Abstract
We demonstrate that among the three monofunctional catalases of Pseudomonas aeruginosa PA14, KatA and, to a lesser extent, KatB, but not KatE, are required for resistance to peroxide and osmotic stresses. KatA is crucial for adaptation to H2O2 stress and full virulence in both Drosophila melanogaster and mice. This dismantling of catalase roles represents a specialized catalytic system primarily involving KatA in responses to adverse environmental conditions.
Catalases are central components of the enzymatic detoxification pathways that prevent the formation of the highly reactive hydroxyl radical (HO.) by decomposing hydrogen peroxide (H2O2) and contribute to a variety of physiological processes involving adaptation and survival mechanisms. Because the toxicity of H2O2 released by phagocytes has been implicated in the host innate immune responses, bacterial pathogens exploit catalytic enzyme systems to survive the host environments. Although the involvement of catalases in virulence mechanisms has been demonstrated in many bacterial pathogens, little is known on the roles of catalases in the pathogenesis of Pseudomonas aeruginosa, an opportunistic human pathogen that is frequently associated with the alveolar surface and is most likely subjected to oxidative stresses within the pulmonary airways (1).
P. aeruginosa is a unique bacterium that has three differentially evolved monofunctional catalase genes, katA, katB, and katE, but no bifunctional catalase (catalase-peroxidase) gene on its genome (25). Like other clade 3 monofunctional catalases, the major catalase, KatA, is H2O2 inducible (4, 13, 18). The expression of a second H2O2-inducible catalase, KatB, that belongs to clade 1 is noteworthy (2, 14). The third catalase, KatE, is one of the clade 2 catalases that are highly conserved among most bacterial species (3, 14). With the exception of a Streptomyces coelicolor catalase (CatB) that plays important roles in osmoprotection and differentiation (5), little else is known about the physiological role of clade 2 catalases.
The purpose of this work is to systematically determine the roles of the three differentially evolved monofunctional catalases in stress responses and survival mechanisms of P. aeruginosa strain PA14 (20). We used nonpolar, unmarked deletion mutants of each catalase and investigated their resistance and adaptation in response to stress conditions in vitro and their virulence in Drosophila melanogaster and mice. We demonstrate that KatA plays an important role in virulence and oxidative and/or osmotic stress responses. In combination with the previously published studies (2, 10, 11, 18, 19, 26), these results suggest the specialized roles of P. aeruginosa catalases in response to environmental stresses and pathogenic interactions as well.
Construction of catalase mutants of P. aeruginosa PA14 and their resistance to H2O2.
The genome sequence of the P. aeruginosa reference strain PAO1 reveals three monofunctional catalase genes, katA, katB, and katE, of different evolutionary origins, clade 3, clade 1, and clade 2, respectively (13, 14). No homologue of bifunctional catalases has been found in P. aeruginosa thus far. Isolation of the gene encoding a manganese-containing nonheme pseudocatalase from lactic acid bacteria (12) and compilation of its homologous sequences from the bacterial genome sequence databases revealed a homologous gene (PA2185 or katM after Mn-catalase) on the 12th variable segment of the PAO1 genome (21).
Most P. aeruginosa strains, including PA14, do not harbor a katM gene (28; Heo and Cho, unpublished). As summarized in Table 1, we have created seven unmarked deletions (katA, katB, katE, katAB, katAE, katBE, and katABE) in strain PA14 to systematically address the potential role of catalases in stress responses and virulence (15).
TABLE 1.
P. aeruginosa strains used in this study
| Genotype | Strain name | Relevant characteristics | Reference or source |
|---|---|---|---|
| Wild type | PA14 | Prototrophic, virulent burn wound isolate | 20 |
| katA | PRL700 | PA14 ΔkatA (0.60-kb deletion of katA) | This study |
| katB | PRL800 | PA14 ΔkatB (0.72-kb deletion of katB) | This study |
| katE | PRL900 | PA14 ΔkatE (2.10-kb deletion of katE) | This study |
| katAB | PRL780 | PRL700 ΔkatB (0.72-kb deletion of katB) | This study |
| katAE | PRL790 | PRL700 ΔkatE (2.10-kb deletion of katE) | This study |
| katBE | PRL980 | PRL900 ΔkatB (0.72-kb deletion of katB) | This study |
| katABE | PRL798 | PRL790 ΔkatB (0.72-kb deletion of katB) | This study |
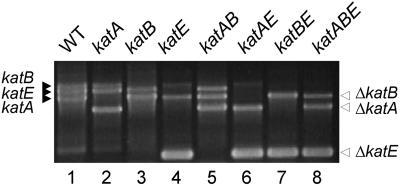
We have verified all the catalase mutants through genetic structure analyses by PCR and Southern hybridization (Fig. 1) (15), expression profiles by total catalase activity staining (15), and growth inhibition on plates containing 100 μM H2O2 (Fig. 2A). All the mutants exhibited doubling times similar to that of the wild type (15). The growth of the katA and to a lesser extent katB but not katE mutants was inhibited by H2O2. The contribution of KatB to H2O2 resistance was more evident in the katAB mutant (Fig. 2A).
FIG. 1.
Creation of catalase mutants. Based on the PAO1 sequences, PCR deletions of each monofunctional catalase were generated and used to create three single (katA, katB, and katE), three double (katAB, katAE, and katBE), and a triple (katABE) mutant in wild-type PA14 (WT) via homologous recombination followed by sacB-dependent segregation as summarized in Table 1. Multiplex PCR using three sets of primers was used to verify the predicted genetic structures of the mutants. The PCR product sizes of the intact genes (designated by the solid arrowhead on the left) for katA, katB, and katE were 2.1, 2.8, and 2.5 kb, respectively, whereas those of deletions (designated by the empty arrowhead on the right) were 1.5, 2.1, and 0.4 kb, respectively. Because the PCR products from the intact katA gene and the deleted katB gene are almost the same size, only two bands were observed for katB and katBE (lanes 3 and 7, respectively).
FIG. 2.
Oxidative stress resistance of catalase mutants. (A) Cells were grown in LB broth at 37°C to an optical density at 600 nm of 0.5. Five 10-fold serial dilutions of the cells in LB broth were spotted onto an LB agar medium containing 100 μM H2O2. (B) Complementation of H2O2-sensitive phenotype of the katA mutant was performed by introducing pUCP18-derived plasmids containing full-length fragments of katA (pUCP-KatA), katB (pUCP-KatB), and katE (pUCP-KatE). Cells were diluted as in A and spotted onto LB agar medium containing 100 μM H2O2 and 200 μg/ml carbenicillin. The numbers (from 106 to 102) indicate the CFU of the cell spots.
The H2O2 sensitivity of the katA mutant was completely restored by introducing the pUCP18 (22)-derived plasmid containing appropriate full-length catalase constructs (Fig. 2B). In contrast, multicopy expression of the katB gene only partially restored the H2O2 resistance of the katA mutant. We conclude that the KatA is most critical in H2O2 resistance, whereas resistance mediated by KatB was only discernible when KatA expression was abolished. In contrast, katE does not affect H2O2 resistance.
KatA is required for adaptation to H2O2 in P. aeruginosa PA14.
In an attempt to investigate the roles of catalases in the adaptation to H2O2 stress, we examined the sensitivity of PA14 to H2O2 in liquid culture. Mid-logarithmic PA14 cells were pretreated with a nonlethal level of H2O2 (1 mM) for 30 min before being exposed to the killing concentration of H2O2 (100 mM). A 30-min treatment time was chosen to investigate the steady-state response rather than the early and acute responses. The viability of cells was determined at 5-min intervals. Less than 0.1% of the unadapted or naive cells remained viable 10 min after exposure to 100 mM H2O2. In contrast, when cells were pretreated with 1 mM H2O2, survival was enhanced more than 1,000-fold (Fig. 3A). The sublethal pretreatment affected the cells' growth and/or survival compared to the control (∼60% survival as shown in Fig. 3B) and is slightly harsher than those previously described in other bacteria (7, 8, 9, 17).
FIG. 3.
Adaptation of catalase mutants to H2O2 stress. (A) PA14 cells were grown in LB broth at 37°C to an optical density at 600 nm of 0.5 and pretreated with 1 mM H2O2 for 30 min. Pretreated (•) and naive (○) cells were then exposed to 100 mM H2O2. Samples taken at designated time points were serially diluted and plated on LB agar to determine the numbers of CFU. The percentages of survival relative to the corresponding untreated cells at the zero time point are shown. Each point represents the mean of five independent experiments. (B) Cells (wild-type, katA, katB, and katAB) were grown in LB at 37°C to an optical density at 600 nm of 0.5, followed by a 30-min pretreatment with 1 mM H2O2. Pretreated cells (empty bar) were then exposed to 100 mM H2O2 for 10 min (solid bar). The percentages of survival relative to the untreated wild-type cells at the zero time point are shown. Each point represents the mean of five independent experiments with error bars representing standard deviations. Values in lanes 4, 7, and 8 are less than 0.1.
We analyzed all the catalase mutants in the adaptation experiment to determine whether catalases participate in the adaptive response to H2O2. The katA mutant was more sensitive to 1 mM H2O2 than the wild type was. Moreover, the katAB mutant was even more sensitive (<10−4 viability) to the pretreatment. Therefore, the residual survival (∼25%) of the katA mutant bacteria by the pretreatment may be attributed to KatB, which is in a good agreement with the results on solid agar culture (Fig. 2A). The H2O2 pretreatment enhanced the cells' resistance and viability against the killing concentration of H2O2, which was completely abolished in the katA mutant. Killing of the pretreated katB and katE mutant bacteria by 1 mM H2O2 was discernible (Fig. 3B and data not shown). This result suggests that the basal and/or inducible expression of KatA, but not KatB, is responsible for the adaptation to H2O2, despite the rapid induction of katB by H2O2 in the presence of functional KatA (19) (data not shown).
It is clear, however, that KatA and KatB have overlapping but distinct roles in oxidative stress responses, since the multicopy KatB failed to fully compensate for the absence of KatA in terms of H2O2 resistance and adaptation (data not shown). The catalytic functions involving both KatA and KatB during normal growth and oxidative stress remain to be further deciphered by combining this result with detailed and systematic gene expression analyses in each catalase mutant background with or without oxidative challenge.
KatA is preponderantly required for osmoprotection in P. aeruginosa PA14.
A minor catalase (CatB) from the actinomycete S. coelicolor is known to be required for resistance to osmotic stress and differentiation (5). We tested whether P. aeruginosa catalase mutants are susceptible to osmotic stresses. As shown in Fig. 4A, KatA was critical in the resistance to KCl treatments (0.8 M and 0.9 M), whereas deletion of katB or katE had no significant effect on salt resistance. However, the different KCl sensitivities of the katA and katAB mutants, depending on the KCl concentration, suggest that KatB may play a minor role in osmoresistance as in H2O2 resistance (Fig. 2).
FIG. 4.
Osmotic stress resistance of catalase mutants. Cells were grown in LB broth at 37°C to an optical density at 600 nm of 0.5. Three 10-fold serial dilutions of the cells in LB broth were spotted onto BDT (Bushnell-Hass minimal salt) agar medium containing 0.8 M or 0.9 M KCl (A) or 32% or 34% sucrose (B). The numbers (105, 104, and 103) indicate the CFU of the cell spots. (C) Complementation of the salt-sensitive phenotype of the katA mutant was performed by introducing pUCP18-derived plasmids (pUCP-KatA and pUCP-KatB) as in Fig. 2A. Dilutions were made in LB broth or in filter (0.2 μm)-sterilized spent culture supernatants from the stationary-phase cultures of either the wild type (+WT s/n) or the katA mutant (+katA s/n) cells and then spotted onto BDT agar medium containing 0.9 M KCl and 200 μg/ml carbenicillin. The numbers indicate the CFU of the cell spots.
Since KCl increases ionic strength as well as osmotic strength, we used a nonionic osmolyte, sucrose, with comparable amounts of KCl (23). As shown in Fig. 4B, sucrose treatments at 32% (∼0.89 M) and 34% (∼0.94 M) exhibited similar results as observed in KCl treatments, uncovering the involvement of KatA in sucrose resistance, although the responses to the two different concentrations were more subtle than those in the KCl treatments, especially in the katA and katAB mutants, indicating the minor role of KatB in this condition.
The sensitive phenotype of the katA mutant was restored by trans complementation with the pUCP18-derived plasmid expressing KatA (Fig. 4C). Unlike H2O2 sensitivity, however, multicopy KatB could not restore growth of the katA mutant on salt-containing media, which may imply differential functions and/or regulations of KatA and KatB in response to osmotic stress.
It is intriguing that the cell-free culture supernatant from the wild-type culture in the stationary growth phase could restore the KCl sensitivity of the katA mutant, although we were not sure whether or not the supplied activities absent in the culture supernatant of the katA mutant were working extracellularly. Further experimentation is needed to unravel how catalases such as P. aeruginosa KatA and S. coelicolor CatB protect against osmotic stresses. Considering that the general stress responses likely require alternative sigma factors (3, 6), it will be of special interest to analyze the gene expression in response to specific and general stress conditions.
KatA is required for virulence in P. aeruginosa PA14.
The in vitro oxidative and osmotic stress phenotypes of catalase mutants are most likely related to the survival pathways, and therefore likely implicated in virulence due to unfavorable conditions P. aeruginosa may encounter in the host environment. We examined whether the P. aeruginosa catalases play a role in host infection using the D. melanogaster model, since it was a simple alternative model host to evaluate P. aeruginosa virulence potentials, as measured by fly mortality and in vivo proliferation of P. aeruginosa (16, 27).
D. melanogaster infection was performed by pricking 2- to 5-day-old adult flies with 50 to 200 CFU of PA14 cells as described previously (16). Mortality was monitored at 25°C for up to 54 h postinfection (Fig. 5). Four catalase mutants (katA, katAB, katAE, and katABE) commonly deficient in KatA exhibited significant virulence attenuations in terms of delayed death kinetics (by more than 10 h) and lower mortality, whereas the remaining three mutants (katB, katE, and katBE) were as virulent as the wild type (Fig. 5A). Reintroduction of the full-length katA gene restored the attenuated virulence of the katA mutant to the wild-type level, whereas katA mutant cells harboring a multicopy plasmid expressing either KatB or KatE were still avirulent (Fig. 5B).
FIG. 5.
Virulence of catalase mutants in D. melanogaster. Fly mortality was determined using groups of 100 flies. Flies were infected with 50 to 200 CFU of the wild-type or mutant bacteria that had been grown in LB broth to an optical density at 600 nm of 3.0 and kept at 25°C. Flies that died within 12 h postinfection were excluded from mortality determination. Mortality studies were repeated at least five times with similar results. Mutants with katA deleted are indicated with solid symbols and those with intact katA are shown as open symbols. Symbols: ×, wild type; ⋄ and ♦, single mutants; ○ and •, double mutants; ▪, triple mutant.
The virulence attenuation of the katA mutant was verified by bacterial proliferation in D. melanogaster (Fig. 6). PA14 cells proliferate almost exponentially in flies, as described by Lee et al. (16), where the linear regression analyses from the 57 data points (from live flies) gave a slope of 0.1734, which is statistically significant (r2 = 0.897). The slope corresponds to a doubling time of 1.736 h. However, not all katA cells proliferate exponentially in flies, unlike the wild type. The bacterial proliferations from 78 live flies were delayed about 6 h, and some infected flies completely cleared the bacteria (Fig. 6B).
FIG. 6.
Bacterial proliferation in D. melanogaster. Batches of 10 flies were infected either with the wild type (A) or the katA mutant (B) as described for Fig. 5. Homogenates of individual infected flies were collected every 6 h up to 48 h postinfection and plated on LB agar to determine the CFU per fly. The CFU determined from live (open symbols) and dead (solid symbols) flies are shown in a log scale, showing a statistically significant linearity (*) only for the wild-type bacteria. The results are representative of three independent experiments. Symbols: ○ and •, wild type; ⋄ and ♦, katA mutant.
The involvement of KatA in virulence was further verified in mammalian hosts, using the mouse peritonitis model as described previously (24). The mice were monitored from 6 to 64 h after intraperitoneal challenge with 5 × 106 CFU of bacterial cells and regarded as dead when they displayed ruffled fur, evidence of dehydration, and nonresponsiveness to stimuli. More than 90% of the mice that had been infected with the wild-type cells died within 36 h in our experimental conditions (Fig. 7). As in D. melanogaster, the katA, katAB, katAE, and katABE mutants were less virulent in the mouse peritonitis model, with ∼40% mice surviving the infection, exhibiting delayed killing (by more than 20 h).
FIG. 7.
Virulence of catalase mutants in mice. LB broth-grown cells (optical density at 600 nm of 3.0) were harvested and washed twice with phosphate-buffered saline (150 mM NaCl, 20 mM phosphate, pH 7.0), and appropriately diluted to reach 5 × 106 CFU in 100 μl of phosphate-buffered saline containing 1% mucin, which helps to induce infection in naive mice as an adjuvant. Groups of 10 anesthetized BALB/c mice (4 to 6 weeks old) were infected intraperitoneally with 100 μl of bacterial suspension. Percentages of survivors over the indicated time points are shown. Each point represents the mean of three independent. Mutants with katA deleted are indicated by solid symbols and those with intact katA are shown asopen symbols. Symbols: ×, wild type; ⋄ and ♦, single mutants; ○ and •, double mutants; ▪, triple mutant.
Conclusion.
These phenotypic analyses of the three monofunctional catalases (KatA, KatB, and KatE) in P. aeruginosa PA14 suggest that the catalytic system of KatA is crucial for oxidative and osmotic stress responses. KatA is also required for adaptation to peroxide stress and for virulence of this bacterium, which is intuitively understandable in that it is critical for stress responses as well as adaptations in vitro that may resemble unfavorable host environments. It is also explainable in part by the regulation of katA, which involves quorum-sensing circuits (11).
The pivotal roles of KatA in virulence mechanisms can be further authenticated and generalized, by investigating its involvement in virulence of other P. aeruginosa strains such as PAO1, since the multifactorial nature of virulence pathways is related with the genetic backgrounds that accounts for different virulence potentials, and its expression and regulation in conjunction with related enzymes and regulators such as RpoS and OxyR.
Acknowledgments
We are grateful to Gee Lau for helpful comments.
This work was supported by grants from the 21C Frontier Microbial Genomics and Applications Center (MG05-0104-05-0) and the Korea Research Foundation (2004-015-C00505) and by a Special Research Grant from Sogang University (2002-3010) to Y.-H. Cho.
Editor: V. J. DiRita
REFERENCES
- 1.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 2.Brown, S. M., M. L. Howell, M. L. Vasil, A. J. Anderson, and D. J. Hassett. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 177:6536-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, Y.-H. 1999. Gene expression and the role of catalases in Streptomyces coelicolor A3(2). Ph.D. thesis. Seoul National University, Seoul, Korea.
- 4.Cho, Y.-H., and J.-H. Roe. 1997. Isolation and expression of the catA gene encoding the major vegetative catalase in Streptomyces coelicolor Müller. J. Bacteriol. 179:4049-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, Y.-H., E.-J. Lee, and J.-H. Roe. 2000. A developmentally regulated catalase required for proper differentiation and osmoprotection of Streptomyces coelicolor. Mol. Microbiol. 35:150-160. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y.-H., E.-J. Lee, B.-E. Ahn, and J.-H. Roe. 2001. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42:205-214. [DOI] [PubMed] [Google Scholar]
- 7.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 8.Demple, B., J. Halbrook, and S. Linn. 1983. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J. Bacteriol. 153:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowds, B. C., P. Murphy, D. J. McConnell, and K. M. Devine. 1987. Relationship among oxidative stress, growth cycle, and sporulation in Bacillus subtilis. J. Bacteriol. 169:5771-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassett, D. J., E. Alsabbagh, K. Parvatiyar, M. L. Howell, R. W. Wilmott, and U. A. Ochsner. 2000. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol. 182:4557-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi, T., Y. Kono, and K. Tanaka. 1996. Molecular cloning of manganese catalase from Lactobacillus plantarum. J. Biol. Chem. 271:29521-29524. [DOI] [PubMed] [Google Scholar]
- 13.Klotz, M. G., and P. C. Loewen. 2003. The molecular evolution of catalytic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 20:1098-1112. [DOI] [PubMed] [Google Scholar]
- 14.Klotz, M. G., G. R. Klassen, and P. C. Loewen. 1997. Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol. Biol. Evol. 14:951-958. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J.-S. 2005. Phenotypic analyses of of monofunctional catalases in Pseudomonas aeruginosa. M.S. thesis. Sogang University, Seoul, Korea.
- 16.Lee, J.-S., S.-H. Kim, and Y.-H. Cho. 2004. Dithiothreitol attenuates the pathogenic interaction between Pseudomonas aeruginosa and Drosophila melanogaster. J. Microbiol. Biotechnol. 14:367-372. [Google Scholar]
- 17.Lee, J.-S., Y.-C. Hah, and J.-H. Roe. 1993. The induction of oxidative enzymes in Streptomyces coelicolor upon hydrogen peroxide treatment. J. Gen. Microbiol. 139:1013-1018. [Google Scholar]
- 18.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palma, M., D. DeLuca, S. Worgall, and L. E. Quadri. 2004. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 186:248-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 21.Robbe-Saule, V., C. Coynault, M. Ibanez-Ruiz, D. Hermant, and F. Norel. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol. Microbiol. 39:1533-1545. [DOI] [PubMed] [Google Scholar]
- 22.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 23.Shortridge, V. D., A. Lazdunski, and M. L. Vasil. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol. 6:863-871. [DOI] [PubMed] [Google Scholar]
- 24.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217-228. [DOI] [PubMed] [Google Scholar]
- 25.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 26.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vodovar, N., C. Acosta, B. Lemaitre, and F. Boccard. 2004. Drosophila: a polyvalent model to decipher host-pathogen interactions. Trends Microbiol. 12:235-242. [DOI] [PubMed] [Google Scholar]
- 28.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]