Abstract
Marine brown algae produce the highly recalcitrant polysaccharide fucoidan, contributing to long-term oceanic carbon storage and climate regulation. Fucoidan is degraded by specialized heterotrophic bacteria, which promote ecosystem function and global carbon turnover using largely uncharacterized mechanisms. Here, we isolate and study two Planctomycetota strains from the microbiome associated with the alga Fucus spiralis, which grow efficiently on chemically diverse fucoidans. One of the strains appears to internalize the polymer, while the other strain degrades it extracellularly. Multi-omic approaches show that fucoidan breakdown is mediated by the expression of divergent polysaccharide utilization loci, and endo-fucanases of family GH168 are strongly upregulated during fucoidan digestion. Enzymatic assays and structural biology studies reveal how GH168 endo-fucanases degrade various fucoidan cores from brown algae, assisted by auxiliary hydrolytic enzymes. Overall, our results provide insights into fucoidan processing mechanisms in macroalgal-associated bacteria.
Subject terms: Enzymes, Bacterial structural biology, Glycobiology, Environmental microbiology, Bacterial physiology
Degradation of the recalcitrant polysaccharide fucoidan, produced by marine brown algae, is carried out by some bacteria through unclear mechanisms. Here, the authors provide insights into fucoidan processing mechanisms by studying two alga-associated bacterial strains, using microscopy, multi-omic, biochemical and structural techniques.
Introduction
Macroalgal forests represent one of the largest and most productive coastal vegetated habitats on Earth, with a net primary production equivalent to the Amazon rainforest1,2. Macroalgae are ubiquitously found in coastal environments, including rocky intertidal habitats, coral reefs, and kelp forests1. Massive accumulations of free-floating macroalgae are also found in the open-ocean, such as the golden tides of Sargassum, which are expanding due to anthropogenic climate change3. The increasing arrival of seaweed tides to the coastline causes rising concerns due to their detrimental economic consequences4. However, macroalgae represent a valuable raw material, and globally, play a major beneficial role in regulating Earth’s climate by fixing atmospheric CO2 and storing carbon in the ocean. It has been estimated that 173 Tg of CO2 are fixed annually by macroalgae, and ca. 80% is sequestered in the deep sea5. Most of the carbon storage is mediated by brown algae2 which contain in their cell wall the highly recalcitrant polysaccharide fucoidan6. This polymer confers macroalgae a considerable resistance to degradation, being currently considered a climate-relevant polymer7. Moreover, fucoidan is gaining increasing attention due to its relevant biological effects, including anti-tumoral, anti-inflammatory, immunomodulatory, and antiviral activities8–10 with beneficial health implications.
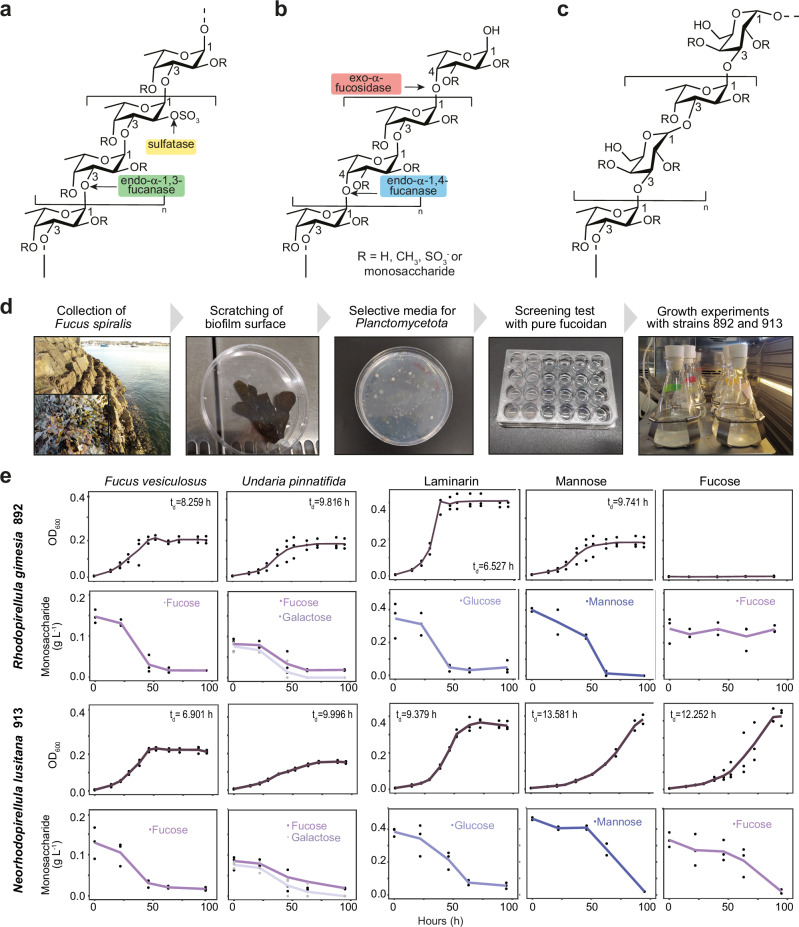
Fucoidans are complex sulfated polysaccharides whose precise chemical structure is mostly unknown since the composition and branching patterns are highly dependent on producing algal species, growth stage and environmental conditions11. Due to its complexity, only a small fraction of the structure of fucoidan extracts from most algae species has been elucidated12–18. Fucoidans can be divided into two main types, homo- and heterofucans, depending on their backbone composition14 (Fig. 1a–c and Supplementary Table 1). The main chain of homofucans is formed exclusively by α-1,3-fucose (Type I; Fig. 1a) or alternating α-1,3/α-1,4-fucose residues (Type II; Fig. 1b) that are extensively decorated with sulfate, ester groups, and other monosaccharides. In contrast, heterofucans contain a main chain composed of various sugar monomers, including glucose, mannose, rhamnose, xylose, and uronic acids, with sulfated fucose being found at branching sites14,19–21. A particular type of heterofucans typically produced by brown algae are galactofucans, composed of β-d-galactose and α-l-fucose in variable ratios14,22,23 (Fig. 1c). The remarkable complexity of fucoidan structures is a major obstacle to advance in our understanding of the specialized degradation pathways of this polysaccharide24, as well as the underlying mechanisms of their therapeutic applications.
Fig. 1. Chemical structure of fucoidans and experimental work with Planctomycetota strains isolated from Fucus spiralis.
Chemical structures of the common backbone chains of Type I (a) and Type II (b) homofucans, including the target site for the fucoidan-processing enzymes. c Chemical structure of the common backbone of the galactofucan (heterofucan) found in Undaria pinnatifida22. d Workflow of the experimental work. e Growth curves and carbohydrate consumption of strains 892 and 913 in different carbon sources (i.e., fucoidan from Fucus vesiculosus and Undaria pinnatifida, laminarin, mannose and fucose). Data points at each time represent three biologically independent experiments. Doubling times (td) have been derived from optical density measurement values (OD600) during exponential phase. Source data are provided as a Source Data file.
The slow turnover of fucoidans in the environment is mediated by a highly specific catalytic machinery encoded in just a few marine microbial taxa25,26. It is commonly assumed that the more complex a polysaccharide is, the greater the number of enzymes needed for its depolymerization27–29. Specifically, the degradation of fucoidan requires a combination of different enzymes, including glycoside hydrolases (GHs), carbohydrate esterases (CEs), and sulfatases (SAs), acting in a coordinated manner (Fig. 1a, b). A recently discovered marine bacterial strain affiliated with Verrucomicrobiota (Lentimonas sp. CC4) contains hundreds of fucoidan-degrading enzymes largely concentrated in a megaplasmid30. Yet, this strain is highly specialized in the digestion of homofucans, being unable to grow on heterofucans produced by macroalgae such as Sargassum or Undaria pinnatifida30. This finding highlights the exquisite specificity of fucoidan-processing enzymes, which are extremely sensitive to the polysaccharide chemical structure.
Identifying the precise catalytic pathways and mechanisms for the breakdown of distinct types of fucoidan is currently a major challenge. It has been largely assumed that exo-acting fucosidases from Carbohydrate Active enZyme31(CAZy) families GH29, GH95 and/or GH141 process the polysaccharide in combination with a set of SAs (e.g., families S1_17, S1_22 and S1_25 according to SulfAtlas database30,32–34) and CEs (e.g., family CE7 according to CAZy database30). These enzymes remove the side chains and negatively charged groups, before endo-acting enzymes (i.e., endo-fucanases or endo-fucoidanases35) can access the backbone structure. Endo-fucanases may hold a key role in the complete digestion of fucoidan, but so far, their biochemical and structural characterization lags behind that of exo-fucosidases35,36. According to the CAZy database31, there are currently four known families of endo-fucanases (GH107, GH168, GH174 and GH187) and a few representatives of these enzyme families have been recently characterized, most of them from marine Flavobacteriia. Among the GH107 family, some members are endo-α-1,4-fucanases that hydrolyze α-1,4-l-fucosidic bonds in sulfated homofucans composed of alternating 1,3- and 1,4-α-l-fucopyranose residues37–39, while others display hydrolytic activity against homogenous α-1,3-l-fucopyranoses in sulfated homofucans40,41. Additionally, endo-α-1,3-l-fucanase activity has been reported for members of the GH168, GH174 and GH187 families42–45. More recently, the enzyme FWf5 from Wenyingzhuangia fucanilytica CZ1127T has been reported as an α-1,4-endo-fucanase belonging to the GH168 family46. However, structural information is still needed in these enzyme families to determine their substrate specificity for the vast diversity of existing fucoidan structures.
Notably, a significant fraction of putative endo-fucanases included in the CAZy database that remain uncharacterized are identified in genomes of marine Planctomycetota, a phylum that is receiving great interest at the ecological and biotechnological level47. Members of Planctomycetota are typically associated with marine sinking particles25,48,49 and the surface of seaweeds50. Indeed, these microorganisms are well-known for their capability to degrade complex sulfated polysaccharides derived from macroalgae51,52. Provided with fascinating cell biology features such as cytosolic compartmentalization53, some Planctomycetota contain bacterial microcompartments (BMC) to metabolize l-fucose residues from algal polysaccharides54. Therefore, there is evidence suggesting that this phylum plays a crucial, yet completely uncharacterized role in fucoidan degradation. Here, we aim to understand the processing mechanisms of recalcitrant fucoidan in Planctomycetota. By combining the isolation of fucoidan-degrading Planctomycetota strains associated with macroalgae and their in-depth analysis by genomics, transcriptomics, proteomics and fluorescent microscopy, we gain new insights into fucoidan processing by marine bacteria. In addition, through enzymatic assays and structural biology, we provide a model on how GH168 endo-fucanases are assisted by auxiliary hydrolytic enzymes to get access and process certain fucoidan cores. Conversely, we show that Rho5174, an endo-fucanase from family GH168 present in Planctomycetota, is able to partially hydrolyze fucoidan from a specific macroalgae source. Finally, we also provide structural insights into the mechanism of substrate recognition for this important enzyme family.
Results
Planctomycetota strains isolated from Fucus spiralis degrade homo- and heterofucoidans
From nearly 100 isolates obtained by seeding biofilm samples of Fucus spiralis in culture media tailored to promote the growth of Planctomycetota (Fig. 1d), we identified 24 isolates affiliated with this phylum. In substrate utilization assays using (i) homofucan from Fucus vesiculosus or (ii) galactofucan from Undaria pinnatifida as single carbon source, only two isolates affiliated to the Pirellulaceae family, 892 and 913, showed a clear increase in OD when growing on both fucoidans (Fig. 1e). Isolate 892 was affiliated with Rhodopirellula gimesia SWK7 (ANIb 97.97%, coverage 86.70%) and isolate 913 to Rhodopirellula lusitana DSM 25457 (ANIb 97.53%, coverage 86.69%, recently reclassified as Neorhodopirellula lusitana). Both isolates were selected for an in-depth study of their mechanisms for fucoidan degradation.
Isolates 892 and 913 (892 and 913 strains, hereinafter) displayed similar growth patterns on both types of fucoidans, achieving higher growth rates in F. vesiculosus as compared to U. pinnatifida (Fig. 1e). Both strains were also able to grow on the highly labile marine polysaccharide laminarin, which is a β(1,3)-glucan with β(1,6)-branches, and the monosaccharide mannose. However, clear differences in growth were found when fucose was added as the sole carbon source. While strain 892 was unable to utilize fucose under our experimental conditions, strain 913 could efficiently use it as a carbon source (Fig. 1e). In general, both strains showed a higher lag phase growing on monosaccharides than on polysaccharides.
We assessed the ability of strains 892 and 913 to consume fucoidan by monitoring the disappearance of its monosaccharides using High-Performance Anion Exchange Chromatography with Pulsed Amperometry Detection (HPAEC-PAD). As depicted in Fig. 1e, the decreasing concentration of monosaccharides in supernatant filtrates was concomitant with the increase in OD during bacterial growth. Both strains, consumed ca. 90% and 77% of fucose contained in fucoidan from F. vesiculosus and U. pinnatifida, respectively. Interestingly, the galactose residue contained in the heterofucan of U. pinnatifida was completely consumed, being undetectable at the end of the growth curves. In addition, both strains processed ca. 85% of glucose contained in laminarin. When using monosaccharides for growth, complete consumption of mannose was observed for strain 892, and ca. 95% consumption was detected for strain 913 after 95 h. Finally, HPAEC-PAD data also confirmed the inability of strain 892 to process fucose, while strain 913 consumed ca. 95% of this monosaccharide during the experiment (Fig. 1e).
Planctomycetota strains 892 and 913 express highly heterogeneous polysaccharide utilization loci for fucoidan degradation
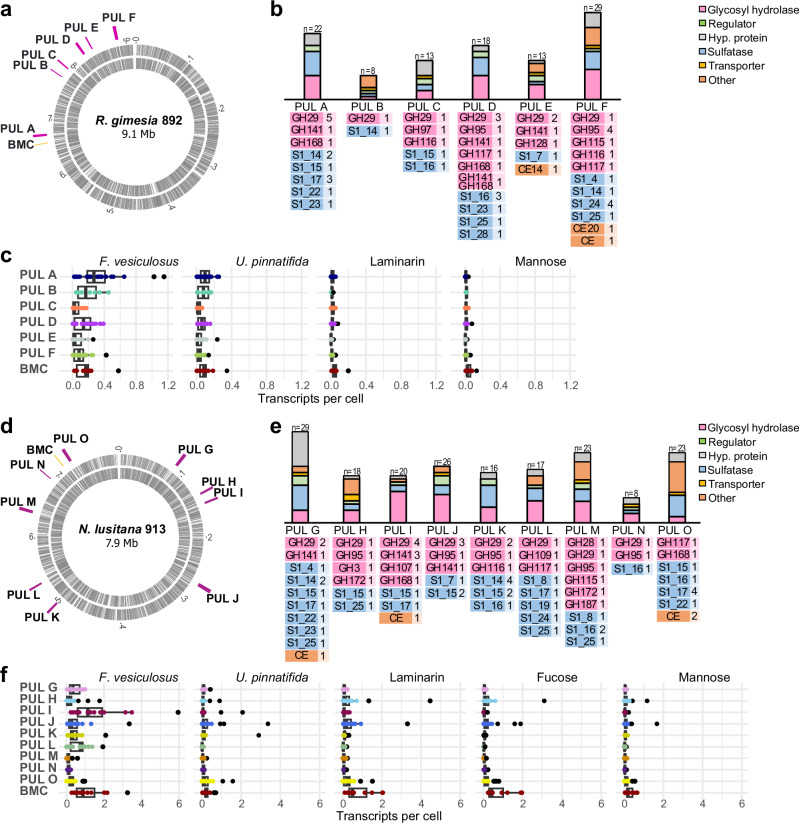
We found 6 and 9 putative fucoidan Polysaccharide Utilization Loci (PULs) in the genomes of strains 892 and 913, respectively. These loci are enriched with exo-fucosidases, endo-fucanases and SAs (Fig. 2), with no evidence of genomic structure similarity between the strains (Fig. 3, Supplementary Datasets 1 and 2). Additionally, we found a genomic region that likely encodes a BMC for fucose metabolism in both strains (Figs. 2; Supplementary Datasets 1 and 2). Interestingly, PUL B from strain 892 also contains genes associated with the metabolism of fucose, such as aldolase, fucose isomerase, and carbohydrate kinase, together with a transporter (Major Facilitator Superfamily, MFS) (Fig. 3, Supplementary Dataset 1). The fucoidan PULs in strain 892 have an average size of 10 Kbp and are co-located with the BMC in the same genomic region (Fig. 2a; PULs A to F). In contrast, the fucoidan PULs in strain 913 have an average size of 50 Kbp and are dispersed across the genome (Fig. 2d; PULs G to O).
Fig. 2. Fucoidan Polysaccharide Utilization Loci (PULs) in Planctomycetota strains and global expression under different carbon sources obtained by quantitative transcriptomics.
Genomic location of putative fucoidan PULs in strains 892 (a) and 913 (d) and functional composition of the PULs (b and e). Besides endo-fucanases (GH107, GH168 and GH187) and exo-fucosidases (GH29, GH95, GH141), all indicated in pink color, we identified enzymes from families GH97 (α-glucosidase/α-galactosidase), GH115 (α-glucuronidase), GH116 (β-glucosidase), GH117 (α-neoagarobiosidase) and GH128 (endo-β−1,3-glucanase/ exo-β−1,3-glucanase) in the PULs of strain 892 (b), and enzymes from families GH3 (β-glucosidase), GH28 (endo-polygalacturonase/exo-polygalacturonase), GH109 (α-N-acetylgalactosaminidase), GH115 (α-glucuronidase), GH116 (β-glucosidase), GH117 (α-neoagarobiosidase), GH172 (α-d-arabinofuranosidase) in the PULs of strain 913 (e). Pannels c and f show mean transcripts per cell of individual genes in each of the fucoidan PULs and the BMC (highlighted in different colors) in strains 892 and 913, respectively, grown on different carbon sources (i.e., fucoidan from Fucus vesiculosus and Undaria pinnatifida, laminarin, mannose and fucose) in three biologically independent experiments. Box plots indicate median (middle line), 25th, 75th percentile (box), and 5th and 95th percentile (whiskers). Individual plotted data points represent the transcripts per cell and black points indicate the outliers. Transcripts per cell were quantified by transcriptomics using internal mRNA standards in three biologically independent replicates. Source data are provided as a Source Data file. BMC: Bacterial microcompartment; GH: glycosyl hydrolase; S: sulfatase; CE: carbohydrate esterase.
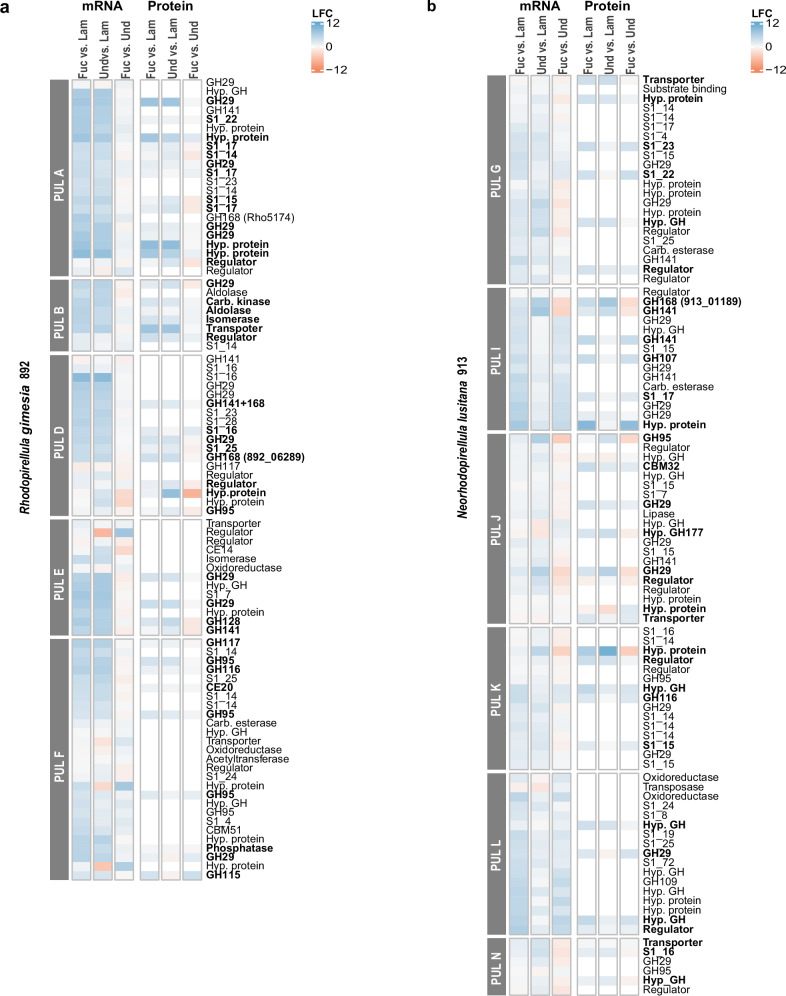
Fig. 3. Differential gene expression at mRNA and protein level in the identified fucoidan PULS from strains 892 and 913.
Log2-fold changes (LFC) of mRNA and protein expression data are based on pairwise comparisons between different carbon sources in strains 892 (a) and 913 (b). Color scale indicates upregulation (blue) or downregulation (red) of transcripts or proteins in the indicated fucoidan vs. other carbon sources. Expression values were quantified in three biologically independent replicates (n = 3). Only PULs where fucosidases were detected at the protein level are shown. Bold letters highlight those proteins that were significantly more abundant in one of the fucoidan sources as compared to laminarin (two-sided Student’s t-test, p value < 0.05). Source data are provided as a Source Data file. Fuc: fucoidan from Fucus vesiculosus; Und: fucoidan from Undaria pinnatifida; Lam: laminarin; GH: glycosyl hydrolase; S: sulfatase; CE: carbohydrate esterase; CBM: carbohydrate binding modules; Hyp: hypothetical; Carb: carbohydrate.
The number of predicted exo-fucosidases located in fucoidan PULs is similar in both strains, with 13 GH29, 5 GH95, and 3 GH141 homologs in strain 892, and 15 GH29, 5 GH95, and 4 GH141 in strain 913 (Fig. 2b, e). The average identity in these enzymes ranged between ca. 25 to 70% (Supplementary Datasets 3 and 4). Interestingly, both strains differ in the composition and number of putative endo-fucanases (Fig. 2b, e). In strain 913, we found enzymes from families GH168 (2), GH107 (1), and GH187 (1) (Fig. 2e), while in strain 892, there are three representatives of the GH168 family, one encoded in PUL A (892_05174) and two in PUL D (892_06282 and 892_06289). Additionally, two homologs of GH187 were found in strain 892 (892_02309 and 892_05521), but they were encoded outside the described PULs (Supplementary Dataset 1). The fucoidan PULs in both strains had a different number of SAs (25 and 38 in strains 892 and 913, respectively), all assigned to the S1 family and distributed in ca. 10 subfamilies. The dominant SA families were S1_14 and S1_16 in strain 892, and S1_14, S1_15 and S1_17 in strain 913 (Fig. 2). Moreover, several fucoidan PULs from both strains contained SAs from families S1_17, S1_22 and S1_25, which are known to be involved in the cleavage of the sulfate group from fucosyl residues33,34.
We used quantitative transcriptomics with internal RNA standards to measure the average expression of the PULs when growing on both types of fucoidan, produced by F. vesiculosus and U. pinnatifida, in comparison to the labile polysaccharide laminarin and the monosaccharides fucose and mannose (Figs. 2c, f, Supplementary Datasets 1 and 2). In strain 892, most GHs within all six putative fucoidan PULs were actively expressed (Supplementary Fig. 1 and Supplementary Dataset 1). Notably, PULs A and D, which contained the three GH168 endo-fucanases, along with PUL B, displayed the highest mean expression values when growing on both types of fucoidan (Wilcoxon test, p-value < 0.05, Fig. 2c, Supplementary Dataset 5). For strain 913, we observed that growth on fucoidan from F. vesiculosus induced the expression of GHs within most fucoidan PULs (Supplementary Fig. 1, Supplementary Dataset 2), with the highest overall expression observed in PUL I, which contains the predicted endo-fucanases GH168 and GH107 (Fig. 2f). The expression of GHs was lower when strain 913 was grown on fucoidan from U. pinnatifida compared to F. vesiculosus (Supplementary Fig. 1), and growth on fucose did not induce the transcription of any fucoidan PULs. This suggests that substrate-recognition and transcription activation for the turnover of fucoidan may be mediated by other components or structural identities that are more complex than its main monosaccharide. Lastly, the BMC compartment was induced in both strains when they were growing on both fucoidan sources, indicating an active metabolism of fucose residues.
Endo-fucanases are strongly upregulated during the degradation of different types of fucoidan
We conducted a gene expression analysis on RNAseq datasets of strains 892 and 913 growing on the different carbon sources (Supplementary Fig. 2). The DEseq2 package was used to identify specific genes that were up- or downregulated by specific carbon sources. We followed the criteria Log2-fold change (LFC) > 2 and p < 0.05 to determine significant changes (Fig. 3, Supplementary Fig. 3, Supplementary Datasets 1 and 2). When both strains were grown on fucoidan compared to laminarin or mannose, we observed upregulation of the expression of most SAs, exo-fucosidases, and all endo-fucanases within the PULs (LFC > 2, p < 0.05; Fig. 3, Supplementary Datasets 1 and 2, Supplementary Fig. 3). Interestingly, in strain 913, out of the 15 exo-fucosidases upregulated by fucoidan compared to laminarin, seven were upregulated only in the presence of F. vesiculosus (families GH29 and GH141) and two were upregulated only in U. pinnatifida (families GH95 and GH141). Moreover, in the same strain, the GH107 homolog was upregulated only in fucoidan from F. vesiculosus (LFC = 4.08, p < 0.05; Supplementary Dataset 2), while the upregulation of the two GH168 endo-fucanases was particularly pronounced in U. pinnatifida (LFC > 5.9).
The proteome analysis of strain 892 revealed that most exo-fucosidases and endo-fucanases located in fucoidan PULs were translated to protein (16 out of 24), including the two GH168 homologs found in PUL D (Fig. 3a). However, only 8 out of 18 SAs were detected, which were assigned to families S1_14, S1_15, S1_16, S1_17, S1_22, and S1_25 (Fig. 3a, Supplementary Dataset 1). We assessed differentially regulated proteins among various carbon sources using Student’s t-test (LFC > 1.5, p < 0.05). We observed a total of 11 GHs and 4 SAs upregulated when strain 892 was grown on fucoidan (from F. vesiculosus or U. pinnatifida) compared to other carbon sources (Fig. 3a, Supplementary Dataset 1). Upon comparison of the two fucoidan types, significant differences were only observed in two GH29 homologs and two galactosidases (GH116 and GH115, LFC > 1.5, p < 0.05; Fig. 3a; Supplementary Dataset 1). In genomic regions outside putative fucoidan PULs, 13 GHs were differentially expressed at the protein level in at least one type of fucoidan compared to other carbon sources. Additionally, 11 SAs were upregulated during growth in U. pinnatifida as compared to F. vesiculosus (LFC > 1.5, p < 0.05), including families S1_4 (3), S1_7 (3) and S1_16 (1), which are involved in removal of sulfated groups linked to a galactosyl residue34. For strain 913, when growing on fucoidan, only 6 out of 38 SAs, and less than half of the exo-fucosidases and endo-fucanases located in PULs (10 from 29) were identified by proteomics (Figs. 3b, Supplementary Dataset 2). Out of these, two endo-fucanases (GH107 and GH168), both located in PUL I, exhibited varying protein expression depending on the type of fucoidan, in agreement with transcriptomic results. The protein GH107 was only upregulated when growing on fucoidan from F. vesiculosus (20-fold change), whereas GH168 was more strongly induced in fucoidan from U. pinnatifida (128-fold change at the protein level), consistent with transcriptomic data. In terms of exo-fucosidases, only four showed differential expression at the protein level between fucoidan sources (LFC > 1.5, p < 0.05). Both types of fucoidan upregulated one SA from subfamily S1_16, while four SAs from subfamilies S1_23, S1_22, S1_17, and S1_15 were induced only in F. vesiculosus.
The endo-fucanase family GH168 is consistently present in Planctomycetota genomes enriched in fucosidases
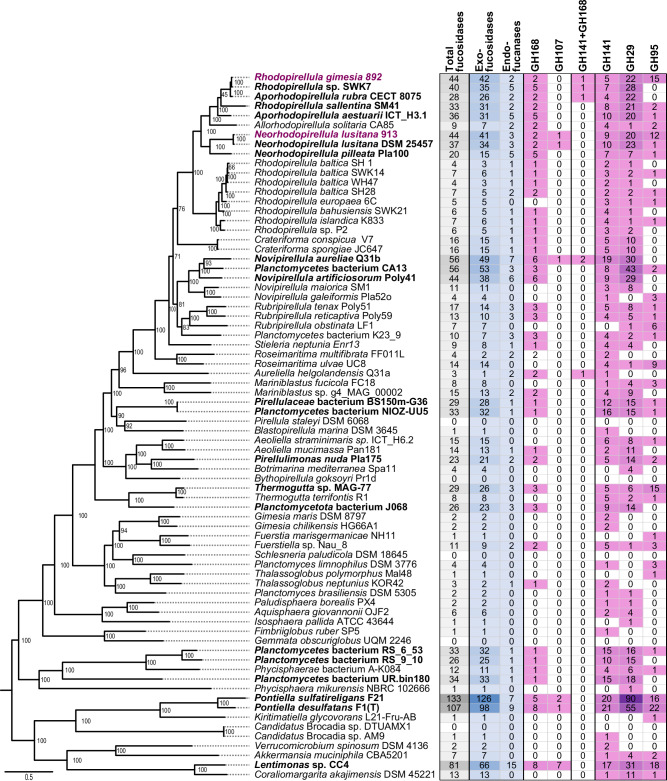
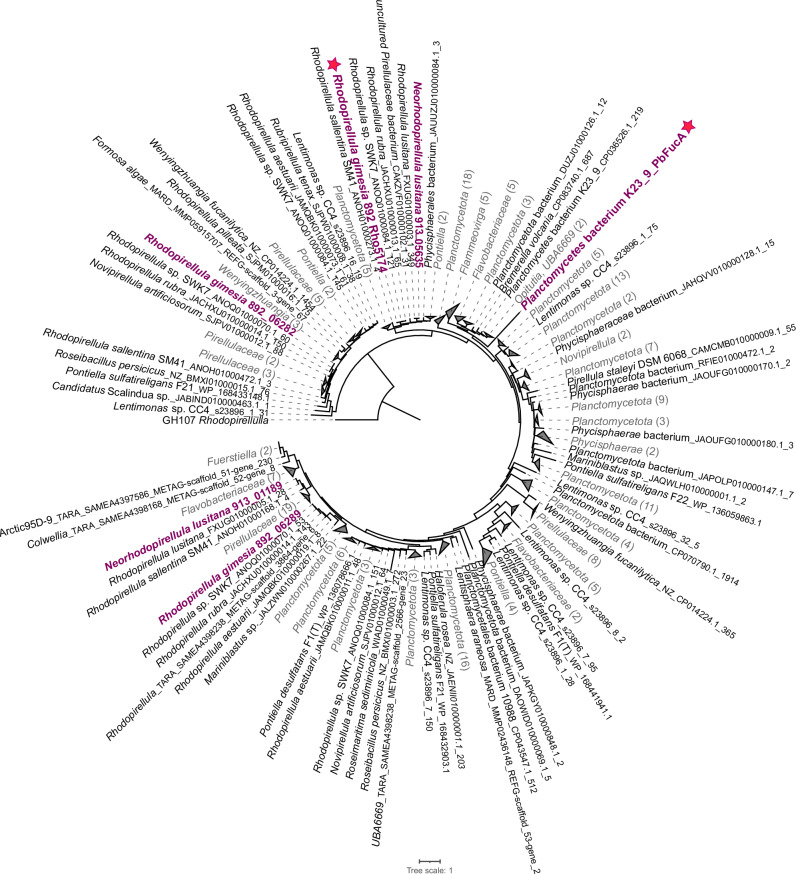
To expand our knowledge on the distribution of fucoidan-degrading enzymes in the phylum Planctomycetota, we performed a screening of enzymes from families GH29, GH95, GH141, GH107 and GH168 in all available genomes at NCBI belonging to this phylum (> 5000 genomes). We found that almost 50% of Planctomycetota genomes analyzed (2735) contained at least one exo-fucosidase, and 16% contained more than 5 homologs of these enzymes. However, endo-fucanases were present in less than 3% of available genomes, and generally in low abundance, indicating that the presence of these enzymes is not a conserved genomic trait (Supplementary Dataset 6). This result confirms a wider distribution of exo-fucosidases than endo-fucanases in bacterial genomes26,30. The presence of exo-fucosidases and endo-fucanases was clearly enriched in the class Planctomycetia, especially within the orders Pirellulales (i.e., some members of Rhodopirellula, Aporhodopirellula, Neorhodopirellula and Novipirellula) and Phycisphaerales (Fig. 4).
Fig. 4. Endo-fucanases and exo-fucosidases detected in selected genomes from Planctomycetota and Verrucomicrobiota phyla.
The recently characterized endo-fucanase families GH187 and GH174 were not included in this comparative analysis since the assignation was not found to be reliable using dbCAN v3.0.7. Taxa enriched in fucosidases, i.e., displaying ≥ 20 fucosidases, are highlighted in bold. Strains 892 and 913 are highlighted in magenta. The phylogenetic tree on the left side shows the taxonomic profiling of the selected genomes, based on the amino acid sequences of 10 core genes aligned using the Phylogenetic Tree Service from BV-BRC database (https://www.bv-brc.org)154 without deletion and duplication allowed, using Candidatus Brocadiia as outgroup. Enzymes with more than one domain have not been considered in the total sum of fucosidases. Source data are provided as a Source Data file.
Members of the genera Aporhodopirellula, Neorhodopirellula and Rhodopirellula (i.e., A. rubra, A. aestuarii, R. sallentina, N. lusitana), with genomes closely related to strains 892 and 913, exhibited a comparable number of exo-fucosidases and endo-fucanases (Fig. 4; Supplementary Dataset 6). Another group of Rhodopirellula genomes (R. baltica, R. europaea, R. islandica) displayed a low number of exo-fucosidases (3–6 copies of each family). However, most of them still contained one homolog of GH168. Notably, among endo-acting fucanases, GH168 was more widely distributed across Planctomycetota as compared to GH107, which was found only in a few genomes (e.g., Neorhodopirellula lusitana, Novipirellula aureliae; Fig. 4; Supplementary Dataset 6). This suggests that GH168 endo-fucanases play a significant role in fucoidan degradation in Planctomycetota. Interestingly, in our expanded analysis, we identified novel putative GH168 enzymes coupled with additional exo-fucosidase domains, but they were less widely distributed than the enzymes comprising a single domain (Fig. 4; Supplementary Dataset 6).
We carried out a phylogenetic analysis of GH168 homologs present in the Planctomycetota genomes and other known fucoidan degraders from Bacteroidota and Verrucomicrobiota phyla (Fig. 5, Supplementary Fig. 4). Our findings reveal a remarkable diversity of GH168 enzymes, exhibiting a wide range of total percent identities, spanning from 18% to 100% (see Supplementary Dataset 7). Highly diverse GH168 homologs were also found at the level of individual genomes, with different copies assigned to distant clusters in the phylogenetic tree. For example, the percent identity among GH168 homologs in strains 892 and 913 ranged between 20–80% (coverage > 50%; Supplementary Dataset 7). Conversely, some GH168 homologs from phylogenetically distant bacteria shared high similarity (>70% identity), suggesting horizontal gene transfer events. This was the case of some endo-fucanases recently characterized from the Flavobacterium Wenyingzhuangia fucanolytica42,43, which shared high similarity (>60%) with some GH168 homologs of members of Verrucomicrobiota (Kiritimatiella, Lentimonas, Pontiella, Roseibacillus,) and Planctomycetota, mainly Pirellulaceae (e.g., Aporhodopirellula, Aureliella, Mariniblastus Neorhodopirellula, Novipirellula, Rubripirellula), including strain 892 (Supplementary Fig. 4).
Fig. 5. Phylogenetic tree of GH168 homologs.
Maximum likelihood phylogenetic tree of GH168 homolog amino acid sequences from all available Planctomycetota genomes at NCBI, selected genomes of Bacteroidota and Verrucomicrobiota, and environmental sequences found in the The Ocean Microbiomics Database gene catalog126. Source data are provided as a Source Data file. The number of collapsed nodes are indicated in parentheses. The GH168 enzymes of strains 892 and 913 are highlighted in magenta. The GH168 enzymes Rho5174 and PbFucA, which are the focus of this study, are highlighted with star symbols.
Insights into fucoidan processing by GH168 enzymes from Planctomycetota
To investigate the enzymatic activity and substrate specificity of various members of the GH168 family in Planctomycetota, we focused on two enzymes: 892_05174 from the newly isolated strain 892 (hereinafter referred to as Rho5174) and PbFucA from a phylogenetically distant member within the Pirellulaceae family, Planctomycetes bacterium K23_9. Both enzymes were expressed and purified to apparent homogeneity. PbFucA (UniProt code A0A517NMB4, residues 35-392; Supplementary Fig. 5) shares 36% sequence identity with Rho5174 and contains a predicted signal peptide (residues 1-34). In contrast, Rho5174 did not show a signal peptide in its N-terminus, according to SignaIP-6.055.
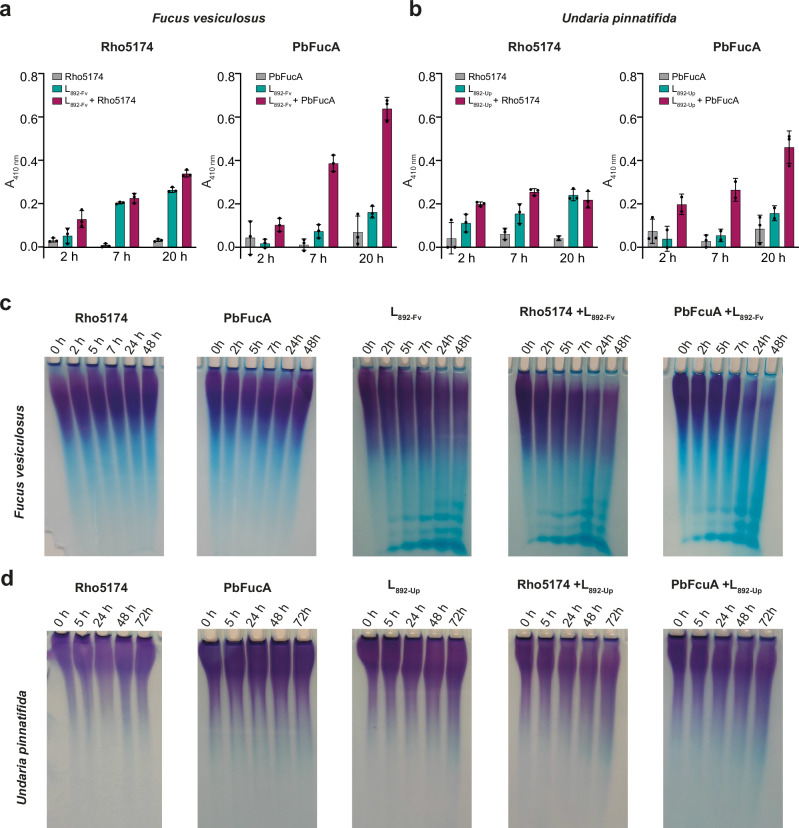
We studied the ability of Rho5174 and PbFucA to hydrolyze fucoidan from F. vesiculosus and U. pinnatifida measuring the released reduced sugar by the p-hydroxybenzoic acid hydrazide (PAHBAH) method30,56. Only residual hydrolytic activity was observed after 20 h incubation under these conditions (Fig. 6a, b). In contrast, the addition of strain 892 lysates previously grown in the presence of fucoidan from F. vesiculosus (L892-Fv) or U. pinnatifida (L892-Up) to the mixtures clearly triggered degradation of the corresponding fucoidans after just 2 h of incubation with PbFucA (Fig. 6a, b). This synergistic effect was less pronounced when Rho5174 and the lysates were incubated with both fucoidans (Fig. 6a, b). Importantly, we observed very reduced or no hydrolytic activity of both enzymes against fucoidan from U. pinnatifida and F. vesiculosus, respectively, when combined with the lysates of strain 892 grown in mannose (L892-man) (Supplementary Fig. 6). Thus, this indicates that Rho5174 and PbFucA can get access and process fucoidan from F. vesiculosus or U. pinnatifida assisted by hydrolytic enzymes of strain 892, which are exclusively expressed in the presence of these fucoidan substrates. Moreover, Rho5174 and PbFucA did not exhibit exo-fucosidase activity against 4-nitrophenyl-α-l-fucopyranoside (pNP-Fuc), confirming that the enzymes exclusively display an endo-fucanase activity (Supplementary Fig. 7).
Fig. 6. Hydrolytic activity of PbFucA, Rho5174 and strain 892 lysates against fucoidan from F. vesiculosus and U. pinnatifida.
Hydrolytic activity of Rho5174 (left) or PbFucA (right) against fucoidan from F. vesiculosus (a) and U. pinnatifida (b). The hydrolytic activity of the enzymes, strain 892 lysate grown in F. vesiculosus (L892-Fv, in panel a) or U. pinnatifida (L892-Up, in panel b), and the combination of enzyme and lysate are colored in gray, cyan, and magenta, respectively. Data represent the mean ± SD derived from three biologically independent experiments (n = 3). c C-PAGE analysis of the hydrolytic activity of Rho5174 and PbFucA, strain 892 lysate grown in F. vesiculosus (L892-Fv), and the combination of enzymes and lysate against fucoidan from F. vesiculosus. d C-PAGE analysis of the hydrolytic activity of Rho5174 and PbFucA, strain 892 lysate grown in U. pinnatifida (L892-Up) and the combination of enzymes and lysate against fucoidan from U. pinnatifida. Source data are provided as a Source Data file.
We performed Carbohydrate PolyAcrylamide Gel Electrophoresis (C-PAGE) to detect degradation products of fucoidan from various species of brown algae, including F. vesiculosus, U. pinnatifida, Cladosiphon okamuranus, Ecklonia maxima and Macrocystis pyrifera, after incubation with Rho5174 or PbFucA. These fucoidans differ in monosaccharide composition, sulfation degree and glycosidic linkage patterns (Supplementary Fig. 8 and Supplementary Table 1). We assessed the hydrolytic activity of the enzymes on these fucoidans, either alone or in combination with cellular lysate L892-Fv. For U. pinnatifida, we performed parallel experiments using the cellular lysate L892-Up (Fig. 6d). Notably, L892-Fv exhibited clear hydrolytic activity on the fucoidan from F. vesiculosus (Fig. 6c), and slightly degraded the fucoidan from E. maxima and M. pyrifera (Supplementary Fig. 9). The combination of L892-Fv with Rho5174 or PbFucA, significantly increased the degradation capability of the latter enzymes against these fucoidans, as evidenced by the attenuation of high molecular weight fucoidan in the gels (Supplementary Fig. 9). This supports the notion that enzymes present in the lysate facilitate the access of Rho5174 and PbFucA to these fucoidan backbones. L892-Fv could not hydrolyze fucoidan from C. okamuranus or U. pinnatifida. However, hydrolysis of fucoidan from U. pinnatifida was detected when using L892-Up, but only at a very low level (Fig. 6d). This was indicated by a smear in the upper part of the C-PAGE gel, suggesting a partial breakdown of this fucoidan source (Fig. 6d). Interestingly, Rho5174 exhibited hydrolytic activity without the assistance of other enzymes against the fucoidan from C. okamuranus, but not against other fucoidans tested (Supplementary Fig. 9), indicating a high selectivity for the fucoidan source. PbFucA could also partially hydrolyze fucoidan from C. okamuranus, but less efficiently than Rho5174 (Supplementary Fig. 9).
Insights into fucoidan recognition and processing of the endo-fucanase GH168 family
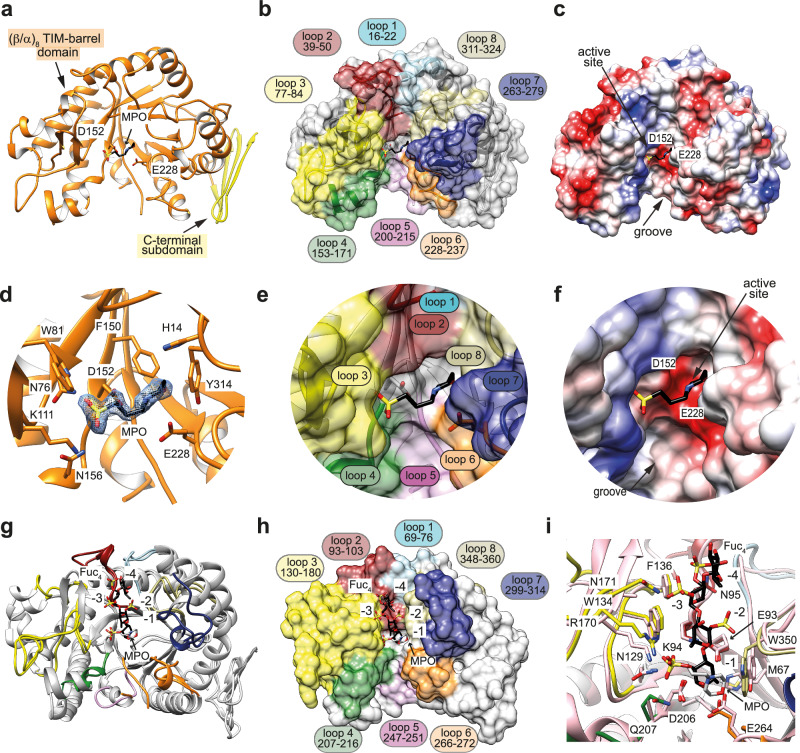
To better understand the structural determinants that govern substrate specificity of the GH168 family of endo-fucanases, we determined the structure of Rho5174 and PbFucA (Fig. 7, Supplementary Figs. 5 and 10-13; Supplementary Table 2; Supplementary Note 1; see “Method” section for further details). The overall protein scaffold and the conformation of both proteins were essentially preserved (PDB codes 9F9V and 8RG3; r.m.s.d. of 1.03 Å from 284 residues, Supplementary Fig. 13). Thus, we refer to Rho5174 to describe the overall structure of both enzymes (see Supplementary Note 1 for PbFucA structure details). Rho5174 contains (i) a catalytic domain adopting a conserved (β/α)8 barrel topology (residues 1-331) and (ii) a small C-terminal subdomain (residues 332-368) comprising a β-sheet conformation composed of 4 antiparallel β-strands, with an overall size of 63 Å × 39 Å × 45 Å (Fig. 7). The catalytic domain and the C-terminal subdomain interact with each other through an extensive contact area of ca. 997 Å2, representing 7% of the total accessible surface of the isolated domains (Fig. 7). A large groove runs at the center of the catalytic domain, containing the active site of the enzyme. This groove is surrounded by the loops connecting β1–α1 (loop 1; residues 16-22, light blue), β2–α2 (loop 2; residues 39–50, dark red), β3–α3 (loop 3; residues 77–84, green), β4–α4 (loop 4; residues 153–171, yellow), β5–β6 (loop 5; residues 200–215, pink), β6–α5 (loop 6; residues 228–237, orange), β7–α6 (loop 7; residues 263–279, dark blue), and β8–α7 (loop 8; residues 311–324, khaki; Fig. 7) that form the fucoidan binding site. The conformation of these loops is largely conserved between Rho5174 and PbFucA, except for loop 3 and loop 5. Loop 3 is longer in Rho5174 than in PbFucA, adopting a β-hairpin conformation. We could not model loop 5 in the Rho5174 structure, most likely due to the higher flexibility of this loop compared to the PbFucA structure (Supplementary Fig. 13).
Fig. 7. The overall structure of Rho5174.
a Cartoon representation showing the general fold and secondary structure organization of Rho5174. Domains, MOPS molecule (MPO) and catalytic residues are annotated. b Surface representation of the Rho5174. Loops that form the main groove of Rho5174 are highlighted in different colors. c Electrostatic surface representations of Rho5174 showing the location of the catalytic site. d–f Close-up view of the catalytic active site of Rho5174 shown as cartoon/stick and the electron density map of MOPS molecule (MPO) shown at 1.0 σ r.m.s.d (d) with annotated loops (e), and as electrostatic surface representation (f). g Superposition of the X-ray crystal structures of Rho5174 with the MPO molecule in gray (PDB code 9F9V) and Fun168A in complex with the tetrasaccharide product (α-l-Fucp-1 → 3-α-l-Fucp2,4(OSO3−)−1 → 3-α-l-Fucp2(OSO3−)−1 → 3-α-l-Fucp2(OSO3−) in black (Fun168A-Fuc4; PDB code 8YA7). Binding subsites for Fuc4 product are annotated. h Surface representation of the crystal structure of Fun168A-Fuc4 (PDB code 8YA7). The MPO molecule in Rho5174, colored in light gray, is superimposed. i Close-up view of the superposition of the fucoidan binding site of Fun168A (loops in different colors) and Rho5174 (colored in pink). The residues of Fun168A interacting with the fucoidan derivative are annotated and fucose residues are numbered according to the binding subsites.
Despite much effort, we were unable to crystallize Rho5174 or PbFucA in complex with fucose or chemically synthesized sodium methyl-α-l-fucopyranoside 2-sulfate (Fuc2SO4)57 (Supplementary Fig. 14). However, we identified one molecule of 3-morpholinopropane-1-sulfonic acid (MOPS) in the central groove of the Rho5174 structure (Fig. 7). The sulfonic acid group of MOPS interacts with the side chains of N76 of β3, W81 and K111 of loop 3 and Q156 of loop 4. The morpholine ring of MOPS makes hydrophobic interactions with H14 of loop 1, F150 of β4, and Y314 of loop 8 (Fig. 7d). Structural comparison of Rho5174 and Fun168A, an endo-1,3-fucanase from the GH168 family whose structure was recently solved in complex with fucotetraose (Fun168A-Fuc4; PDB code 8YA758;) revealed that the MOPS molecule in Rho5174 occupies the equivalent position to Fuc (-1) in the Fun168A-Fuc4 complex (Fig. 7g–i). Specifically, the sulfate group attached to the O2 position of Fuc makes a salt bridge with the side chain of R170 (K111 in Rho5174) in loop 3 and hydrogen bonds with the side chains of N129 and W134 (N76 and W81 in Rho5174) also in loop 3. Fuc (-1) makes hydrophobic interactions with W350 (Y314 in Rho5174) in loop 8 (Fig. 7h). Altogether, the experimental data support a common binding mode for Fuc (-1) in Rho5174 and Fun168A. It is worth noting that Fun168A hydrolyzes the α-1,3 bonds between Fucp(2OSO3-) and α-l-Fuc of sulfated fucoidans from Isostichopus badionotus and Thelenota ananas43 (Supplementary Fig. 15a), which are two species of sea cucumbers.
A double-displacement catalytic mechanism has been initially proposed for Fun168A, with D206 and E264 being critical residues for this enzyme, since the mutants D206E and E264Q were inactive42. In Fun168A, the distance between D206 and the atom C1 of the fucosyl residue at the 1 subsite was 3.0 Å, indicating that D206 serves as a nucleophile. In addition, the distance between E264 and the atom O1 was 2.7 Å, indicating that E264 functions as the general acid/base residue58. Interestingly, in the central groove of Rho5174, the MOPS molecule is located near the predicted catalytic residues D152 and E228 (Fig. 7a, d). These two residues are strictly conserved in both Rho5174 and PbFucA structures (D152 and E228, in Rho5174, D184 and E254, in PbFucA) and other members of the GH168 family, including Fun168D from W. funcanilytica CZ112743 (Supplementary Fig. 15c). Fun168D is an α-1,3 fucanase that acts between α-l-Fucp(2OSO3-) and α-l-Fucp(2OSO3-) of fucoidan from I. badionotus and between α-l-Fucp(2OSO3-) and α-l-Fucp(2,4OSO3-) of fucoidan from Holothuria tubulosa, supporting a common catalytic mechanism (Supplementary Fig. 16).
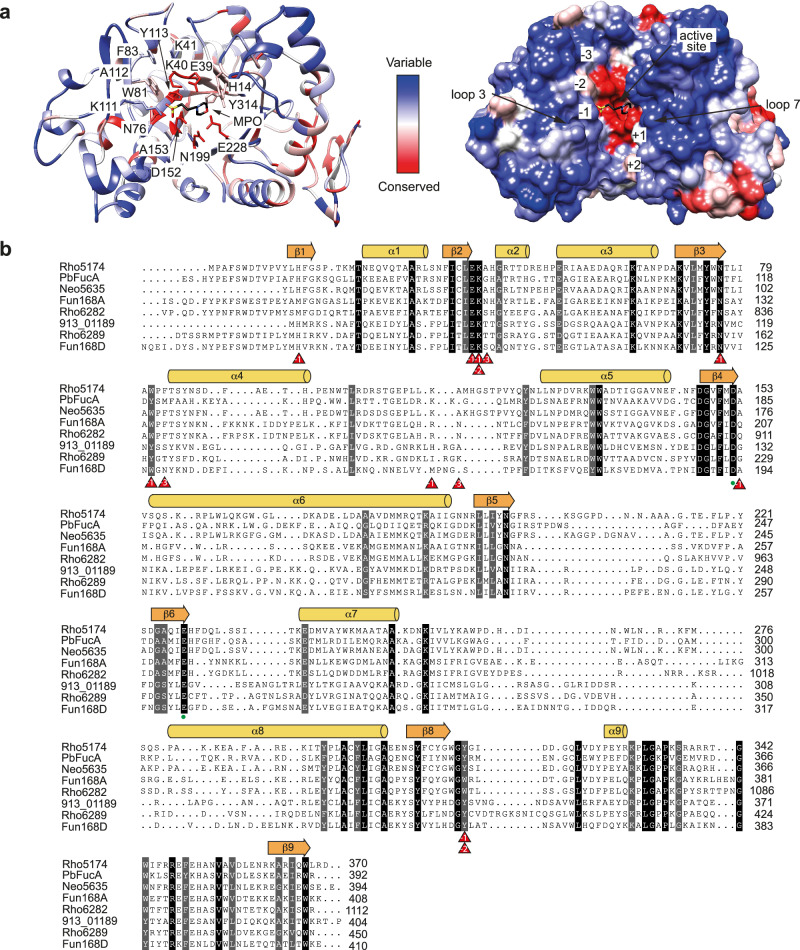
In an extended analysis of the GH168 enzymes from strains 892 and 913, the Alphafold-predicted structures revealed a similar overall topology to Rho5174 and PbFucA, with a GH168 domain followed by a small β-sandwich subdomain (Supplementary Fig. 17). Sequence alignment and structural comparison analysis of crystal structures of Rho5174, PbFucA, Fun168A-Fuc4 and the predicted structures of Fun168D (Supplementary Dataset 8) and other GH168 enzymes from strains 892 and 913 highlight that not only the catalytic residues (D152 and E228 in Rho5174), but also those forming subsites -1 and -2 are well conserved among them (Fig. 8 and Supplementary Fig. 17). However, the residues comprising subsites -3, +1, and +2 are highly diverse among the enzymes, leading to cavities of different sizes and shapes. This is exemplified in the Fun168A-Fuc4 crystal structure by N95 (A41 in Rho5174 and G81 in PbFucA), and N171 (A112 in Rho5174 and G149 in PbFucA), which interact with the sulfate from O2 and O4 of Fuc (-3), respectively.
Fig. 8. Structural basis of GH168 family substrate specificity.
a Structural comparison analysis of Rho5174 with X-ray crystal structures of PbFucA, Fun168A, and the Alphafold models of GH168 enzymes from strains 892 and 913, and Fun168D from W. funcanilytica CZ1127T. Conserved and variable residues are colored in red and blue, respectively. b Structure weighted sequence alignment of GH168 enzymes from 892 and 913 strains, PbFucA from P. bacterium K23_9 and Fun168A and Fun168D from W. funcanilytica CZ1127T. Catalytic residues are highlighted with green circles. Residues interacting with fucose moieties in the X-ray crystal structure of FunA168A-Fuc4 are highlighted with red triangles.
Interestingly, Rho5174 showed a high sequence identity with a GH168 homolog in strain 913, 913_05635 in PUL_O (76% identity, Supplementary Table 3, Supplementary Fig. 17). This implies that Rho5174 and 913_05635 (Supplementary Dataset 9) might hydrolyze similar fucoidan fragments in both strains, showing a longer loop 3 with respect to other members, which contains instead a β-hairpin motif (Supplementary Fig. 17). Conversely, the enzymes 892_06289 (PUL D; Supplementary Dataset 10) and 913_01189 (PUL I; Supplementary Dataset 11), which share almost 49% of sequence identity and display a shorter loop 3 and a longer loop 7 than other GH168 enzymes, were detected by proteomics in both strains when growing on fucoidan. In the latter enzyme-predicted structures, loop 7 also adopts a β-hairpin as in the Fun168D AlphaFold model (Supplementary Fig. 17 and Supplementary Fig. 15d–e). It is worth noting that Fun168A cannot accept α-l-Fucp(2OSO3−) at subsite +1 as Fun168D does, most likely due to these differences in loops 3 and 7.
Finally, the enzyme 892_06282 in strain 892 (PUL D; Supplementary Dataset 12) exhibits an additional hydrolytic domain belonging to the GH141 family at the N-terminus (Supplementary Fig. 17), representing a novel multi-domain enzyme. It should be noted that, to date, the two members of the GH141 family that have been biochemically characterized showed α-fucosidase activity on pectins59 and endo-xylanase activity60. However, this family of enzymes has also been identified in several fucoidan processing bacteria24,30, suggesting a key role in the hydrolysis of this polysaccharide. The enzymes 892_06282 and Fun168A show 64% sequence identity and shorter loops 3 and 7 than the other enzymes (Supplementary Table 3; Supplementary Fig. 17).
Planctomycetota strains 892 and 913 strains exhibit different mechanisms of fucoidan uptake and degradation
In addition to the catalytic machinery to process fucoidan, the cellular mechanisms employed by marine bacteria to uptake and digest this polysaccharide remain unexplored. To study the uptake of fucoidan by strains 892 and 913 by fluorescence microscopy, we synthesized a fluorescently labeled fucoidan analog from F. vesiculosus, i.e., FITC-Fucoidan. Both strains featured an oval cellular shape with ca. 2 µm diameter and typical planctomycetal morphology, including budding events and a marked nucleoid. In experiments exposing mid-exponential growing cells to FITC-Fucoidan, both strains showed a high ability for polymer uptake, i.e., ca. 80% of bacterial cells analyzed colocalized with the signal of FITC-Fucoidan by using Nile Red as cellular envelope marker (Fig. 9a, b). A Rhodopirellula strain isolated from the same niche but with no fucoidan degradation capability (strain 1062) was used as a negative control. Under the same experimental conditions, no FITC-fluorescence was observed in strain 1062, thus ruling out any unspecific binding of FITC-Fucoidan to the cells (Fig. 9a).
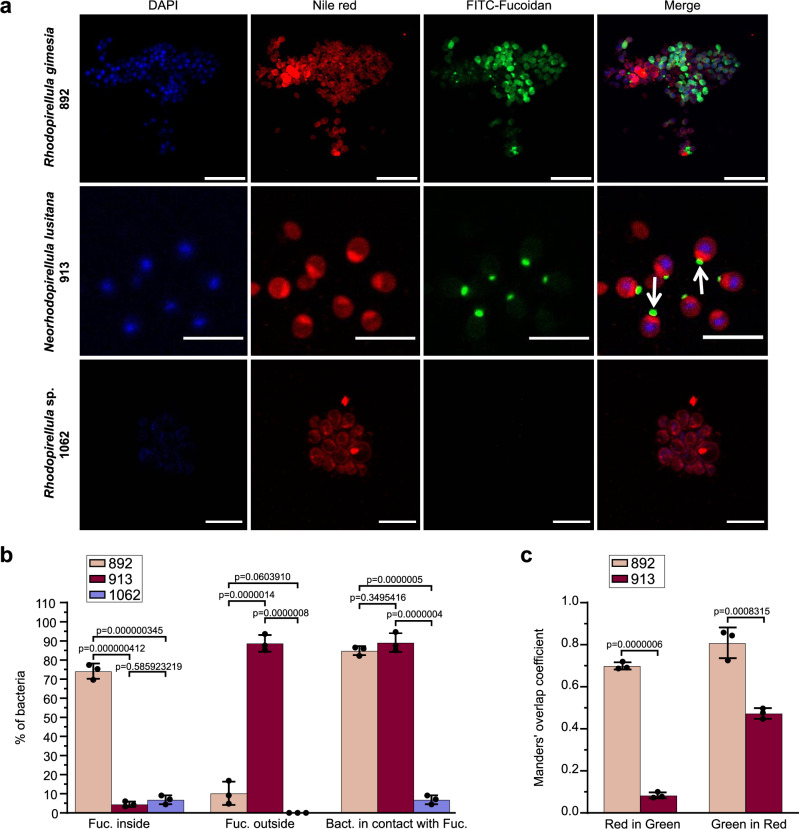
Fig. 9. Visualization of Planctomycetota cells consuming FLA-fucoidan.
a Strains 892, 913, and 1062 were grown in the presence of FITC-Fucoidan (0.05%) fixed and visualized by confocal microscopy. Bacterial cellular envelope and nucleoid region were labeled using Nile Red (3 µg ml−1) and DAPI (1 µg ml−1), respectively. Scale bar = 10 μm b Quantification of cells with fucoidan detected intra- and extracellularly and in contact with the bacterial envelope. c Colocalization results between FITC-Fucoidan and Nile red. Orange bar (M1): fraction of Nile red (red signal) in FITC-Fucoidan (green signal) and dark red bar (M2): fraction of FITC-Fucoidan (green) in Nile RED (red). Manders’ overlap coefficients were calculated employing the JaCoP plug-in for ImageJ software. Data represent the mean ± SD derived from three biologically independent experiments (n = 3) and p-values of one-way ANOVA Tukey´s multiple comparison test are indicated. Each dot represents a single experiment. Source data are provided as a Source Data file.
The closely related strains 892 and 913 exhibited remarkably different patterns of fucoidan uptake (Fig. 9). In the case of strain 892, the analysis of the FITC-Fucoidan fluorescent signal clearly indicated an internalization of the polysaccharide (>75% of cells had the fluorescent signal inside the cell). However, in the case of strain 913, the FITC-Fucoidan was mainly visualized outside the cells (>80% of analyzed cells) but in close contact with the cell membrane, mostly concentrated in a cellular pole. In some cases, we observed that the cells were placed nearby along a fiber of FITC-Fucoidan, seemly attached to the polysaccharide, or growing in rosette-like structures with the FITC-Fucoidan concentrated in the center (Figs. 9; Supplementary Fig. 18). Manders analysis indicated that in strain 892, FITC-Fucoidan internalization takes place through different bacterial cell envelope areas (i.e., high colocalization values for FITC-Fucoidan vs. Nile red and vice versa, Fig. 9c). However, for strain 913, our analysis showed that while FITC-Fucoidan strongly colocalized with the Nile red signal (Fig. 9c; high green in red colocalization value), very low colocalization of Nile red with FITC-Fucoidan (low red in green colocalization value) was observed. This result supports that, unlike strain 892, internalization of FITC Fucoidan in strain 913 takes place in a single area of the cell envelope. A two-phase membrane segregation with a highly hydrophobic phase in close contact with the FITC-Fucoidan was observed in strain 913, suggesting an accumulation of invaginations of the inner membrane at that site (Fig. 9, white arrows). Since Nile red fluorescence strongly depends on the hydrophobic environment of the lipid membrane, we speculate that the observed membrane heterogeneity could correspond to the generation of a highly hydrophobic phase where lipids and membrane proteins required for oligosaccharide breakdown and intracellular transport concentrate.
Overall, the visualization results are consistent with the idea that strains 892 and 913 have contrasting feeding strategies. Supporting this notion, the analysis of the proteomic samples during the growth experiments indicated different expression patterns of membrane transporters and secretory systems in both strains. In strain 913, up to 86 membrane transporters and 28 proteins related to secretion systems were upregulated at the proteomic level when exposed to fucoidan from F. vesiculosus, with average fold-change ratios ranging between 4 and 128, respectively, as compared to laminarin and mannose. From those, a total of 15 transporters were induced by growth on fucose. This result indicates that a common set of transporters are expressed during growth on fucoidan from F. vesiculosus and its primary monosaccharide fucose, reinforcing the idea that free fucose is liberated during the extensive external digestion of fucoidan in strain 913. By contrast, a low number of transporters and secretory proteins were induced by growth on fucoidan in strain 892 (i.e., only 4 transporters and 4 proteins related to the secretory pathway), even if both strains contained a similar inventory of proteins with these functions in their genomes (Supplementary Fig. 19). These results are fully aligned with the observations made by fluorescence microscopy, pointing to two independent degradation and internalization mechanisms: while cells from strain 913 process fucoidan externally, cells from strain 892 mainly perform internal digestion of this polysaccharide.
Discussion
The marine polysaccharide fucoidan produced by brown algae exhibits a remarkable recalcitrance. This enigmatic feature is at the core of its climate relevance, as it promotes the accumulation and long-term sequestration of carbon in the ocean7. So far, only a narrow diversity of microbial taxa have shown the capacity to degrade fucoidan, mainly affiliated with Bacteroidota and Verrucomicrobiota, which are found in fucoidan-rich environments such as sediments, marine particles and the phycosphere of microalgae like diatoms24–26,61. Even if fucanolytic enzymes have been detected in metagenomes from brown algae62, the epiphytic macroalgal microbiome remains an unexplored niche for uncovering mechanisms of fucoidan degradation. Such gap of knowledge may critically limit our capacity to obtain accurate estimates of the carbon effectively sequestered by macroalgae, which results from the balance of algal production and microbial degradation processes63,64. The two fucoidan-degrading marine strains isolated here were associated with a biofilm sample from the macroalgae F. spiralis. These strains are affiliated to the closely related genera Rhodopirellula and Neorhodopirellula65 from the phylum Planctomycetota, which are consistently found on the surface of diverse brown macroalgae51,66–70. Thus, they represent ecologically relevant models for studying the mechanisms of fucoidan degradation in this important, yet so far overlooked, environment. Strikingly, despite being closely related at the phylogenetic level and sharing a common niche, both strains exhibited a different genomic arrangement of fucanolytic enzymes and contrasting fucoidan feeding mechanisms. Such differences likely relate to niche partitioning processes that may sustain their coexistence in the algal biofilm62, as reported in marine bacteria during phytoplankton blooms27,61,71.
Two non-exclusive mechanisms have been proposed for bacterial feeding on complex carbohydrates in markedly different environments such as the human gut and marine waters72–74. In the classical external degradation mechanism, a myriad of enzymes with distinct substrate specificities are secreted outside the cell in soluble form or attached to the cell-surface, to perform the degradation in the surrounding environment75. Thus, the uptake of externally hydrolyzed products is not restricted to the enzyme-producing bacteria, but also to other members of the microbial community, enhancing cross-feeding events76. In the alternative selfish mechanism, bacteria perform a partial external depolymerization of high-molecular-weight carbohydrates leading to the formation of debranched oligosaccharides, which are selectively imported into the cell to complete their metabolism27,77. Our results by confocal microscopy revealed that strain 892 consumed fucoidan from F. vesiculosus through a selfish mechanism since the polysaccharide was mainly visualized inside the cell. Interestingly, fucose, the main monosaccharide of the homofucan, did not sustain its growth, which is a characteristic phenotype for selfish bacteria28,77. On the other hand, the feeding mechanism of strain 913 on fucoidan from F. vesiculosus was consistent with an extensive external degradation, including (i) the up-regulation of a wide range of transporters and the Type II Secretion system, (ii) the ability to grow on the monosaccharide fucose, and (iii) the visualization of fucoidan mainly outside but in close proximity to the cells, suggesting that it is retained in the cellular envelope. This is an outstanding observation, since the few previous visualizations available on free-living marine bacteria consuming fucoidan have only shown degradation through a selfish mechanism74, and external hydrolytic activity of fucosidases in environmental samples has been rarely reported27,77. Our co-localization analysis suggests that in contrast to the pattern observed in strain 892, in strain 913 fucoidan concentrates in a single defined focus along the cellular envelope paired with a higher-ordered region (i.e., higher Nile red fluorescence) in contact with the polysaccharide. Since cellular membranes are known to phase separate into high and low-ordered nanodomains, it is tempting to speculate that strain 913 generates a membrane phase compartmentalization with a higher-ordered nanodomain in contact with the polysaccharide containing all the required machinery to externally degrade and transport hydrolysis products through the bacterial envelope.
The genomic architecture of the enzymatic repertoire for fucoidan degradation in strains 892 and 913 also showed clear differences. While in strain 913 the fucoidan PULs were scattered across the entire genome, most of the fucanolytic PULs in strain 892 clustered together in a genomic region with a similar size (ca. 1 Mb, PULs B to F) to the reported fucoidan-degrading megaplasmid found in Lentimonas CC4, affiliated with Verrucomicrobiota30. This invites the hypothesis that this region may have been integrated into the genome of strain 892 from a giant extrachromosomal element. Despite the differences at the genomic level, both strains showed the ability to grow efficiently on homofucan and galactofucan sharing a core fucoidan degradative pathway encoded by exo-fucosidases from families GH29, GH95, and GH141, the endo-fucanase GH168 and SAs S1_17, S1_22 and S1_25. Interestingly, the proteomic investment for fucoidan turnover in strains 892 and 913 was much lower as compared to the highly specialized fucoidan degrader Lentimonas CC4, whose core degradative pathway involved more than 100 exo-fucosidases and SAs, as revealed by proteomics30. While approximately 60% of these enzymes were differentially regulated in two structurally related homofucans30, Lentimonas CC4 seems to be unable to grow on heterofucans or even labile polysaccharides, suggesting a high degree of specialization. On the other hand, strains 892 and 913 digested up to 80% of two markedly different fucoidans (homofucan and galactofucan) by upregulating a reduced set of enzymes, i.e., a total of 13 to 15 enzymes, including exo-fucosidases, endo-fucanases and SAs (Supplementary Table 4), indicating a higher flexibility in fucoidan degradation. The differential expression of selected variants of these enzyme families in strain 913 when growing on both fucoidan structures suggests differences in their substrate recognition, especially for the endo-fucanases of families GH107 and GH168.
We reveal that enzymes of family GH168 are consistently present in Planctomycetota genomes enriched in fucosidases, suggesting their importance in fucoidan degradation. Our hydrolytic experiments showed that Rho5174 and PbFucA, both GH168 endo-fucanases, were able to increase the degradation rate of fucoidan from F. vesiculosus, and to a lesser extent from E. maxima and M. pyrifera, when combined with the cellular lysate of strain 892. Both enzymes required the action of other enzymes with complementary activities/specificities present in this fucoidan-degrading bacteria, likely including SAs, CEs, and exo-fucosidases. These findings support the notion that the latter enzymes hydrolyze the heavily chemically decorated structures that protect the glycosidic linkages of the fucoidan backbone as a first degradative step. Afterwards, the endo-fucanases can access the core and accelerate the polysaccharide degradation30.
The enzyme Rho5174 was also able to hydrolyze fucoidan from C. okamuranus on its own, whereas the cellular lysate of strain 892 could not. However, this hydrolysis was rather weak, suggesting the need of other enzymes to efficiently degrade this polysaccharide. Fucoidan from C. okamuranus mainly consists of α-1,3-fucose (type I fucoidan), partially sulfated at position O4, and has a high percentage of acetylation and glucuronic acid30,78. In contrast, fucoidan from F. vesiculosus is composed of alternating α-1,3/α1,4-fucose (type II fucoidan) with O2 sulfation30,79. The fact that Rho5174 was transcriptionally upregulated when strain 892 was grown on F. vesiculosus, but not detected at the proteomic level suggests post-transcriptional regulatory mechanisms that depend on the fucoidan source. The ability of Rho5174 to partially initiate the hydrolysis of fucoidan from C. okamuranus indicates that distinct sources of fucoidan may follow substantially different degradation pathways. It is worth noting that other GH168 enzymes that have been characterized so far, belonging to the flavobacterium W. fucanilytica CZ1127T, can also initiate the degradation of fucoidans from sea cucumbers without the action of other enzymes42,43. Future work will be needed to determine the specific linkage and cleavage sites of Rho5174 and PbFucA.
Our structural comparison of GH168 family of enzymes revealed that they share a conserved (β/α)8 barrel catalytic domain and a common binding site for fucoidan, which runs in the center of the enzyme. Yet, we identified unique structural elements (loops 3, 4 and 7) that vary among different enzymes of the same strain and might partially define the substrate specificity of each endo-fucanase80–83. Such structural variability, related to distinct fucoidan recognition for each enzyme, may allow bacteria to process different chemical structures of the polysaccharide backbone, increasing the overall fucoidan degradation rate. Interestingly, several enzymes of the GH168 family found in both strains (Rho5174 and 913_05635, or 892_06289 and 913_01189) share these unique elements that form the binding site for fucoidan, which would lead to the recognition of similar fragments of this polymer. However, we also found some highly conserved residues that interact with O2 sulfated of Fuc (-1) and Fuc (-2) in the crystal structure of Fun168A-Fuc458. This suggests that these enzymes could recognize at least O2 sulfated fucose at these subsites, as is the case of the preferred substrates of Fun168A and Fun168D42,43. Beyond the relevance of fucoidan at the biogeochemical level, the study of the substrate specificity of new endo-fucanases and other fucoidan-degrading enzymes will be fundamental to obtain chemically defined fucoidan fragments and establishing the relationship between structure and biological activity on this glycan. Several studies revealed the benefits of fucoidan in human health, including immunomodulation, antioxidant, anti-allergic, antitumor, antiviral, anti-inflammatory, and hepatoprotective effects84–91. However, most of the earlier studies were performed using non-homogeneous fucoidan structures, varying in molecular weight, branching, linkages, and degree of sulfation and decoration11,92,93. Thus, advancing our understanding of substrate specificity of fucoidan degrading enzymes will be critical to produce low molecular weight, homogeneous and less complex fucoidan fragments, to apply this knowledge in the field of biomedicine.
The diversity of enzymes expressed for fucoidan degradation reflects the structural complexity of this polymer. However, such complexity, per se, does not explain their bioaccumulation in the environment. Other complex polysaccharides such as xylan94,95, pectins59, or α-mannans28,96 are effectively degraded by environmental microorganisms. Similarly, in other systems, such as the mucus barrier of the human gut, commensal bacteria continuously adapt to digest new dietary glycans regardless of their complexity97–101. The strong fucanolytic capacity of the strains shown here, investing a rather small enzymatic repertoire, also challenges the rationale behind the high recalcitrance of this polymer and invites the hypothesis that other processes are regulating the fucoidan degradative process. The microbiota found on the surface of macroalgae is typically host-specific102–104, which has been proposed to be related to the polysaccharide composition of the algae cell wall50,105. The gelatinous matrix composed of fucoidan provides a stable environment promoting interactions and coexistence with commensal bacteria105. The relationship between the macroalgae and their core associated community is tight and shapes fundamental roles in the algal life cycle, considering them as a sole organism termed “holobiont”106. A comparable functional role can be assigned to the mucus barrier in the human digestive tract, which is densely colonized by microbes and is considered another holobiont system107,108. Some members of the intestinal microbiota, such as Akkermansia municiphila or Bacteroides thetaiotaomicron, modulate the decorations of the glycoproteins composing the mucus barrier using a set of specialized GHs. However, these glycoproteins are used as a preferred carbon source only after long starvation periods, presumably because this implies the autodigestion of their habitat109,110. We hypothesize that a similar process can operate in the brown algae holobiont system. Some microorganisms with the ability to process fucoidan, such as members of the Pirellulaceae family, may act as primary colonizers, allowing the establishment of a bacterial community into the algal surface. Their distinct fucoidan-feeding mechanisms and enzymatic content would determine the subsequent succession of microbial communities in the macroalgae, as proposed in the context of the degradation of other polymers74,111,112. Moreover, the selective processing of structural units of fucoidan, such as sulfation or other decorations, may change the physical properties of the environment and shape the spatial distribution of the microbiome113. Nevertheless, the induction of the complete fucoidan degradative pathway should be tightly regulated at the community level, as their activation would compromise the stability of their environmental niche. Understanding the mechanisms of interaction and sensitivity to environmental change in this tightly regulated system should be a priority for future quantification of carbon fluxes mediated by macroalgae, which need to be reframed under the holobiont concept63,64.
Methods
Materials
Fucoidan from F. vesiculosus (F5631) and U. pinnatifida (F8315) for growth test in minimal medium and hydrolytic experiments were purchased from Sigma-Aldrich. Fucoidan from F. serratus (YF09360), F. vesiculosus (YF57714), D. potatorum (YF157165), E. maxima (YF157166), M. pyrifera (YF145109) and C. okamuranus (YF146834) for hydrolytic experiments were purchased from Biosynth. Fucoidan from U. pinnatifida (F8315) for hydrolytic experiments, tyramine and fluorescein isotiocyanate (FITC) were purchased from Sigma-Aldrich.
Isolation of Planctomycetota strains from Fucus spiralis
Samples of F. spiralis were collected in September 2021 from an intertidal zone of the Bidasoa estuary, near Hondarribia harbor (43.363036 N, 1.785446 E, Basque Country, Spain) and stored in natural seawater at 4 °C overnight before starting the isolation procedure. For the selective isolation of Planctomycetota strains from the surface of F. spiralis, a biofilm suspension was prepared by carefully scraping 15 g of algae into 5 mL of sterile natural seawater supplemented with the fungicide cycloheximide (20 mg L−1). An aliquot of this suspension was plated directly into solid media selective for Planctomycetota114 containing natural seawater (80%), ampicillin (200 mg L−1), streptomycin (200 mg L−1), and four different variations of carbon and energy sources: N-acetylglucosamine (1 g L−1), a crude extract of fucoidan from F. serratus (YF09360, Biosynth, 2 g L−1), a mixture of crude fucoidan from F. serratus (YF09360, Biosynth, 2 g L−1) and N-acetylglucosamine (A4106; Sigma-Aldrich, 1 g L−1), and a complex medium containing d-glucose (0.25 g L −1), Bacto peptone (BD Biosciences, 0.25 g L−1), BD Bacto yeast extract (BD Biosciences; 0.25 g L−1) and N-acetylglucosamine (A8625; Sigma-Aldrich 1 g L−1). All carbon sources were sterilized by filtration through 0.22 µm pore-size filters and added to the medium after autoclaving. Noble agar (BD Biosciences) was employed as gellifying agent and pH was adjusted to 8.0.
Plates were inoculated with 20 µL of the biofilm suspension at two different dilutions (10−1 and 10−2) in duplicates and incubated for 1 month at 25 °C. A selection of colonies with different phenotypic traits was examined by light microscopy using a Olympus microscope BX60 with a Olympus cellSens software. Colonies were selected for purification when microscopic analysis showed that cells displayed oval shape, budding growth or the formation of rosettes, which are characteristic morphologies for Planctomycetota. Pure cultures were re-inoculated into rich liquid media containing glucose, bactopeptone and levan extract (complex culture medium, see above), and stored in 20% glycerol at −80 °C. A random set of 20 isolates were selected for amplification of partial 16S rRNA genes (using primers 27 f and 1492r115) and Sanger sequencing by STAB VIDA.
Growth test in fucoidan minimal medium (MM)
To identify putative fucoidan-degrading strains, selected cultures were inoculated in minimal media (MM) supplemented with pure fucoidan ( ≥ 95%) as sole carbon source. In the MM, natural seawater was substituted for 250 ml 4x concentrated artificial seawater (46.94 g L−1 NaCl, 7.84 g L−1 Na2SO4, 21.28 g L−1 MgCl2·6H2O, 2.86 g L−1 CaCl2·2H2O, 0.384 g L−1 NaHCO3, 1.384 g L−1 KCl, 0.192 g L−1 KBr, 0.052 g L−1 H3BO3, 0.08 g L−1 SrCl2·6H2O and 0.006 g L−1 NaF) before autoclaving. Then, the medium was supplemented with 0,50 g L−1 KH2PO4, 0,40 g L−1 NH4Cl and 0.005 g L−1 BactoTM casamino acids (223050; Thermo Fisher). As a carbon source, pure fucoidan from U. pinnatifida (≥95% purity, F8315; Sigma–Aldrich) and F. vesiculosus (≥95% purity, F8190; Sigma–Aldrich) were tested at a final concentration of 0.5 g L−1. Isolates were inoculated into 1 mL filter-sterilized fucoidan-containing MM in 24-well plates and incubated for 7 days (25 °C, 85 rpm). At the initial and final times, we obtained measurements of optical density of the cultures in a microplate reader (Biotech Synergy).
Growth experiments with Planctomycetota isolates 892 and 913
The Planctomycetota isolates 892 and 913 were cultured in triplicate 500 mL Erlenmeyers flasks containing 200 mL of MM medium with 0.05% (wt/vol) fucoidan from F. vesiculosus (F8190; Sigma–Aldrich), fucoidan from U. pinnatifida (F8315; Sigma–Aldrich); laminarin from Laminaria digitata (L9634-1G; Sigma–Aldrich), l-fucose (A16789.03; Thermo Fisher) and mannose (A10842.14; Thermo Fisher). Pre-cultures grown on the same media were used to set the starting optical density to 0.005 (OD 600 nm). Flasks were incubated at 25 °C with continuous shaking at 85 rpm.
To compare growth patterns under different carbon sources, samples were taken in regular intervals of ca. 8 h during 5 days for measuring OD600 and substrate utilization using the phenol-sulfuric acid method (see below). Samples for transcriptomic and proteomic analyzes were collected at mid-exponential phase when OD600 was close to 0.08 (i.e., after 20 to 40 h). Briefly, duplicate samples (40 mL) were centrifuged at 10,000 × g and 25 °C. Cells were flash-frozen in liquid nitrogen and stored at −80 °C until analysis. Samples of the supernatant were filtered using 0.22-µm pore-size syringe filters and stored at −80 °C to analyze carbohydrate consumption.
Carbohydrate quantification in bacterial supernatants using High-Performance Anion Exchange Chromatography with Pulsed Amperometry Detection (HPAEC-PAD)
The monosaccharide content in samples collected during the growth experiments was determined by the HPAEC-PAD analysis technique. Briefly, 2 mL of supernatants were centrifuged before sample preparation at 19,000 × g for 10 min at 4 °C followed by filtration through 0.22 µm pore syringe-filter (13 mm). Then, samples were diluted 1:10 with MQ water and 0.5 mL of this solution was added to 2 mL of 1.25 M HCl to perform the acid hydrolysis of the polysaccharide. The reaction was incubated for 20 h at 100 °C and neutralized by evaporation with nitrogen gas at 60 °C. Finally, samples were rehydrated with MQ water, diluted 1:20 (final sample dilution 1:1,000) to avoid interference from salts and filtered again through 0.22 µm pore syringe-filter (13 mm). Carbohydrates were analyzed by high-performance anion exchange chromatography (HPAEC) with pulsed amperometric detection (PAD) using an ICS 5000+ ion chromatography system (Dionex, Thermo Fisher Scientific). Monosaccharides were separated using a precolumn and an analytical column Dionex CarboPac PA210-Fast-4 µm and detected using an ED 50 electrochemical cell equipped with a combined pH-Ag/AgCl reference electrode and a gold working electrode. Dionex EGC (Eluent Generator Cartidge) 500 KOH with Anion Trap Column continuously regenerated (Dionex CR-ATC500) was used as eluent source. An isocratic method of 18 mM KOH for 15 min, at a flow rate of 0.1 mL min−1, was used for the separation of monosaccharides followed by and additional period to clean-up the columns (up to 60 mM KOH). Finally, the eluent was reduced again to 18 mM KOH for re-equilibration of the columns. Chromatogram time was set to 35 min. The injection volume was set to 2.5 µL. Calibration curves for fucose, galactose, mannose, glucose, rhamnose and xylose were performed by multi-elemental standards of different concentrations in the range of 0.01 to 1 ppm, from 1000 ppm standards of each carbohydrate. Five levels of calibration standards were run to generate a standard curve. System control and data acquisition were performed with Chromeleon v 7.2 SR5 software (Dionex). The determination via HPAEC-PAD was performed in the Phytotron SGiker Service from the University of Basque Country (UPV/EHU, Spain).
Synthesis of FITC-Fucoidan
Fucoidan from F. vesiculosus (Sigma-Aldrich) was dissolved in phosphate buffer (pH 8.0). Tyramine (2.91 mmol, 400 mg) and NaBH3CN (2.39 mmol, 150 mg) were added to the mixture and incubated at 37 °C for 96 h with continuous stirring. The mixture was centrifuged at 11,500 × g for 10 min, and the supernatant was mixed with anhydrous ethanol and crystallized at -20 °C. Finally, the product was precipitated by centrifugation and evaporated to dryness in vacuo. Next, the synthesized fucoidan-tyramine compound was dissolved in NaHCO3 (0.5 mol L−1, pH 8.5), mixed with fluorescein isotiocyanate (FITC) (0.064 mmol, 25 mg) and incubated at 37 °C for 24 h. The mixture was centrifuged at 11,500 × g for 10 min, and the supernatant was added to a solution of anhydrous ethanol and crystallized at -20 °C. The crystallized product was collected by centrifugation and freeze-dried. Finally, free FITC contaminants were removed by dialysis (membrane molecular mass cut-off: 10 kDa) for 24 h. After collecting the liquid inside the dialysis bag, the FITC-Fucoidan was lyophilized, yielding 155 mg (60%) of pure FITC-Fucoidan.
Visualization of fucoidan uptake by strains 892 and 913 using confocal microscopy
Cells from strains 892 and 913 were grown on MM with fucoidan from F. vesiculosus at 0.05%. A bacterial suspension (OD600 = 0.1) was diluted (1:20) into 1 mL of fresh MM medium containing fluorescein-labeled fucoidan (0.05%). After 40 h of incubation at 25 °C and 85 rpm in 24-well plates, cells were stained with Nile Red (Sigma-Aldrich, 3 µg mL−1 final concentration) and DAPI (Thermo Fisher, 1 µg mL−1 final concentration) for 30 and 20 min, respectively. Then, cells were washed twice with MM medium at 19,000 × g for 10 min and fixed with paraformaldehyde (4%, 10 min). Samples were washed again with PBS and resuspended in 50 µL of PBS. Samples were mounted on agarose pads (1%) on microscope slides, covered with VectaShield and observed in a confocal microscope Zeiss LSM800.
Quantification of bacterial interaction with fucoidan
The type of interaction with fucoidan from different bacterial strains was determined by an experienced observer blind to sample identity. A total of 100 cells were counted for each experimental condition. Bacteria were divided into two classes: those that had fucoidan inside and those that had fucoidan outside. Additionally, all bacteria that displayed physical contact with the compound, either internally or externally, were considered ‘in contact with fucoidan”. Fucoidan and Bacterial Membrane Colocalization assay between the fluorophore Nile Red and FITC-Fucoidan was quantified as the Manders´ overlap coefficients using the JaCoP plug-in for ImageJ software116.
DNA extraction, whole-genome sequencing and assembly
Cultures were grown in 25 mL of liquid rich media for 5 days and pelleted. Cell pellets were washed twice with TE buffer, resuspended in 100 µL, and digested using lysozyme (1 mg mL−1, 37 °C, 1 h) and proteinase K (100 µg mL−1, 55 °C, overnight). DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega). Whole-genome sequencing was performed by combining Illumina MiSeq (2 x 150 pb) and GridION (Oxford Nanopore). Library preparation and sequencing were carried out at CNAG-CRG (National Center for Genomic Analysis, Spain). For each sequenced isolate, genomic sequence reads derived using Nanopore GridION and paired end sequence reads derived using Illumina MiSeq, were hybrid assembled using Spades v3.13.1117. Contigs with length > 5 Kbp were selected for subsequent analyzes (see Supplementary Table 5 for assembly details).
Genome annotation of Planctomycetota strains 892 and 913 and comparison with other reference genomes
Genes were predicted using Prokka v 1.14.6118 and annotated with InterProScan v.5.57-90.0119 using the databases Pfam120, TIGRFAMs121, CDD122, SMART123 and SUPERFAMILY124. GHs in the genomes were identified using dbCAN v3.0.7125. Three different tools implemented in dbCAN for CAZyme annotation were used, (i) HMMER search against annotated CAZYme domain boundaries, (ii) DIAMOND search against the CAZy pre-annotated CAZyme sequence database and (iii) HMMER for dbCAN-sub database, using default parameters. SAs were classified into families and subfamilies using SulfAtlas database based on the highest-scoring BLAST hit34.
For identifying putative PULs for fucoidan utilization in the genomes, we searched for regions enriched in CAZymes from families GH29, GH95, GH107, GH141 or/and GH168, co-localized with SAs and regulators. The presence of transporters (such as ABC transporters, Major Facilitators Transporters) was also considered, but not mandatory for the identification of putative PULs. In the absence of a reliable method to detect GH187 and GH174 family members using dbCAN v3, homologs of these enzymes were identified in our strains by using BLASTP against the available sequences of these enzyme families in the CAZY database (Nov 22, 2023).
Phylogenetic diversity of GH168 endo-fucanases in Planctomycetota
To assess the phylogenetic distribution of fucosidases, all genomes from the phylum Planctomycetota available at NCBI (5,202, accessed September 2023), NCBI RefSeq genomes from the phylum Verrucomicrobiota (530) and selected genomes from the phylum Bacteroidota (135) were retrieved and analyzed using the same methodology explained above, considering only hits by HMMER and at least one additional tool (DIAMOND and/or HMMER for dbCAN-sub) as implemented in dbCAN v3125. The aminoacidic sequences from the detected GH168 homologs were used to build a custom database. To identify additional GH168 family members from environmental samples, we performed a local BLASTP against The Ocean Microbiomics Database gene catalog126 using our GH168 custom database, selecting hits with >30% identity and >70% coverage. To confirm the identity of the latter GH168 hits, a second annotation with dbCAN v3.0.7125 was performed.
For assessing the phylogenetic relationship among GH168 enzymes, all the detected GH168 amino acid sequences from Planctomycetota, Bacteroidota and Verrucomicrobiota phyla and those from the environmental gene catalog were aligned using MAFFT v7127. Ambiguously aligned regions were removed with trimAl v1.4128, using a gap score cut-off of 0.95 and unaligned sequences were manually discarded by inspection of the alignment with MSAViewer129. A maximum likelihood tree was generated using RAxML v8.2.12130 with the PROTGAMMAUTO model. The resultant phylogenetic tree was visualized and annotated with iTol131. The homology of all retrieved GH168 proteins was assess using a reciprocal best match approach using the rbm.rb script implemented in the Enve-omics collection132.
Transcriptomic analysis in carbohydrate degradation assays with strains 892 and 913
Frozen cell pellet samples (45 mL) reserved for RNA extraction were immediately resuspended in 10 mL of RNA lysis buffer to obtain a clarified solution. A total of 5 mL of the lysate was spiked with internal RNA standards obtained by in vitro transcription of genomic templates of Saccharolobus solfataricus (formerly Sulfolobus solfataricus) P2 (NBCI Taxon ID 273057) for quantitative benchmarked transcriptomics133. Five standards were spiked individually to each sample at a conc. of ca. 20 pg µL−1. Then, RNA was extracted using the MirVana miRNA isolation Kit (Thermo Fisher) following the manufacturer’s recommendations. Samples were treated with Turbo DNAse-free kit (Thermo Fisher) and RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.) and Qubit™ RNA High Sensitivity (Thermo Fisher). RNA quality and integrity were checked using an Agilent RNA 6000 Nano Bioanalyzer (Agilent Technologies, Inc. 2001).
The RNA sequencing library was generated from 200-500 ng total RNA with the Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus Microbiome kit (Illumina) following the manufacturer’s recommendations. The cDNA libraries were sequenced as 50-bp paired-end reads on an Illumina NovaSeq 6000 platform (CNAG, Spain). Raw sequences were trimmed using Trimmomatic134, with Sliding Window:50:35 and MINLEN:50 parameters, and reads passing quality thresholds were paired. rRNA sequences were removed using SortMeRNA v4.3.6135, and Bowtie2 v2.4.2136 was used with the “–non-deterministic” parameter to map the remaining reads to the assembled genomes of the Planctomycetota strains. Read count tables were obtained with HTSeq137 using –stranded = reverse, -a 10 and -m intersection-nonempty parameters. To identify RNA internal standard reads of S. solfataricus, the same procedure was performed using the genome S. solfataricus. Individual transcript abundance (Ta) of protein-coding genes in each RNA sample was calculated as described in ref. 133, using Eq. (1),
| 1 |
where Ta corresponds to the estimated number of transcripts of an individual protein-coding gene, Ts corresponds to the number of reads assigned to the corresponding protein-coding gene, Sa corresponds to the number of molecules of internal RNA standards from S. solfataricus spiked to the RNA sample, and Ss corresponds to the number of reads assigned to S. solfataricus internal standards. To calculate transcript abundance per cell, Ta values were divided by total cell abundance in the corresponding sample. The number of cells was determined by flow cytometry using a BD Accuri™ C6 Plus Flow Cytometer in samples preserved with glutaraldehyde (0.25% final concentration) and stained with Sybr Green I (Thermo Fisher; 10X final concentration). Differential expression under different experimental conditions was analyzed using DESeq2 v1.24138 in R v4.2.1139, discarding genes with less than 10 transcripts in all samples prior to normalization, and implemented without automatic independent filtering. Patters in gene expression were visualized with heatmaps created with the R package ComplexHeatmap v2.12.1140. To assess the differences in the overall PUL expression, a pairwise Wilcoxon rank-sum test was performed using the R package rstatix.
Proteomic analysis in carbohydrate degradation assays with strains 892 and 913
Cell pellets were washed in 50 mM of Tris-HCl pH 6.8 at 4 °C before sample analysis. Sample preparation and analysis were conducted in the Proteomic Facility from the CIC bioGUNE (Derio, Spain). Samples were submitted to LC-MS label-free analysis using a hybrid trapped ion mobility spectrometry–quadrupole time of flight mass spectrometer (timsTOF Pro with PASEF, Bruker Daltonics, Billerica, MA, United States) coupled online to an EVOSEP ONE chromatograph (Evosep, Odense, Denmark). Samples were incubated and digested following the filter-aided sample preparation (FASP) protocol141. Trypsin was added to a trypsin:protein ratio of 1:50, and the mixture was incubated overnight at 37 °C, dried out in a RVC2 25 speedvac concentrator (Christ), and resuspended in 0.1% FA. Peptides were desalted and resuspended in 0.1% FA using C18 stage tips (Millipore). Digested sample (200 ng) was loaded in an EVOSEP ONE chromatograph (Evosep, Odense, Denmark) and resolved with a 44 min run (30 SPD protocol). Data was acquired using the standard DDA PASEF-standard_1.1sec_cycletime method, that scans between 100–1,700 m/z, and 0.60–1.60 v.s/cm2 using a ramp time of 100 ms. The column was heated to 50 °C using an oven. Protein identification and quantification were carried out using PEAKS Studio software using default settings (Bioinformatics solutions). Searches were carried out against a database consisting of strains 892 and 913 protein entries, with precursor and fragment tolerances of 20 ppm and 0.05 Da. Data (protein area) was loaded onto Perseus platform142 and further processed (log2 transformation, imputation) before statistical analysis (two-sided Student’s t-test). Proteins with a p value < 0.05 were considered for further analyzes. Differentially abundant proteins were considered with a LFC ≥ 1.5 and a p-value ≤ 0.05 in at least one condition.
Cloning, expression, and purification of 892_058729 (Rho5174) and PbFucA
The full-length gene encoding Rho5174 and PbFucA, from strain 892 and Planctomycetes bacterium K23_9 respectively, were cloned into a pET29a vector using NdeI and HindIII sites (ATG:biosynthetics GmbH, Mezhausen, Germany). The predicted signal peptide from PbFucA sequence (UniProt code: A0A517NMB4, residues 35-392)143 has been removed. Escherichia coli BL21(DE3) cells were transformed with the corresponding plasmid and grown in LB medium supplemented with 50 μg mL−1 kanamycin at 37 °C and 200 rpm. Cell cultures were induced at OD600 of 0.6 – 0.8 by adding 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). After 16 h at 18 °C, cells were harvested by centrifugation at 5,000 × g for 30 min at 4 °C and resuspended in 125 mL of buffer A (50 mM Tris-HCl, pH 7.5, 500 mM NaCl) containing protease inhibitors (A32955; Thermo Fisher) and 3 µL of Benzonase (71205; Merck). Cells were lysed by sonication in ice (30 cycles of 10 s pulses with 60 s cooling intervals between the pulses, and 60% amplitude) at 4 °C. The lysate was centrifuged at 20,000 × g for 40 min at 4 °C. The supernatant was filtered through 0.22 µm filters and then applied into a HisTrap HP column (5 mL, Cytiva) previously equilibrated with buffer A. The elution was performed in a ÄKTATM Prime Plus system (GE healthcare, Uppsala, Sweden) using a linear gradient from 0% to 100% of buffer B (50 mM Tris pH 7.5, 500 mM NaCl, 500 mM imidazole) for 40 min at 3.5 mL min−1. The purity of the protein was evaluated by SDS-PAGE (TruePAGE Precast Gels, Merck) and protein bands were visualized by Coomassie brilliant blue. For enzymatic activity experiments, the fractions were further purified by size-exclusion chromatography (SEC) using a Superdex 75 10/300 increased column and 50 mM Tris pH 7.5, 150 mM NaCl as running buffer. The eluted protein was concentrated, flash-frozen, and stored at -80 °C. For X-ray crystallography experiments, the fractions were purified by SEC using the same column and 20 mM Tris-HCl pH 7.5 as running buffer.
Preparation of lysates of Planctomycetota strains 892 and 913
Cells from 200 mL culture of Planctomycetota strains grown in F. vesiculosus (Sigma-Aldrich), U. pinnatifida (Sigma-Aldrich), and mannose at a final OD600 between 0.15- 0.19 were centrifuged at 16,000 × g. Cell pellets were resuspended in 12 mL of 50 mM Tris-HCl, pH 7.5, 500 mM NaCl containing a pill of protease inhibitors (A32955; Thermo Fisher), and 2 µL of Benzonase (71205; Merck). Cells were lysed by sonication (18 cycles of 10 s pulses with 59 s pulses of cooling intervals and 10% amplitude) at 4 °C. The lysate was centrifugated at 30,000 × g for 30 min at 4 °C. The lysates were flash-frozen and stored at -80 °C for fucoidan degradation activity assays.
Fucoidan degradation activity assays using the p-hydroxybenzoic acid hydrazide (PAHBAH) method
For enzymatic hydrolytic experiments (n = 3), a solution of 1 mg mL−1 of fucoidan from different algal species was incubated with Rho5174 and PbFucA at a final concentration of 2 and 1 mg mL−1, respectively, in 50 mM Tris, 150 mM NaCl pH 7.5 to a final reaction volume of 100 µL. It is worth mentioning that we were not able to concentrate PbFucA without precipitation to assay more than 1 mg mL−1 of the enzyme in the reactions. For the lysate hydrolytic experiments, a solution of 1 mg mL−1 of fucoidan from different algae species was incubated with lysates from strain 892 (1:5, v/v) in 50 mM Tris pH 7.5, 150 mM NaCl to a final reaction volume of 100 µL. For enzyme and lysate from strain 892 hydrolytic experiments, a solution of 1 mg mL−1 of fucoidan from different algae species was incubated with each enzyme at a final concentration indicated above, and lysate from strain 892 (1:5, v/v) in 50 mM Tris, 150 mM NaCl pH 7.5 to a final reaction volume of 100 µL. The Rho5174 and PbFucA reactions were incubated at 37°C and 25°C, respectively, for 0, 2, 7 and 20 h for each experiment. PbFucA precipitates at higher temperatures than 25 °C. We stopped the reactions by denaturation of the enzyme at 98 °C for 5 min. The reducing sugar equivalents of the samples were quantified with the para-hydroxybenzoic acid (PAHBAH) assay56. Briefly, the PAHBAH working reagent was freshly prepared at a ratio of 1:9 solution A (0.3 M 4-hydroxybenzhydrazide, 0.6 M HCl) to solution B (48 mM trisodium citrate, 10 mM CaCl2, 0.5 M NaOH). The working reagent was added to the reaction sample (1:10). This mixture was heated for 10 min at 99 °C under constant mixing at 300 rpm. Next, the absorbance of the mixture was measured at 410 nm in 200 plate reader Infinite® M Plex microplate reader (TECAN) to quantify the reduced sugar in the reaction. The results of each reaction were normalized to 0.
Exo-fucosidase activity assays using 4-nitrophenyl-α-l-fucopyranoside
We evaluated the exo-fucosidase activity of Rho5174 and PbFucA against 4-nitropheyl-α-l-fucopyranoside (pNP-Fuc). 50 µM of each enzyme was incubated with 5 mM pNP-Fuc in 0.1 M citrate-phosphate buffer along a pH gradient of 5.0, 6.0, 7.0 and 8.0 at 37 °C for 24 h in a final volume 75 µL following described protocols80,144. The reaction was stopped using 75 µL of 0.4 M of phosphate buffer pH 10.4. The p-nitrophenol released was measured spectrophotometrically at 405 nm Infinite 200 plate reader Infinite® M Plex microplate reader (TECAN) and evaluated using a standard curve of p-nitrophenol. We used FucOB as positive control using the described conditions80.
Enzymatic assays for Carbohydrate Polyacrylamide Gel Electrophoresis (C-PAGE)
For enzymatic hydrolytic experiments, a solution of 5 mg mL−1 of fucoidan from different algal species was incubated with Rho5174 and PbFucA at a final concentration of 2 and 1 mg mL−1, respectively, in 50 mM Tris, 150 mM NaCl pH 7.5. It is worth mentioning that we were not able to concentrate PbFucA without precipitation to assay more than 1 mg mL−1 of the enzyme in the reactions. For the lysate hydrolytic experiments, a solution of 5 mg mL−1 of fucoidan from different algae species was incubated with lysates from strain 892 (1:5, v/v) in 50 mM Tris pH 7.5, 150 mM NaCl to a final reaction volume of 100 µL. For enzyme and lysate from strain 892 hydrolytic experiments, a solution of 5 mg mL−1 of fucoidan from different algae species was incubated with each enzyme at a final concentration indicated above, and lysate from strain 892 (1:5, v/v) in 50 mM Tris, 150 mM NaCl pH 7.5 to a final reaction volume of 100 µL. The reactions were incubated at 37°C and 25 °C, respectively, for 0, 2, 5, 7, 24, 48 or 72 h for each experiment. PbFucA precipitates at higher temperatures than 25 °C. We stopped the reactions by denaturation of the enzyme at 98 °C for 5 min. Samples were stored at -20 °C for further analysis by C-PAGE in duplicate assays. Briefly, 2 µL of loading buffer (0.02% phenol red in 20% glycerol) was added to 10 µL of reaction. C-PAGE was performed on a 20%(w/v) acrylamide/bisacrylamide electrophoresis gel in 100 mM Tris–borate buffer pH 8.8 at 30 mA and 120 V for 1 h. The gel was stained with 0.05% Alcian blue 8GX (Panreac, Spain) in 2% acetic acid for 1 h, followed by destained in water for 20 min. Subsequently, the gel was stained with 0.01% O-toluidine blue (Sigma-Aldrich, Germany) in 50% aqueous ethanol and 1% acetic acid was applied for 30 min and distained in water for 1 h.
Rho5174 and PbFucA crystallization and data collection
Rho5174 was crystallized by mixing 0.25 µL of a protein solution at 10 mg mL−1 in 20 mM Tris-HCl pH 7.5 and 10 mM of sodium methyl-α-l-fucopyranoside 2-sulfate (Fuc2SO4; Supplementary Fig. 14)57 with 0.25 µL of Morpheus screening condition G8, Molecular Dimensions (0.1 M sodium HEPES:MOPS acid pH 7.5; 0.1 M sodium formate, 0.1 M ammonium acetate 0.1 M sodium citrate tribasic dihydrate; 0.1 M potassium sodium tartrate tetrahydrate, 0.2 M sodium oxamate; 9.25% v/v MPD; 9.25% PEG 1000; 9.25% w/v PEG 3350). Crystals grew in 2–3 days. PbFucA was crystallized in three crystal forms, referred thereafter as PbFucA-1, PbFucA-2 and PbFucA-3. The first crystal form, PbFucA-1, was obtained by mixing 0.25 µL of a protein solution at 5.4 mg mL−1 in 20 mM Tris-HCl pH 7.5 with 0.25 µL of JCSG+ screening condition B4, Molecular Dimensions (100 mM HEPES pH 7.5 10% w/v PEG 8000). Crystals grew in 2–3 days. The PbFucA-2 crystal form was achieved by random microseeding of PbFucA-1 crystals, 0.25 µL of protein solution at 5.4 mg mL−1 in 20 mM Tris-HCl pH 7.5 and 2 mM fucose was mixed with 0.05 µL of PbFucA-1 seeds and 0.20 µL of Morpheus screening condition E6, Molecular Dimensions (0.12 M ethylene glycols, 0.1 M buffer system 2 pH 7.5, 30% v/v precipitant mix 2). Crystals grew in 2–3 days. The PbFucA-3 crystal form was obtained by mixing 0.25 µL of protein solution at 5.4 mg mL−1 in 20 mM Tris-HCl pH 7.5 and 2 mM fucose and 0.25 µL of Morpheus screening condition D10, Molecular Dimensions (0.12 M alcohols, 0.1 M buffer system 3 pH 8.5, 30% v/v precipitant mix 2). All crystals were transferred to a cryo-protectant solution containing 30% glycerol and frozen under liquid nitrogen, except for Rho5174 crystals that grew in a cryo-protectant solution. Complete X-ray diffraction datasets were collected at the beamline BL13 XALOC (Alba Synchrotron, Spain) for Rho5174 and beamlines I24 for PbFucA-1 and PbFucA-3 and i04 for the PbFucA-2 (Diamond Light Source (DLS), Oxfordshire, UK). Rho5174 crystallized in triclinic space group P1 at 1.6 Å (PDB code 9F9V, Supplementary Table 2). The PbFucA-1 form crystallized in the monoclinic space group P1211 with six molecules in the asymmetric unit (MAU) and diffracted to a maximum resolution of 2.2 Å (PDB code 8RG3, Supplementary Table 2). The PbFucA-2 form crystallized in the orthorhombic space group of P212121 with 2 MAU at a resolution of 1.4 Å (PDB code 8RG4, Supplementary Table 2). The PbFucA-3 form crystallized in the tetragonal space group P41212 with 2 MAU to a maximum resolution of 2.1 Å (PDB code 8RG5, Supplementary Table 2). All datasets were integrated and scaled with XDS following standard procedures145.
Rho5174 and PbFucA structures determination and refinement
Structure determination of Rho5174 and PbFucA-1 crystal form was carried out by molecular replacement methods implemented in Phaser146 and the PHENIX suite147 and using the Alphafold model as a template148. Structure determination of PbFucA-2 and PbFucA-3 were carried out by molecular replacement using the crystal structure of PbFucA-1 chain A as template model. The final manual building was performed with Coot149 and refinement with phenix-refine. The structure was validated by MolProbity150. Data collection and refinement statistics are presented in Supplementary Table 2. Molecular graphics and structural analyzes were performed with the UCSF Chimera package151.
Structural analysis and sequence alignments
Structure-based sequence alignment analysis was performed using Chimera151. Z- score values were produced by using DALI. Domain interface analysis was performed using PISA152. Conserved and similar residues were labeled using the Multiple Align Show server (www.bioinformatics.org/SMS/multi_align.html).
Alphafold models of GH168 enzymes from strains 892 and 913, and Fun168D
The Alphafold-predicted structures were obtained using ColabFold153 and the full-length sequence of 892_06282 (Supplementary Dataset 12) and 892_06289 (Supplementary Dataset 10) from strain 892, 913_01189 (Supplementary Dataset 11) and 913_05635 (Supplementary Dataset 9) from strain 913 and Fun168D (Supplementary Dataset 8; UniProt code: A0A1B1Y6G8).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the Basque Government, including Elkartek projects MOSAIC and EMOTION with grant numbers KK-2021-00034 (LAS, MEG, BT) and KK-2022/00107 (LAS, MEG, BT, FXC), project BIOMATRIX (LAS), and grant number IT1625-22 (FXC). Additional funding was obtained by the MICINN/FEDER EU grant PID2019-105649RB-I00 (MEG), the MICINN grants PID2021-125469NB-C33 (LAS), PID2021-122177NA-I00 (BT), PID2020-117405GB100 (FXC), National Institute of Health grant R01AI149297 (MEG), La Caixa Foundation grant LCF/BQ/DR19/11740011 (MGA), and, as appropriate, by “ERDF A way of making Europe” by the “European Union” or by the “European Union NextGenerationEU/PRTR”. BT was supported by a “Ramón y Cajal” fellow, FXC was supported by Fundacion Ramon Areces (grant number CIVP20S11276) and UA and JOI were supported by a PhD grant of the Basque Government PhD. We thank support of A. Lanzén for assistance in bioinformatics and J. Franco and JM Garmendia for providing samples of F. spiralis. We acknowledge Diamond Light Source (proposals mx28360) and ALBA synchrotron beamline BL13-XALOC (2023087729) for providing access to synchrotron radiation facilities. The authors thank for technical and human support provided by SGIker, Phytotron Service of UPV/EHU during the HAEPAC-PAD analysis, and the Proteomic Facility from the CIC bioGUNE (Derio, Spain) for sample preparation and data analysis. This is contribution number 1244 of AZTI, Marine Research, Basque Research and Technology Alliance (BRTA).
Author contributions
F.X.C., M.E.G., B.T., and L.A.S. conceived the project. C.P.C., A.M.M., R.L., O.T., U.A., M.G.A., M.L., A.M.G., I.G.A. and B.R.C. performed the experiments. C.P.C. performed the isolation of bacterial strains. C.P.C., U.A., and R.L. performed the experimental work with bacterial strains and collected samples for all associated analyzes. R.L. carried out the bioinformatic analysis of multi-omics data, and C.P.C., R.L., and L.A.S. interpreted the results. A.M.M. and M.G.A. expressed, purified, and crystallized proteins, and collected and processed X-ray data. E.M. and J.L.C. synthesized sodium methyl-α-l-fucopyranoside 2-sulfate (Fuc2SO4). A.M.M., M.G.A., M.E.G., and B.T. solved and analyzed the crystal structure and performed structural and sequence analysis. A.M.M., A.M.G., and I.G.A., and B.R.C. carried out enzymatic assays, and A.M.M., I.G.A., B.R.C., M.E.G., and B.T. interpreted the activity results. C.P.C., J.A.N.G., and J.O.I. performed FITC-Fucoidan experiments. O.T. and M.L. analyzed microscopy results and synthesized FITC-Fucoidan. C.P.C., A.M.M., R.L., O.T., U.A., M.G.A., M.L., I.G.A., B.R.C., F.X.C., M.E.G., B.T. and L.A.S. analyzed the results. C.P.C., A.M.M., R.L., F.X.C., M.E.G., B.T. and L.A.S. wrote the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The atomic coordinates and structure factors have been deposited with the Protein Data Bank (PDB), accession code 8RG3 (PbFucA-1), 8RG4 (PbFucA-2), 8RG5 (PbFucA-3) and 9F9V (Rho5174). The whole genome sequencing datasets and transcriptome sequencing data generated in this study have been deposited in the European Nucleotide Archive (ENA) under Bioproject accession number PRJEB71092 and Zenodo. The genome assemblies of 892 and 913 strains generated in this study have been deposited under accession number GCA_963859505.1 and GCA_963859645.1 respectively. Proteomic data has been deposited to the ProteomeXchange Consortium via the PRIDE repository with the dataset identifier PXD057797. The crystal structures have been deposited in PDB under the following accession codes: 8RG3, 8RG4, 8RG5, and 9F9V. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Carla Pérez-Cruz, Alicia Moraleda-Montoya, Raquel Liébana.
Contributor Information
Francesc-Xabier Contreras, Email: xabier.contreras@ehu.eus.
Marcelo E. Guerin, Email: mrccri@ibmb.csic.es
Beatriz Trastoy, Email: beatriz.trastoy@gmail.com.
Laura Alonso-Sáez, Email: lalonso@azti.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55268-w.
References
- 1.Pessarrodona, A. et al. Global Seaweed Productivity. Sci. Adv. 8, eabn2465 (2022). [DOI] [PMC free article] [PubMed]
- 2.Duarte, C. M. et al. Global estimates of the extent and production of macroalgal forests. Glob. Ecol. Biogeogr.31, 1422–1439 (2022). [Google Scholar]
- 3.Wang, M. et al. The great Atlantic Sargassum belt. Science365, 83–87 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Smetacek, V. & Zingone, A. Green and golden seaweed tides on the rise. Nature504, 84–88 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Krause-Jensen, D. & Duarte, C. M. Substantial role of macroalgae in marine carbon sequestration. Nat. Geosci.9, 737–742 (2016). [Google Scholar]
- 6.Deniaud-Bouët, E. et al. Chemical and enzymatic fractionation of cell walls from Fucales: Insights into the structure of the extracellular matrix of brown algae. Ann. Bot.114, 1203–1216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck-Wiese, H. et al. Fucoid brown algae inject fucoidan carbon into the ocean. Proc. Natl Acad. Sci.120, e2210561119 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park, H. Y. et al. Anti-inflammatory effects of fucoidan through inhibition of NF-κB, MAPK and Akt activation in lipopolysaccharide-induced BV2 microglia cells. Food Chem. Toxicol.49, 1745–1752 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Chen, L.-M. et al. Oligo-Fucoidan prevents IL-6 and CCL2 production and cooperates with p53 to suppress ATM signaling and tumor progression. Sci. Rep.7, 11864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradhan, B. et al. A state-of-the-art review on fucoidan as an antiviral agent to combat viral infections. Carbohydr. Polym.291, 119551 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ale, M. T. & Meyer, A. S. Fucoidans from brown seaweeds: an update on structures, extraction techniques and use of enzymes as tools for structural elucidation. RSC Adv.3, 8131–8141 (2013). [Google Scholar]
- 12.Ponce, N. M. A. & Stortz, C. A. A comprehensive and comparative analysis of the fucoidan compositional data across the phaeophyceae. Front. Plant Sci.11, 556312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, H. T. T. et al. Novel enzyme actions for sulphated galactofucan depolymerisation and a new engineering strategy for molecular stabilisation of fucoidan degrading enzymes. Mar. Drugs16, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zayed, A., Avila-Peltroche, J., El-Aasr, M. & Ulber, R. Sulfated galactofucans: an outstanding class of fucoidans with promising bioactivities. Mar. Drugs20, 412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopplin, G. et al. Structural characterization of fucoidan from laminaria hyperborea: assessment of coagulation and inflammatory properties and their structure-function relationship. ACS Appl. Bio Mater.1, 1880–1892 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Wang, L. et al. Structural characterization of a fucoidan from Ascophyllum nodosum and comparison of its protective effect against cellular oxidative stress with its analogues. Int. J. Biol. Macromol.239, 124295 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Yang, Y., Hu, T., Li, J., Xin, M. & Zhao, X. Structural characterization and effect on leukopenia of fucoidan from Durvillaea antarctica. Carbohydr. Polym.256, 117529 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Sichert, A. et al. Ion-exchange purification and structural characterization of five sulfated fucoidans from brown algae. Glycobiology31, 352–357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, J., Zhang, Q., Zhang, Z., Zhang, H. & Niu, X. Structural studies on a novel fucogalactan sulfate extracted from the brown seaweed Laminaria japonica. Int. J. Biol. Macromol.47, 126–131 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Bilan, M. I. et al. Sulfated polysaccharides of the Vietnamese brown alga Sargassum aquifolium (Fucales, Sargassaceae). Carbohydr. Res.449, 23–31 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Imbs, T. I., Shevchenko, N. M., Semenova, T. L., Sukhoverkhov, S. V. & Zvyagintseva, T. N. Compositional heterogeneity of sulfated polysachcharides synthesized by brown alga Costaria costata. Chem. Nat. Compd.47, 96–97 (2011). [Google Scholar]
- 22.Chang, C. C., Cheng, J. J., Lee, I. J. & Lu, M. K. Purification, structural elucidation, and anti-inflammatory activity of xylosyl galactofucan from Armillaria mellea. Int. J. Biol. Macromol.114, 584–591 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Ke, S. et al. Structural characterization of sulfated galactofucan from Undaria pinnatifida and its effect on type 2 diabetic mice. J. Oceanol. Limnol.41, 300–313 (2023). [Google Scholar]
- 24.Orellana, L. H. et al. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J.16, 630–641 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reintjes, G., Heins, A., Wang, C. & Amann, R. Abundance and composition of particles and their attached microbiomes along an Atlantic Meridional Transect. Front. Mar. Sci.10, 1–15 (2023). [Google Scholar]
- 26.Vidal-Melgosa, S. et al. Diatom fucan polysaccharide precipitates carbon during algal blooms. Nat. Commun.12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnosti, C. et al. The biogeochemistry of marine polysaccharides: sources, inventories, and bacterial drivers of the carbohydrate cycle. Ann. Rev. Mar. Sci.13, 81–108 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Cuskin, F. et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature517, 165–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reisky, L. et al. A marine bacterial enzymatic cascade degrades the algal polysaccharide Ulvan. Nat. Chem. Biol.15, 803–812 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Sichert, A. et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol.5, 1026–1039 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Drula, E. et al. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res50, D571–D577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silchenko, A. S. et al. Fucoidan sulfatases from marine bacterium Wenyingzhuangia Fucanilytica CZ1127T. Biomolecules8, 1–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silchenko, A. S. et al. Discovery of a fucoidan endo-4O-sulfatase: Regioselective 4O-desulfation of fucoidans and its effect on anticancer activity in vitro. Carbohydr. Polym.271, 118449 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Stam, M. et al. SulfAtlas, the sulfatase database: state of the art and new developments. Nucleic Acids Res51, D647–D653 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barzkar, N. et al. A recent update on Fucoidonase: source, Isolation methods and its enzymatic activity. Front. Mar. Sci.10, 1–13 (2023). [Google Scholar]
- 36.Wu, H., Owen, C. D. & Juge, N. Structure and function of microbial α- fucosidases: a mini review. Essays Biochem67, 399–414 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silchenko, A. S. et al. Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiology27, 254–263 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Zueva, A. O. et al. Expression and biochemical characterization of two recombinant Fucoidanases from the marine bacterium Wenyingzhuangia Fucanilytica CZ1127. T. Int. J. Biol. Macromol.164, 3025–3037 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Ropartz, D. et al. In-depth structural characterization of oligosaccharides released by GH107 Endofucanase MfFcnA reveals enzyme subsite specificity and sulfated fucan substructural features. Glycobiology32, 276–288 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Tran, V. H. N. et al. The endo-α(1,3)-fucoidanase Mef2 releases uniquely branched oligosaccharides from Saccharina Latissima fucoidans. Mar. Drugs20, 305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, C. et al. Overexpression and biochemical characterization of a truncated endo-α (1 → 3)-fucoidanase from Alteromonas sp. SN-1009. Food Chem.353, 129460 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Shen, J., Chang, Y., Zhang, Y., Mei, X. & Xue, C. Discovery and characterization of an endo-1,3-fucanase from marine bacterium Wenyingzhuangia Fucanilytica: a novel glycoside hydrolase family. Front. Microbiol.11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen, J. et al. Characterization of a novel endo-1,3-fucanase from marine bacterium Wenyingzhuangia Fucanilytica reveals the presence of diversity within glycoside hydrolase family 168. Carbohydr. Polym.318, 121104 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Liu, G. et al. Characterization of an endo-1,3-fucanase from marine bacterium Wenyingzhuangia Aestuarii: The first member of a novel glycoside hydrolase family GH174. Carbohydr. Polym.306, 120591 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Shen, J. et al. Discovery of a catalytic domain defines a new glycoside hydrolase family containing endo-1,3-fucanase. Carbohydr. Polym.323, 121442 (2024). [DOI] [PubMed] [Google Scholar]
- 46.Silchenko, A. S. et al. The discovery of the fucoidan-active Endo-1→4-α-l-fucanase of the GH168 family, which produces fucoidan derivatives with regular sulfation and anticoagulant activity. Int. J. Mol. Sci. 25, 218 (2024). [DOI] [PMC free article] [PubMed]
- 47.Kallscheuer, N. & Jogler, C. The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol. Adv.53, 107818 (2021). [DOI] [PubMed] [Google Scholar]
- 48.DeLong, E. F., Franks, D. G. & Alldredge, A. L. Phylogenetic diversity of aggregate‐attached vs. free‐living marine bacterial assemblages. Limnol. Oceanogr.38, 924–934 (1993). [Google Scholar]
- 49.Wiegand, S., Jogler, M. & Jogler, C. On the maverick planctomycetes. FEMS Microbiol. Rev.029, 739–760 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Lage, O. M. & Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol.5, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faria, M. et al. Planctomycetes attached to algal surfaces: Insight into their genomes. Genomics110, 231–238 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Wegner, C. E. et al. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genomics9, 51–61 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Boedeker, C. et al. Determining the bacterial cell biology of Planctomycetes. Nat. Commun.8, 14853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erbilgin, O., McDonald, K. L. & Kerfeld, C. A. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl. Environ. Microbiol.80, 2193–2205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol.40, 1023–1025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lever, M. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem.47, 273–279 (1972). [DOI] [PubMed] [Google Scholar]
- 57.Leder, I. G. Synthesis of the sodium salts of methyl α-l-fucopyranoside 2-, 3-, and 4-sulfates. J. Carbohydr. Chem.12, 95–103 (1993). [Google Scholar]
- 58.Chen, G. et al. Structural investigation of Fun168A unraveling the recognition mechanism of endo-1,3-fucanase towards sulfated fucan. Int. J. Biol. Macromol.271, 132622 (2024). [DOI] [PubMed] [Google Scholar]
- 59.Ndeh, D. et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature544, 65–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinze, S. et al. Identification of endoxylanase XynE from Clostridium thermocellum as the first xylanase of glycoside hydrolase family GH141. Sci. Rep.7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Priest, T., Heins, A., Harder, J., Amann, R. & Fuchs, B. M. Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group. ISME J.16, 1570–1582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song, W., Wemheuer, B., Steinberg, P. D., Marzinelli, E. M. & Thomas, T. Contribution of horizontal gene transfer to the functionality of microbial biofilm on a macroalgae. ISME J.15, 807–817 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Loos, L. M., Eriksson, B. K. & Falcão Salles, J. The macroalgal holobiont in a changing sea. Trends Microbiol27, 635–650 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Trevathan-Tackett, S. M. et al. A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol.3, 1509–1520 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Sreya, P. et al. Revisiting the taxonomy of the genus Rhodopirellula with the proposal for reclassification of the genus to Rhodopirellula Sensu Stricto, Aporhodopirellula gen. nov., Allorhodopirellula gen. nov. and Neorhodopirellula gen. nov. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol.116, 243–264 (2023). [DOI] [PubMed] [Google Scholar]
- 66.Wiegand, S. et al. Analysis of bacterial communities on North Sea macroalgae and characterization of the isolated planctomycetes adhaeretor mobilis gen. Nov., sp. nov., roseimaritima multifibrata sp. nov., rosistilla ulvae sp. nov. and rubripirellula lacrimiformis sp. nov. Microorganisms9, 1–33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bondoso, J. et al. Epiphytic Planctomycetes communities associated with three main groups of macroalgae. FEMS Microbiol. Ecol.93, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bengtsson, M. M., Sjøtun, K. & Lanzén, A. & Øvreås, L. Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J.6, 2188–2198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bengtsson, M. M. & Øvreås, L. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol10, 261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lage, O. M. & Bondoso, J. Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol.78, 366–375 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Avcı, B., Krüger, K., Fuchs, B. M., Teeling, H. & Amann, R. I. Polysaccharide niche partitioning of distinct Polaribacter clades during North Sea spring algal blooms. ISME J.14, 1369–1383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnosti, C. Fluorescent derivatization of polysaccharides and carbohydrate-containing biopolymers for measurement of enzyme activities in complex media. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.793, 181–191 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Hehemann, J. H. et al. Single cell fluorescence imaging of glycan uptake by intestinal bacteria. ISME J.13, 1883–1889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reintjes, G., Arnosti, C., Fuchs, B. M. & Amann, R. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J.11, 1640–1650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Traving, S. J., Thygesen, U. H., Riemann, L. & Stedmon, C. A. A model of extracellular enzymes in free-living microbes: which strategy pays off? Appl. Environ. Microbiol.81, 7385–7393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pontrelli, S. et al. Metabolic cross-feeding structures the assembly of polysaccharide degrading communities. Sci. Adv.8, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giljan, G. et al. Selfish bacteria are active throughout the water column of the ocean. ISME Commun.3, 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim, S. J., Wan Aida, W. M., Schiehser, S., Rosenau, T. & Böhmdorfer, S. Structural elucidation of fucoidan from Cladosiphon Okamuranus (Okinawa Mozuku). Food Chem.272, 222–226 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Lahrsen, E., Schoenfeld, A.-K. & Alban, S. Size-dependent pharmacological activities of differently degraded fucoidan fractions from Fucus vesiculosus. Carbohydr. Polym.189, 162–168 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Anso, I. et al. Turning universal O into rare Bombay type blood. Nat. Commun. 14, 1765 (2023). [DOI] [PMC free article] [PubMed]
- 81.Trastoy, B. et al. Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases. Nat. Commun.14, 1705 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.García-Alija, M. et al. Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE. Nat. Commun.13, 1137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trastoy, B. et al. Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides. Nat. Commun.11, 899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deniaud-Bouët, E., Hardouin, K., Potin, P., Kloareg, B. & Hervé, C. A review about brown algal cell walls and fucose-containing sulfated polysaccharides: Cell wall context, biomedical properties and key research challenges. Carbohydr. Polym.175, 395–408 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Maruyama, H., Tamauchi, H., Hashimoto, M. & Nakano, T. Suppression of Th2 immune responses by Mekabu fucoidan from Undaria Pinnatifida Sporophylls. Int. Arch. Allergy Immunol.137, 289–294 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Chen, B. R. et al. Immunomodulation and mechanisms of fucoidan from Cladosiphon Okamuranus Ameliorates atopic dermatitis symptoms. Int. J. Biol. Macromol.189, 537–543 (2021). [DOI] [PubMed] [Google Scholar]
- 87.Mizuno, M., Fujioka, A., Bitani, S., Minato, K. I. & Sakakibara, H. Anti-allergic activity of fucoidan can be enhanced by coexistence with quercetin. Int. J. Mol. Sci.23, 12163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oliveira, C., Neves, N. M., Reis, R. L., Martins, A. & Silva, T. H. A review on fucoidan antitumor strategies: From a biological active agent to a structural component of fucoidan-based systems. Carbohydr. Polym.239, 116131 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Lahrsen, E., Liewert, I. & Alban, S. Gradual degradation of fucoidan from Fucus vesiculosus and its effect on structure, antioxidant and antiproliferative activities. Carbohydr. Polym.192, 208–216 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Cao, L. M. et al. Antitumor activity of fucoidan: a systematic review and meta-analysis. Transl. Cancer Res.10, 5390–5405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao, Y. et al. Fucoidan extracted from Undaria pinnatifida: source for nutraceuticals/functional foods. Mar. Drugs16, 321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flórez-Fernández, N., Torres, M. D., González-Muñoz, M. J. & Domínguez, H. Potential of intensification techniques for the extraction and depolymerization of fucoidan. Algal Res30, 128–148 (2018). [Google Scholar]
- 93.Wang, P. et al. Analysis of structural heterogeneity of fucoidan from Hizikia fusiforme by ES-CID-MS/MS. Carbohydr. Polym.90, 602–607 (2012). [DOI] [PubMed] [Google Scholar]
- 94.Pereira, G. V. et al. Degradation of complex arabinoxylans by human colonic Bacteroidetes. Nat. Commun.12, 459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dutschei, T. et al. Marine bacteroidetes enzymatically digest xylans from terrestrial plants. Environ. Microbiol. 25, 1713–1727 (2023) [DOI] [PubMed]
- 96.Chen, J. et al. Alpha- and beta-mannan utilization by marine Bacteroidetes. Environ. Microbiol.20, 4127–4140 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Hehemann, J.-H. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature464, 908–912 (2010). [DOI] [PubMed] [Google Scholar]
- 98.Pluvinage, B. et al. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat. Commun.9, 1043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hehemann, J., Kelly, A. G., Pudlo, N. A., Martens, E. C. & Boraston, A. B. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Nati. Acad. Sci. USA109, 19786–19791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pudlo, N. A. et al. Diverse events have transferred genes for edible seaweed digestion from marine to human gut bacteria. Cell Host Microbe30, 314–328.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mathieu, S. et al. Ancient acquisition of ‘alginate utilization loci’ by human gut microbiota. Sci. Rep.8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lachnit, T., Blümel, M., Imhoff, J. F. & Wahl, M. Specific epibacterial communities on macroalgae: Phylogeny matters more than habitat. Aquat. Biol.5, 181–186 (2009). [Google Scholar]
- 103.Lachnit, T., Meske, D., Wahl, M., Harder, T. & Schmitz, R. Epibacterial community patterns on marine macroalgae are host‐specific but temporally variable. Environ. Microbiol.13, 655–665 (2011). [DOI] [PubMed] [Google Scholar]
- 104.Marzinelli, E. M. et al. Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol.17, 4078–4088 (2015). [DOI] [PubMed] [Google Scholar]
- 105.Singh, R. P. & Reddy, C. R. K. Seaweed-microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol.88, 213–230 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Egan, S. et al. The seaweed holobiont: Understanding seaweed-bacteria interactions. FEMS Microbiol. Rev.37, 462–476 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Wang, B. X., Wu, C. M. & Ribbeck, K. Home, sweet home: how mucus accommodates our microbiota. FEBS J.288, 1789–1799 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Postler, T. S. & Ghosh, S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab.26, 110–130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wardman, J. F., Bains, R. K., Rahfeld, P. & Withers, S. G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol.20, 542–556 (2022). [DOI] [PubMed] [Google Scholar]
- 110.Pudlo, N. A. et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. MBio6, 01282–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Enke, T. N. et al. Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol.29, 1528–1535 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Datta, M. S., Sliwerska, E., Gore, J., Polz, M. F. & Cordero, O. X. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun.7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McCallum, G. & Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol.22, 105–118 (2023). [DOI] [PubMed] [Google Scholar]
- 114.Wiegand, S. et al. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol.5, 126–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lane, D. J. & Collins, M. L. Current methods for detection of DNA/Ribosomal RNA Hybrids. Rapid Methods Autom. Microbiol. Immunol. 54–75 (1991).
- 116.Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc.224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol.19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 119.Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res.49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haft, D. H. et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res.41, D387–D395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marchler-Bauer, A. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pandurangan, A. P., Stahlhacke, J., Oates, M. E., Smithers, B. & Gough, J. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res47, D490–D494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zheng, J. et al. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res51, W115–W121 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paoli, L. et al. Biosynthetic potential of the global ocean microbiome. Nature607, 111–118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Katoh, K., Misawa, K., Kuma, K. I. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yachdav, G. et al. MSAViewer: interactive javascript visualization of multiple sequence alignments. Bioinformatics32, 3501–3503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodriguez-R, L. M. & Konstantinidis, K. T. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ4, e1900v1 (2016).27123377 [Google Scholar]
- 133.Gifford, S., Satinsky, B. & Moran, M. A. Quantitative Microbial Metatranscriptomics. in Methods in Molecular Biology (eds. Paulsen, I. T. & Holmes, A. J.)1096, 213–229 (Humana Press, 2014). [DOI] [PubMed]
- 134.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Anders, S., Pyl, P. T. & Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed]
- 139.R Core Team. R: A language and environment for statistical computing. (2022). R Foundation for Statistical Computing, Vienna. https://www.R-project.org.
- 140.Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 143.Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol.37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 144.Megson, Z. et al. Characterization of an α-fucosidase from the periodontal pathogen Tannerella forsythia. Virulence6, 282–292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kabsch, W. XDS. Acta Crystallogr. Sect. D. Biol. Crystallogr.66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr.40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr.66, 213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 150.Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res32, W615–W619 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 152.Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol.372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 153.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olson, R. D. et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res.51, D678–D689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The atomic coordinates and structure factors have been deposited with the Protein Data Bank (PDB), accession code 8RG3 (PbFucA-1), 8RG4 (PbFucA-2), 8RG5 (PbFucA-3) and 9F9V (Rho5174). The whole genome sequencing datasets and transcriptome sequencing data generated in this study have been deposited in the European Nucleotide Archive (ENA) under Bioproject accession number PRJEB71092 and Zenodo. The genome assemblies of 892 and 913 strains generated in this study have been deposited under accession number GCA_963859505.1 and GCA_963859645.1 respectively. Proteomic data has been deposited to the ProteomeXchange Consortium via the PRIDE repository with the dataset identifier PXD057797. The crystal structures have been deposited in PDB under the following accession codes: 8RG3, 8RG4, 8RG5, and 9F9V. Source data are provided with this paper.