Abstract
Chlamydiae are obligately intracellular pathogens which cause infections associated with a broad range of diseases in both livestock and humans. In addition, a large proportion of animals may become persistently infected asymptomatic carriers and serve as reservoirs for other animals which also shed these potential zoonotic pathogens. Reducing the chlamydial load of animals is therefore of major importance, and since large-scale antibiotic treatment is neither desired nor feasible, alternative means of prevention are needed. Here we performed a study comparing the efficacy of a probiotic strain of Enterococcus faecium on the reduction of both the rate of natural infection and the shedding of chlamydiae in swine. The presence of Chlamydiaceae was detected by species-specific PCR of fecal samples of sows taken at three times prior to the birth of piglets. Piglets delivered from chlamydia-positive sows in either the control or the probiotic group were also examined for the frequency of chlamydiae at various ages. Eighty-five percent of the piglets from the control group were found to be chlamydia positive, whereas chlamydiae were found in only 60% of piglets from the probiotic group, results confirmed by fluorescence in situ hybridization and immunohistology, which showed higher rates of infection in the control group. In addition to the reduced frequency of chlamydia-positive piglets in the probiotic group, the time of appearance of positive samples was delayed. To our knowledge, these data show for the first time that a probiotic strain of E. faecium can reduce the rate of carryover infections of piglets by obligate intracellular pathogens.
Members of the order Chlamydiales are obligately intracellular parasites of eukaryotic cells, displaying a unique developmental cycle alternating between extracellular infectious elementary bodies (EB) and intracellular replicating reticulate bodies (RB). In humans, chlamydiae are the leading cause of preventable blindness, sexually transmitted disease, and pneumonia and have also been linked to cardiovascular disease (21, 23, 35, 53, 54). Several chlamydial species also are responsible for a variety of clinically and economically important diseases in livestock and domestic animals. In swine, chlamydiae are associated with a broad range of diseases, including abortion and delivery of weak piglets (70, 71), orchitis, epididymitis, and urethritis in boars (56), pericarditis, polyarthritis, and polyserositis in piglets (69), conjunctivitis (48), pneumonia (30), and pseudomembranous or necrotizing enteritis (38, 45). Outbreaks of chlamydiosis in pigs have been reported for industrial animal stocks in Eastern European countries, where the habitat of the chlamydiae involved appeared to be the intestinal tract (25, 39, 47, 62, 64). Persistent infections of the gut result in intermittent shedding of chlamydiae into the environment, and due to the high tenacity of these organisms, infected feces becomes a source of infection for other animals, as well as for humans (42, 62).
Chlamydia suis (formerly a porcine serovar of Chlamydia trachomatis), Chlamydia pecorum, and Chlamydophila abortus (formerly Chlamydia psittaci serovar 1) have all been isolated from the porcine gut (28, 58, 66, 72). Chlamydia spp. show greater than 80% sequence identity within the genes encoding their 16S and/or 23S rRNAs (16). Based on 16S-23S ribosomal intergenic spacer signature sequences, C. suis is highly related (99%) to the human C. trachomatis species, and C. suis isolate S-45 is identical to human genital C. trachomatis serovar D (16). Both species also show identical or near-identical VS4 epitopes, encoded by the ompA gene (15). Indeed, monoclonal antibodies (MAbs) to VS4 of C. trachomatis and chlamydial serogroup B MAbs, routinely used to serotype human C. trachomatis, can lead to cross-reactions with C. suis, leading to species misidentification (15).
The zoonotic potential of C. suis for humans remains an important unresolved question. Case reports have described the transmission of C. abortus from small ruminants to humans and have been associated with abortion in infected humans as well as health-related problems of pig, cattle, and sheep farmers (9, 44). Several studies revealed a prevalence of chlamydiae in swine ranging from 12 to 30%, with the frequency of detection on individual farms ranging from 0 to 70% (28, 66, 72). Chlamydiae are found localized predominantly in the large intestine, where chlamydial inclusions are situated in the cytoplasm of enterocytes (11, 24, 46, 48, 72). C. suis isolates recovered from the intestines of diarrheic pigs were shown to cause intestinal lesions and diarrhea in gnotobiotic pigs at infective doses from 105 to 106 inclusion-forming units (IFU)/ml (47). In young, weaned pigs, the same infective doses do not result in diarrhea, but they do result in intestinal lesions and persistent intestinal infections (46). Challenge infections of mice with C. trachomatis (MoPn) have shown that cell-to-cell transmission, systemic dissemination, and autoinoculation with infectious fluids also contribute to chlamydial spread (42). The key importance of intestinal chlamydial infections is therefore not necessarily their enteropathogenicity but rather the frequent persistent infections, conferring carrier status.
For livestock, antiinfectives and vaccines are well-established means to reduce infections. However, the use of antiinfectives, especially as feed supplements, has been criticized due to the development of antibiotic resistances, and vaccination against porcine chlamydiae is not well established. In addition to hygienic measures, the use of probiotics as feed supplements for livestock animals is increasing in importance. Probiotics are microorganisms which confer beneficial effects in the prevention and treatment of certain pathological conditions (27), and the probiotic principle has been suggested to be useful for the treatment of intestinal disorders and for enhancing the gut mucosal barrier function (1, 33, 55). The protective effects of probiotics against intestinal infections were shown both with animals and with in vitro cell culture models (6, 18, 19, 32, 63, 67). However, while many beneficial effects of probiotics for both animals and humans have been described, the mode of action of probiotics remains largely unclear (49).
To our knowledge, no studies to date have examined the influence of probiotics on infections by obligate intracellular bacteria. We therefore determined the effect of an Enterococcus faecium probiotic strain (NCIMB 10415), as a representative of the autochthonous gut flora of pigs, on the rate of chlamydial infection in swine as a possible means to decrease infections of newborn piglets. The choice of this probiotic bacterium was also based on prior studies reporting beneficial effects, leading to its licensing by the European Union as a feed supplement for animals. Furthermore, we examined the suitability of the E. faecium probiotic strain as a prophylactic measure for the prevention of carryover infections by monitoring the rate of infection by chlamydiae in newborn piglets from naturally chlamydia infected sows.
MATERIALS AND METHODS
Animals and design of the study.
Hybrid Landrace × Duroc sows were randomly assigned to two groups, consisting of an untreated control group receiving normal feed and a second group of sows receiving feed supplemented with Cylactin LBC ME10 (probiotic group), a microencapsulated preparation of Enterococcus faecium SF68 (NCIMB 10415), at a concentration of 50 mg/kg (sows) or 100 mg/kg (piglets) feed. The concentration of viable Enterococcus faecium bacteria in the preparations was determined to be 9 × 109 CFU/g. The duration of application was 13 weeks for sows and 8 weeks for piglets. Sows were fed a diet supplemented with Cylactin LBC or an unsupplemented diet, beginning 24 days after mating. Suckling piglets of both the probiotic and control groups had access to prestarter feed ad libitum from days 14 to 28. After weaning (day 28), the starter diet for piglets was with or without supplementation according to the treatment group.
Samples.
Three independent fecal samples from each of a total of 22 sows were taken prior to the birth of piglets to detect carriers. DNA from this sample matrix was prepared using the QIAmp DNA Stool kit (QIAGEN, Germany) according to the manufacturer's instructions. Tissue and fecal samples isolated from euthanized piglets were used for determination of colonization rates by PcR determinations with DNA from the ileum, colon descendens, and feces. DNA extractions were done by using the QIAamp DNA Stool kit or the QIAamp DNA mini-kit (QIAGEN, Germany) according to the instructions of the manufacturer. Only piglets delivered from chlamydia-positive carrier sows were examined.
Cultivation.
Chlamydiae were propagated in the permanent Buffalo green monkey (BGM) kidney epithelial cell line. Cells were cultured in Eagle's minimal essential medium (EMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% vitamins, 1% l-glutamine, 1% nonessential amino acids, 2.5 μg/ml amphotericin, and 50 μg/ml gentamicin (all components from Biochrom, Germany) and were incubated at 37°C under 5% CO2. After infection of cells, the cell culture medium was modified to contain 2% FCS and 2 μg/ml cycloheximide.
For cultivation of chlamydiae, feces or pooled mucosal samples from the colon ascendens and colon descendens were used. Samples were placed in 2SP transport medium (0.2 M sucrose, 0.02 M phosphate buffer, pH 7.2) containing 50 μg/ml gentamicin, 10 μg/ml amphotericin B, 25 μg/ml vancomycin, and 20% FCS, transported on ice, and processed within 24 h. Preparation of infectious chlamydiae from feces and mucosa was as follows. The matrix was disrupted with glass beads (diameter, 1 mm) in a BeadBeater (BioSpec, Bartlesville, Okla.) for 2 min on ice. Cellular debris was removed from the homogenates by centrifugation at 500 × g and 4°C for 10 min, and the resulting supernatant was centrifuged again for 20 min at 1,500 × g and 4°C. Supernatants were centrifuged for 45 min at 30,000 × g for sedimentation of EB. The sediment was resuspended in 2 ml EMEM and used for infection of cultured cells. For each specimen, 0.2 ml inoculum was placed on 24-h-old BGM monolayers on glass coverslips (13 mm) in 24-well microtiter plates (Corning, Schiphol, The Netherlands). Plates were centrifuged at 2,500 × g and 37°C for 1 h. After 2 h of incubation, the inoculum was removed by a wash with phosphate-buffered saline and was replaced by fresh EMEM. After 3 and 5 days of incubation, duplicate coverslips were fixed with methanol and stained for the presence of characteristic chlamydial inclusions (22). Two wells were harvested by the freeze-thaw method (65) for a second passage/infection. If no inclusions were detected within five passages, the specimen was considered negative.
Genus- and species-specific PCR.
Genus and species-specific nested PCR targeting the ompA gene was used for detection of chlamydiae in swine as described by Schiller et al. (58) and modified by Sachse and Hotzel (52). The principle of the nested amplifications and species differentiation is as follows. In the first amplification step (outer primer pair, 191CHOMP and CHOMP371), a genus-specific product is generated. The second step of PCR involves one genus-specific inner primer (201CHOMP or CHOMP336s) and one species-specific primer (TRACH269 or 218PSITT/204PECOR). Primer pairs 204PECOR and CHOMP336s (C. pecorum), 218PSITT and CHOMP336s (C. abortus; formerly C. psittaci serovar 1), and 201CHOMP and TRACH269 for C. suis (formerly a porcine serovar of C. trachomatis) were used.
Oligonucleotide primers are listed in Table 1. The sensitivity of the nested PCR has been reported as less than 1 IFU/ml (51, 52). DNA controls were prepared by proteinase K digestion from cell culture monolayers infected with either C. suis S45, C. abortus B577, C. psittaci (avian), or C. pecorum LW613 (kindly provided by M. M. Wittenbrink, Institut für Veterinärbakteriologie, Zürich, Switzerland) as described by Meijer et al. (37). Cross-contamination was prevented by the use of aerosol-resistant pipette tips in all pre-PCR steps. Samples were considered positive when the species-specific amplicon of the expected size was visible on 2% agarose gels after electrophoresis and ethidium bromide staining. This PCR protocol was performed with samples from 22 sows (66 samples) for detection of carrier status and with samples from 40 piglets (120 samples). For some samples, species diagnosis was performed by PCR for the 16S rRNA signature sequence using primers 16SIGF and 16SIGR. Primers 16SF and 16SR were used to generate the 16S rRNA genes of the cultivated isolates. The ompA gene was amplified from the same isolates by using primers CTU and TGLY.
TABLE 1.
Sequences of primers for detection of chlamydiae
| Primer | Sequence (5′-3′)a | Reference |
|---|---|---|
| 191CHOMP | GCIYTITGGGARTGYGGITGYGCIAC | 52 |
| CHOMP371 | TTAGAAICKGAATTGIGCRTTIAYGTGIGCIGC | 52 |
| 201CHOMP | GGIGCWGMITTCCAATAYGCICARTC | 52 |
| CHOMP336s | CCRCAAGMTTTTCTRGAYTTCAWYTTGTTRAT | 52 |
| 218PSITT | GTAATTTCIAGCCCAGCACAATTYGTG | 52 |
| TRACH269 | ACCATTTAACTCCAATGTARGGAGTG | 52 |
| 204PECOR | CCAATAYGCACAATCKAAACCTCGC | 52 |
| 16SIGF | CGGCGTGGATGAGGCAT | 15 |
| 16SIGR | TCAGTCCCAGTGTTGGC | 15 |
| 16SF | GCGTGGATGAGGCATGCAA | 16 |
| 16SR | GGAGGTGATCCAGCCCCA | 16 |
| CTU | ATGAAAAAACTCTTGAAATCGG | 10 |
| TGLY | GGCTACAGCTCTACCATTGA | 10 |
Degenerate nucleotides: K = G or T; M = A or C; R = A or T; Y = C or T; I = inosine.
Immunohistochemistry (IHC).
Tissue samples (ileum, colon ascendens, and colon descendens) were fixed in 4% paraformaldehyde and processed to 4- to 6-μm-thick paraffin sections. For immunohistochemical staining, a genus-specific mouse MAb (clone AC-1; Progen, Heidelberg, Germany) directed against the lipopolysaccharide of chlamydiae (working dilution, 1:200) was used and labeled with streptavidin-biotin using the DAKO ChemMate Detection kit according to the manufacturer's instructions (DAKO Diagnostics, Hamburg, Germany). The number of infected enterocytes of intact tissue per mm of lamina muscularis mucosae was calculated using a histological software program (AxioVision; Zeiss, Oberkochen, Germany).
FISH.
Tissue samples from the ileum, colon ascendens, and colon descendens were washed in phosphate-buffered saline and fixed in 4% paraformaldehyde at 4°C. Tissues were embedded in polymerizing resin (Technovit 8100; Heraeus Kulzer, Wehrheim, Germany) at 4°C according to the vendor's instructions. Polymerized resin blocks were stored at 4°C for preparation of semithin sections (4 μm). For detection and differentiation of chlamydiae, an oligonucleotide-probe set as described by Poppert et al. (43) was used (Table 2). Fluorescence in situ hybridization (FISH) and buffers have been described elsewhere (4). The eubacterial probe EUB338, targeting the 16S rRNA gene sequence of most members of the domain Bacteria, was used as a positive control in all hybridization experiments. DAPI (4′,6′-diamidino-2-phenylindole) staining was also included in the control reagents for labeling of both prokaryotic and eukaryotic DNA.
TABLE 2.
Oligonucleotide probe sequences specific for the Chlamydiales
| Probe | Target organisms | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Chls-0523 | Chlamydiales | CCTCCGTATTACCGCAGC | 43 |
| Chlae-0574 | Chlamydiaceae | CTTTCCGCCTACACGCCC | 43 |
| Chla-0232 | Chlamydia | TAGCTGATATCACATAGA | 43 |
| Chlph-0583 | Chlamydophila | CTAACTTTCCTTTCCGCC | 43 |
| Ct-0623 | C. trachomatis (human) | ATTAGATGCCGACTCGGG | 43 |
| EUB338 | Eubacteria | GCTGCCTCCCGTAGGAGT | 3 |
From all piglets, at least four sections were investigated in four different combinations of oligonucleotide probes. Prewarmed hybridization solution (40 μl) was mixed with 200 ng of the probes and applied to the tissue sections. After incubation in the dark for 4 h in a humidified chamber at 46°C, slides were washed with prewarmed wash buffer (48°C) for 10 min. Slides were rinsed with distilled water, air dried in the dark, coated with Vectashield antifade reagent (Vector Laboratories), and mounted for epifluorescence microscopy.
DNA sequencing.
PCR products were purified using the High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Sequencing was performed by AGOWA (Berlin, Germany). Nucleotide sequence alignments were performed using the BLAST program.
Statistical analysis.
Statistical analysis of the detection frequencies of chlamydiae from specimens of piglets in the two groups was performed using the χ2 test using SPSS 12.0 for Windows.
Nucleotide sequence accession numbers.
Accession numbers for the sequenced DNA regions submitted to the EMBL nucleotide sequence database are AY661794 to AY661798 for the 16S rRNA gene and AY687638 to AY68763 for the ompA gene. Sequenced PCR products of genus-specific nested-PCR products were assigned accession numbers AY687635 to AY687639, and the 16S rRNA signature sequence was assigned accession numbers AY686469 to AY686473.
RESULTS
Detection of chlamydia carrier status of sows.
To determine the chlamydia carrier status of sows, DNA was prepared from fecal samples of 22 sows from either the control or the probiotic group and was used for species-specific PCRs. To increase the probability of identifying positive samples despite intermittent fecal shedding, three independent samples from each sow were examined. The presence of chlamydiae in fecal samples ranged from 27% (6/22) for single-time-point detection to 73% (16 chlamydia-positive sows out of 22) when three samples were investigated. The species diagnosis from the second, species-specific step of the nested PCRs verified the presence of C. trachomatis in all 16 sows. The C. trachomatis-specific primer TRACH 269 targets the ompA gene and detects both human C. trachomatis species and C. suis (formerly a porcine serovar of C. trachomatis).
Detection of chlamydiae in piglets from sows with carrier status by nested PCR.
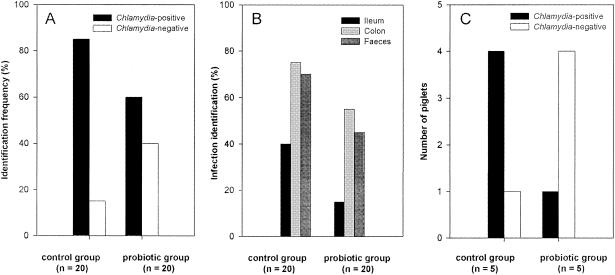
Having established the chlamydia infection status of sows in both the control and the probiotic-fed group, we then determined the frequency of carryover infections in piglets belonging to 10 sows with identifiable carrier status. Of the two groups, four piglets from each sow were examined for chlamydial infection at the ages of 14, 28, 35, and 56 days. Of a total of 40 piglets examined, 29 of 40 (73%) were found to be chlamydia positive, with a positive sample distribution among 11 of 40 (28%) for the ileum, 26 of 40 (65%) for the colon descendens, and 23 of 40 (58%) for the feces. Piglets from the control group showed a frequency of chlamydia-positive samples of 17/20 (85%), in contrast to 12/20 (60%) in the probiotic group (Fig. 1A). The frequency of detection of chlamydiae was also greater for all three sample types in the control group (Fig. 1B). The reduced frequency of chlamydiae in the probiotic group (3/20; 15%) was most apparent in ileal samples relative to the control group (8/20; 40%). Clear differences were also apparent between the two groups in colon and fecal samples, with the probiotic group showing reduced frequencies relative to the control group of 20% and 25%, respectively. A significant reduction in the frequency (P = 0.05) of chlamydia-positive samples was seen at the age of 14 days, when only one of five piglets from the probiotic group tested positive, compared to four out of five from the control group (Fig. 1C). False negative results were reduced to a minimum by the excellent sensitivity of the nested PCR and the use of specialized DNA extraction kits. The 100% correlation of four independent methods supports the conclusion that the PCR results reflected the actual infection rate in the piglets.
FIG. 1.
Effects of probiotic supplementation on the frequency and distribution of chlamydial isolates in piglets of chlamydia-positive sows. (A) Total-DNA samples from the ileum, colon descendens, and feces isolated from piglets at 14, 21, 35, and 52 days postpartum were subjected to nested PCR for the presence of chlamydiae. Piglets from the control group showed a frequency of chlamydia-positive samples of 17/20 (85%), in contrast to 12/20 (60%) in the probiotic-fed group (P = 0.073). (B) Distribution of chlamydiae in ileal, colon descendens, and fecal samples. The reduced frequency of chlamydiae in the probiotic group (3 of 20; 15%) was most apparent in ileal samples relative to the frequency in the control group (8 of 20; 40%). Clear reductions in the frequency of chlamydiae were also apparent in colon samples (75% and 55% in control and probiotic piglets, respectively) and fecal samples (70% and 50%, respectively) (ileum, P = 0.073; colon, P = 0.103; feces, P = 0.326). (C) At the age of 14 days, only one of five piglets from the probiotic group was positive for chlamydiae, compared to four out of five from the control group (P = 0.05).
Based on the species diagnosis from sows, carryover infections of piglets were most likely to have been due to Chlamydia suis infection. To verify that these were carryover infections rather than infections due to a susceptibility to other environmental species, the PCR products from six randomly selected piglets from different sows were sequenced. Both the ompA and 16S rRNA gene signature sequences were compared. The sequences of the ompA gene were 92% to 93% identical with that of C. suis, and the 16S rRNA gene sequence was 99% to 100% identical with that of C. suis R22, consistent with the isolates identified in chlamydia-positive sows.
Detection of chlamydia-infected piglets from sows with carrier status by cultivation.
In an effort both to establish pure cultures and to verify the infectivity of fecal and intestinal isolates, fecal and pooled mucosal samples from the colon ascendens and colon descendens from chlamydia-positive piglets were seeded on chlamydia-susceptible cultured BGM cells and cultivated for five passages before the final evaluation. In cell culture, chlamydiae were observed in the second passage from five mucosal samples and three fecal samples. The ompA and 16S rRNA PCR products generated from DNA extracted from cell culture supernatants were sequenced and showed 99% identity to the 16S rRNA gene of C. suis R22 and 92% to 94% identity to the ompA gene of C. suis S45. The sequences for the 16S rRNA and ompA genes from two piglets of the same sow were identical. Specimens of three animals were excluded from the evaluation due to cross-contamination by other bacteria. These results verified the infectivity of the chlamydia isolates identified by the PCR-based methods, and the infective fecal isolates further confirmed the carrier status of the piglets.
Detection of chlamydiae in piglets from sows with carrier status by immunohistochemistry.
The chlamydial infection status of piglets was also verified by PCR-independent means. Sections of paraformaldehyde-fixed, paraffin-embedded tissue from the colon ascendens and colon descendens from 12 randomly selected piglets from either the control or the probiotic group were immunohistochemically labeled with genus-specific mouse MAbs against chlamydial lipopolysaccharide. A total of 7 out of 12 animals in both the control and probiotic-fed groups showed positive results by use of antibodies (Fig. 2 and 3).
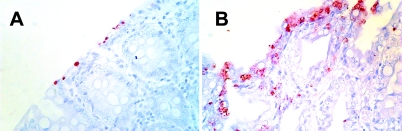
FIG. 2.
Immunohistology of colon ascendens (A) and colon descendens (B) samples from a 35-day-old piglet. A genus-specific monoclonal antibody against chlamydial lipopolysaccharide was used to label chlamydial inclusions in enterocytes. Magnification, ×400.
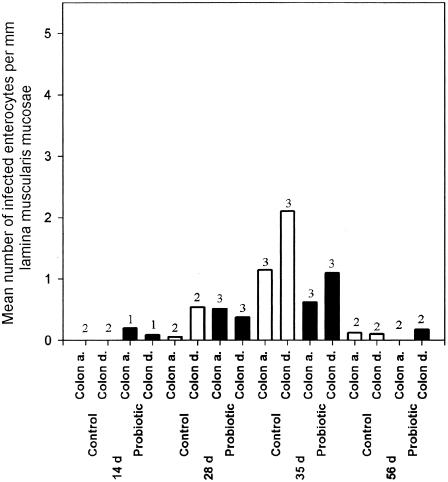
FIG. 3.
Probiotic supplementation reduces the in vivo spread of chlamydiae in enterocytes. Age-dependent distribution of infected enterocytes per mm lamina muscularis mucosae in control (open bars) and probiotic-fed (filled bars) piglets. Shown are the average infected enterocytes for regions of the colon ascendens (Colon a.) and colon descendens (Colon d.) in each group at different ages (in days). Data shown are for a maximum of three piglets of the indicated age postpartum from each group. The number of piglets sampled is given above each bar. Only determinations from PCR-positive piglets were used for calculation of means.
Semiquantitative analyses of the frequency of chlamydia-infected enterocytes per mm lamina propria mucosae showed large differences in the degree of infection when the mean values for individuals were compared. There were clear differences between the probiotic and control groups in both the colon ascendens and the colon descendens in 35-day-old piglets (Fig. 3). At this age the infection rate of enterocytes was high. In both groups, however, the colon descendens showed the largest number of infected enterocytes. Chlamydiae were detected only in the cytoplasm of enterocytes and never below the lamina propria mucosae. When the results of the PCR determinations were compared to those of the immunohistological examinations, among 24 animals examined, 4 that were identified as chlamydia positive by PCR could not be verified immunohistochemically. In contrast, all chlamydia-positive animals identified immunohistologically were also found to be positive by PCR.
Examination of intestinal tissue from piglets for chlamydial infection by FISH.
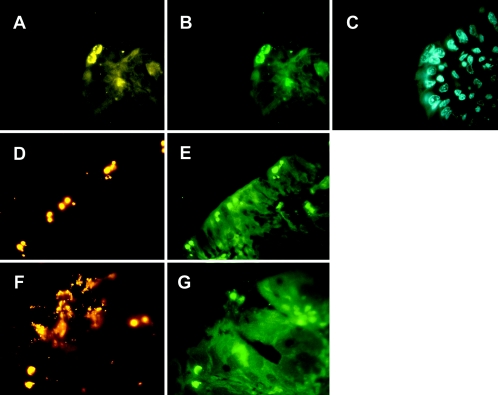
To validate the results of the immunohistochemical determinations, sections of resin-embedded tissue from the ileum, colon ascendens, and colon descendens from identical piglets were also hybridized with chlamydia oligonucleotide probes Chls-0523, Chlae-0574, Chla-0232, Chlph-0583, and Ct-0623 (Table 2), as well as a universal eubacterial probe, EUB338, which detects nearly all bacteria (2). No hybridization signals were observed by using probe Chlph-0583 or Ct-0623, specific for Chlamydophila or C. trachomatis, respectively. Combined with the results from both PCR and sequencing, the results shown in Fig. 4 were therefore consistent with the conclusion that all chlamydia-positive piglets were infected with C. suis. Furthermore, the use of multiple probes verified the utility of FISH for discrimination of both genus- and species-specific infections by chlamydiae. Of 12 animals examined per study group, 6 showed positive hybridization signals in the control group compared to 5 from the probiotic-fed group. For the three regions of the intestine examined for each piglet, 10 out of a total of 36 sections showed chlamydial inclusions in the control group compared with 8 of 36 in the probiotic group.
FIG. 4.
In situ detection of chlamydiae in the intestine of a 35-day-old piglet by FISH. (A through C) Visualization of chlamydiae in the colon ascendens by using the Cy3-labeled probe Chls-523 (A), the fluorescein-labeled probe Chlae-574 (B), or control staining by DAPI (C). (D through G) In situ identification of chlamydiae in the colon descendens by Chls-523 (D), Chlae-574 (E and F), or the Cy3-labeled probe EUB338 (G). (A through C) Original magnification, ×1,000; (D through G) original magnification, ×400.
DISCUSSION
The observations in this study of reduced numbers of chlamydia-positive piglets from infected sows in the probiotic-treated group, including reduced tissue localizations and infected enterocytes and a delayed time of appearance of positive samples, suggest that the probiotic E. faecium strain reduces both the rate and the severity of carryover infections of piglets by chlamydiae.
Consistent with other studies (28, 66), we confirmed a high level of persistent chlamydia infections in sows in a conventional swine herd (16/22; 63% chlamydia positive). For the detection of chlamydiae in fecal samples, multiple assays were imperative, as shown by the different results obtained between single and triplicate sample testing of sows using PCR-based methods. Only piglets of sows with chlamydia carrier status were followed up for the presence of chlamydial infection. The most consistent and reproducible results for the presence of chlamydiae in piglets were obtained with tissues of the large intestine (65%) compared to small-intestine tissues (ileum), where only 28% of samples were found positive. The chlamydial inclusions observed in enterocytes consisted of both single inclusions and apparently proliferating clusters (Fig. 2 and 4), also confirming results from other studies (11, 45).
For diagnostic purposes, the cultivation of chlamydiae is unsuitable, particularly for C. suis, which shows a very low success rate (5, 59). Additional problems include coinfection of intestinal samples with mycoplasmas; control measures include the use of cycloheximide and other antibiotics for transport and processing of samples, but these can also reduce the propagation and proliferation of chlamydiae. PCR-based detection methods are sensitive, able to detect less than 1 IFU in cell culture or tissues (52). The specificity of PCR is further validated by sequencing of the PCR products and by 16S rRNA gene signature sequence PCR. In this study, all supporting validation methods resulted in the species diagnosis of C. suis. While IHC is an excellent tool for visualization of the localization of chlamydiae in tissues, the lower sensitivity of IHC compared to PCR is understandable, since single chlamydial inclusions could not be visualized and tissue section examinations are time-consuming. Here, we have used IHC only as a validation method for tissue sections from which extracted DNA was used in PCRs. While FISH with 16S rRNA sequences specific for different taxonomic levels provided a valuable tool for visualization in situ, the method is limited, because only metabolically active chlamydial (RB) forms are detected. However, this difference provides information distinguishing between environmental contamination and genuine infection status.
In the herd of sows examined, only C. suis was identified. The transmission of chlamydiae to piglets most likely results from carryover infections from sows to piglets through contact with infectious feces. This would be consistent with the sequence results indicating that piglets of one sow showed the same isolate of C. suis (see nucleotide accession numbers AY687630, AY687631, AY661795, and AY661795). These observations suggest a transmission model in which oral uptake of infectious chlamydiae leads to initial colonization of enterocytes of the large intestine. A weakened immune response resulting from stress or other factors, or infection by more-virulent strains of chlamydiae, would lead to ascending infection and manifestation in the small intestine. An outcome of colonization resulting in latent infections is suggested by the observation that none of the infected piglets in this study showed clinical signs of enteric infections, as previously observed in challenge trials with chlamydiae in gnotobiotic pigs (47). However, during the course of the study, we noted that many of the sows that tested positive for C. suis showed a high rate of reduced fertility, resulting in lower-than-average litter numbers (unpublished observations).
At least three mechanisms are possible whereby the probiotic Enterococcus faecium strain could reduce the infection rate in piglets: (i) inhibition or reduction of infection of host cells, (ii) reduced proliferation within infected cells, and/or (iii) a more rapid clearance of chlamydia-infected cells. Attachment of chlamydiae to host cells is at least partially dependent on an interaction with the glycosaminoglycan receptor (26). Competition for binding to the glycosaminoglycan receptor is possible, similar to the antagonism for binding of the intestinal mucus receptor (80-kDa protein, involved with glycoprotein) shown for Enterococcus faecium strain 18C23 and K88-positive enterotoxigenic Escherichia coli in a model of porcine small-intestine mucus (29). Reduced entry could also result from antimicrobial agents produced by E. faecium, which may act against chlamydiae (7, 14, 50). However, a recent study found that none of the eight known bacteriocin (enterocin) genes are present in the strain of E. faecium used in this study, and the strain shows no antimicrobial activity against gram-negative bacteria (20; unpublished observations).
The second possibility, i.e., a reduction in chlamydial replication and proliferation within host cells, is less clear. Very little is known about the interactions between probiotics and intestinal epithelial cells. A recent study, however, suggested that secreted products of probiotic Lactobacillus strains can lead to activation of the eukaryotic transcriptional activator NF-κB (8). Prior studies have also suggested competitive interactions between probiotics and epithelial cells (13, 34, 36). It is therefore possible that probiotic bacteria could influence any number of the previously observed interactions between chlamydiae and their host cells (12, 31, 60, 61). Because chlamydiae are obligately intracellular pathogens, it is possible that changes in host cell gene expression patterns due to probiotic bacterial interactions may have indirect consequences on the replication of chlamydiae.
The third possibility could be a more rapid elimination of chlamydia-infected cells by an elevated immune response, either specific or innate, resulting from probiotic treatment (40, 41). However, in parallel studies on immune cell populations of piglets, including those reported here and additional piglets from the same study, no immediate indications of changes in protective immune cell populations or antibody production were observed (57). How probiotic bacteria can affect or prevent infection by enteropathogens through effects on the immune response remains unclear; the answer may be related to changes in gene expression and/or cytokine production indirectly affecting immune cell functions rather than to their populations per se. Clearly, further work will be required to answer this question.
Finally, the observed reductions in chlamydial infection of intestinal epithelia may be the result of indirect effects of the E. faecium probiotic strain on the makeup of the intestinal flora. Probiotics may support a more rapid colonization of the intestine with a healthy, commensal bacterial flora in piglets, most likely “inherited” from the sows. Prior studies with germ-free mice have shown that commensal bacteria enhance development of both the intestinal epithelia and the gastrointestinal lymphoid system (17, 68). In this manner, a balanced microbial population would support the inherent defense mechanisms of a healthy intestinal tract, resulting in better control of intestinal pathogens. With regard to chlamydial infections, the rate of infection of epithelial cells per se may not be affected, but accelerated development (and turnover) of the epithelia combined with a more developed intestinal tract would help keep the rate of further epithelial cell infections low. Further studies, including in vitro studies, from this ongoing project should help clarify many of these remaining questions.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG) grant FOR 438/1-1 and a fellowship from the Studienstiftung des deutschen Volkes to M.P.
We are grateful to M. Filter (Institut für Molekularbiologie und Biochemie, Charité-Universitätsmedizin Berlin) for statistical evaluation of the data.
Editor: J. B. Bliska
REFERENCES
- 1.Alvarez-Olmos, M. I., and R. A. Oberhelman. 2001. Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin. Infect. Dis. 32:1567-1576. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen, A. A. 1994. Presented at the 25th Annual Meeting of the American Association of Swine Practitioners.
- 6.Asahara, T., K. Shimizu, K. Nomoto, T. Hamabata, A. Ozawa, and Y. Takeda. 2004. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect. Immun. 72:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audisio, M. C., G. Oliver, and M. C. Apella. 2001. Effect of different complex carbon sources on growth and bacteriocin synthesis of Enterococcus faecium. Int. J. Food Microbiol. 63:235-241. [DOI] [PubMed] [Google Scholar]
- 8.Bai, A. P., Q. Ouyang, W. Zhang, C. H. Wang, and S. F. Li. 2004. Probiotics inhibit TNF-α-induced interleukin-8 secretion of HT29 cells. World J. Gastroenterol. 10:455-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazala, E., and J. Renda. 1992. Latent Chlamydia infections as the cause of health disorders in swine, cattle and sheep breeders in Czechoslovakia. Berl. Munch. Tierarztl. Wochenschr. 105:145-149. [PubMed] [Google Scholar]
- 10.Bush, R. M., and K. D. Everett. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51:203-220. [DOI] [PubMed] [Google Scholar]
- 11.Chae, C., D. S. Cheon, D. Kwon, O. Kim, B. Kim, J. Suh, D. G. Rogers, K. D. Everett, and A. A. Andersen. 1999. In situ hybridization for the detection and localization of swine Chlamydia trachomatis. Vet. Pathol. 36:133-137. [DOI] [PubMed] [Google Scholar]
- 12.Coombes, B. K., D. L. Johnson, and J. B. Mahony. 2002. Strategic targeting of essential host-pathogen interactions in chlamydial disease. Curr. Drug Targets Infect. Disord. 2:201-216. [DOI] [PubMed] [Google Scholar]
- 13.Dahan, S., G. Dalmasso, V. Imbert, J. F. Peyron, P. Rampal, and D. Czerucka. 2003. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect. Immun. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elmer, G. W., and L. V. McFarland. 2001. Biotherapeutic agents in the treatment of infectious diarrhea. Gastroenterol. Clin. N. Am. 30:837-854. [DOI] [PubMed] [Google Scholar]
- 15.Everett, K. D. 2000. Chlamydia and Chlamydiales: more than meets the eye. Vet. Microbiol. 75:109-126. [DOI] [PubMed] [Google Scholar]
- 16.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 17.Falk, P. G., L. V. Hooper, T. Midtvedt, and J. I. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez, M. F., S. Boris, and C. Barbes. 2003. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 94:449-455. [DOI] [PubMed] [Google Scholar]
- 19.Filho-Lima, J. V., E. C. Vieira, and J. R. Nicoli. 2000. Antagonistic effect of Lactobacillus acidophilus, Saccharomyces boulardii and Escherichia coli combinations against experimental infections with Shigella flexneri and Salmonella enteritidis subsp. typhimurium in gnotobiotic mice. J. Appl. Microbiol. 88:365-370. [DOI] [PubMed] [Google Scholar]
- 20.Foulquie Moreno, M. R., R. Callewaert, B. Devreese, J. Van Beeumen, and L. De Vuyst. 2003. Isolation and biochemical characterisation of enterocins produced by enterococci from different sources. J. Appl. Microbiol. 94:214-229. [DOI] [PubMed] [Google Scholar]
- 21.Gerbase, A. C., J. T. Rowley, and T. E. Mertens. 1998. Global epidemiology of sexually transmitted diseases. Lancet 351(Suppl. 3):2-4. [DOI] [PubMed] [Google Scholar]
- 22.Gimenez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 23.Grayston, J. T., S. P. Wang, C. C. Kuo, and L. A. Campbell. 1989. Current knowledge on Chlamydia pneumoniae, strain TWAR, an important cause of pneumonia and other acute respiratory diseases. Eur. J. Clin. Microbiol. Infect. Dis. 8:191-202. [DOI] [PubMed] [Google Scholar]
- 24.Guscetti, F., R. Hoop, I. Schiller, L. Corboz, T. Sydler, and A. Pospischil. 2000. Experimental enteric infection of gnotobiotic piglets with a Chlamydia psittaci strain of avian origin. J. Vet. Med. B 47:561-572. [DOI] [PubMed] [Google Scholar]
- 25.Guscetti, F., I. Schiller, T. Sydler, L. Corboz, and A. Pospischil. 1998. Experimental Chlamydia psittaci serotype 1 enteric infection in gnotobiotic piglets: histopathological, immunohistochemical and microbiological findings. Vet. Microbiol. 62:251-263. [DOI] [PubMed] [Google Scholar]
- 26.Hackstadt, T. 1999. Cell biology, p. 101-138. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 27.Havenaar, R., and J. H. J. Huis in't Veld. 1992. Probiotics: a general review, p. 151-170. In B. J. B. Wood (ed.), The lactic acid bacteria. Chapman & Hall, London, United Kingdom.
- 28.Hoelzle, L. E., G. Steinhausen, and M. M. Wittenbrink. 2000. PCR-based detection of chlamydial infection in swine and subsequent PCR-coupled genotyping of chlamydial omp1-gene amplicons by DNA-hybridization, RFLP-analysis, and nucleotide sequence analysis. Epidemiol. Infect. 125:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, L. Z., R. R. Marquardt, and X. Zhao. 2000. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Appl. Environ. Microbiol. 66:4200-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielstein, P., H. Stellmacher, F. Horsch, and J. Martin. 1983. Chlamydia infection of swine. 1. Experimental chlamydia pneumonia of swine. Arch. Exp. Veterinarmed. 37:569-586. (In German.) [PubMed] [Google Scholar]
- 31.Krull, M., A. C. Klucken, F. N. Wuppermann, O. Fuhrmann, C. Magerl, J. Seybold, S. Hippenstiel, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 162:4834-4841. [PubMed] [Google Scholar]
- 32.Lievin-Le Moal, V., R. Amsellem, A. L. Servin, and M. H. Coconnier. 2002. Lactobacillus acidophilus (strain LB) from the resident adult human gastrointestinal microflora exerts activity against brush border damage promoted by a diarrhoeagenic Escherichia coli in human enterocyte-like cells. Gut 50:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilly, D. M., and R. H. Stillwell. 1965. Probiotics: growth-promoting factors produced by microorganisms. Science 147:747-748. [DOI] [PubMed] [Google Scholar]
- 34.Lu, L., and W. A. Walker. 2001. Pathologic and physiologic interactions of bacteria with the gastrointestinal epithelium. Am. J. Clin. Nutr. 73:1124S-1130S. [DOI] [PubMed] [Google Scholar]
- 35.Mabey, D. C., A. W. Solomon, and A. Foster. 2003. Trachoma. Lancet 362:223-229. [DOI] [PubMed] [Google Scholar]
- 36.Mack, D. R., S. Michail, S. Wei, L. McDougall, and M. A. Hollingsworth. 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276:G941-G950. [DOI] [PubMed] [Google Scholar]
- 37.Meijer, A., G. J. Kwakkel, A. de Vries, L. M. Schouls, and J. M. Ossewaarde. 1997. Species identification of chlamydia isolates by analyzing restriction fragment length polymorphism of the 16S-23S rRNA spacer region. J. Clin. Microbiol. 35:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nietfeld, J. C., B. H. Janke, P. Leslie-Steen, D. J. Robison, and D. H. Zeman. 1993. Small intestinal Chlamydia infection in piglets. J. Vet. Diagn. Investig. 5:114-117. [DOI] [PubMed] [Google Scholar]
- 39.Nietfeld, J. C., P. Leslie-Steen, D. H. Zeman, and D. Nelson. 1997. Prevalence of intestinal chlamydial infection in pigs in the Midwest, as determined by immunoperoxidase staining. Am. J. Vet. Res. 58:260-264. [PubMed] [Google Scholar]
- 40.Perdigon, G., S. Alvarez, M. Rachid, G. Aguero, and N. Gobbato. 1995. Immune system stimulation by probiotics. J. Dairy Sci. 78:1597-1606. [DOI] [PubMed] [Google Scholar]
- 41.Perdigon, G., M. Locascio, M. Medici, A. Pesce de Ruiz Holgado, and G. Oliver. 2003. Interaction of bifidobacteria with the gut and their influence in the immune function. Biocell 27:1-9. [PubMed] [Google Scholar]
- 42.Perry, L. L., and S. Hughes. 1999. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect. Immun. 67:3686-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poppert, S., A. Essig, R. Marre, M. Wagner, and M. Horn. 2002. Detection and differentiation of chlamydiae by fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:4081-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pospischil, A., R. Thoma, M. Hilbe, P. Grest, D. Zimmermann, and J. O. Gebbers. 2002. Abortion in humans caused by Chlamydophila abortus (Chlamydia psittaci serovar 1). Schweiz. Arch. Tierheilkd. 144:463-466. (In German.) [DOI] [PubMed] [Google Scholar]
- 45.Pospischil, A., and R. L. Wood. 1987. Intestinal Chlamydia in pigs. Vet. Pathol. 24:568-570. [DOI] [PubMed] [Google Scholar]
- 46.Rogers, D. G., and A. A. Andersen. 2000. Intestinal lesions caused by a strain of Chlamydia suis in weanling pigs infected at 21 days of age. J. Vet. Diagn. Investig. 12:233-239. [DOI] [PubMed] [Google Scholar]
- 47.Rogers, D. G., and A. A. Andersen. 1996. Intestinal lesions caused by two swine chlamydial isolates in gnotobiotic pigs. J. Vet. Diagn. Investig. 8:433-440. [DOI] [PubMed] [Google Scholar]
- 48.Rogers, D. G., A. A. Andersen, A. Hogg, D. L. Nielsen, and M. A. Huebert. 1993. Conjunctivitis and keratoconjunctivitis associated with chlamydiae in swine. J. Am. Vet. Med. Assoc. 203:1321-1323. [PubMed] [Google Scholar]
- 49.Rolfe, R. D. 2000. The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130:396S-402S. [DOI] [PubMed] [Google Scholar]
- 50.Saavedra, L., M. P. Taranto, F. Sesma, and G. F. de Valdez. 2003. Homemade traditional cheeses for the isolation of probiotic Enterococcus faecium strains. Int. J. Food Microbiol. 88:241-245. [DOI] [PubMed] [Google Scholar]
- 51.Sachse, K., E. Grossmann, C. Jager, R. Diller, and H. Hotzel. 2003. Detection of Chlamydia suis from clinical specimens: comparison of PCR, antigen ELISA, and culture. J. Microbiol. Methods 54:233-238. [DOI] [PubMed] [Google Scholar]
- 52.Sachse, K., and H. Hotzel. 2003. Detection and differentiation of chlamydiae by nested PCR. Methods Mol. Biol. 216:123-136. [DOI] [PubMed] [Google Scholar]
- 53.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed] [Google Scholar]
- 54.Saikku, P., M. Leinonen, L. Tenkanen, E. Linnanmaki, M. R. Ekman, V. Manninen, M. Manttari, M. H. Frick, and J. K. Huttunen. 1992. Chronic Chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki Heart Study. Ann. Intern. Med. 116:273-278. [DOI] [PubMed] [Google Scholar]
- 55.Salminen, S., E. Isolauri, and E. Salminen. 1996. Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie Leeuwenhoek 70:347-358. [DOI] [PubMed] [Google Scholar]
- 56.Sarma, D. K., M. K. Tamuli, T. Rahman, B. R. Boro, B. C. Deka, and C. K. Rajkonwar. 1983. Isolation of chlamydia from a pig with lesions in the urethra and prostate gland. Vet. Rec. 112:525. [DOI] [PubMed] [Google Scholar]
- 57.Scharek, L., J. Guth, K. Reiter, K. D. Weyrauch, D. Taras, P. Schwerk, P. Schierack, M. F. G. Schmidt, L. H. Wieler, and K. Tedin. 2005. Influence of a probiotic Enterococus faecium strain on development of the immune system of sows and piglets. Vet. Immunol. Immunopathol. 105:151-161. [DOI] [PubMed] [Google Scholar]
- 58.Schiller, I., R. Koesters, R. Weilenmann, B. Kaltenboeck, and A. Pospischil. 1997. Polymerase chain reaction (PCR) detection of porcine Chlamydia trachomatis and ruminant Chlamydia psittaci serovar 1 DNA in formalin-fixed intestinal specimens from swine. Zentbl. Veterinarmed. B 44:185-191. [DOI] [PubMed] [Google Scholar]
- 59.Schiller, I., A. Schifferli, P. Gysling, and A. Pospischil. 2004. Growth characteristics of porcine chlamydial strains in different cell culture systems and comparison with ovine and avian chlamydial strains. Vet. J. 168:74-80. [DOI] [PubMed] [Google Scholar]
- 60.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 62.Shewen, P. E. 1980. Chlamydial infection in animals: a review. Can. Vet. J. 21:2-11. [PMC free article] [PubMed] [Google Scholar]
- 63.Silva, A. M., E. A. Bambirra, A. L. Oliveira, P. P. Souza, D. A. Gomes, E. C. Vieira, and J. R. Nicoli. 1999. Protective effect of bifidus milk on the experimental infection with Salmonella enteritidis subsp. typhimurium in conventional and gnotobiotic mice. J. Appl. Microbiol. 86:331-336. [DOI] [PubMed] [Google Scholar]
- 64.Storz, J., and B. Kaltenboeck. 1993. The Chlamydiales, p. 27-64. In Z. Woldehiwet and M. Ristic (ed.), Rickettsial and chlamydial diseases of domestic animals. Pergamon Press, Oxford, United Kingdom.
- 65.Suchland, R. J., W. M. Geisler, and W. E. Stamm. 2003. Methodologies and cell lines used for antimicrobial susceptibility testing of Chlamydia spp. Antimicrob. Agents Chemother. 47:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szeredi, L., I. Schiller, T. Sydler, F. Guscetti, E. Heinen, L. Corboz, E. Eggenberger, G. E. Jones, and A. Pospischil. 1996. Intestinal Chlamydia in finishing pigs. Vet. Pathol. 33:369-374. [DOI] [PubMed] [Google Scholar]
- 67.Todoriki, K., T. Mukai, S. Sato, and T. Toba. 2001. Inhibition of adhesion of food-borne pathogens to Caco-2 cells by Lactobacillus strains. J. Appl. Microbiol. 91:154-159. [DOI] [PubMed] [Google Scholar]
- 68.Umesaki, Y., and H. Setoyama. 2000. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2:1343-1351. [DOI] [PubMed] [Google Scholar]
- 69.Willigan, D. A., and P. D. Beamer. 1955. Isolation of a transmissible agent from pericarditis of swine. J. Am. Vet. Med. Assoc. 126:118-122. [PubMed] [Google Scholar]
- 70.Wittenbrink, M. M., X. Wen, N. Bohmer, G. Amtsberg, and A. Binder. 1991. Bacteriologic studies of the occurrence of Chlamydia psittaci in organs of swine and in aborted swine fetuses. Zentbl. Veterinarmed. B 38:411-420. (In German.) [PubMed] [Google Scholar]
- 71.Woollen, N., E. K. Daniels, T. Yeary, H. W. Leipold, and R. M. Phillips. 1990. Chlamydial infection and perinatal mortality in a swine herd. J. Am. Vet. Med. Assoc. 197:600-601. [PubMed] [Google Scholar]
- 72.Zahn, I., L. Szeredi, I. Schiller, U. Straumann Kunz, E. Burgi, F. Guscetti, E. Heinen, L. Corboz, T. Sydler, and A. Pospischil. 1995. Immunohistochemical determination of Chlamydia psittaci/pecorum and C. trachomatis in the piglet gut. Zentbl. Veterinarmed. Berlin 42:266-276. (In German.) [PubMed] [Google Scholar]