Abstract
Porphyromonas gingivalis, one of the causative agents of adult periodontitis, attaches and forms biofilms on substrata of Streptococcus gordonii. Coadhesion and biofilm development between these organisms requires the interaction of the short fimbriae of P. gingivalis with the SspB streptococcal surface polypeptide. In this study we investigated the structure and binding activities of the short fimbriae of P. gingivalis. Electron microscopy showed that isolated short fimbriae have an average length of 103 nm and exhibit a helical structure with a pitch of ca. 27 nm. Mfa1, the major protein subunit of the short fimbriae, bound to SspB protein, and this reaction was inhibited by purified recombinant Mfa1 and monospecifc anti-Mfa1 serum in a dose-dependent manner. Complementation of a polar Mfa1 mutant with the mfa1 gene restored the coadhesion phenotype of P. gingivalis. Hence, the Mfa1 structural fimbrial subunit does not require accessory proteins for binding to SspB. Furthermore, the interaction of Mfa1 with SspB is necessary for optimal coadhesion between P. gingivalis and S. gordonii.
Porphyromonas gingivalis, a gram-negative anaerobe, is recognized as one of the primary pathogens in severe manifestations of adult periodontitis (33). P. gingivalis colonizes the dental plaque biofilm that accumulates on the supragingival and subgingival tooth surfaces (37, 43). Dental plaque is complex and dynamic, and the initial colonizers comprise predominantly gram-positive commensals such as Streptococcus gordonii and related streptococci, as well as Actinomyces species (14, 21, 30). Later biofilm inhabitants such as P. gingivalis are capable of binding to the antecedent organisms, and these attachment mechanisms are thought to drive the temporal and spatial development of pathogenic plaque (14, 17, 30, 32, 35). In addition, the coadhesion of P. gingivalis with primary colonizing bacteria such as streptococci may be important in the invasion of dentinal tubules by P. gingivalis (19).
P. gingivalis adherence to S. gordonii is multimodal and involves at least two distinct sets of adhesins and receptors. The long (major) fimbriae of P. gingivalis are predominantly comprised of the FimA protein and interact with glyceraldehyde 3-phosphate dehydrogenase on the streptococcal surface (20). Subsequent accretion of P. gingivalis into a mixed species biofilm requires an additional interaction between the short (minor) fimbriae and the Ssp major surface proteins on the streptococcal surface (16). The short fimbriae of P. gingivalis have been described independently by two groups (8, 27) as 0.1 to 0.5 μm in length and antigenically and genetically distinct from the long fimbriae (FimA) that can extend up to 3 μm (8, 27, 47). The short fimbriae are comprised predominantly of the Mfa1 protein. The possible contribution of minor fimbrial components to structure and function has not been investigated, although the mfa1 gene was shown to be cotranscribed with the downstream gene PG0179 (4).
The Ssp proteins are members of the antigen I/II family of streptococcal surface proteins that are highly conserved across all the human oral streptococcal species (2, 13). However, despite the high degree of structural similarity, P. gingivalis can discriminate between antigen I/II proteins from different species. In particular, P. gingivalis adheres to SspA and SspB proteins of S. gordonii but not to the antigen I/II homologue of Streptococcus mutans, SpaP. This species-specific interaction is determined by a discrete domain designated BAR (SspB adherence region), that spans amino acid residues 1167 to 1250 of SspB (2, 5). Within this domain, the amino acids asparagine at position 1182 and valine at 1185 appear to confer a unique structural determinant that is required for binding and is not conserved in S. mutans SpaP (5).
In this study, we have examined the morphology of the short fimbriae and defined the role of the Mfa1 protein as a receptor for SspB BAR. Purified recombinant Mfa1 (rMfa) and antibodies to this protein competitively inhibited P. gingivalis-S. gordonii binding. Furthermore, rMfa bound in a dose-dependent manner to the SspB BAR peptide. Complementation of a P. gingivalis Mfa1-deficient mutant with full-length mfa1 in trans restored binding activity of the P. gingivalis strain for S. gordonii. Thus, the Mfa1 major structural subunit protein of the short fimbriae of P. gingivalis is responsible for binding to the S. gordonii SspB BAR domain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are listed in Table 1. P. gingivalis 33277 and derivatives were grown anaerobically at 37°C in Trypticase soy broth supplemented, per liter, with 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione. When necessary, gentamicin, erythromycin, or tetracycline was added to the medium at a final concentration of 200, 10, or 5 μg/ml, respectively. Solid medium was prepared by supplementation with 5% sheep blood and 1.5% agar. S. gordonii DL1 was cultured under static conditions in Trypticase peptone broth supplemented with 5 g of yeast extract per liter and with 0.5% glucose as a carbon source. Escherichia coli strains were grown in Luria-Bertani broth containing, when necessary, the antibiotics ampicillin (100 μg/ml) or trimethoprim (200 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| S. gordonii DL-1 | Laboratory stock; 28 | |
| P. gingivalis | ||
| 33277 | Type strain from ATCC | Laboratory stock |
| SMF1 | Derivative of 33277 with an insertional inactivation of the mfa1 gene; Emr | 16 |
| cSMF1 | SMF1 containing pT-MFA, complemented strain; Emr Tcr | This study |
| KDP98 | Derivative of 33277 with an insertional inactivation of the fimA gene; Emr | 42 |
| E. coli | ||
| DH5α | F φ80dlacZΔ(lacZYA-argF)U169 endA1 supE44 recA1 relA1 | BRL |
| BL21(DE3)pLysS | Host for pET30 vector | Novagen |
| Plasmids | ||
| pET30 | pET expression vector | Novagen |
| pET-MFA | pET-30 with the mfa1 open reading frame | This study |
| pT-COW | Shuttle vector plasmid; Amr Tcr in E. coli; Tcr in P. gingivalis Mob+ Rep+ | N. Shoemaker |
| pT-MFA | pT-COW containing a 2.5-kb fragment containing the upstream and coding region of the mfa1 gene | This study |
Resistance to erythromycin (Emr), tetracycline (Tcr), and ampicillin (Amr) is indicated. Mob+, mobilizable; Rep+, ability to replicate in P. gingivalis.
Recombinant proteins and antiserum.
Recombinant Mfa1 protein was produced by PCR amplification of the mfa1 coding sequences on the P. gingivalis 33277 chromosomal DNA using primers designed from the TIGR genome sequence. The amplification product was cloned into the pET-30 expression system (Novagen) with the resulting plasmid encoding the full-length Mfa1 protein containing a C-terminal penta-histidine tag. After induction in E. coli, rMfa was purified by chromatography over an Ni2+ metal chelation resin and by elution with imidazole. Purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and purified rMfa was used to produce monospecific rabbit antibodies by Covance (Denver, PA). Synthetic BAR peptide representing residues 1167 to 1193 of SspB was synthesized by BioSynthesis (Lewisville, TX). Purity of peptide was assessed by high-pressure liquid chromatography to be >95%.
Coprecipitation.
SspB protein was used to coprecipitate its P. gingivalis binding partner (Mfa1) essentially as described previously (4). SspB protein (5) was incubated with biotin-labeled P. gingivalis cell extract for 2 h at room temperature, followed by incubation with SspB antibodies (5) at room temperature for 30 min. Protein A-Sepharose beads were added and reacted at 4°C for 1 h. The beads were recovered by centrifugation at 2,500 × g for 5 min, washed four times in phosphate-buffered saline (PBS) buffer, and resuspended in 100 μl of SDS-PAGE sample buffer. The suspension was subjected to SDS-PAGE and immunoblotting. The blots were visualized with avidin-peroxidase conjugate and 3,3′-diaminobenzidine tetrahydrochloride or with Mfa antibodies. Controls included omission of SspB protein, omission of SspB antibodies, and the use of preimmune serum.
Protein identification.
Protein bands were excised from Coomassie blue-stained SDS-PAGE gels, and sequencing was performed using in situ proteolytic digestion, tandem mass spectrometry, and the SEQUEST database search program as previously described (3). Briefly, a Finnigan TSQ 7000 triple quadrupole mass spectrometer coupled to a capillary high-pressure liquid chromatograph was used to generate collision-induced dissociation daughter ion mass spectra (MS2) from electrospray-derived parent ions representing intact tryptic fragments from proteins contained in the gel band. The MS2 mass spectra were searched against an in-house-modified version of the P. gingivalis W83 open reading frame database that contained additional GenBank sequences derived from strain 33277. All high-scoring SEQUEST hits (6, 44) against the P. gingivalis database (3, 39) were verified by manual interpretation of the MS2 mass spectra (12).
Complementation of the mfa1 gene.
Shuttle vector plasmid pT-COW (7) (kindly provided by N. Shoemaker, University of Illinois) was used for complementation. The encoding region of mfa1 along with 820 bp of upstream sequence was amplified by high-fidelity PCR using ProofStart DNA polymerase (QIAGEN, Inc.), and the PCR product was cloned into pT-COW. The recombinant plasmid, pT-MFA, was introduced by conjugation into the Mfa1-deficient mutant, P. gingivalis SMF1 (16), as described previously (29), to create P. gingivalis cSMF1. After conjugation, erythromycin- and tetracycline-resistant transconjugants were selected, and plasmid identity was confirmed by restriction analysis.
Immunoblotting of P. gingivalis.
Expression of Mfa was investigated by immunoblotting. Lysates of 5 × 106 P. gingivalis cells were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Antisera to rMfa or to P. gingivalis 33277 whole cells (45) (1:10,000) were used as probes with peroxidase-conjugated secondary antibody (1:3,000). Antigen-antibody binding was developed with 0.05% diaminobenzidine tetrahydrochloride.
ELISA.
The level of cell surface Mfa1 protein was determined by enzyme-linked immunosorbent assay (ELISA) after adsorption of P. gingivalis strains onto Maxisorp plates (Nunc). Briefly, P. gingivalis cells were harvested, washed with PBS, and fixed with 0.5% formalin in PBS at 4°C overnight. After three washes with PBS, 105 cells were added to each well for 2 h at 4°C. After being washed to remove unbound bacteria, P. gingivalis cells were reacted with rMfa antibodies (1:10,000) followed by peroxidase-conjugated secondary antibody (1:3,000), each for 1 h at 37°C. Antigen-antibody binding was determined by a colorimetric reaction using the 3,3′,5,5′-tetramethylbenzidine liquid substrate (Sigma Aldrich) in a Bio-Rad Benchmark Microplate reader at 655 nm. Antiserum to P. gingivalis 33277 whole cells was used as control to compare the cell numbers fixed in each well.
Interbacterial binding assay.
Adherence of P. gingivalis to S. gordonii was determined with a nitrocellulose blot assay as described previously (5, 18). Briefly, S. gordonii cells were suspended in PBS, and 108 bacteria were deposited on nitrocellulose paper in a dot blot apparatus. After adsorption of the streptococci, the filter was washed three times in PBS containing 0.1% Tween 20 (PBS-T) and incubated for 1 h at room temperature with [3H]thymidine-labeled P. gingivalis (5 × 10−4 mean cpm/cell) suspended in PBS-T. After being washed with PBS-T to remove unbound organisms, the experimental areas of the filter were excised, and bound P. gingivalis was quantitated by scintillation spectroscopy. For antibody inhibition experiments, radiolabeled P. gingivalis cells were incubated with antiserum for 1 h at 37°C prior to incubation with immobilized streptococci. No visible aggregates of P. gingivalis cells were observed by phase microscopy after reaction with the anti-Mfa serum. Inhibition of coadhesion by purified rMfa protein was determined by exposing the adsorbed S. gordonii cells to rMfa for 1 h at 37°C prior to the addition of labeled P. gingivalis. Controls for nonspecific inhibition were preimmune sera and recombinant P. gingivalis SerB protein (a putative serine phosphatase) that was produced by the same procedure as rMfa.
Protein binding assay.
BAR peptide (0 to 30 μg/ml) was immobilized on Xenobind (Xenopore Corporation) covalent binding microwell plates overnight at room temperature according to the manufacturer's protocol. The plate was subsequently washed with PBS containing 0.1% Triton X-100 (PBS-X) and blocked for 2 h with 10% fetal bovine serum at 37°C. After the plate was washed with PBS-X, purified rMfa protein (20 μg/ml) was added and incubated for 2 h at room temperature. The plate was washed again with PBS-X, and the primary anti-Mfa antibody (1:5,000) was incubated for 1.5 h at room temperature. The reaction was developed using goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate and o-phenylenediamine dihydrochloride peroxidase substrate (Sigma Aldrich). Reactions were stopped with 1 M H2SO4 and analyzed using a Perkin Elmer Wallac Victor 3 microplate reader at 405 nm. Control binding of rMfa to fetal bovine serum was subtracted from values obtained with BAR.
Purification of Mfa1.
Native Mfa1 protein (75 kDa) was purified essentially as described previously (48). Because Mfa1 is not easily sheared off from the cell surface, purification was carried out from fractions of broken cells of a FimA-deficient mutant, KDP98, using gel filtration in the presence of SDS. To avoid breaking fimbriae, bacterial cells were disrupted in a French pressure cell by three passes at 100 MPa in the presence of 10 μg/ml of RNase and DNase without sonication. The degree of purity and identity of Mfa1 were verified by SDS-PAGE and Western blotting with antiserum against the rMfa protein.
Electron microscopy.
Purified Mfa1 at 1.8 mg of protein per ml in 10 mM Tris-HCl, pH 7.5, 0.01% SDS, and 0.01% NaN3 was diluted and negatively stained with 2% phosphotungstic acid (pH 7.4) and observed under a JEM 1010 transmission electron microscope (JEOL). Micrographs were digitized with a resolution of 2 pixels/nm, and electrically expanded images were used to measure the physical dimensions of 100 and 50 different fibers for length and pitch, respectively. The width of thicker and thinner portions was determined by measurement of 20 sites for each region.
RESULTS
Pysical characteristics of the short fimbriae.
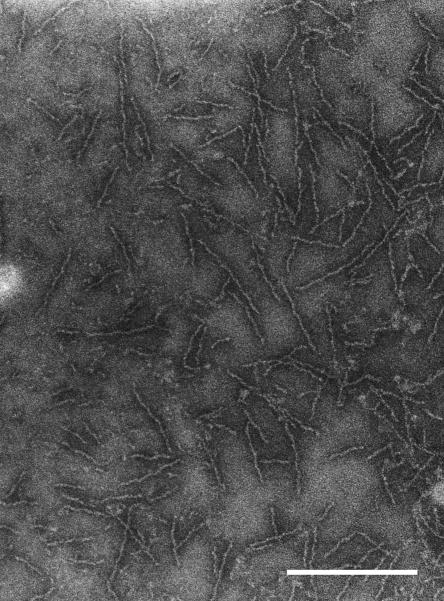
In order to examine the structure of the short fimbriae without contamination by long fimbriae, short fimbriae were purified from the FimA mutant strain KDP98. In addition, sonication was avoided during the isolation process to minimize shearing of the fimbrial structures. The fimbrial preparation was analyzed by electron microscopy, and a micrograph is shown in Fig. 1. The length of the purified short fimbriae, as determined from measurements of 100 fibers, ranged from 60 to 200 nm, consistent with previous observations (8). The majority of the fibers were 80 to 120 nm long, with an average of 103 nm. The short fimbriae displayed a helical structure, with a pitch of ca. 27 nm, and presented a thicker portion ca. 6.5 nm wide along with a thinner portion ca. 3.5 nm wide.
FIG. 1.
Electron micrograph of purified short fimbriae. The short fimbrial preparations were purified from P. gingivalis KDP98 and negatively stained with 2% phosphotungstic acid. Bar, 0.2 μm.
Amino acid sequence analysis of SspB ligand.
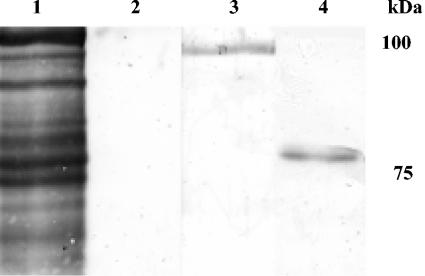
Previous study (4) of the P. gingivalis ligand for SspB showed that SspB coprecipitated a protein of ca. 100 kDa from a P. gingivalis surface extract. Amino acid sequencing of the 100-kDa protein identified it as Mfa1. In addition, disruption of the mfa1 gene resulted in loss of coadhesion and biofilm formation with S. gordonii. However, as disruption of mfa1 has a polar effect on the downstream cotranscribed gene PG0179, the contribution of Mfa1 to binding and biofilm development could not be determined. Furthermore, the predicted size of Mfa1 (61 kDa) combined with that of the cotranscribed PG0179 protein (37 kDa) would produce a composite protein of approximately 100 kDa. To clarify the identity of the 100-kDa protein recovered in the coprecipitation experiments and to determine the roles of the Mfa1 and PG0179 proteins in coadhesion and biofilm development, a series of experimental approaches was undertaken. We first repeated the pull-down assay described by Chung et al. (4) and coprecipitated P. gingivalis outer membrane proteins with SspB and anti-SspB antibodies. The resulting band was confirmed as containing Mfa1 by immunoblotting with Mfa antibodies (Fig. 2). This SspB-binding component of P. gingivalis was then subjected to more complete amino acid sequence analysis by tandem mass spectrometry of tryptic fragments. Sequences were obtained from peptides that covered more than 50% of Mfa1, including a peptide with a C terminus. No sequence was obtained that was derived from PG0179 or any other P. gingivalis protein. Thus, the SspB ligand would appear to be Mfa1 alone and is not a translational fusion or other composite protein. We conclude that the discrepancy in the sizes of Mfa1 observed by different methodologies results from the extended period of boiling required for complete denaturation of Mfa1 (Fig. 2, compare lanes 3 and 4). Mfa1 has a predicted molecular mass of 61 kDa; however, reported sizes based on SDS-PAGE do vary considerably, from 67 to 75 kDa (11, 27); thus, aberrant migration of this protein is not unusual. Moreover, multimers of Mfa1 subunits can form and require heating at over 100°C for more than 5 min to dissociate (8). In our coprecipitation assays, boiling for 10 min was required to generate a band of approximately 75 kDa (Fig. 2).
FIG. 2.
Blot analysis of P. gingivalis extract coprecipitated with SspB. Lane 1, biotinylated P. gingivalis extract only developed with avidin-peroxidase; lane 2, control without SspB antibodies in the coprecipitation reaction; lane 3, P. gingivalis extract coprecipitated with SspB, boiled in sample buffer for 5 min and developed with Mfa antibodies; lane 4, P. gingivalis extract coprecipitated with SspB, boiled in sample buffer for 10 min and developed with Mfa antibodies. Molecular mass markers are at right.
Inhibition of coadhesion between P. gingivalis and S. gordonii by Mfa1 antibody and recombinant protein.
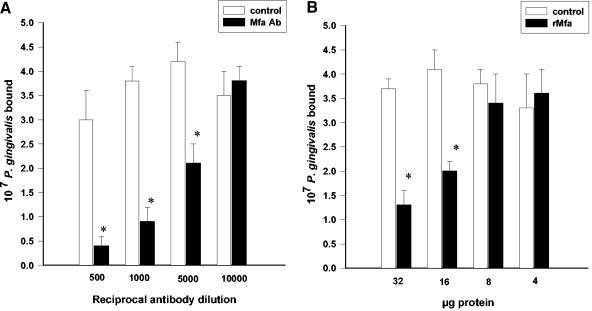
To examine the role of Mfa1 protein in the interbacterial interaction between whole cells of P. gingivalis and S. gordonii, antiserum to the rMfa protein (Fig. 3A) or purified rMfa protein itself (Fig. 3B) was tested for the ability to inhibit coadhesion. rMfa antibodies were able to impede the binding between P. gingivalis and S. gordonii by up to 88% in a dose-dependent manner. Furthermore, rMfa protein demonstrated dose-dependent inhibition of coadhesion up to 65%. These data indicate that the Mfa1 protein can engage Ssp proteins on the streptococcal cell surface and that this interaction is necessary for optimal adhesion of P. gingivalis to S. gordonii.
FIG. 3.
Recombinant Mfa1 protein or rMfa antibodies inhibit P. gingivalis-S. gordonii coadhesion. (A) Tritiated P. gingivalis cells (108) were reacted with rMfa antiserum or with preimmune serum (control) prior to incubation with streptococcal cells adsorbed to a nitrocellulose membrane. (B) Adsorbed S. gordonii cells were reacted with rMfa or control (SerB) protein prior to exposure to tritiated P. gingivalis cells (108). Error bars indicate standard deviations (n = 3). Asterisk, statistically significant in comparison to control (P < 0.01; t test).
Characteristics and binding properties of the complemented strain, P. gingivalis cSMF1.
To confirm that Mfa1 mediates P. gingivalis-S. gordonii coadhesion without the involvement of accessory proteins, an Mfa1-deficient mutant of P. gingivalis (SMF1) was complemented in trans with the intact mfa1 gene carried on plasmid pT-COW under the control of the endogenous mfa1 promoter. Expression of Mfa1 in the complemented strain cSMF1 was confirmed by immunoblot analysis. Figure 4A shows that cSMF1 expresses the Mfa1 protein and associated degradation products. Scanning densitometry and image analysis with Kodak 1D software revealed that the major anti-Mfa reactive band was 58% more intense in cSMF1 compared to the parental strain. Thus, expression of Mfa1 in the complemented strain is higher than in the parental strain. The most likely explanation for the increase in expression is that mfa is present on a multicopy plasmid in the recombinant strain. Localization of Mfa1 to the cell surface was determined by ELISA after fixation of P. gingivalis strains on 96-well plates. As shown in Fig. 4B, cSMF1 cells showed significantly higher (P < 0.001; t test) reactivity with anti-Mfa serum compared to preimmune serum. The ELISA data also confirmed greater (P < 0.001; t test) expression of Mfa1 in the complemented strain compared to the parent. In contrast, parent, mutant, and complemented mutant strains showed equivalent reactivity with whole P. gingivalis antiserum, indicating that equivalent numbers of bacteria were deposited and retained on the plates. Similar ELISA data were obtained with P. gingivalis cells cultured to mid-log and early stationary phases of growth (not shown). In an interbacterial binding assay, levels of cSMF1 binding to S. gordonii were comparable to those of the parental strain (Fig. 4C). The presence of higher levels of Mfa1 on the cSMF1 strain, thus, did not result in greater binding activity, possibly as a consequence of steric constraints or suboptimal presentation of recombinant protein. The Mfa1-deficient strain showed only low levels of accumulation on S. gordonii, consistent with previous reports (16). Thus, expression of mfa1 in a Mfa1-deficient background restores the ability of P. gingivalis to bind to S. gordonii. Furthermore, as the complemented strain is deficient in Pg0179 (due to the polar effect of the mutation of the chromosomal mfa1), these data provide corroborating evidence that the short fimbrial subunit itself is responsible for binding and does not require the presence of protein PG0179.
FIG. 4.
P. gingivalis strain cSMF1, complemented in trans with mfa1, expresses Mfa1 protein which is located on the cell surface and confers a coadhesion phenotype on the organism. (A) Immunoblot of whole-cell lysates. Lanes 1, strain 33277; lanes 2, SMF1 (Mfa1−); lanes 3, cSMF1 probed with antiserum to rMfa or to 33277 whole cells (1:10,000). Size standards (kDa) are shown in lane M. (B) ELISA of fixed cells of P. gingivalis strains 33277, SMF1, or cSMF1 probed with preimmune serum, antiserum to rMfa, or antiserum to 33277 whole cells. (C) Coadhesion of tritiated cells of P. gingivalis stains 33277, SMF1, or cSMF1 with S. gordonii cells adsorbed to a nitrocellulose membrane. Plot is the result of a curve-fitting algorithm using SigmaPlot V8. Error bars represent standard deviation (n = 3).
Binding of rMfa to BAR peptide.
SspB, the streptococcal ligand of Mfa1, possesses a discrete region, designated BAR, that spans amino acid residues 1167 to 1250 and is sufficient to promote adherence of P. gingivalis cells, without interacting with FimA (5, 16). To determine if rMfa recognized and bound to BAR, we examined the interaction of purified rMfa with an immobilized synthetic peptide representing the BAR sequence. As shown in Fig. 5, rMfa was able to bind to the synthetic BAR peptide in a dose-dependent fashion. Binding exhibited second-order kinetics, with half-maximal binding occurring at an input BAR concentration of approximately 2.5 μg/ml. This result is similar to the kinetics of binding of intact P. gingivalis cells to BAR (half-maximal binding at 2.0 μg/ml) previously reported (5). Thus, rMfa is capable of interacting with the coadhesion epitope of SspB, consistent with its role as the binding partner for SspB in P. gingivalis-S. gordonii coadhesion.
FIG. 5.
rMfa binds to synthetic BAR peptide in an ELISA format. Immobilized BAR was reacted with rMfa (20 μg/ml) and binding was detected with antiserum to rMfa (1:5,000). Plot is the result of a curve-fitting algorithm using SigmaPlot v8. Error bars represent standard deviations (n = 3). OD405nm, optical density at 405 nm.
DISCUSSION
Colonization of the oral cavity by P. gingivalis necessitates adherence to available surfaces such as the preexisting plaque biofilm on tooth surfaces. Consistent with this constraint, P. gingivalis can attach to a variety of common oral species including Fusobacterium nucleatum and S. gordonii (14, 17). Indeed, in vivo studies have shown that introduction of P. gingivalis into the mouths of human volunteers results in the organism locating almost exclusively on streptococcal-rich preformed plaque (32). S. gordonii is a major component of early plaque on the supragingival tooth surface and can also be found subgingivally (31, 34, 43). P. gingivalis cells can adhere to S. gordonii in vitro and accumulate into biofilm microcolonies on substrata of S. gordonii (16). Initial attachment of P. gingivalis to streptococcal cells involves interactions of the long fimbrial protein, FimA (15), with streptococcal glyceraldhehyde-3-phosphate dehydrogenase (20). While disruption of the fimA gene results in approximately a twofold reduction of coadhesion with S. gordonii, a FimA-deficient mutant of P. gingivalis retains the capability to form mixed species biofilms with S. gordonii in the absence of shear forces (16, 19). In contrast, disruption of the ssp genes of S. gordonii reduces coadhesion and ablates biofilm formation (16). Thus, coadhesion mediated through the S. gordonii Ssp proteins appears to be the predominant adhesive requirement for mixed species biofilm formation. Previous studies have demonstrated that the S. gordonii SspB protein binds to the short fimbriae of P. gingivalis (4). However, this report was inconclusive as regards the participation of accessory fimbrial components in the binding event. In particular, data from Chung et al. (4) suggested that the product of the PG0179 gene, which is immediately downstream of, and cotranscribed with, the structural subunit gene mfa1, may be associated with the short fimbriae and involved in binding to SspB. However, the results presented in the current study demonstrate that it is the Mfa1 protein that is necessary and sufficient for binding to SspB. This conclusion is based on the following observations: (i) purified recombinant Mfa1 protein can compete with P. gingivalis cells for binding sites on S. gordonii and impede coadhesion; (ii) monospecific antibodies to rMfa can inhibit P. gingivalis-S. gordonii coadhesion; (iii) in contrast, an Mfa1-deficient mutant that is also deficient in expression of the PG0179 gene is deficient in binding to S. gordonii, and complementation of this mutant with mfa1 in trans restores binding activity to wild-type levels; (iv) rMfa can bind to the BAR peptide that constitutes the adhesin domain of SspB. Moreover, the finding that coadhesion occurs between the complemented strain cSMF1 and S. gordonii, and the ability of rMfa to bind to the BAR domain of SspB, indicates that Mfa1 is the naturally occurring ligand of SspB. As SspB-dependent coadhesion is involved in the initiation of the events that lead to mixed species biofilm formation (16), it is possible that engagement of SspB by Mfa1 may initiate a signal transduction pathway within P. gingivalis that culminates in an adaptive response that allows the organism to adopt a “biofilm-ready” phenotype. Participation of additional signaling factors such as AI-2 is then required for biofilm formation to occur (22).
The Mfa1 protein was originally identified as a 75-kDa protein that copurified with the long fimbriae. Subsequently, the 75-kDa protein was purified as an immunodominant surface antigen forming a large, stable complex with an apparent molecular mass of about 2,000 kDa that was assumed (in the absence of morphological observation) to be a globular outer membrane or surface protein complex (48). Two independent groups later proposed the existence of short fimbriae, which they called minor and PgII fimbriae, with subunit molecular masses of 67 kDa and 72 kDa, respectively (8, 27). Little is known about the morphology of these fimbriae, however, in part because purification is difficult in the presence of the long fimbriae. Nonetheless, it is now well established that the 75-kDa protein, Mfa1 (67 kDa), and PgII (72 kDa) are the same polypeptides, based on their identical N-terminal amino acid sequences and extensive similarity of primary amino acid sequence deduced from the gene sequences (9, 26, 41). Furthermore, the short fimbriae comprised of Mfa1 are distinct from a third fimbrial type consisting of a 53-kDa protein (1). In this study, electron micrographs were taken of the short fimbriae purified from a FimA-deficient strain. The short fimbriae were slightly wider and much shorter (width, ca. 6.5 nm; average length, 103 nm) than the FimA fimbriae (width, ca. 5 nm; length, up to 3 μm) although both appear to exhibit a helical structure. The short fimbriae are also more uniform in length, consistent with the elution pattern of an almost symmetrical peak from gel filtration during purification (48), whereas the long FimA fimbriae elute as a much broader peak under similar conditions (data not shown). The fact that the short fimbriae were difficult to shear off the surface of bacteria without sonication, in contrast to the FimA fimbriae, is also consistent with fimbriae of a short type. Indeed, the short fimbriae were distributed to the soluble as well as the membrane fractions when cells were broken. Furthermore, a residual population remained in the peptidoglycan fraction even after intensive SDS extraction (23; unpublished data), indicating that they may bind or penetrate through the peptidoglycan layer. Such a location could potentially allow the short fimbriae to function as a part of signal transduction pathway following binding of SspB to the exposed surface regions of the molecule; however, more research is required to address this possibility.
Complementation of gene disruptions in P. gingivalis has been achieved in only a limited number of studies, mostly with proteolytic enzymes (25, 29) and with the response regulator fimR (24). In the case of surface fimbrial adhesins, Takahashi et al. (36) successfully expressed the long fimbriae on the surface of heterologous P. gingivalis stains that produce serologically distinct FimA. In this situation, with both recombinant and native FimA present, the recombinant fimbriae interfered with the adhesive activity of the host strain. In the current study we utilized the shuttle vector plasmid pT-COW (7, 24) for complementation of a P. gingivalis strain deficient in short fimbriae. Expressed Mfa protein derived from this plasmid was transported to the surface of P. gingivalis and displayed functional activity. As this shuttle vector replicates in P. gingivalis 33277, this system may prove to be applicable to the complementation in trans of mutations in other P. gingivalis genes for surface proteins.
Both the short and long fimbriae of P. gingivalis are comprised of a major structural subunit. Studies have suggested that proteins PG2134 and PG2135, encoded by genes downstream of fimA (PG2132), may be minor components of long fimbriae (40, 46). It is not known if the short fimbriae are comprised solely of Mfa1 or also contain minor components. It remains possible that PG0179, which is cotranscribed with Mfa1, could be a short fimbrial component or could be necessary for optimal presentation of the short fimbriae; however, PG0179 does not contribute to binding to S. gordonii. In addition to their distinct physical properties, and their individual streptococcal receptor specificity, the long and short fimbriae have other contrasting biological properties. The short fimbriae are more active in promoting bone resorption in rats and elicit a secreted cytokine profile from macrophages distinct from that of the long fimbriae (9, 10). Adherence of FimA and Mfa1 mutants to epithelial cells is also dissimilar (38). While FimA has been shown to bind to integrins on the gingival epithelial cell surface (45), the binding receptor for Mfa1 is unknown.
In conclusion, the short fimbriae of P. gingivalis are ca. 6.5 nm wide and on average 103 nm long. The short fimbrial subunit protein Mfa1 is the naturally occurring binding partner for the SspB streptococcal adhesin, and interaction between SspB and Mfa1 is necessary for the optimal coadhesion between P. gingivalis and S. gordonii.
Acknowledgments
This work was supported by grant DE12505 from the NIDCR (R.L.); by Grants-in-Aid for Scientific Research (C) (15591957 to F.Y. and 15561958 to Y.M.) from the Japan Society for the Promotion of Science; and by the AGU High-Tech Research Center Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan (F.Y. and Y.M.).
We thank H. Xie for helpful advice and T. Atsumi and J. Iwami for analyzing electron micrographs and for their technical assistance.
Editor: V. J. DiRita
REFERENCES
- 1.Arai, M., N. Hamada, and T. Umemoto. 2000. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol. Lett. 193:75-81. [DOI] [PubMed] [Google Scholar]
- 2.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W., K. E. Laidig, Y. Park, K. Park, J. R. Yates III, R. J. Lamont, and M. Hackett. 2001. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst 126:52-57. [DOI] [PubMed] [Google Scholar]
- 4.Chung, W. O., D. R. Demuth, and R. J. Lamont. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68:6758-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectra of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 7.Gardner, R. G., J. B. Russell, D. B. Wilson, G. R. Wang, and N. B. Shoemaker. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 62:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamada, N., H. T. Sojar, M.-I. Cho, and R. J. Genco. 1996. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 64:4788-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada, N., K. Watanabe, M. Arai, H. Hiramine, and T. Umemoto. 2002. Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis. Oral Microbiol. Immunol. 17:197-200. [DOI] [PubMed] [Google Scholar]
- 10.Hiramine, H., K. Watanabe, N. Hamada, and T. Umemoto. 2003. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol. Lett. 229:49-55. [DOI] [PubMed] [Google Scholar]
- 11.Hongyo, H., S. Kokeguchi, H. Kurihara, M. Miyamoto, H. Maeda, S. Takashiba, and Y. Murayama. 1998. Comparative study of two outer membrane protein genes from Porphyromonas gingivalis. Microbios 95:91-100. [PubMed] [Google Scholar]
- 12.Hunt, D. F., J. R. Yates III, J. Shabanowitz, S. Winston, and C. R. Hauer. 1986. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 83:6233-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkinson, H. F., and R. J. Lamont. 1997. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 8:175-200. [DOI] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamont, R. J., C. A. Bevan, S. Gil, R. E. Persson, and B. Rosan. 1993. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 8:272-276. [DOI] [PubMed] [Google Scholar]
- 16.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 17.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 18.Lamont, R. J., and B. Rosan. 1990. Adherence of mutans streptococci to other oral bacteria. Infect. Immun. 58:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love, R. M., M. D. McMillan, Y. Park, and H. F. Jenkinson. 2000. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon binding specificity of streptococcal antigen I/II adhesin. Infect. Immun. 68:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 22.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami, Y., T. Masuda, M. Imai, J. Iwami, H. Nakamura, T., Noguchi, and F. Yoshimura. 2004. Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol. Immunol. 48:561-569. [DOI] [PubMed] [Google Scholar]
- 24.Nishikawa, K., and F. Yoshimura. 2001. The response regulator FimR is essential for fimbrial production of the oral anaerobe Porphyromonas gingivalis. Anaerobe 7:255-262. [Google Scholar]
- 25.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa, T., H. Mori, K. Yasuda, and M. Hasegawa. 1994. Molecular cloning and characterization of the genes encoding the immunoreactive major cell-surface proteins of Porphyromonas gingivalis. FEMS Microbiol. Lett. 120:23-30. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa, T., K. Yasuda, K. Yamada, H. Mori, K. Ochiai, and M. Hasegawa. 1995. Immunochemical characterisation and epitope mapping of a novel fimbrial protein (Pg-II fimbria) of Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 11:247-255. [DOI] [PubMed] [Google Scholar]
- 28.Pakula, R., and W. Walczak. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31:125-133. [DOI] [PubMed] [Google Scholar]
- 29.Park, Y., B. Lu, C. Mazur, and B. C. McBride. 1997. Inducible expression of a Porphyromonas gingivalis W83 membrane-associated protease. Infect. Immun. 65:1101-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 31.Scannapieco, F. A., L. Solomon, and R. O. Wadenya. 1994. Emergence of human dental plaque and host distribution of amylase-binding streptococci. J. Dent. Res. 73:1627-1635. [DOI] [PubMed] [Google Scholar]
- 32.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 34.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 35.Stinson, M. W., K. Safulko, and M. J. Levine. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi, Y., H. Yoshimoto, D. Kato, N. Hamada, M. Arai, and T. Umemoto, T. 2001. Reduced fimbria-associated activities of Porphyromonas gingivalis induced by recombinant fimbrial expression. FEMS Microbiol. Lett. 195:217-222. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, S., Y. Murakami, K. Seto, K. Takamori, M. Yosida, K. Ochiai, S. Watanabe, and S. Fujisawa. 2003. The detection of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in the supragingival plaque of children with and without caries. Pediatr. Dent. 25:143-148. [PubMed] [Google Scholar]
- 38.Umemoto, T., and N. Hamada. 2003. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J. Periodontol. 74:119-122. [DOI] [PubMed] [Google Scholar]
- 39.Wang, T., Y. Zhang, W. Chen, Y. Park, R. J. Lamont, and M. Hackett. 2002. Reconstructed protein arrays from 3D HPLC/tandem mass spectrometry and 2D gels: complementary approaches to Porphyromonas gingivalis protein expression. Analyst 127:1450-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe, K., T. Onoe, M. Ozeki, Y. Shimizu, T. Sakayori, H. Nakamura, and F. Yoshimura. 1996. Sequence and product analyses of the four genes downstream from the fimbrilin gene (fimA) of the oral anaerobe Porphyromonas gingivalis. Microbiol. Immunol. 40:725-734. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, K., T. Takasawa, F. Yoshimura, M. Ozeki, M. Kawanami, and H. Kato. 1992. Molecular cloning and expression of a major surface protein (the 75-kDa protein) of Porphyromonas (Bacteroides) gingivalis in Escherichia coli. FEMS Microbiol. Lett. 92:47-56. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe-Kato, T., J. Hayashi, Y. Terazawa, C. I. Hoover, K. Nakayama, T. Noguchi, and F. Yoshimura. 1998. Isolation and characterization of transposon-induced mutants of Porphyromonas gingivalis deficient in fimbriation. Microb. Pathogen. 24:25-35. [DOI] [PubMed] [Google Scholar]
- 43.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722-732. [DOI] [PubMed] [Google Scholar]
- 44.Yates, J. R., III, J. K. Eng, A. L. McCormack, and D. Schieltz. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426-1436. [DOI] [PubMed] [Google Scholar]
- 45.Yilmaz, Ö., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 46.Yoshimura, F., Y. Takahashi, E. Hibi, T. Takasawa, H. Kato, and D. P. Dickinson. 1993. Proteins with molecular masses of 50 and 80 kilodaltons encoded by genes downstream from the fimbrilin gene (fimA) are components associated with fimbriae in the oral anaerobe Porphyromonas gingivalis. Infect. Immun. 61:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshimura, F., K. Takahashi, Y. Nodasaka, and T. Suzuki. 1984. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 160:949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimura, F., K. Watanabe, T. Takasawa, M. Kawanami, and H. Kato. 1989. Purification and properties of a 75-kilodalton major protein, an immunodominant surface antigen, from the oral anaerobe Bacteroides gingivalis. Infect. Immun. 57:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]