Abstract
The cis-regulatory elements encoded in an mRNA determine its stability and translational output. While there has been a considerable effort to understand the factors driving mRNA stability, the regulatory frameworks governing translational control remain more elusive. We have developed a novel massively parallel reporter assay (MPRA) to measure mRNA translation, named Nascent Peptide Translating Ribosome Affinity Purification (NaP-TRAP). NaP-TRAP measures translation in a frame-specific manner through the immunocapture of epitope tagged nascent peptides of reporter mRNAs. We benchmark NaP-TRAP to polysome profiling and use it to quantify Kozak strength and the regulatory landscapes of 5’ UTRs in the developing zebrafish embryo and in human cells. Through this approach we identified general and developmentally dynamic cis-regulatory elements, as well as potential trans-acting proteins. We find that U-rich motifs are general enhancers, and upstream ORFs and GC-rich motifs are global repressors of translation. We also observe a translational switch during the maternal-to-zygotic transition, where C-rich motifs shift from repressors to prominent activators of translation. Conversely, we show that microRNA sites in the 5’ UTR repress translation following the zygotic expression of miR-430. Together these results demonstrate that NaP-TRAP is a versatile, accessible, and powerful method to decode the regulatory functions of UTRs across different systems.
Subject terms: RNA decay, RNA quality control
The cis-regulatory elements encoded in the sequence and structure of mRNAs determine their translational output. Here the authors develop NaP-TRAP, an assay to measure translation in a frame-specific manner, revealing dynamic regulatory elements in zebrafish embryos and HEK293T cells.
Introduction
Life requires spatial and temporal control of protein expression. The protein output of a given transcript reflects the integration of translation and mRNA stability. While there has been a considerable effort to understand the factors driving mRNA synthesis1, maturation2, and decay3–6, the regulatory frameworks governing translational control remain elusive7,8. Cis-regulatory elements encoded in the sequence and structure of mRNAs modulate these processes. Although these elements are distributed throughout the transcript, they are often concentrated in the regions upstream and downstream of the main coding sequence, the 5’ and 3’ untranslated regions (UTRs) respectively9,10. Given that initiation is the rate-limiting step of translation, there has been a particular interest in understanding the regulatory elements found in 5’ UTRs11–14. These elements include internal ribosome entry sites (IRESs)15, G-quadruplexes16, iron response elements (IREs)17, pyrimidine-rich elements (TOP18, PRTE19, CERT20), as well as upstream Open Reading Frames (uORFs)21–23. Regulatory elements function by recruiting trans-factors to the mRNA. Differences in the pool of cellular RNA binding proteins (RBPs), translation initiation factors and ribosome components have been shown to influence the regulatory potential of these elements11.
To investigate translation control, several studies have utilized ribosome profiling24,25. While this method accurately quantifies the translation efficiency of individual genes, its capacity to characterize the function of cis-regulatory elements is limited. The generation of ribosome-protected fragments decouples the translation measurement of a given mRNA from its cognate untranslated regions26. Thus, measurements of translation efficiency often reflect the amalgamation of several isoforms of a given gene, each with a unique set of regulatory elements. This process is complicated further by the fact that endogenous transcripts contain multiple regulatory elements that exert differential and often competing effects on translation.
Massively parallel reporter assays (MPRAs) are particularly suited to address these challenges. MPRAs measure the abundance and/or translation efficiency of thousands of reporters simultaneously. These assays have improved our understanding of translation control and mRNA stability. Previous studies have characterized Kozak strength27, uORFs28–30, IRESs31,32, codon optimality33,34, RNA structure35, microRNA binding sites36, cytoplasmic polyadenylation37, and the effects of variation in human UTRs38–41. These assays have also identified novel sequence motifs driving translation and decay4,42,43. Despite these insights, conclusions based on MPRAs have been limited by the methods used to measure translation: growth-selection44, fluorescent-cell sorting (FACS)28,45, polysome profiling39,41, Translating Ribosome Affinity Purification (TRAP-seq)46, and direct analysis of ribosome targeting (DART)43 (Table 1). In MPRAs where reporters are encoded in DNA, measurements of translation can be confounded by additional layers of transcriptional regulation. Furthermore, in polysome fractionation and TRAP-seq, which quantify the number of ribosomes on each transcript through sucrose gradient fraction and immunocapture of epitope-tagged ribosomes, respectively, inactive ribosomes, ribosomes translating out of frame, or ribosomes translating ORFs outside of the coding sequence may skew translation measurements. Additionally, the methodological complexity of these assays has reduced the use of translation-based MPRAs across diverse systems.
Table 1.
Comparing NaP-TRAP to existing translation-based MPRA methods
| Method | System | Input | Specialized Equipment | Multi-Frame | Frame-specific | Translation Read-out |
|---|---|---|---|---|---|---|
| Growth-selection | Cell lines | DNA | None | No | Yes | Steady-State |
| FACS | Cell lines | DNA / RNA | Flow-cytometer | No | Yes | Steady-State |
| DART | Cell lysate | RNA | Ultracentrifuge | No | No | Equilibrium |
| Polysome profiling | Model organisms | DNA / RNA | Ultracentrifuge | No | No | Instantaneous |
| TRAP-seq | Model organisms | DNA / RNA | None | No | No | Instantaneous |
| NaP-TRAP | Model organisms | DNA / RNA | None | Yes | Yes | Instantaneous |
NaP-TRAP was developed to quantify the translation of thousands of reporters simultaneously in a frame-specific manner. In contrast to existing methods, NaP-TRAP can be adapted to dynamic model systems (e.g. the developing zebrafish embryo) and does not require specialized equipment or a large amount of source material to measure translation.
Here, we develop NaP-TRAP (Nascent Peptide Translating Ribosome Affinity Purification), a novel method to measure in-frame translation through the immunocapture of nascent peptides. More specifically, by including an N-terminal FLAG tag in the coding sequences of reporter mRNAs, we enrich for reporters in a manner proportional to the number of active ribosomes translating the main ORF in frame. First, we benchmark NaP-TRAP to polysome profiling and use reporter assays of increasing complexity to validate this method. Second, using NaP-TRAP we quantify the Kozak strength in zebrafish by measuring the translation of thousands of reporters simultaneously. Third, we assess the regulatory potential of endogenous 5’ UTR sequences in the developing zebrafish embryo and HEK293T cells. Lastly, through this approach, we identify common and developmentally modulated motifs that regulate translation in vertebrates as well as potential effector RBPs known to bind these sequences. In doing so we demonstrate that NaP-TRAP is an accessible, versatile, and quantitative method, which has the capacity to measure translation control across multiple systems and identify regulatory sequences in vivo.
Results
NaP-TRAP measures translation via the immunocapture of nascent chains
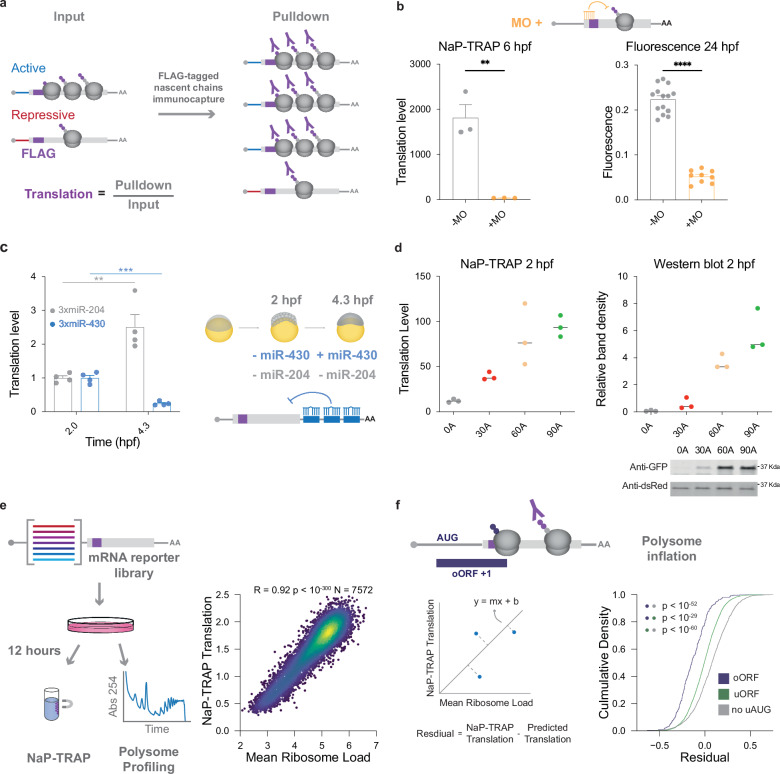
Cis-regulatory elements encoded in mRNAs modulate translation efficiency. To measure the regulatory potential of these elements we developed a novel massively parallel reporter assay (MPRA), NaP-TRAP (Nascent Peptide Translating Ribosome Affinity Purification). We reasoned that we could enrich for reporters in a manner proportional to their translation efficiency through the immunocapture of FLAG-tagged nascent chain complexes immobilized by cycloheximide treatment (Fig. 1a). To this end, we measured translation as a ratio of the amount of a reporter mRNA in the pulldown relative to its input. This approach decouples translation measurements from differences in mRNA abundance.
Fig. 1. NaP-TRAP measures translation through immunocapture.
a Schematic detailing the NaP-TRAP method. FLAG-tagged nascent chain complexes of reporter mRNAs are enriched via an anti-FLAG immunoprecipitation. Translation is measured as a ratio of reporter reads in the pulldown relative to the input. b NaP-TRAP derived translation at 6 hpf (left, N = 3 replicates; 25 embryos per replicate) and fluorescence measurements at 24 hpf (right, - MO N = 14 embryos, +MO N = 9 embryos) in the presence (+ MO) or absence (- MO) of a translation blocking morpholino (two-sided unpaired t-test: ** p < 0.005, **** p < 0.0001; NaP-TRAP + /− MO, p = 0.0036; Fluorescence +/− MO p < 10−4; error bars = SEM). c NaP-TRAP derived translation values for 3xFLAG-GFP-3xmiR-430 (blue) and 3xFLAG-GFP-3xmiR-204 (gray) at 2 hpf (N = 4 replicates; 25 embryos per replicate) and 4.3 hpf (N = 4 replicates; 25 embryos per replicate) (two-sided unpaired t-test: ** p < 0.01, *** p < 0.005; miR-204 2hpf vs 4 hpf, p = 0.0071; miR-430 2hpf vs 4 hpf = 0.0001; error bars = SD). Schematic representation of repression of translation exercised by miR-430, but not miR-204 at 4.3 hpf. d NaP-TRAP derived translation values at 2 hpf (left, N = 3 replicates; 25 embryos per replicate) and quantification of band intensity of western blot and example immunoblot (right, N = 3 replicates; 5 embryos per replicate) of GFP reporters with poly-A tails of 0, 30, 60 and 90 As in the presence of a DsRED injection control. e In HEK293T cells, NaP-TRAP derived translation values are strongly correlated with mean ribosome load (polysome profiling) for a 5’ UTR mRNA reporter library (two-sided Pearson’s R = 0.92, N = 7572 reporters). f A cumulative density plot of residuals (NaP-TRAP translation – predicted translation) for reporters containing oORFs (N = 247 reporters), uORFs (N = 3834 reporters), and no upstream start codons (N = 1993 reporters) (p-values were calculated using a two-sided Mann-Whitney U-test; oORF vs noAUG p < 10−52; oORF vs uORF p < 10−29; uORF vs noAUG p < 10−60).
To evaluate the capacity of NaP-TRAP to measure translation we performed three reporter assays. First, we measured the effect of a translation-blocking morpholino. We co-injected 3xFLAG-GFP and DsRed mRNAs into single-cell zebrafish embryos in the presence or absence of a translation-blocking morpholino, targeting the start codon of 3xFLAG-GFP and performed NaP-TRAP at 6 hpf. To measure the amount of 3xFLAG-GFP reporter mRNA present in the input and pulldown fractions of each anti-FLAG immunoprecipitation, we employed reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Given that the DsRed reporter mRNA was neither enriched by the anti-FLAG immunoprecipitation nor targeted by the translation-blocking morpholino, we utilized the relative abundance of the DsRed reporter mRNA in each fraction to normalize NaP-TRAP derived translation values across experimental conditions. Using NaP-TRAP we observed a significant decrease in translation in the presence of the morpholino at 6 hours post fertilization (hpf) (Fig. 1b). These results are consistent with the translational repression in morpholino-injected embryos observed by the decrease in fluorescence (GFP/DsRed) at 24 hpf (Fig. 1b).
Next, we tested the capacity of NaP-TRAP to capture dynamic translation control. In the developing zebrafish embryo, the microRNA miR-430 is one of the first zygotically transcribed genes. At 4.3 hpf, the expression of miR-430 results in the translation repression, deadenylation, and eventual decay of targeted mRNAs47,48. To quantify this effect, we measured the translation of 3xFLAG-GFP reporters with partial complementarity to two different microRNAs in their 3’ UTRs, miR-430 (3xFLAG-GFP-3xmiR-430) and miR-204 (3xFLAG-GFP-3xmiR-204). We selected miR-204 target sites as a control given that this microRNA is not expressed in the early embryo. Using NaP-TRAP we measured the translation of mRNA reporters at 2 and 4.3 hpf, before and after miR-430 expression. As described above we normalized translation values across experimental conditions using a DsRed control mRNA. We observed that the translation of the miR-430 reporter decreased ~4.3-fold after the expression of miR-430, while the control miR-204 reporter increased ~2.5 fold (Fig. 1c), consistent with the role of miR-430 in translational repression and the global increase in translation observed during the early stages of development49.
Third, poly-A tail length has been shown to be correlated with translation during early development48,50. To test whether NaP-TRAP quantifies the level of translation, we compared the translation of 3xFLAG-GFP mRNA reporters with different poly-A tail lengths (0, 30, 60, and 90 adenosines respectively), using DsRed mRNA as an injection control. We observed that increasing the poly-A tail length increases translation as measured by NaP-TRAP and western blot (Fig. 1d). Together these results demonstrate that: (1) NaP-TRAP measures the translation of individual reporters targeting general and developmentally dynamic cis-regulatory elements and (2) NaP-TRAP derived translation values are quantitative and correlate with protein output. Further, these experiments highlight the versatility of the method as they demonstrate the capacity of NaP-TRAP to quantify cis-regulation mediated by the 5’ and 3’ UTRs, as well as the poly-A tail.
NaP-TRAP measures translation in a frame-specific manner
To evaluate NaP-TRAP’s performance as an MPRA, as well as benchmark the approach against an established method, we performed NaP-TRAP and polysome profiling on a 5’ UTR mRNA reporter library in HEK293T cells (11,088 reporters, Supplementary Table 3). To this end, we transfected the in vitro transcribed mRNA library into HEK293T cells and performed NaP-TRAP and polysome profiling at 12 hours post-transfection (hpt) (Supplementary Fig. 1a-b). To limit the effect of out-of-frame translation, we incorporated stop codons in frames +1 and +2 early in the coding sequence of 3xFLAG GFP. We observed a strong correlation between Mean Ribosome Load (MRL) and NaP-TRAP-derived translation values (R2 = 0.85), suggesting that NaP-TRAP measures quantitatively the number of translating ribosomes on individual reporters (Fig. 1e).
To demonstrate the capacity of NaP-TRAP to measure frame-specific translation we adopted three different approaches. First, we performed a linear regression comparing MRL (polysome profiling) to NaP-TRAP translation and measured the residual of reporters containing upstream open reading frames (uORFs and oORFs, overlapping upstream open reading frames). The residual is the difference between the experimentally determined NaP-TRAP translation and the translation value calculated based on the linear regression (experimental—predicted translation; Fig. 1f). We observed a negative shift in the residuals of reporters containing oORFs and reporters containing uORFs when compared to reporters containing no upstream start codons (Fig. 1f, Mann-Whitney U test, p < 10−52 and p < 10−60 respectively). This shift towards negative values suggests that polysome profiling MRL values for reporters containing upstream start codons are inflated relative to NaP-TRAP-derived translation values. This result highlights the importance of measuring translation in a frame-specific manner compared to the number of bound ribosomes. Second, to evaluate the capacity of NaP-TRAP to measure frame-specific translation we selected 12 reporters containing upstream start codons for which the two methods diverged and measured their protein output using nano- and firefly luciferase in HEK293T cells at 12 hpt. NaP-TRAP-derived translation values correlated more strongly with dual luciferase activity than MRL (R2 = 0.81 and R2 = 0.65, respectively, Supplementary Fig. 1c-e). Finally, to measure translation in three frames simultaneously, we encoded FLAG, HA, and MYC tags into frames 1, 2, and 3, respectively of a GPF luciferase reporter. Using a degenerated oligo reporter containing uORFs and oORFs in each frame, we were able to detect translation in frames 2 and 3 in zebrafish in the absence of frame 1 translation, indicating that NaP-TRAP can be designed to detect frame-specific translation (Supplementary Fig. 2). Taken together these results demonstrate that: (1) NaP-TRAP is a quantitative MPRA method and (2) the frame specificity of NaP-TRAP derived translation values results in a more accurate measurement of protein output than MRL for reporters with out-of-frame translation.
Using NaP-TRAP to investigate Kozak strength in the developing zebrafish embryo
Next, we designed a library to quantify the regulatory potential of the Kozak sequence in the developing zebrafish embryo. We elected to measure Kozak strength for two reasons: (1) the Kozak sequence is a strong determinant of translation initiation and (2) Kozak strength is often inferred based on the frequency of these sequences in the genome51,52. To generate a library of diverse Kozak sequences, we incorporated six random nucleotides upstream and one random nucleotide downstream of the AUG of the 3xFLAG-GFP reporter (Fig. 2a)53. We injected the in vitro transcribed mRNA library into single-cell zebrafish embryos and measured the translation using NaP-TRAP at 6 hpf (Supplementary Fig. 3a). We identified active and repressive Kozak sequences, and generated position weight matrices for the top and bottom 10% of reporters based on their translation (Fig. 2a). We observed that active Kozak sequences were enriched in C’s and A’s, whereas repressive Kozak sequences were enriched in U’s and G’s.
Fig. 2. NaP-TRAP measures the effect of Kozak strength on translation.
a A schematic detailing the Kozak library containing six random nucleotides upstream and one nucleotide downstream of the start codon of 3xFLAG-GFP (top left). Histogram of NaP-TRAP translation values at 6 hpf (bottom left). Sequence logos for the top and bottom 10% of reporters based on translation (right)63. b, c Cartoon of random forest regression model feature generation through the one-hot encoding of reporter sequences (b). Scatterplot comparing the model’s prediction to the experimentally derived translation values of a test set of reporters (two-sided Pearson’s R; N = 785 reporters) (c). d Permuted feature importance derived from random forest regression model (N = 10 repeats, error bars = SD). Nucleotide positions in purple correlate negatively with translation, whereas positions in blue correlate positively with translation (two-sided Spearman Rank Correlation Coefficient). e Comparison of translation measurements at 6 hpf and an in silico-derived Kozak score based on the frequency of Kozak sequences in the transcriptome (two-sided Pearson’s R; N = 2616 reporters; p < 10−56)26 f, g. The translation of seven reporters with a Kozak score between 295-305 was measured using a dual luciferase-based assay (f). Plot comparing relative luciferase activity (Nano luciferase / Firefly luciferase), and NaP-TRAP translation values (two-sided Pearson’s R; N = 3 replicates; 5 embryos per replicate, p < 0.0033; error bars = SEM) (g).
To examine the effect of nucleotide identity and position on translation, we employed a random forest regression model (RFM). Model features were generated based on nucleotide identities and positions within the Kozak sequence (Fig. 2b). Prior to training the model, we divided the data into a test and a training set, 30% and 70% of reporters, respectively, and used a 5-fold cross-validation to optimize model parameters. The predicted values of the model were correlated with the translation of the test set (R = 0.73, Fig. 2c); thus, we used this model to predict the translation of all possible Kozak sequences (Supplementary Fig. 3b, Supplementary Table 2). To identify the features informing the predictive power of the model, we performed a permuted feature importance analysis. Briefly, we shuffled the identities of the features of the model one at a time and measured the effect of the change on the predictive power of the model. At 6 hpf bases G, G, U, and G in positions −5, −4, −3, and −2 were the most repressive features, whereas C, A/C, and A in positions −4, −3, +4 were the most activating features (Fig. 2d and Supplementary Fig. 3c).
Lastly, we compared NaP-TRAP translation values to an in silico-derived metric, the Kozak score. In zebrafish, Kozak strength has been previously inferred based on the frequency of a given Kozak sequence within the transcriptome. This metric associates Kozak sequence abundance with increased translation initiation52. Our experimentally derived translation values challenge this hypothesis as we observed a weak correlation between NaP-TRAP and Kozak score (R = 0.31, Fig. 2e). To validate this conclusion, we identified sequences that had similar Kozak scores (300 + /− 5) yet exhibited different NaP-TRAP derived translation values and measured their translation using a dual luciferase reporter assay (Fig. 2f). The luciferase translation values correlated strongly with NaP-TRAP (Pearson’s R = 0.92; Fig. 2g). This result not only highlights the importance of measuring Kozak strength experimentally, but also demonstrates that NaP-TRAP derived translation values correlate well with protein production. Taken together, these results demonstrate that NaP-TRAP can be employed as a MPRA. By measuring the translation of thousands of reporters simultaneously, we quantify and model Kozak strength in a vertebrate system.
Investigating the regulatory potential of endogenous 5’ UTRs
Next, we investigated the regulatory activity of endogenous 5’ UTRs during the early stages of embryogenesis. Prior to zygotic genome activation, the translation of maternally supplied mRNAs drives development54,55. Given that initiation is the rate-limiting step of translation, 5’ UTRs provide a mechanism for maternally supplied mRNAs to modulate their protein output. To study the cis-regulatory elements encoded in these mRNAs, we designed an 11,088-sequence synthetic oligo library (124-nt long; Table S3) by tiling the 5’-UTRs of 1725 zebrafish genes every 25 nucleotides (Fig. 3a). To specifically investigate the regulatory role of 5’ UTR elements in translation and identify novel regulators, we chose to maintain a constant 5’ UTR length, Kozak sequence, and poly-A tail length in the reporter mRNAs, as these factors are well-established modulators of translation initiation in developing embryos. We injected the in vitro transcribed library into single-cell zebrafish embryos and measured translation at 2 hpf and 6 hpf using NaP-TRAP. We selected these timepoints as these occur before and after zygotic genome activation. Through this approach we sought to understand how the expression of zygotic genes modulates translation via 5’ UTRs. To quantify relative changes in translation, we added an internal spike-in at the RNA extraction step (Fig. 3a). Translation values were strongly correlated across replicates (Pearson’s R > = 0.90; Supplementary Figs. 4a,b and 5a,b) and with protein abundance at both timepoints (dual luciferase assay; 2 hpf Pearson’s R 0.68, 6 hpf Pearson’s R 0.86, Supplementary Fig. 4c-d). We observed a mean increase in translation between 2 hpf and 6 hpf of ~1.9 fold, consistent with the gradual de-repression of translation during the first 24 hours of development (Fig. 3b and Supplementary Fig 5c)49,56.
Fig. 3. Using NaP-TRAP to predict the translation of zebrafish 5’ UTRs.

a A schematic detailing global changes in translation during the maternal-to-zygotic transition in the developing zebrafish embryo (top left). The 5’ UTR library was generated by tiling the 5’ UTRs of 1725 zebrafish genes (bottom left). The NaP-TRAP workflow with the addition of spike-ins at the RNA extraction step (right). b Comparison of translation values between 2 and 6 hpf of the 5’ UTR library (two-sided Mann-Whitney U-test; p < 10−100; N = 8529 reporters). Minima and maxima of the box represent the first and third quartiles, respectively, whereas the whiskers correspond to 1.5x the interquartile range (Q1-Q3). The center of the box is the median. c Schematic detailing random forest regression model feature selection (k-mer counts 1-6 nt and features characterizing upstream ORFs). d–e Scatterplot comparing each model’s prediction to the experimentally derived translation values of a test set of reporters (test set = 30% of reporters used; N = 2559 reporters; two-sided Pearson’s R) (d, e). f, g The correlation between the top 12 features and translation (measured using permuted feature importance Supplementary Fig. 4e-f; blue refers to a positive correlation and purple refers to a negative correlation; two-sided Spearman Rank Correlation Coefficient).
Next, we utilized a random forest regression model (RFM) trained on sequence elements and features characterizing upstream AUGs (uAUGs) to predict translation (Fig. 3c and Supplementary Fig. 5d-i). To prevent model over-fitting, we divided our data into test and training sets (30% and 70% of reporters respectively) and used a 5-fold cross-validation during model training. The predictions of the models correlated with the test set at both timepoints (R = 0.74; Fig. 3d, e). Using a permuted feature importance analysis, we identified the number of upstream uORFs as well as the Kozak strength of uORFs and out-of-frame oORFs (ooORFs) as the features contributing most significantly to the predictive power of the model (Supplementary Fig. 4e-f). We observed that U-repeats activated translation and G/GG sequences and uORFs suppressed translation at both timepoints. Interestingly, we also observed that Cs suppressed translation at 2 hpf, whereas at 6 hpf C and CU sequences activated translation (Fig. 3f, g). Given that G-richness is an important repressive feature in both RFMs (Fig. 3f, g), we elected to investigate the role of structure, Minimum Free Energy (MFE), in translation57. At both timepoints we observe a moderate correlation between MFE and translation (2 hpf R = 0.28; 6 hpf R = 0.25), suggesting a role for structure in translation repression (Supplementary Fig. 6a,e). Lastly, given the role of upstream open reading frames in the feature importance analysis, we repeated the random forest analysis on reporters that contained no upstream start codons (Supplementary Fig. 4g-l). In the absence of upstream start codons, the importance and prominence of C- and CU-sequences increased at 6 hpf (Supplementary Fig. 4i,l). Altogether these results demonstrate the following: (1) uAUGs are the most prominent repressive elements during early embryogenesis, and (2) 5’ UTR regulatory landscapes are dynamic during development.
Identifying sequences driving differential translation
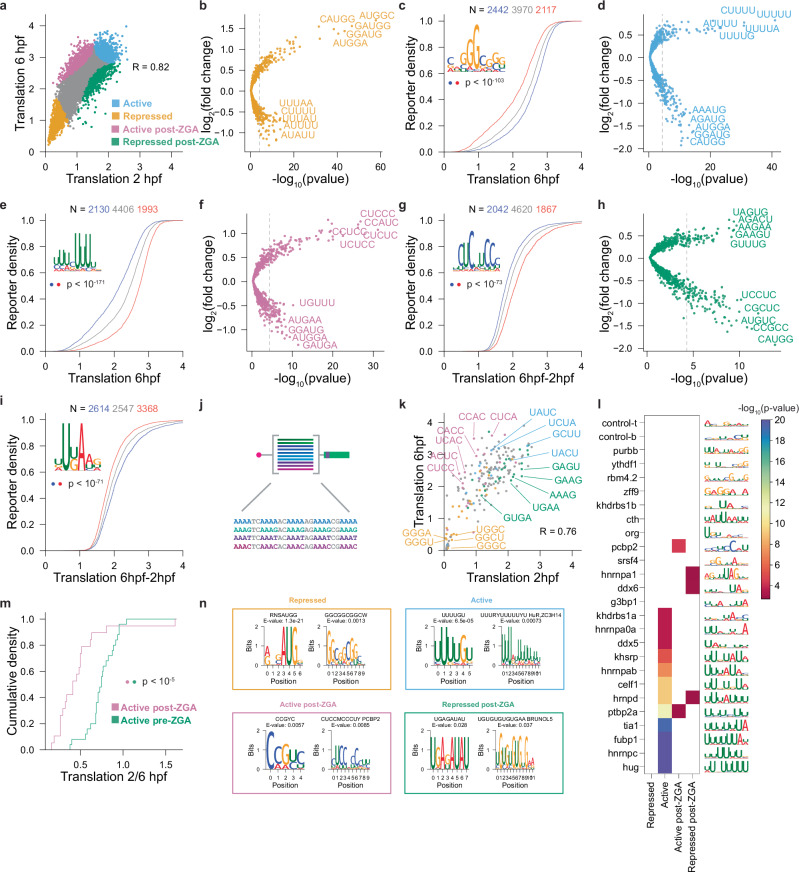
To identify the sequence elements modulating differential translation during development, we divided the 5’ UTR reporters into four groups based on their relative translation at 2 and 6 hpf: (1) repressed, (2) active, (3) repressed post-ZGA, and (4) active post-ZGA (Fig. 4a and Supplementary Fig. 5j) and performed a differential pentamer enrichment analysis on each group. Repressed reporters were significantly enriched in upstream ORFs and GC-rich pentamers and depleted in U-rich tracks (p > 10−5 hypergeometric test, Fig. 4b and Supplementary Fig 5k). Conversely, active reporters were enriched in U-rich pentamers (UUUUU, CUUUU, UUUUA, GUUUU) and depleted in upstream start codons (Fig. 4d and Supplementary Fig. 5l). Reporters that were more highly translated after genome activation were enriched in C-rich pentamers (CUCUC, CUCCC, CCAUC, CCUCC) and depleted in U-repeats (Fig. 4f and Supplementary Fig. 5m). In contrast, reporters that were repressed post-ZGA were enriched in AG/UG-rich sequences (UAGUG, UAUUG, AAGAA, AGACU), as well as sequence motifs complementary to the seed site of the microRNA miR-430 (GCACU and GCACUU; Table S3) and depleted in uORFs and pyrimidine repeats (Fig. 4h and Supplementary Fig. 5n). Conversely, we observed that sequences enriched in those motifs (top 20%, red) were differentially translated compared to a group of sequences depleted in those motifs (bottom 20%, blue) (Fig. 4c, e, g, i). For example, sequences enriched in GGCGGG were repressed at 2 and 6 hpf (p value < 10−128 and p value < 10−103, Mann-Whitney U-test, Fig. 4c), while sequences enriched in UUUUU were highly translated at 2 and 6 hpf (p value < 10−230 and p-value < 10−171, Mann-Whitney U-test, Fig. 4e).
Fig. 4. NaP-TRAP captures general and dynamic 5’ UTRs cis-regulation.
a The translation of the 5’ UTR library at 2 hpf and 6 hpf using NaP-TRAP (two-sided Pearson’s R; N = 8,529 reporters). Reporters were divided into four groups based on their translation rank at both time-points: repressed, active, active post-ZGA, and repressed post-ZGA (orange, blue, pink, and green). b–i Fold-change enrichment and depletion for all pentamers in each group from (a) relative to the reporter library (one-sided hypergeometric test to calculate significance; Bonferroni corrected p-value threshold, p < 5 * 10−6; repressed (b), active (d), active post-ZGA (f), and repressed post-ZGA (h)). Using hierarchical clustering sequence motifs were generated from enriched pentamers. The cumulative distribution of translation for reporters enriched (red, top 20%) and depleted (blue, bottom 20%) in a representative motif were plotted (middle 80% gray). The significance in the difference of the distributions of translation was determined using a two-sided Mann-Whitney U-test ((c), p < 10−103; (e), p < 10−171; (g), p < 10−73; (i), p < 10−71). j Schematic detailing the library of all possible tetramer repeats separated by dinucleotide spacers. k Translation measurements of the validation library at 2 and 6 hpf (two-sided Pearson’s R; N = 195 reporters). Tetrameric reporters were labeled based on whether their repeat was enriched in the reporter groups described above (a). l Heat map showing the significance of the overlap between eCLIP motifs for RBPs expressed in the early embryo (top 10) and the motifs enriched in each class in panel (a) (one-sided hypergeometric test). m Cumulative distribution plot comparing the translation (2hpf / 6 hpf) of validation reporters identified as active post ZGA (pink) and repressed post ZGA (green) (two-sided Mann-Whitney U-test: p < 10−5). n Using STREME, motifs were generated from the reporter groups described above (a). The information content of representative motifs was plotted: repressed (b), active (d), active post-ZGA (e), and repressed post-ZGA (f). Displayed E-values reflect the output of STREME. Tomtom was utilized to compare motifs to a database of human RBP motifs (E-value < 0.05).
To independently validate the temporal regulation of these elements, we generated a library of 5’ UTR sequences containing all possible tetramer repeats separated by dinucleotide spacers and measured translation using NaP-TRAP at 2 and 6 hpf (Fig. 4j). The distributions of reporters encoding repressed, repressed post-ZGA, and active post-ZGA tetramers largely recapitulated the distributions of their respective groups in the 5’ UTR library (Fig. 4k, Supplementary Fig. 7b-e Supplementary Table 3 and Supplementary Table 4). Consistent with this, reporters enriched in “active motifs post-ZGA” revealed a larger increase in translation compared to those enriched in “repressed motifs post-ZGA” (Fig. 4m). For some motifs (e.g. U-rich sequences in the active group), we observed a different behavior in the tetramer library, an effect that may stem from the differences in the motif length. Indeed, the motif analyses presented above identified that poly-U tracts of 6-8 Us are associated with highly translated reporters (Fig. 4e, l, n). Altogether, these results identify sequence motifs in 5’UTRs that modulate general and dynamic translation during embryonic development.
Finally, we compared motifs enriched in each group to the RNA binding motifs preferentially bound by RBPs expressed in the early zebrafish embryo5 and an RBP database58 using STREME and Tomtom59,60. We found a significant enrichment for HuR binding sites among the constitutively active motifs and PCPB2 among the active post-ZGA motifs (Fig. 4n, l and Supplementary Fig. 7a). These results reveal 5’ UTR elements with differential regulatory activity on translation during early embryogenesis and identify potential trans-factors participating in this dynamic regulation.
miR-430 binding sites in the 5’ UTR suppress translation
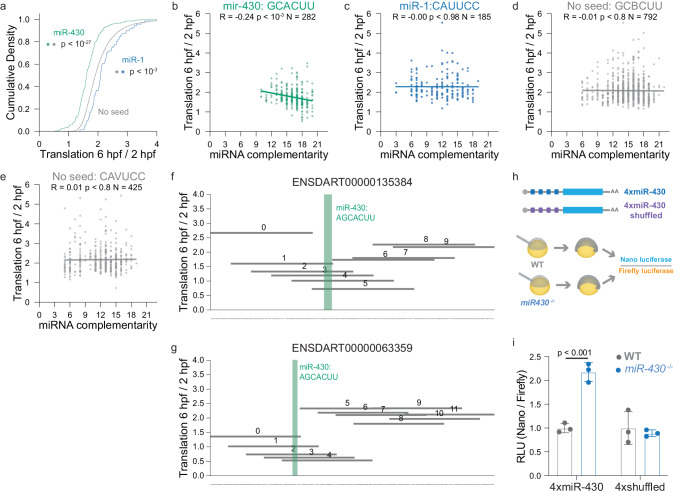
miR-430 is one of the first zygotically transcribed genes in the developing zebrafish embryo47. The enrichment of miR-430 seeds in reporters that were repressed after ZGA suggests that miR-430 can target mRNAs through 5’ UTR binding to regulate their temporal expression during the maternal-to-zygotic transition. To determine whether this repressive effect is specific for miR-430, we compared the translation between 2 hpf and 6 hpf for reporters containing seeds for miR-430 or miR-1, a microRNA that is not expressed in the early stages of development. We observed a significant decrease in the translation of miR-430 containing reporters (6 vs 2 hpf) relative to the translation of reporters lacking either seed (miR-430: p < 10−27, miR-1: p < 0.001, Mann-Whitney U test). In contrast, miR-1 control reporters were not significantly decreased (Fig. 5a).
Fig. 5. Zygotic expression of miR-430 inhibits translation of mRNAs with 5’ UTR seed sites.
a Cumulative distributions of translation (6 hpf / 2 hpf) for reporters containing miR-430 (GCACUUA, GCACUUU, AGCACUU; N = 212 reporters) or miR-1 (ACAUUCC, CAUUCCA; N = 71 reporters) heptamers, labeled in green and blue respectively. Reporters labeled in gray contain neither seed sequence (N = 8,242 reporters) (two-sided Mann-Whitney U test, p < 10−27 miR-430 reporters vs reporters with no seed; two-sided Mann-Whitney U test, p < 0.001 miR-1 reporters vs reporters with no seed). b, c The effect of the number of complementary bases to miR-430 ((b), N = 282 reporters) or miR-1 ((c), N = 185 reporters) on the translation of reporters containing seed sequences at 6 hpf versus 2 hpf (two-sided Pearson’s R; (b) p < 10−3, (c) p < 0.98). d–e The effect of the number of complementary bases to miR-430 ((d), N = 792 reporters) and miR-1 ((e), N = 425 reporters) on translation for reporters containing seeds with a single mismatch at 6 hpf versus 2 hpf (two-sided Pearson’s R; (d) p < 0.8, (e) p < 0.8). f, g Plots showing the delta translation (Translation 6 hpf / 2hpf) of reporter fragments that tile the zebrafish 5’ UTR (ENSDART00000135384 / usp9 (f), ENSDART00000063359 / ucp2 (g)). The seed sites of miR-430 (AGCAUUU) are labeled in green. h, i Schematic detailing dual luciferase assay measuring the inhibitory effect of miR-430 binding sites in the 5’ UTR (h). 4xmiR-430-nanoluc and 4xmiR-430-shuffled-nanoluc were injected in wild-type and miR-430 knockout embryos. Relative Luciferase Activity (RLU) values were normalized to each reporter (N = 3 replicates; 5 embryos per replicate; two-sided unpaired t-test; (i) p < 0.001; error bars = SD).
To investigate the mechanism driving miR-430-mediated translation repression, we measured the degree of complementarity between the microRNA and the targeted reporter. Translation of the miR-430 reporters was negatively correlated to miRNA-5’ UTR complementarity (Fig. 5b). Whereas translation of the miR-1 control reporters or miR-430 reporters with a single mismatch (CAUUCC, GCBCUU) were not significantly correlated with miRNA-5’ UTR complementarity (Fig. 5c–e), suggesting that microRNA complementarity is correlated with translation repression and that the seed is crucial for this interaction.
To evaluate whether miR-430 expression drives the translation repression of 5’ UTRs with seed sites, we constructed two nano-luciferase reporters: 4xmiR-430-nanoluc and 4xmiR-430-MUT-nanoluc, where the miR-430 seed (GCACUU) has been mutated (GCUCUA). We injected these reporter mRNAs together with a firefly luciferase control mRNA into wild-type and miR-430−/− mutant embryos and measured relative luciferase activity at 6 hpf (Fig. 5h). We observed a ~2fold decrease in the relative luciferase activity of the 4xmiR-430 reporter in the wild-type condition when compared to the mutant (two-tailed t-test; p-value < 10−5). In contrast, we observed no significant difference in luciferase activity between the shuffled reporters when comparing the mutant and wild-type embryos (Fig. 5i). Consistent with these results, this effect is also observed at the level of individual endogenous 5’ UTRs containing a miR-430 seed, where reporter fragments containing the seed are more repressed post-ZGA compared to other fragments (Fig. 5f, g). Taken together these findings demonstrate that the zygotic expression of miR-430 represses the translation of mRNAs with 5’ UTR seed sites and suggest that miRNA target sites in the 5’ UTR can provide significant translational repression in vivo.
The developmentally dynamic role of C-rich motifs
Our analyses also indicated that C-rich pentamers were significantly enriched in reporters that increase translation after ZGA (Fig. 4f). To explore this observation further, we measured the translation of reporters enriched and depleted in each feature (k-mers ≤ 4) for reporters lacking upstream open reading frames. We observed that the k-mers C, CC, CCC, and UCC were some of the most repressive features at 2 hpf, yet activated translation at 6 hpf (Fig. 6a). To determine the role of C-rich sequences in translation regulation, we compared the translation of four wild-type (wt) and four mutant (mut) reporters (C > U and U > C). For the wt reporters two were enriched in Us and highly translated, while the other two were enriched in Cs and were repressed pre-ZGA and active following ZGA (Fig. 4a). We observed that mutating Cs to Us in the C-rich reporters enhanced translation at 2hpf (Fig. 6c), whereas mutating Us to Cs repressed translation (Fig. 6d). In contrast, this effect was largely reduced at 6 hpf (Fig. 6e, f). These results support the findings of the differential enrichment analysis and indicate that there is a translational switch driven by C-rich sequences following ZGA (Fig. 6b). Altogether, these results validate novel developmentally dynamic mechanisms of 5’ UTR-mediated translation control, demonstrating the importance of quantifying cis-regulation in non-steady state systems.
Fig. 6. The dynamic effect of C-richness on translation.
a Plot comparing feature rank at 2 and 6 hpf (k-mers ≤ 4). At each time point feature rank was determined by calculating the mean difference in translation between non-uAUG reporters enriched and depleted (top and bottom 20%) in each feature (red: repressive at 2 hpf, blue: active at 2 hpf, purple: features that were repressive at 2 hpf and then active at 6 hpf). Features were only included if there was a significant difference in mean translation at each timepoint (Bonferroni corrected t-test). b Proposed models explaining how the maternal-to-zygotic transition can alter the effect of cis-regulatory elements in the 5’ UTR (see Fig. 4 g, l–n). c–f The translation measurements of four wild-type (wt) and four mutant (mut) reporters using NaP-TRAP at 2 hpf and 6 hpf. For the wt reporters, U’s in two active (U-rich) reporters were mutated to C’s (U > C), whereas C’s in two active-post ZGA (C-rich) reporters were mutated to U’s (C > U. Wt reporters are colored gray, whereas mut reporters are labeled red (N = 3 replicates; 25 embryos per replicate; two-sided unpaired t-test, (c) sich211 p < 10−6, atp6v0 p < 10−6; (d) igf1ra p = 0.0673, asun p = 0.0094 (e) sich211 p = 0.0076, atp6v0 p = 0.356 (f) igf1ra p = 0.763, asun p = 0.793; error bars = SD).
NaP-TRAP investigates 5’ UTRs mediated translation control in human cells
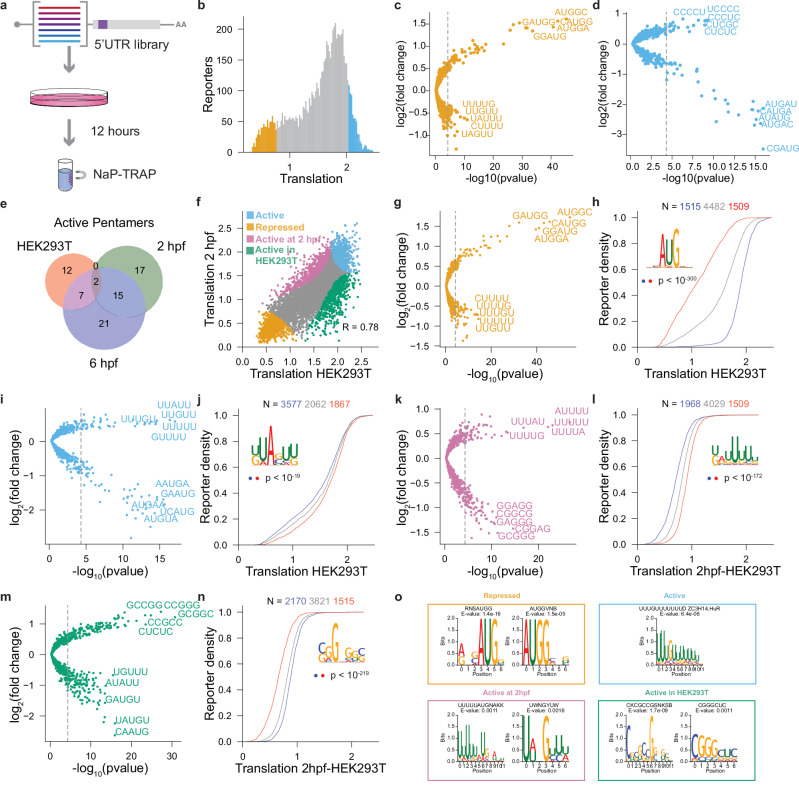
Lastly, we were interested in understanding the regulatory landscape of 5’ UTRs in human cell lines. To this end, we transfected the in vitro transcribed mRNA 5’ UTR library into HEK293T cells and measured translation at 12 hours post-transfection (hpt) using NaP-TRAP (Fig. 7a, b). Replicates were strongly correlated (Pearson’s R ≥ 0.92, Fig. Supplementary Fig. 8 a-c). In HEK293T cells, repressed reporters were enriched in AUG-containing motifs and depleted in U-rich motifs (Fig. 7c), whereas active reporters were enriched in C-rich pentamers and depleted in AUG-containing motifs (Fig. 7d), consistent with those observed in zebrafish at 6 hpf (Fig. 7e and Supplementary Fig. 8d). To identify general and species-specific regulatory elements, we performed a differential enrichment analysis comparing the human and the zebrafish data (Fig. 7f and Supplementary Fig. 8e-i, 2 hpf and 6 hpf, respectively). We divided the reporters into four groups: (1) repressed, (2) active, (3) active in zebrafish, and (4) active in HEK293T cells and identified pentamers enriched and depleted in each group (Fig. 7 g, i, k, m respectively). Using hierarchal clustering, we generated sequence motifs from the enriched pentamers in each group. Sequences enriched in those motifs (top 20%, red) are differentially translated compared to a group of sequences depleted in those motifs (bottom 20%, blue) (Fig. 7h, j, l,n). Our results reveal that uAUGs are general repressors of translation (Fig. 7g,h and Supplementary Fig. 8f) and U-rich motifs are general activators of translation (Fig. 7i, l and Supplementary Fig. 8g,l). In contrast, reporters that are active only in HEK293T cells are enriched in G-rich k-mers, whereas reporters active at 2 hpf or 6 hpf are depleted in these motifs (Fig. 7m and Supplementary Fig. 8i). Finally, we employed a random forest model to predict translation (R = 0.80; Supplementary Fig. 8j). Consistent with the results observed in zebrafish, Kozak strength and number of upstream AUGs were the most predictive features of translation in HEK293T cells, exhibiting a strong negative correlation with translation (Supplementary Fig. 8 k,l). All together these results demonstrate that (1) NaP-TRAP is a robust method that can measure translation across multiple model systems, (2) upstream AUGs are a dominant driver of 5’ UTR mediated translation control in HEK293T cells, and (3) U-rich motifs are general activators of translation.
Fig. 7. Investigating translation control in HEK293T cells.
a Schematic detailing NaP-TRAP in HEK293T cells. Cells were transfected with the 5’ UTR library using lipid nanoparticles. Translation was measured at 12 hpt. b–d The distribution of translation values in HEK293T cells. The top and bottom 10% of reporters based on their translation values are labeled in orange and blue (N = 7506 reporters) (b). A differential enrichment analysis identified pentamers enriched and depleted in repressed (c) and active (d) reporters, blue and orange, respectively (one-sided hypergeometric test with a Bonferroni corrected p-value, p < 5 * 10−6). e Venn-diagram comparing the pentamers enriched in active reporters in HEK293T cells and zebrafish embryos at 2 and 6 hpf. f Translation values at 2 hpf in zebrafish compared to translation values in HEK293T cells (two-sided Pearson’s R). Reporters were divided into four groups based on their translation rank at each condition: active (blue), repressed (orange), active in zebrafish at 2 hpf (pink) and active in HEK293T cells (green) (N = 7506 reporters). g–n Fold-change enrichment and depletion for all pentamers in each group from (f) relative to the reporter library (one-sided hypergeometric test to calculate significance; Bonferroni corrected p-value threshold, p < 5 * 10−6; repressed (g), active (i), active at 2hpf (k), and active in HEK293T (m)). Using hierarchical clustering sequence motifs were generated from enriched pentamers. The cumulative distribution of translation for reporters enriched (red, top 20%) and depleted (blue, bottom 20%) in representative motifs were plotted (middle 80% gray). The significance in the difference of the distributions of translation was determined using a two-sided Mann-Whitney U-test ((h) p < 10−300, (j) p < 10−19, (l) p < 10−172, p < 10−219). o Using STREME motifs were generated from the reporter groups described above (f). The information content of representative motifs was plotted: repressed (g), active (i), active in the developing zebrafish embryo at 2 hpf (k), and active in HEK293T cells (m). Displayed E-values reflected the output of STREME. Tomtom was utilized to compare motifs to a database of human RBP motifs. RBPs were displayed above their corresponding motif (E-value < 0.05).
Discussion
Here, we develop a novel MPRA method, NaP-TRAP, and demonstrate its capacity to measure translation quantitatively. We use NaP-TRAP to characterize cis-regulatory elements in the 5’ and 3’ UTRs, the effect of poly-A tail length, the strength of the Kozak sequence, and the regulatory landscape of the 5’ UTRs of mRNAs in developing zebrafish embryos and HEK293T cells. Using NaP-TRAP we have identified general and developmentally dynamic cis-regulatory elements, as well as characterized global changes to translation associated with early embryogenesis.
NaP-TRAP is an accessible, versatile, and quantitative MPRA method to measure translation. By enriching for reporters through the immunocapture of epitope-tagged nascent chain complexes, NaP-TRAP measures translation in a manner that is proportional to the number of ribosomes actively translating the tagged ORF. This approach is particularly important in systems with a low basal level of translation (e.g., the early stages of vertebrate embryogenesis49 and neurons61) and results in translation measurements that reflect protein output (Fig. 1d, Fig. 2g, and Supplementary Figs. 1d,e and 4c,d). In contrast to existing approaches, NaP-TRAP does not require a large amount of input material or specialized equipment, making the method adaptable to a wide range of model systems, different cell types, and physiological states (Table 1). Further, by injecting or transfecting in vitro transcribed mRNA reporter libraries, NaP-TRAP eliminates the confounding effects of transcriptional regulation associated with MPRAs that introduce reporters as DNA. When quantifying the regulatory potential of the 5’ UTR, transcriptional bias may be more pronounced given the region’s proximity to the promoter sequence.
In this study, we have benchmarked NaP-TRAP to polysome profiling. While we observe a strong correlation between NaP-TRAP-derived translation values and MRL (Fig. 1e, f), for reporters containing uORFs and oORFs, we demonstrate that NaP-TRAP-derived translation values correlate more strongly with protein output than MRL (Supplementary Fig. 1c-e). We attribute this difference to the fact that NaP-TRAP measures frame-specific translation as only the nascent chains of ribosomes translating the main ORF are epitope-tagged and thereby immunocaptured, whereas in polysome profiling and other TRAP methods, inactive ribosomes or ribosomes translating outside of the main open reading frame may affect the measurements of translation.
Previous studies have assumed that the frequency of a given Kozak sequence within the genome correlates strongly with its effect on translation52. While this hypothesis has been challenged by MPRAs performed in cell culture, to date we lack measurements of the effect of Kozak strength on translation in zebrafish27. To demonstrate the capacity of NaP-TRAP to quantify the translation of thousands of mRNA reporters simultaneously, we measured the Kozak strength in zebrafish embryos. In support of this approach, we observed a weak correlation between Kozak strength and an in silico-derived Kozak score (Fig. 2e). While our approach has identified conserved activators and repressors (−3 A and −3/−2 U/G) of translation, we have also characterized novel Kozak sequences that differ from the zebrafish or vertebrate consensus Kozaks, including the activating effect of adenosine in the +4 position. Further, we have utilized our random forest regression model to predict the Kozak strength of all zebrafish Kozak sequences (positions −6 to +4). (Table S2). This prediction will improve our annotation of Kozak strength in the zebrafish, as well as our capacity to modulate protein output.
Development requires dynamic spatial and temporal control of translation. Using NaP-TRAP we have identified 5’ UTR cis-regulatory elements that differentially regulate translation in development. For example, we show that the zygotically expressed microRNA miR-430 represses the translation of reporters containing miR-430 seeds in the 5’ UTR. Our results are consistent with the in vitro observations by Lytle et al. in the 5’ UTR62, and support a physiological role for miRNA-mediated regulation of 5’ UTRs during developmental transitions in vivo. We have also identified a translation switch driven by C-rich motifs63. At 2 hpf C-rich k-mers suppress translation, whereas at 6 hpf these k-mers activate translation. Poly-C and poly-pyrimidine-rich elements in the 5’ UTR (PRTE19 and CERT20) have been shown to increase translation in a transcript-specific manner. While it is unclear if similar mechanisms are at play during zebrafish development, we propose two potential mechanisms to explain this novel 5’ UTR mediated translation control (Fig. 6b). First, the expression or loss of a trans-acting factor that binds C-rich tracts may drive differential translation. Although we have identified poly-C-binding protein 2 (PCBP2) as a potential binder of C-rich pentamers enriched in reporters that are active post-ZGA, future work is needed to characterize the role of PCBPs in development. It is important to recognize that members of the poly-pyrimidine tract-binding protein family (PTBP) and components of the EIF3 complex could also function as trans-acting factors as these RBPs have been shown to activate translation by binding C-rich tracts in the 5’ UTR64,65. Interestingly, components of the EIF3 complex have been implicated in the realization of lineage-specific translation programs in early embryogenesis66.
Second, competition between transcripts for a limited pool of trans-acting factors can affect the potency of a given cis-regulatory element. We observe that the relative importance of U-rich sequences depends on the global rate of translation. When translation is low, the prevalence of U-rich sequences is a prominent predictor of translation (Supplementary Fig. 5 d-f). In contrast, as translation increases, the relative importance of U-rich sequences declines, whereas the relative importance of uORFs increases (Supplementary Fig. 5g-i). We propose that this dynamic reflects a change in the availability of the translation initiation machinery49. We speculate that when the supply of ribosomes is limited, the capacity of the 5’ UTR to recruit ribosomes drives translation. In contrast, as the ribosome pool increases, the effect of 5’ UTRs on ribosome recruitment diminishes (Fig. 6b). Consistent with this model, difference in the composition of ribosomes may further limit the pool of available ribosomes or alter the potency of different regulatory sequences67. This competition model can also be employed to explain the differential effect of C-rich tracts on translation. In the early embryo, reporters enriched in U-repeats may recruit the limited supply of ribosomes more efficiently than reporters enriched in C’s. As the supply of translation machinery increases, the effect of competition diminishes, resulting in the efficient initiation of C-rich 5’ UTRs (Fig. 6b). Repressive trans-acting factors may amplify the effect of competition, as in the absence of scanning 40S ribosomes, these factors can be more readily recruited to the 5’ UTR. The strong correlation between translation efficiency and poly-A tail length exemplifies this model50. Prior to gastrulation the pool of PAPBC1 (poly(A) binding protein cytoplasmic 1) is limited, driving competition between transcripts for PAPBC1. This competition is eliminated following gastrulation as the relative abundance of PAPBC1 and active ribosomes increases following zygotic genome activation, resulting in the decoupling of poly-A tail length and translation efficiency.
C-rich motifs are associated with translation activation in both HEK293T cells and zebrafish post-ZGA, whereas these motifs are repressive before genome activation. This observation adds to a growing body of evidence, which suggests that the early embryo employs a unique translation regulatory regime37,48,49,63,68. NaP-TRAP also identifies conserved regulators of translation across both systems whereby poly-U repeats activate translation and uORFs repress translation. Our motif enrichment analyses identified several U-rich binding RBPs, including Tia1, Fubp, HNRNPC, and HuR, expressed in zebrafish and human cell lines (Fig. 4l). Consistent with our results, recent studies from Reimão-Pinto et al. have also identified HuR binding motifs in zebrafish 5’ UTRs that undergo dynamic translation63. In addition, work from Zinshteyn et al has demonstrated in vitro that yeast EIF4G binds U-repeats greater than five nucleotides, driving translation activation69. While we do not know if EIF4G recruitment modulates translation activation in our experiments, the fact that U repeats are activators of translation across timepoints and experimental systems, suggests that this observation may be driven by a component of the canonical translation initiation machinery.
Despite these similarities, in HEK293T cells we observe two major differences in 5’ UTR mediated translation regulation: (1) G-rich motifs are not associated with translation repression, and (2) that there is no correlation between NaP-TRAP translation and structure (Minimum Free Energy; Fig. 7n, o and Supplementary Fig. 6i). Previous studies have demonstrated in human cell lines that G-rich elements and structure in the 5’ UTR repress translation28. These observations coupled with the fact that human 5’ UTRs are more GC-rich than zebrafish70, suggest that the human translation machinery is equipped to overcome repressive G-rich and structural elements in zebrafish 5’ UTRs. Given the well-characterized effect of temperature on RNA structure, it is an intriguing possibility that these differences in translation regulation of G-rich elements may be related to differences in physiological body temperature between zebrafish and humans (28 C and 37 C, respectively). Future work could explore this phenomenon by measuring the translation of 5’ UTRs of multiple species across model systems and temperature ranges.
Lastly, it is important to quantify translation in a frame-specific manner, given the dominant repressive effect and the prevalence of uORFs in vertebrates22. As a proof of principle, we have expanded NaP-TRAP to measure translation in multiple frames simultaneously (Supplementary Fig 2). Through this approach, we can capture the complex interaction between multiple open reading frames within a single transcript and examine how they affect each other’s expression. This application of NaP-TRAP along with other multi-frame approaches (Fig. 8a–f) will enable the field to identify the regulatory potential of UTRs across numerous cell types and cellular states as well as determine how sequence variation in non-coding regions affects gene output in human health and disease.
Fig. 8. The NaP-TRAP method can be adapted to study the translation of multiple ORFs simultaneously in vivo.
a NaP-TRAP is an accessible, versatile, and quantitative method that measures the translation of thousands of reporters simultaneously through the immunocapture of FLAG-tagged nascent peptides. b Through the over-expression of an HA-tagged RBP. NaP-TRAP can be employed in conjunction with an RNA immunoprecipitation experiment to measure the effect of RBP recruitment on translation. c–f NaP-TRAP quantifies translation in a frame-specific manner. Through the incorporation of additional epitope tags in frames 2,3 or ORFs outside of the main open reading frame, the NaP-TRAP method can be utilized to: (1) measure out-of-frame translation in the main ORF (c), (2) detect IRES sequences in an unbiased manner through the use of a bicistronic reporter (d), (3) identify frameshifting elements (e), and (4) quantify stop codon readthrough (f).
Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Antonio J. Giraldez (antonio.giraldez@yale.edu).
Materials availability
Plasmids generated in this study are available from the Lead Contact on request.
Zebrafish maintenance and mating
Wild-type zebrafish embryos were obtained through natural mating of TU-AB strain of mixed ages (5-18 months). Mating pairs were randomly chosen from a pool of 60 males and 60 females allocated for each day of the month. Fish lines were maintained following the International Association for Assessment and Accreditation of Laboratory Animal Care research guidelines and approved by the Yale University Institutional Animal Care and Use Committee (IACUC).
HEK293T cells
HEK293T (ATCC®) cells were grown in a media consisting of Dulbecco’s Modified Eagle Medium (DMEM) (ThermoFisher Scientific #10569010), 10% heat-inactivated Fetal Bovine Serum (FBS) (ThermoFisher Scientific #16140071), 20 mM HEPES (ThermoFisher Scientific #15630080), 2 mM L-Glutamine (ThermoFisher Scientific #25030081), 1x Penicillin/Streptomycin (ThermoFisher Scientific #15140122) at 37 °C with 5% CO2. mRNA transfections were performed using Lipofectamine™ MessengerMAX™ Transfection Reagent (ThermoFisher Scientific #LMRNA008) in accordance with the manufacture’s protocol. See Supplementary Methods S1 for a detailed protocol.
Method details
NaP-TRAP reporter controls
To enable nascent chain immunocapture, a 3xFLAG tag was incorporated after the first 18 nucleotides of GFP-3xAID* (auxin-inducible domain) using an In-Fusion® HD Cloning kit (Takara #638946) (F: 3xFLAG_inf_fwd; R: AID_inf_rev). While AID* domains were included in the initial NaP-TRAP vector to enable future use of an auxin-inducible degron system, this system was not utilized in this study. The vector also included an SP6 promoter sequence and SV40 poly-adenylation signal. NaP-TRAP reporter and dsRED control (plasmid pCS2 + - DsRED) mRNAs were generated using a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340) from linearized reporter plasmids via NotI-HF® (NEB #R3189L) restriction enzyme digest. In vitro transcribed mRNAs were purified using a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to injection.
To validate NaP-TRAP, two reporter experiments were performed. First, to quantify morpholino-mediated translation repression using NaP-TRAP, 100 pg of 3xFLAG-GFP-3xAID mRNA and 75 pg of dsRED mRNA were injected into single-cell zebrafish embryos in the presence or absence of a morpholino targeting the start codon of 3xFLAG-GFP (250 μM, GFP-MO 5’- ACAGCTCCTCGCCCTTGCTCACCAT-3’, Gene Tools LLC). Embryos were collected at 6 and 24 hpf (25 embryos per NaP-TRAP replicate). Embryos collected for NaP-TRAP were flash-frozen in liquid nitrogen prior to sample processing. NaP-TRAP was performed at 6 hpf, whereas immunofluorescence was measured at 24 hpf. Images were quantified using ImageJ71. Second, to assess the capacity of NaP-TRAP to measure microRNA-mediated repression in the 3’ UTR, two additional reporters were generated: (1) 3xFLAG-GFP-3xmiR-430 and (2) 3xFLAG-GFP-3xmiR-204. Three binding sites of either miR-430 or miR-204 were cloned into the 3’ UTR of 3x-FLAG-GFP-3xAID*, using an In-Fusion® HD Cloning kit (Takara #638946) (F: FGFP_inf_fwd; R: 3xmir430_inf_rev and 3xmir204_inf_rev, respectively). Single-cell embryos were injected with 20 pg of both the 3xFLAG-GFP-3xmiR-430 and 3xFLAG-GFP-3xmiR-204 mRNAs, as well as 160 pg of DsRED mRNA. Twenty-five embryos per replicate were collected and flash-frozen in liquid nitrogen at 2 and 4.3 hpf.
To test if NaP-TRAP is able to capture the translational regulatory activity provided by poly(A) tail length in the early embryos, NaP-TRAP experiments with reporters harboring differential poly(A) tail length were performed. The 3xFLAG-GFP vector was amplified using a forward primer against SP6 and reverse primer against the SV40 poly(A) signal containing the differential number of untemplated Ts at the 5’-end (0, 30, 60, or 90). The amplicons were generated by performing a PCR (SP6_F and either 0 A/30 A/60 A/90A_R primers) using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 18 cycles at 98 °C, 62 °C, and 72 °C for 15, 20, and 30 seconds, respectively. The amplicons were gel purified using Zymo Gel DNA recovery (Zymo, D4008) and recovered with DNA clean and concentrator kit (Zymo, D4014). PCR products were used for in vitro mRNA synthesis using mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340) generating NaP-TRAP reporters with various poly(A) tail length. Before injection, the embryos were dechorinated using Pronase (Sigma, P8811-1G) and 1-cell stage embryos were injected with a mix of 40 pg of NaP-TRAP reporter with a specific poly(A) tail length and 40 pg of DsRed control. Twenty-five embryos per replicate were collected and flash-frozen in liquid nitrogen at 2 hpf for a total of three replicates per condition. Translation was then measured by NaP-TRAP and qPCR as mentioned below.
*For methods detailing NaP-TRAP and qPCR translation measurements see sections: (1) NaP-TRAP (Nascent Peptide Translating Ribosome Affinity Purification) and (2) NaP-TRAP qPCR analysis, respectively.
Primer sequences:
(SP6_fwd: GCTTGATTTAGGTGACACTATA, 0A_rev: GTTGTTGTTAACTTGTTTATTGC, 30A_rev: TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGTTGTTGTTAACTTGTTTATTGC, 60A_rev: TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGTTGTTGTTAACTTGTTTATTGC, 90A_rev: TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGTTGTTGTTAACTTGTTTATTGC)
To validate the NaP-TRAP values of reporters with differential poly(A) tail length, western blot experiments were performed. Specifically, 1-cell embryos were injected with a mix of 40 pg of NaP-TRAP reporter with a specific poly(A) tail length and 40 pg of DsRed control as for the NaP-TRAP experiment. At 2 hpf, 30 embryos were mechanically deyolked using P20 tips (Rainin, 30389225) and cells were collected in 1.5 ml tube and kept on ice until all the embryos were collected. Tubes were then spun at 1100 x g for 5 min at 4 C and the excess water removed carefully to not lose the embryonic cell pellets. The cells pellets were lysed in 20 µL of 1X RIPA buffer with gentle vortexing. After cells lysis, the tubes were spun at 16,000 x g for 15 minutes at 4 C and the supernatant was transferred in a new 1.5 mL tube. While the deyolking of early-stage embryos is essential to eliminate yolk lipids and proteins for clean gel migration, achieving reproducibility in this step can be challenging. As a result, standard protein quantification methods may not accurately reflect the number of proteins from the embryonic cells due to the variability in yolk content. Hence, to load the same amount of embryonic cell lysate, a first 5 µL protein samples were loaded on SDS-PAGE gel (BioRAD, 4561084) and ran at 120 volt until the 10 kDa band of the pre-stained ladder (BioRAD, 1610374) reached the bottom. The proteins were transferred from the gel to 0.45 um LF-PVDF membrane (Biorad, 1620260) using a Transblot Turbo (BioRAD, 1704150) and following the manufacturer’s protocol. The membrane was then probed against the DsRed loading control, using anti-DsRED (1:5000) antibody (SCBT, sc-390909) with an overnight incubation at 4 C. In the next morning, the membrane was washed with 20 mL of 1x TBST for 10 minutes for a total of three washes. Next, the membrane was incubated with anti-Mouse (Licor, 926-68070) antibodies for 1 hour at room temperature. The membrane was developed and imaged using an Odyssey Classic Imager (Licor, 9120). The band intensities were quantified using ImageJ71 and used to adjust the sample loading volume for a second SDS-PAGE gel. After adjusting the loading volume of each sample to ensure equal amount of DsRed per sample, proteins were loaded onto a second SDS-PAGE gel. The proteins were then resolved and transferred using the same protocol as for the first gel. The LF-PVDF membrane was probed with anti-DsRed (1:5000) antibody (SCBT, sc-390909) and anti-GFP (1:2000) antibody (Sigma Aldrich, SAB4701015) overnight at 4 C. The membrane was processed and imaged using the same protocol and instrument as the first membrane. The band density was measured using ImageJ71 and graph was plotted using Prism GraphPad.
NaP-TRAP reporter library assembly
To eliminate excess cytoplasmic mature 3xFLAG-GFP protein that may otherwise compete with nascent peptide for immunocapture, a C-terminal PEST domain was incorporated into the NaP-TRAP reporter plasmid using an In-Fusion® HD Cloning Kit (Takara #638946). The 3xFLAG-GFP vector was amplified from the 3xFLAG-GFP-3xAID* plasmid (F: GFP_dd1_inf_fwd, R: GFP_dd1_inf_rev), whereas the PEST domain insert was generated using PCR overlap extension (F: PEST_fwd, R: PEST_rev). For the sake of brevity, 3x-FLAG-GFP-PEST is referred to as 3xFLAG-GFP in the text and figures of this manuscript unless stated otherwise.
NaP-TRAP reporter libraries were constructed using three different PCR reactions:
First, the common coding sequence and 3’ UTR of the reporters were amplified from the 3xFLAG-GFP plasmid, using a forward primer targeting the N-terminus of GFP and a reverse primer targeting the 3’ end of the 3’ UTR (F: 3xFLAG_GFP_fwd, pA-R: pcr_II_pA_rev, sv40-R: sv40_rev). For the Kozak library, a forward primer targeting the sequence immediately downstream of the variable Kozak sequence was used (F: ntrapK_GPF_fwd). All PCR amplicons were gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L).
Second, reporter libraries were amplified from 1 ng of single-stranded DNA oligo pools using KAPA 2X HiFi HotStart ReadyMix (Roche #7958935001) for 10-20 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds, respectively (F: SP6_II_adapt, R: GFP-aug_rev). For initial Kozak library generation see Random Kozak Library section. Each product was PCR purified using a DNA Clean & Concentrator-5 (Zymo #D4014).
Third, to generate a template for in vitro transcription, a PCR overlap extension was performed between the reporter library and the purified 3xFLAG-GFP-PEST amplicon using KAPA HiFi HotStart ReadyMix (Roche #7958935001) (0.1-1 ng of template DNA). After 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds respectively, primers targeting the 5’ SP6 promoter sequence and the 3’ end of the 3xFLAG-GFP-PEST amplicons were added (F: SP6_II_adapt, pA-R: pCS2_3utr_60 A, sv40-R: sv40_rev) followed by an additional 20 cycles with the same conditions. Unless stated explicitly, reporter libraries contained a 60A tail (pA-R: pCS2_3utr_60A). For libraries with an SV40 polyadenylation signal, a reverse primer targeting the 3’ end of the SV40 poly-adenylation signal was used for the amplification of 3xFLAG-GFP-PEST and the assembly of reporter library (R: SV40_rev).
Lastly, templates for in vitro transcription were gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L). Reporter mRNAs were generated using a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). In vitro transcribed mRNAs were purified using a Monarch® RNA Cleanup Kit (NEB #T2040L). All libraries were injected into single-cell zebrafish embryos at 20 pg per embryo.
Random Kozak library
The random Kozak library consisted of an I5 Illumina adaptor (5’- CCCTACACGACGCTCTTCCGATCT-3’) followed by the 5’ UTR of Xenopus beta-globin, seven random nucleotides, six upstream and one downstream of the start codon, and the N-terminus of 3xFLAG GFP. The Kozak library was generated by performing a PCR overlap extension using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds respectively (F: kozak_ntrap_fwd, R: kozak_7_nt_rev, 1 µL of each primer at 100 uM).
Zebrafish 5’UTR library
A custom single-stranded DNA oligo pool consisting of 11,088 oligos was ordered from GenScript (12 K oligo pool). Each 170 nt oligo contained an Illumina I5 (5’- CCCTACACGACGCTCTTCCGATCT-3’) adaptor sequence, a 124-nucleotide variable region, and 22 nt region with homology to the Kozak sequence and N-terminus of 3xFLAG-GFP (5’-GTAAACATGGTGAGCAAGGGCG-3’). The variable region of the 5’ UTR library was generated using a custom script, tiling the 5’ UTRs of 1,775 maternally supplied genes and six IRES sequences (human AQP4, human MYT2, human NRF, human XIAP, EMCV and crTMV) in 124 nucleotide segments every 25 nucleotides.
Validation library (Tetramer repeats)
A custom single-stranded DNA oligo pool consisting of 256 oligos was ordered from Twist Bioscience as part of a 12 K oligo pool. The design of the common regions of library were identical to that of the zebrafish 5’ UTR library. The variable region (124 nucleotides) consisted of repeats of all possible tetramers. Each repeat occurred 21 times and was separated by a dinucleotide spacer. The dinucleotide spacers were repeated in a pattern across the variable region (TC, AC, AG, CG). These dinucleotide spacers were selected to prevent the creation of unintended upstream ORFs.
NaP-TRAP multi-frame library
The NaP-TRAP multi-frame library consisted of an I5 Illumina adaptor (5’- CCCTACACGACGCTCTTCCGATCT-3’) followed by a 5’ UTR containing three potential ORFs in frames 1, 2 and 3, respectively, and a CDS consisting of 3xFLAG-HA-MYC-GFP-Nano-Luciferase. Using two degenerate oligos, a 5’ UTR library, consisting of different combinations of no ORFs, uORFs and oORFs, was assembled through PCR overlap extension, using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds respectively. The overlap product was purified using a DNA Clean & Concentrator-5 (Zymo #D4014).
MF_ORF_fwd – 5’- CCCTACACGACGCTCTTCCGATCTACAGCACGATSGGGGATCTTCAGCTTTMACTGTTCAAGACACTCGATCAT-3’
MF_ORF_rev – 5’-
CTCCTCGCCCTTGCTGACSATGGTGGCGGCGTKAAAGAGCAATACCCCCSATCGACTTTGGATKATGTACAGACTTCSATGATCGAGTGTCTTGAACAGT-3’
3xFLAG-HA-MYC-GFP-Nano-Luciferase was synthesized as a gene fragment from Twist Bioscience. To measure translation in all frames simultaneously, stop codons were eliminated from frames 2 and 3 of the CDS. Epitope tags were designed as three repeats of FLAG-HA and MYC. Single nucleotide spacers were placed in between epitope each tag to modulate the frame each of the epitopes were expressed in. The fragments were assembled using PCR overlap extension, using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 120 seconds respectively. After which primers were added to amplify the assembled fragments (F:mf_gfp_fwd, R: pcr_II_pA_rev) for an addition 10 cycles using the same cycling parameters. The product was then gel purified with a Monarch® DNA Gel Extraction Kit (NEB #T1020L).
Finally, the multi-frame 5’ UTR and 3xFLAG-HA-MYC-GFP-Nano-Luciferase gene fragments were assembled through PCR overlap, using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 120. Forward and reverse primers were added to amplify the assembled reporter library for an additional 10 cycles to add an SP6 promoter sequence and a hard encoded 60 A tail respectively (F: SP6_II_adapt, R: pCS2_3utr_60A). Following the PCR the library was gel extracted using a Monarch® DNA Gel Extraction Kit (NEB #T1020L) and the reporter mRNAs were in vitro transcribed using a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340) following manufacturer’s protocol. Reporter mRNAs were purified using a Monarch® RNA Cleanup Kit (NEB #T2040L) and injected into single-cell zebrafish embryos at 20 pg per embryo.
NaP-TRAP spike-ins
The design of the spike-in reporters was identical to that of the zebrafish 5’ UTR and tetramer validation libraries. Each spike-in reporter contained a 20-nucleotide identifier (see below). NaP-TRAP spike-ins were generated by performing a PCR overlap extension using KAPA HiFi HotStart ReadyMix (Roche #7958935001) (F: ntrap_sp1_fwd, ntrap_sp2_fwd, ntrap_sp3_fwd, ntrap_sp4_fwd, ntrap_sp5_fwd; R: ntrap_sp_rev) (1 µL of each primer at 100 µM) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds, respectively. Amplicons were gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L). Next, the purified amplicons were cloned into the 3xFLAG-GFP-PEST vector using In-Fusion® HD Cloning (Takara #638946) (vector F: ntrap_spike_inf_fwd, R: ntrap_spike_inf_rev). To generate spike-in mRNAs, plasmids were amplified using KAPA HiFi HotStart ReadyMix (Roche #7958935001) (F: Sp6-Il-adapt, R: pCS2_3utr_60A) for 20 cycles at 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds, respectively. Products were then gel purified with a Monarch® DNA Gel Extraction Kit (NEB #T1020L) and then in vitro transcribed with a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). mRNAs were purified with a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to use. Spike-ins were pooled and added at the RNA extraction step at varying amounts of 1, 5, 25, 50, and 125 fg for the 5’ UTR library.
Spike-in #1 TGACGTGGAAGTCGGTCAAG
Spike-in #2 GTCCAGAGACAAAGTCCGGG
Spike-in #3 CACGAGGAGGAACCAGTGAC
Spike-in #4 CTGTTGTTGTGTGAAGGGCG
Spike-in #5 GCTCTCGGTCTCGGAAGAAG
Zebrafish injection
Following zebrafish mating embryos were dechorionated using Pronase (Sigma-Aldrich #10165921001). The embryos were then injected with 20 pg of in vitro transcribed mRNA at the single-cell stage. Post injections, embryos were incubated at 28 C for 2 to 6 hours post fertilization. Stage matched embryos (64-cell and shield respectively) were flash-frozen in liquid nitrogen. For the Kozak, zebrafish 5’ UTR, and the validation libraries 50 embryos were collected per replicate. For the multi-frame library, 75 embryos with collected per replicate. Frozen embryos were lysed in 500 µL of lysis buffer (see below).
HEK293T RNA transfection
For NaP-TRAP HEK293T cells were plated on 6-well plates coated with poly-d-lysine 24 hours prior to transfection, (200,000-300,000 cells per well). In accordance with the manufactures protocol, the cells were transfected with 2 ug of reporter library per well the using Lipofectamine™ MessengerMAX™ (ThermoFisher Scientific # LMRNA008). For polysome profiling, HEK293T cells were grown on 10 cm2 cell culture plates coated with poly-d-lysine (1-2 million cells per plate). Given the larger number of cells per replicate, cells were transfected with 10 ug of reporter library per plate. After one hour at 37 C, the cells were washed with 1x DPBS and fed with fresh pre-warmed media. Post twelve hours of transfection, HEK293T cells were treated with cycloheximide at a final concentration of 100 ug/mL for 10 minutes at 37 C and washed on ice three times with 1 mL of ice-cold PBS (with 100 ug/mL cycloheximide) prior to lysis. Next, 500 uL of lysis buffer (see below) was added to each well/plate. HEK293T cells were mechanically scrapped using a cell scraper, prior to lysate collection.
Representation of reporter mRNAs
For each NaP-TRAP experiment, 20 pg of mRNA reporter library were injected into each embryo. Given that the mRNA reporters are ~1.2 kb long and the molecular weight of a single base is around 340 daltons, the molecular weight of each reporter mRNA is estimated to be ~408 kilodaltons. Injecting 20 pg of mRNA should result in roughly 29.5 million mRNAs or in the case of the zebrafish 5’ UTR library (11,088 reporters) 2662 mRNAs per reporter per embryo. Given that 50 embryos per replicate were collected, there is estimated to be roughly 133,118 copies of each reporter at the start of each experiment. In the zebrafish 5’ UTR library experiments, at 6 hpf over 75% of reporters passed the QC (100 unique reads in the input), suggesting that injecting 20 pg per embryo provided sufficient coverage.
NaP-TRAP (Nascent Peptide Translating Ribosome Affinity Purification)
For a more detailed protocol, see Methods S1- A Detailed Protocol for NaP-TRAP.
Prior to performing the NaP-TRAP, the following buffers were prepared: 10x salt buffer (150 mM Tris 7.4, 1 M NaCl, 100 mM MgCl2), bead wash buffer (1x salt buffer, 2 mM DTT, 1% triton-X 100), lysis buffer (1x salt buffer, 1% triton-X 100, 2 mM DTT, 100 µg/mL Cycloheximide, 40 U/mL RNaseOUT™ Recombinant Ribonuclease Inhibitor (ThermoFisher Scientific #10777019), 1x cOmplete™ EDTA-free Protease Inhibitor (Roche #11873580001)), and NaP-TRAP wash buffer (lysis buffer + 400 mM NaCl).
Zebrafish embryo and HEK293T cell lysates were incubated for 10 minutes at 4 °C. The lysate was passed through a 25-guage needle (5-10 times). Next, the sample was centrifuged at 16,000 g for 5 minutes at 4 °C. The supernatant was transferred to a new Eppendorf tube and 2 µL of DNase I (NEB #M0303L) was added. After a 15 minutes incubation at 4 °C, the samples were diluted to 1 mL using additional lysis buffer and 75 µL of lysate was collected from each sample to serve as an input.
To capture tagged nascent chains of reporter mRNAs, an immunoprecipitation using anti-FLAG magnetic beads was performed. Magnetic beads were purchased from three suppliers during the course of the study due to supply chain issues (ANTI-FLAG® M2 Magnetic Beads, Millipore® #M8823; Anti-Flag Magnetic Beads, BioTools LLC #B26102; Pierce™ Anti-DYKDDDDK Magnetic Agarose, Thermo Fisher Scientific #88836). For the zebrafish and human cell experiments 10 µL and 20 µL of beads (binding capacity of >0.8 mg of FLAG peptide / mL) were utilized per replicate, respectively. The magnetic beads were washed with 800 µL of bead wash buffer three times prior to being added to the lysis solution. Upon addition of the lysate sample to magnetic beads, tubes were placed on a rotator at 4 °C for 2 hrs. Following incubation and bead capture, the beads were washed with 800 µL of NaP-TRAP wash buffer three times. The beads (pulldown) and the inputs were then resuspended in 1 mL of Trizol (Invitrogen #15596-018). For the zebrafish 5’ UTR and tetramer validation libraries, spike in reporters were added. RNA extractions were performed in accordance with the manufacture’s protocol. RNA pellets were resuspended in 11 µL of nuclease-free H2O.
Multi-frame NaP-TRAP
The protocol of multi-frame NaP-TRAP mirrors that of single-frame NaP-TRAP with the exception of the pulldown step. More specifically, following the collection of the input, the cell lysate was divided into three equal parts for the immunoprecipitation of FLAG, HA, and MYC respectively. Each fraction was diluted to 1 mL with lysis buffer. Next, for each replicate 10 μl of Anti-FLAG (Pierce™ Anti-DYKDDDDK Magnetic Agarose, Thermo Fisher Scientific #88836), Anti-HA (Pierce™ Anti-HA Magnetic Beads # 88836), and Anti-MYC (Pierce™ Anti-c-Myc Magnetic Beads #88842) beads were washed and then added to their respective fractions. As described above the beads were incubated with the lysate for 2 hours at 4 C prior to being washed three times with 800 μl of NaP-TRAP wash buffer. The beads (pulldown) and the inputs were then resuspended in 1 mL of Trizol (Invitrogen #15596-018) and spike in reporters were added. RNA extractions were performed in accordance with the manufacture’s protocol. RNA pellets were resuspended in 11 µL of nuclease-free H2O.
Polysome profiling
HEK293T cells were incubated with 100 μg/ml cycloheximide for 10 min at 37 °C to arrest translation. After washing with prechilled 1x DPBS supplemented with 100 μg/ml cycloheximide, cells were lysed in NaP-TRAP lysis buffer by manual scrapping using a cell scraper. Cell lysates for polysome profiling were prepared following the same protocol as done for NaP-TRAP until the immunocapture stage. 1 ml of cell lysates were layered onto 10–60% sucrose gradients, prepared with lysis buffer, in a thin-walled ultracentrifuge tube and centrifuged in a Beckman SW‐40Ti rotor at 155,000 g. for 3 h. Gradients were fractionated in a Teledyne ISCO fractionator using >60% sucrose chase solution. Absorbance was monitored at 254 nm to obtain the polysome profile and a total of twenty-seven 500 μl fractions were collected at a flow rate of 1 ml/min. Fractions corresponding to the single monosome or polysome peak were pooled postcollection prior to RNA extraction.
RNA was extracted using TRIzol (Thermo Fisher Scientific) reagent. Briefly, 500 μl was added to each fraction and vortexed. 100 µl of chloroform was added and the mixture was vortexed and then incubated for 2-3 min at room temperature. Fractions were spun at 12,000 g. for 30 min and the aqueous phase was transferred to a fresh Eppendorf tube. To the aqueous phase, 250 μl of isopropanol and 1 μl of GlycoBlue was added and vortexed. After incubating at room temperature for 10 mins, the samples were centrifuged at 14,000 g for 30 mins. The RNA pellets were washed with 80% ethanol, dried and resuspended in 11 ul of nuclease-free water.
Library preparation for next-generation sequencing
For a detailed protocol see Methods S1- A Detailed Protocol for NaP-TRAP.
Reverse transcription primers (4 µM total) targeting the N-terminus of 3xFLAG GFP were added to purified input and pulldown RNAs (5’-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-CTAC-10N UMI-TAAC-6 nt sample barcode-GCGTCATGGTCTTTGTAGTCCTC-3’; see NaP-TRAP barcode RT primers in Supplementary Table 1). These primers contained a 6 nucleotide sample barcode and a 10 nucleotide Unique Molecular Identifier (UMI) to allow for demultiplexing and read deduplication, as well as a 3’ I7 Illumina adaptor sequence. To increase library complexity the primer pairs were staggered by 1 nt (e.g., ntrap_RT_b1.1, ntrap_RT_b1.2). Reverse transcription was performed using the Superscript III kit (Invitrogen #18080044) in accordance with manufacturer’s instructions. Reverse transcription reactions were performed at 55 °C. cDNA from replicates were pooled and purified by adding AMPure XP Reagent (Beckman Coulter #A63881) at 1.8x of the original sample volume. Illumina I5 and I7 forward and reverse primers containing a 10-nucleotide index (see below) were utilized to amplify cDNA libraries via PCR (Kappa Polymerase Master Mix). To reduce the number of PCR duplicates 12-18 cycles were utilized. Amplicons were purified by adding AMPure XP Reagent (Beckman Coulter #A63881) at 0.9x the volume of the PCR reaction. Libraries were sequenced on Illumina NovaSeq 6000 platform.
Illumina I5 primer:
5’- AATGATACGGCGACCACCGAGATCTACAC-10 nt index-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’
Illumina I7 primer:
5’- CAAGCAGAAGACGGCATACGAGAT-10 nt index-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’
NaP-TRAP-qPCR analysis
Following NaP-TRAP, cDNA was synthesized using random hexamers (ThermoFisher Scientific #N8080127) and SuperScript™ III Reverse Transcriptase (Invitrogen #18080044) following the manufacturer’s protocol. Translation values were determined by qPCR using the Power SYBR™ Green PCR Master Mix (Applied Biosystems #4367659). Levels of reporters in the input and pulldown were normalized using the 2−∆∆Ct method72, where the NaP-TRAP reporter was the target and DsRed the control. Translation was calculated as a ratio of fold enrichment in the pulldown relative to the input.
Primers utilized in qPCR:
-
-
Translation blocking morpholino (GFP_qpcr_fwd, GFP_qpcr_rev)
-
-
MicroRNA seeds in 3’ UTR (3xmir-430_qpcr_fwd, 3xmir-204_qpcr_fwd, T7_qpcr_rev)
-
-
U/C rich reporters (igf1ra_wt_qPCR_fwd, igf1ra_mut_qPCR_fwd, asun_wt_qPCR_fwd, asun_mut_qPCR_fwd, sich211_wt_qPCR_fwd, sich211_mut_qPCR_fwd, atp6v0_wt_qPCR_fwd, atp6v0_mut_qPCR_fwd, GFP_qPCR_5p_rev)
-
-
DsRed (dsRED_qpcr_fwd, dsRED_qpcr_rev)
Dual luciferase assays
To validate NaP-TRAP translation measurements, single-cell zebrafish embryos were co-injected with mRNAs encoding nano and firefly luciferase (0.5 pg of nano / 19.5 pg of firefly). Embryos were collected at either 2 hpf or 6 hpf and frozen in liquid nitrogen (5 embryos per replicate). Nano and firefly luciferase activities were measured using the Nano-Glo® Dual-Luciferase® Reporter Assay System (Promega #N1610). Firefly luciferase activity was utilized to normalize nano luciferase measurement across reporters.
NaP-TRAP NanoLuc reporter construction
Firefly luciferase was amplified from a pCS2 + -Fluc plasmid using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 20 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 50 seconds (F: SP6_ext_fwd, R: SV40_60A_rev), respectively. Products were gel purified with a Monarch® DNA Gel Extraction Kit (NEB #T1020L) and then in vitro transcribed with a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). mRNAs were purified with a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to injection.
We substituted NanoLuc luciferase (amplified from a gBlocks™ Gene Fragment, Integrated DNA Technologies IDT) for GFP in the 3xFLAG-GFP-PEST vector using In-Fusion® HD Cloning (Takara #638946) (3xFLAG-PEST vector F: PEST_fwd, R: FLAG_rev_inf; NanoLuc insert F: FLAG-Nluc_inf_fwd, R: Nluc_inf_rev). Note: The PEST domain was included in NanoLuc constructs to reduce the half-life of the NanoLuc protein and thereby improve the sensitivity of the assay. To generate NanoLuc reporters 3xFLAG-NanoLuc-PEST was amplified using KAPA HiFi HotStart ReadyMix (Roche #7958935001) (F: 3xFLAG_GFP_fwd R: pcr_II_pa_rev) and gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L).
Kozak NanoLuc reporters
To construct Kozak NanoLuc reporters, 3xFLAG-NanoLuc (amplicon from the previous section) was amplified with seven different forward primers (ntrap_k1_fwd, ntrap_k2_fwd, ntrap_k3_fwd, ntrap_k4_fwd, ntrap_k5_fwd, ntrap_k6_fwd, and ntrap_k7_fwd) and a common reverse primer (pcr_II_pa_rev) using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 20 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 45 seconds, respectively. Products were gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L). To add an SP6 promoter sequence and a 60 A hard-encoded tail, the purified product (1 ng) was amplified using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 20 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 45 seconds, respectively (F: Sp6-Il-adapt, R: pCS2_3utr_60A). Products were gel purified with a Monarch® DNA Gel Extraction Kit (NEB #T1020L) and then in vitro transcribed with a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). mRNAs were purified with a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to injection. Note, the input of one of the replicates of the Kozak reporter K4 was below the read filter cutoff in the NaP-TRAP Kozak Library experiment.
Generating NanoLuc reporters using PCR overlap
The 5’ UTRs of the Zebrafish 5’ UTR library validation reporters and 4xmiR-430 NanoLuc Reporters were generated using PCR overlap (see below for forward and reverse primers), using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds, respectively (1 µL of each primer at 100 µM). The extension products were then gel purified using a Monarch® DNA Gel Extraction Kit (NEB #T1020L). In contrast, the 5’ UTRs of the NanoLuc reporters, which were used to benchmark NaP-TRAP to polysome profiling (Supplementary Fig. S1d-e) were purchased as gene fragments from Twist Biosciences.
Full-length reporters were constructed by performing a PCR overlap extension of the 5’ UTR products with the NanoLuc amplicon described in the Cloning NanoLuc section using KAPA HiFi HotStart ReadyMix (Roche #7958935001). After 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 40 seconds, respectively, primers targeting the SP6 promoter sequence (F: Sp6-Il-adapt), and the 3’ end of the nanoLuc amplicon were added (R: pCS2_3utr_60A). The PCR reaction was continued for an additional 20 cycles at the same cycle conditions. Products were gel purified with a Monarch® DNA Gel Extraction Kit (NEB #T1020L) and then in vitro transcribed with the mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). mRNAs were purified with a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to injection.
Polysome validation Supplementary Fig. 1 (R1 = ENSDART00000104444_9, R2 = ENSDART00000054416_18, R3 = ENSDART00000079144_6, R4 = ENSDART00000132526_5, R5 = ENSDART00000147616_4, R6 = ENSDART00000156579_17, R7 = ENSDART00000091683_7, R8 = ENSDART00000146852_0, R9 = ENSDART00000061736_2, R10 = ENSDART00000006419_2, R11 = ENSDART00000035538_2, R12 = ENSDART00000067843_4).
Zebrafish 5’ UTR library validation reporters (R13: slc16a1_fwd / slc16a1_rev, R14: spred1_fwd / spred1_rev, R15: eif4ebp2_fwd / eif4ebp2_rev, R16: fth1a_fwd / fth1a_rev, R17: atp6v0_wt_fwd / atp6v0_wt_rev, R18: wdr41_fwd / wdr41_rev, R19: slc38a7_fwd / slc38a7_rev, R20: igf1ra_fwd / igf1ra_rev, R21: atp2b4_fwd / atp2b4_rev).
4xmiR-430 NanoLuc- (WT-F: 4xmiR-430-fwd, WT-R: 4xmiR-430-rev; MUT-F: 4xmiR-430-MUT-fwd, MUT-R: 4xmiR-430-MUT-rev).
C and U rich reporters
C- and U-rich wild-type and mutant reporters were generated through PCR overlap extension (Primers used: igf1ra_wt_fwd, igf1ra_wt_rev; igf1ra_mut_fwd, igf1ra_mut_rev; asun_wt_fwd, asun_wt_rev; asun_mut_fwd, asun_mut_rev; sich211_wt_fwd, sich211_wt_rev; sich211_mut_fwd, sich211_mut_R atp6v0_wt_fwd, atp6v0_wt_rev; atp6v0_mut_F atp6v0_mut_R) using KAPA HiFi HotStart ReadyMix (Roche #7958935001) for 10 cycles of 98 °C, 60 °C, and 72 °C for 15, 20, and 30 seconds, respectively (1 µL of each primer at 100 µM). The extension products were then gel purified using a using Monarch® DNA Gel Extraction Kit (NEB #T1020L).
Extension products were cloned into the 3xFLAG-GFP-3xAID* vector using In-Fusion® HD Cloning (Takara #638946) (vector F: 3xFLAG_GFP_F R: I5_inf_rev). Plasmids were digested with NotI-HF® (NEB #R3189L) and in vitro transcribed with a mMESSAGE mMACHINE™ SP6 transcription kit (Invitrogen™ #AM1340). mRNAs were purified with a Monarch® RNA Cleanup Kit (NEB #T2040L) prior to injection.
Quantification and statistical analysis
Read trimming and reporter mapping
Reporter libraries were sequenced on an Illumina NovaSeq 6000 platform. Sequencing data were stored using LabxDB73. Paired-end reads were trimmed and demultiplexed using ReadKnead (https://github.com/vejnar/ReadKnead). Barcodes identifying replicates and UMIs were extracted from read two.
Kozak library
Using ReadKnead the 5’ common region of the Kozak reporters were removed. Next, we extracted the Kozak sequence (NNNNNNATGN) and discarded sequences that did not include ATG in positions 7-10. We then utilized a custom script to eliminate PCR duplicates. UMIs were considered identical if they had a Hamming Distance less than 2. For the Kozak library, reporters were counted. Reporters with indels in the Kozak sequence or reporters without an AUG in the appropriate position were eliminated.
Zebrafish 5’ UTRs
For the zebrafish 5’ UTR and validation libraries, reads were mapped to a library-specific index using Bowtie274. PCR duplicates were eliminated using UMIs. UMIs were considered identical if they had a Hamming Distance less than 2.
Calculation of NaP-TRAP-derived translation values
Reads for each experiment were normalized by dividing the read counts of each reporter by the sum of the total number of reads mapped to the spike-ins. In the 5’ UTR reporter library (pA and sv40 in zebrafish) spike-in #3 was eliminated from the analysis, because in some of the samples the read counts for spike-in #3 did not correlate with amount of spike-in added. In the absence of spike-ins, read counts were normalized based on the total number of mapped reads per replicate (reads per million, RPM). Translation values were calculated as a ratio of reads in the pulldown relative to reads in the input. Translation values were only included in the downstream analyses if the input contained greater than or equal to 100 unique reads across all replicates.
Translation values were calculated as a ratio of reads in the pulldown relative to reads in the input. Translation values were only included in the downstream analyses if the input contained greater than or equal to 100 unique reads across all replicates.
Calculation of mean ribosome load for polysome profile
Read counts across all fractions sequenced were normalized to account for differences in the number of reads sequenced across the different fractions. Normalized reads were derived by dividing each read count by the total number of mapped reads in that fraction. To get a normalized measure of distribution of reads across the polysome fractions, normalized read counts for each reporter was divided by the total normalized reads obtained for that reporter across all the fractions collected. Finally, the mean ribosome load (MRL) for a reporter was calculated as the sum of the product of polysome profile normalized read counts with the corresponding ribosome count. (1 for 80S, 2 for disome, etc.).
Calculation of translation residual
To determine whether there were systemic differences between NaP-TRAP-derived translation values and MRL, a linear regression was performed (y = NaP-TRAP translation, x = MRL). To examine the bias of different subsets of reporter mRNAs, the residual was calculated (residual = NaP-TRAP derived translation – predicted translation). Difference in the culminative distributions of translation residuals were evaluated using a Mann-Whitney U-test for the following reporter mRNAs; reporters containing: (1) oORFs, (2) uORFs, and (3) no upstream ORFs.
Random forest regression models
Random forest regression models were employed to predict translation (scikit-learn 1.3; RandomForestRegressor)75. For the Kozak library, features were generated by one-hot encoding positions -6 to -1 and position +4 of each reporter sequence. In contrast, for the 5’ UTR library, k-mer counts (1-6 nucleotides) and uORF features were generated using a custom python script. For the 5’ UTR library, features were filtered by calculating the Spearman Rank Correlation Coefficient (SPR) between each feature and translation prior to model training. Features that had a correlation greater than 0.05 or less than -0.05 were included in the random forest model.
To prevent model overfitting, the data were divided randomly into two groups: a test and training set, comprised of 30% and 70% of the reporters, respectively. To optimize model parameters, the training data were divided into five different groups of equal size and a 5-fold cross-validation was performed. The following parameters were optimized using an exhaustive grid search (n_estimators: 20, 100, 200; max_features: 10, 20 and 30 percent of supplied features; max_depth: 3, 5, 7 and min_samples_split: 2, 4, 8). Bootstrapping was employed to select samples used to train each tree. The predictive power of the model was assessed using the test set. It should be noted that the random forest cannot predict translation values higher than the maximum value of the training set. Hence the sharp cut-offs in Fig. 3e and Supplementary Fig. 8j. Lastly, a permuted feature importance analysis was performed to identify the features with the greatest predictive power.
Quantifying the effect of the structure of translation
The Minimum Free Energy (MFE, structure) of reporter mRNAs was determined using the Vienna RNAFold program.57 Given that in silico RNA structure predictions are more accurate for shorter RNAs and the fact that the coding sequence and 3’ UTR of reporter mRNAs are identical, the first 200 nucleotides of each reporter were utilized to determine MFE. To investigate the effect of structure on translation the Pearson Correlation Coefficient was measured between translation (2 hpf and 6 hpf) and MFE. Reporters were divided further into groups based on whether they contained: no upstream start codons, uORFs, out-of-frame oORFs, and in frame oORFs. For each group the correlation between structure and translation was determined.
Differential motif enrichment analysis
Reporters were ranked based on their translation at 2 and 6 hpf. Using the sum and difference of these rankings across timepoints, four groups of reporters were generated: (1) repressed, (2) active, (3) repressed post-ZGA (active in HEK293T cells), and (4) active post-ZGA (active in zebrafish). Repressed and active reporters constituted the top and bottom 10% of reporters based on the sum of their ranks at 2 and 6 hpf, respectively, whereas the repress post-ZGA and active post-ZGA, were the top and bottom 10% of groups based on the difference between their ranks at 2 and 6 hpf. A differential motif enrichment analysis was performed on each group. Fold enrichment values were determined by dividing the count of each k-mer in the reporter group by the count of the k-mer in the library, whereas the significance of the fold-change was determined using a hypergeometric test (Bonferroni corrected p-value threshold).
To generate motifs from the kmers enriched in each of the reporter groups defined above, we performed hierarchical clustering (scipy 1.14.0 cluster.heirarchy.linkage) using the UPGMA (unweighted pair group method with arithmetic mean) method76. Distances between kmers were computed using a modified hamming distance metric, which minimized the hamming distance between two kmers by sliding one kmer over the other. Kmers were clustered into motifs. Clusters were combined if their average distance was less than 2.5. Clusters were assembled into motifs based on the position which minimized their modified hamming distance. The information content of motifs was plotted using a custom script informed by the source code of LogoMarker77.
To determine the regulatory potential of these motifs, reporters in the zebrafish 5’ UTR library enriched and depleted in each motif were identified (top and bottom 20%). Enrichment and depletion of motifs were based on the sum of counts of kmers that were clustered to form each motif. A Mann-Whitney U-test was utilized to determine whether there was a significant difference in the distributions in the translation at 2 hpf, 6 hpf, and in HEK293T cells, as well as the delta translation (translation 6 hpf / translation 2 hpf, translation 2hpf / HEK293T, and translation 6 hpf / HEK293T) for reporters enriched and depleted in each motif.
Using Zebrafish eCLIP data to identify candidate RBPs
To identify candidate RBPs, hexamers significantly enriched in the reporters groups described above were compared to the top 10 hexamers enriched in eCLIP experiments from a previous study investigating the regulatory potential of RBPs highly expressed in the early stages of zebrafish development5. To identify the top 10 motifs enriched for each of the RBPs tested, the rank of hexamers across each of the replicates were summed. The top 10 hexamers with the lowest summed rank, which were not enriched in the control groups of eCLIP experiments, were utilized for further analysis. A hypergeometric test was performed to determine whether the overlap between hexamers enriched in reporter groups described above and each of the RBPs was significant (Bonferroni corrected p-value threshold).
Using STREME and Tomtom to identify motifs and candidate RBPs modulating translation
STREME was also employed to identify motifs modulating differential translation59. Reporter sequences (the insert sequence) for each of the groups described above were utilized as the input, whereas all reporters which passed QC served as the control. STREME generated motifs with a minimum size of 5 and a maximum size of 12 nucleotides (meme-streme --minw 5 --maxw 12 --thresh 0.05 --rna --align right). Motifs were deemed significant if they had an E-value less than 0.05. Each of the significant motifs identified by STREME were compared to a database of human RBPs58 using Tomtom60. Candidate RBPs were assigned to motifs if they had an E-value was less than 0.05.
miRNA complementarity analysis
miR-430 (GCACUU) and miR-1 (CAUUCC) seeds were identified in the reporter library. The Vienna RNAcofold program57 was utilized to measure the complementarity between the section of the reporter mRNA (20 nt upstream and 7 nt downstream of the 5’ end of seed site) and the microRNA. For miR-430 the miRNA species with the highest complementary was selected for downstream analysis. Reporters with multiple seed sequences were excluded from the downstream analysis. The relationship between complementarity and change in translation (Translation 6 hpf / 2 hpf) was determined by calculating the Pearson’s Correlation Coefficient. To assess the importance of the microRNA seed. The complementarity analysis described above was repeated for reporters containing a seed mismatch for miR-430 (GCBCUU) and miR-1 (CAVUCC).
miR-430a: 5’-UAAGUGCUAUUUGUUGGGGUAG-3’
miR-430b: 5’-AAAGUGCUAUCAAGUUGGGGUAG-3’
miR-430c: 5’-UAAGUGCUUCUCUUUGGGGUAG-3’
miR-1-1 / mir-1-2: 5’-UGGAAUGUAAAGAAGUAUGUAU-3’
Feature rank analysis
To generate the feature rank plot (Fig. 6a), features (k-mer counts of four nucleotides or fewer) were ranked based on the mean difference in translation between reporters enriched and depleted in the feature, top and bottom 20% respectively, at 2 and 6 hpf. Given the prominent effect of upstream open reading frames on translation, reporters with uORFs were excluded from the analysis. Features were only included in the analysis if there was a significant mean difference in translation at either timepoint (Mann-Whitney U test with Bonferroni corrected p-value threshold).
Statistical analyses
The plots and statistical analyses in Figs. 1b–d, 5i and 6c–f were generated using GraphPad Prism. All other analyses unless otherwise stated were performed using custom scripts written in Python 3. Plots were generated using the Matplotlib package78. Venn diagrams were generated using Matplot-venn (https://github.com/konstantint/matplotlib-venn). Statistical analyses were performed using the SciPy76 and NumPy79 packages, whereas the random forest analysis was performed using the scikit-learn package75. Feature and experimental data were stored using an SQLite database (https://www.sqlite.org/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank C. Castaldi, E. Sykes, B. Sullivan, and B. De Kumar from the Yale Center for Genome Analysis for sequencing support; S. Dube, T. Gerson and D. Karpel for technical help; D. Musaev, L. Weiss, S. Hiley, and I. Carmi for feedback on the manuscript; and all members of the Giraldez lab for feedback and support. We would like to acknowledge our funding sources: National Institutes of Health grant R01 HD100035 (A.J.G.), National Institutes of Health grant R35 GM122580 (A.J.G.), National Institutes of Health grant R00 HD093873 (J.-D.B.), and National Institutes of Health grant R35 GM146883 (J.-D.B).
Author contributions
J.-D.B. conceived the NaP-TRAP principle. E.C.S., J.-D.B., and A.J.G. designed the project. E.C.S. and J.-D.B performed the experiments. E.C.S. developed the pipeline to process and analyze NaP-TRAP data. C.E.V. advised E.C.S. on computational methods and supplied reading trimming and demultiplexing software. E.C.S. and A.J.G. wrote the paper. S.K and H.L. contributed experimental analysis and intellectual input. A.G. performed the poly-A tail reporter assay, whereas N.N. performed the sucrose gradient fractionation. J.-D.B., S.K., H.L., and A.G. provided feedback on the paper.
Peer review
Peer review information
Nature Communications thanks Maria Barna and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The sequencing data generated in this study have been deposited in the NCBI under the BioProject ID PRJNA1188270. The processed count tables from the sequencing data are available in Supplementary Data 2-4 or on the Giraldez Lab datahub (https://www.giraldezlab.org/data/).
Code availability
All scripts used to process and analyze NaP-TRAP data will be released at the time of publication (https://github.com/ecstrayer/nap-trap_paper).
Competing interests
E.C.S., J.-D.B., S.K., and A.J.G are inventors on a provisional patent application filed by Yale University with the US patent office covering the NaP-TRAP method and the sequences described here. A.J.G. is founder of and has equity an equity interest in RESA Therapeutics, Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jean-Denis Beaudoin, Email: Jdbeaudoin@uchc.edu.
Antonio J. Giraldez, Email: Antonio.giraldez@yale.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55274-y.
References
- 1.de Boer, C. G. et al. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol.38, 56–65 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogard, N., Linder, J., Rosenberg, A. B. & Seelig, G. A Deep Neural Network for Predicting and Engineering Alternative Polyadenylation. Cell178, 91–106.e123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yartseva, V., Takacs, C. M., Vejnar, C. E., Lee, M. T. & Giraldez, A. J. RESA identifies mRNA-regulatory sequences at high resolution. Nat. Methods14, 201–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabani, M., Pieper, L., Chew, G. L. & Schier, A. F. A Massively Parallel Reporter Assay of 3’ UTR Sequences Identifies In Vivo Rules for mRNA Degradation. Mol. Cell68, 1083–1094 e1085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vejnar, C. E. et al. Genome wide analysis of 3’ UTR sequence elements and proteins regulating mRNA stability during maternal-to-zygotic transition in zebrafish. Genome Res. 29, 1100–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litterman, A. J. et al. A massively parallel 3’ UTR reporter assay reveals relationships between nucleotide content, sequence conservation, and mRNA destabilization. Genome Res. 29, 896–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buccitelli, C. & Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Teixeira, F. K., & Lehmann, R. Translational Control during Developmental Transitions. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a032987 (2019). [DOI] [PMC free article] [PubMed]
- 9.Mignone, F., Gissi, C., Liuni, S., & Pesole, G. Untranslated regions of mRNAs. Genome Biol 3, REVIEWS0004. 10.1186/gb-2002-3-3-reviews0004 (2002). [DOI] [PMC free article] [PubMed]
- 10.Gebauer, F., Preiss, T. & Hentze, M. W. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb. Perspect. Biol.4, a012245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leppek, K., Das, R. & Barna, M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. cell Biol.19, 158–174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. cell Biol.11, 113–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey, J. W. B., Sonenberg, N., & Mathews, M. B. Principles of Translational Control. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a032607 (2019). [DOI] [PMC free article] [PubMed]
- 15.Pelletier, J., Kaplan, G., Racaniello, V. R. & Sonenberg, N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5’ noncoding region. Mol. Cell Biol.8, 1103–1112 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bugaut, A. & Balasubramanian, S. 5’-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res40, 4727–4741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casey, J. L. et al. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science240, 924–928 (1988). [DOI] [PubMed] [Google Scholar]
- 18.Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truitt, M. L. et al. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell162, 59–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chew, G. L., Pauli, A. & Schier, A. F. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun.7, 11663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnstone, T. G., Bazzini, A. A. & Giraldez, A. J. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J.35, 706–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvo, S. E., Pagliarini, D. J. & Mootha, V. K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA106, 7507–7512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc.7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingolia, N. T., Hussmann, J. A., & Weissman, J. S. Ribosome Profiling: Global Views of Translation. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a032698 (2019). [DOI] [PMC free article] [PubMed]
- 26.Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet15, 205–213 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Noderer, W. L. et al. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol.10, 748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia, L. et al. Decoding mRNA translatability and stability from the 5’ UTR. Nat. Struct. Mol. Biol.27, 814–821 (2020). [DOI] [PubMed] [Google Scholar]
- 29.May, G. E. et al. Unraveling the influences of sequence and position on yeast uORF activity using massively parallel reporter systems and machine learning. Elife12. 10.7554/eLife.69611 (2023). [DOI] [PMC free article] [PubMed]
- 30.Lin, Y. et al. Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Res47, 9358–9367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weingarten-Gabbay, S. et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science351. 10.1126/science.aad4939 (2019). [DOI] [PubMed]
- 32.Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol.41, 262–272 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina-Munoz, S. G. et al. Crosstalk between codon optimality and cis-regulatory elements dictates mRNA stability. Genome Biol.22, 14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamble, C. E., Brule, C. E., Dean, K. M., Fields, S. & Grayhack, E. J. Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell166, 679–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauger, D. M. et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl Acad. Sci. USA116, 24075–24083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vainberg Slutskin, I., Weingarten-Gabbay, S., Nir, R., Weinberger, A. & Segal, E. Unraveling the determinants of microRNA mediated regulation using a massively parallel reporter assay. Nat. Commun.9, 529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang, K., Ly, J. & Bartel, D. P. Control of poly(A)-tail length and translation in vertebrate oocytes and early embryos. Dev. Cell59, 1058–1074.e1011 (2024). [DOI] [PubMed] [Google Scholar]
- 38.Schuster, S. L. et al. Multi-level functional genomics reveals molecular and cellular oncogenicity of patient-based 3’ untranslated region mutations. Cell Rep.42, 112840 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim, Y. et al. Multiplexed functional genomic analysis of 5’ untranslated region mutations across the spectrum of prostate cancer. Nat. Commun.12, 4217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griesemer, D. et al. Genome-wide functional screen of 3’UTR variants uncovers causal variants for human disease and evolution. Cell184, 5247–5260.e5219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sample, P. J. et al. Human 5’ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol.37, 803–809 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, W. et al. Massively parallel functional annotation of 3’ untranslated regions. Nat. Biotechnol.32, 387–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niederer, R. O., Rojas-Duran, M. F., Zinshteyn, B. & Gilbert, W. V. Direct analysis of ribosome targeting illuminates thousand-fold regulation of translation initiation. Cell Syst.13, 256–264.e253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuperus, J. T. et al. Deep learning of the regulatory grammar of yeast 5’ untranslated regions from 500,000 random sequences. Genome Res27, 2015–2024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dvir, S. et al. Deciphering the rules by which 5’-UTR sequences affect protein expression in yeast. Proc. Natl Acad. Sci. USA110, E2792–E2801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell135, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giraldez, A. J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science312, 75–79 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Bazzini, A. A., Lee, M. T. & Giraldez, A. J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science336, 233–237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leesch, F. et al. A molecular network of conserved factors keeps ribosomes dormant in the egg. Nature613, 712–720 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang, K., & Bartel, D. P. The molecular basis of coupling between poly(A)-tail length and translational efficiency. Elife10. 10.7554/eLife.66493 (2021). [DOI] [PMC free article] [PubMed]
- 51.Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell44, 283–292 (1986). [DOI] [PubMed] [Google Scholar]
- 52.Grzegorski, S. J., Chiari, E. F., Robbins, A., Kish, P. E. & Kahana, A. Natural variability of Kozak sequences correlates with function in a zebrafish model. PLoS One9, e108475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozak, M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol.196, 947–950 (1987). [DOI] [PubMed] [Google Scholar]
- 54.Vastenhouw, N. L., Cao, W. X., & Lipshitz, H. D. The maternal-to-zygotic transition revisited. Development146. 10.1242/dev.161471 (2019). [DOI] [PubMed]
- 55.Lee, M. T. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature503, 360–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodland, H. R. Changes in the polysome content of developing Xenopus laevis embryos. Dev. Biol.40, 90–101 (1974). [DOI] [PubMed] [Google Scholar]
- 57.Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol.6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature499, 172–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey, T. L. STREME: accurate and versatile sequence motif discovery. Bioinformatics37, 2834–2840 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L. & Noble, W. S. Quantifying similarity between motifs. Genome Biol.8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biever, A., et al. Monosomes actively translate synaptic mRNAs in neuronal processes. Science367. 10.1126/science.aay4991 (2020). [DOI] [PubMed]
- 62.Lytle, J. R., Yario, T. A. & Steitz, J. A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl Acad. Sci. USA104, 9667–9672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reimao-Pinto, M. M., Castillo-Hair, S. M., Seelig, G., & Schier, A. F. The regulatory landscape of 5’ UTRs in translational control during zebrafish embryogenesis. bioRxiv. 10.1101/2023.11.23.568470 (2023).
- 64.Pan, X., et al. 5’-UTR SNP of FGF13 causes translational defect and intellectual disability. Elife10. 10.7554/eLife.63021 (2021). [DOI] [PMC free article] [PubMed]
- 65.Komatsu, K. R. et al. RNA structure-wide discovery of functional interactions with multiplexed RNA motif library. Nat. Commun.11, 6275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujii, K. et al. Controlling tissue patterning by translational regulation of signaling transcripts through the core translation factor eIF3c. Dev. Cell56, 2928–2937.e2929 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue, S. et al. RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature517, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beaudoin, J. D. et al. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol.25, 677–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zinshteyn, B., Rojas-Duran, M. F. & Gilbert, W. V. Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA23, 1365–1375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babendure, J. R., Babendure, J. L., Ding, J. H. & Tsien, R. Y. Control of mammalian translation by mRNA structure near caps. RNA12, 851–861 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Vejnar, C. E. & Giraldez, A. J. LabxDB: versatile databases for genomic sequencing and lab management. Bioinformatics36, 4530–4531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn Res12, 2825–2830 (2011). [Google Scholar]
- 76.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tareen, A. & Kinney, J. B. Logomaker: beautiful sequence logos in Python. Bioinformatics36, 2272–2274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hunter, J. D. Matplotlib: A 2D graphics environment. Comput Sci. Eng.9, 90–95 (2007). [Google Scholar]
- 79.Harris et al. Array programming with NumPy. Nature585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon request. The sequencing data generated in this study have been deposited in the NCBI under the BioProject ID PRJNA1188270. The processed count tables from the sequencing data are available in Supplementary Data 2-4 or on the Giraldez Lab datahub (https://www.giraldezlab.org/data/).
All scripts used to process and analyze NaP-TRAP data will be released at the time of publication (https://github.com/ecstrayer/nap-trap_paper).