Abstract
The NS1 binding protein, known for interacting with the influenza A virus protein, is involved in RNA processing, cancer, and nerve cell growth regulation. However, its role in stress response independent of viral infections remains unclear. This study investigates NS1 binding protein’s function in regulating stress granules during oxidative stress through interactions with GABARAP subfamily proteins. We find that NS1 binding protein localizes to stress granules, interacting with core components, GABARAP proteins, and p62, a protein involved in autophagy. In cells lacking NS1 binding protein, stress granule dynamics are altered, and p62 ubiquitination is increased, suggesting impaired stress granule degradation. Overexpression of NS1 binding protein reduces p62 ubiquitination. In amyotrophic lateral sclerosis patient-derived neurons, reduced NS1 binding protein and p62 disrupt stress granule morphology. These findings identify NS1 binding protein as a negative regulator of p62 ubiquitination and a facilitator of GABARAP recruitment to stress granules, implicating it in stress granule regulation and amyotrophic lateral sclerosis pathogenesis.
Subject terms: Organelles, Molecular biology, Autophagy
The NS1 binding protein (NS1-BP) is involved in RNA processing, cancer, and nerve cell growth regulation. Here, the authors show that NS1-BP regulates stress granule (SG) dynamics under oxidative stress by interacting with GABARAPs and p62. Reduced NS1-BP in amyotrophic lateral sclerosis (ALS) neurons affects SG morphology, linking it to ALS.
Introduction
Autophagy is cellular recycling, breaking down damaged proteins and organelles for energy and cellular repair1. It aids adaptation to stress conditions, including nutrient shortage, oxidative stress, and infection2,3. Regulated by different signals, autophagy maintains cell health, preventing toxic build-up.
Conversely, stress granules (SGs) are dynamic structures that form in cell cytoplasm in response to stress, including heat shock, viral infection, or oxidative stress4. Comprising RNA and RNA-binding proteins, they store mRNA temporarily, shielding it from degradation. When stress subsides, SGs aid mRNA translation and regulate gene expression and stress response. SGs are rapidly assembled primarily by key nucleating proteins G3BP1 and G3BP2, which facilitate the recruitment of other RNA-binding proteins and mRNAs into SGs5,6 and disassembled by various chaperons, valosin-containing protein (VCP), or RNA helicase7,8. Some specific or prolonged stress conditions also facilitate SG degradation.
Over the past two decades, approximately 100 proteins have been identified in the SG interactome9–11. Some with low complexity regions enable multiple interactions with various binding partners12. SGs serve as signaling platforms, aiding the coordination of cellular processes during stress, including apoptosis, cell growth, and metabolic control13. Dysregulated SG dynamics can lead to the accumulation of persistent SGs, which sequester essential RNA-binding proteins and mRNAs, disrupting cellular homeostasis14–16, potentially inducing neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS)17,18.
Autophagy and SGs interact closely. Autophagy, activated during stress, clears damaged or aggregated proteins, including those in SGs2, regulating their assembly and disassembly8,19,20. SGs components, including G3BP1, TIA-1 and TDP-43, have been shown to undergo autohagic degradation under prolonged stress conditions or when SG disassembly is impaired21,22. This interplay enables cells to adapt to stress and maintain cellular homeostasis.
Recent evidence associates SGs with autophagy, suggesting a molecular connection9,22,23. Autophagy components, including VCP, p62/SQSTM1 (hereafter referred to as p62), mammalian ATG8s (mATG8s), and Unc-51-like kinase 1/2 (ULK1/2), play roles in the regulation or clearance of SGs. For example, VCP is known to regulate selective autophagy, which can facilitate the clearance of SGs that persist under certain stress conditions or fail to disassemble properly8,23. Additionally, ULK1/2, associated with autophagy, regulate SG disassembly through phosphorylation and activation of VCP/p9722. Moreover, SG homeostasis is influenced by tripartite motif-containing protein 21 (TRIM21)-mediated ubiquitination of Ras-GTPase-activating protein (GAP)-binding protein 1 (G3BP1) and autophagy-dependent SG elimination24. Among autophagy regulators of SGs, p62 stands out as a multifunctional protein. It aids in degrading ubiquitinated proteins via autophagy and participates in various signaling pathways such as nutrient sensing, oxidative stress, infection, immunity, and inflammation25. Acting as an autophagic receptor/adaptor, p62 interacts with ubiquitinated cargo via its ubiquitin association (UBA) domain and recruits them to the growing autophagosome membrane through its LC3-interacting (LIR) motif26–28. Recent research indicates that p62-droplets also facilitate autophagosome formation and counter oxidative stress29. Additionally, the C9ORF72-p62 complex aids in the autophagic clearance of SG via arginine methylation19.
Proteomic analyses have revealed a network of proteins associated with SGs, including mATG8 proteins, such as LC3B and GABARAP, key constituents of these granules9. These proteins interact with LIR motif-containing proteins, including p62, acting as adaptors/receptors, and recruit selective cargos or autophagic machinery proteins to autophagosomes during selective autophagy30,31. However, the specific mechanisms by which p62 and mATG8 proteins selectively recognize SGs and regulate autophagic SG clearance remain unclear. Identifying the components in SGs that interact with p62 and mATG8 proteins and how they recruit autophagic machinery for autophagosome formation under various stress conditions is essential. Recent findings indicate that various types of selective autophagy employ receptors/adaptors with LIR motifs to recognize and tether specific substrates to phagophores. Recently, LC3 subfamily- or GABARAP subfamily-selective LIR motifs have been identified, along with several proteins that bind to either the LC3 subfamily or GABARAP subfamily, aiming to characterize their function in selective autophagy32,33.
Therefore, this study aims to investigate NS1-BP, a protein characterized as containing an LIR motif that specifically interacts with GABARAP subfamily proteins; however, not with LC3 subfamily proteins. Co-immunoprecipitation (co-IP) and liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis revealed NS1-BP interaction with SG components, including Ataxin2, Poly(A)-binding protein (PABP), T-cell-restricted intracellular antigen-1 (TIA-1), and G3BP1. NS1-BP was observed to localize to GABARAP-containing SGs during oxidative stress. Furthermore, it strongly associates with p62, acting as an adaptor/receptor for selective autophagy in an oxidative stress-dependent manner. NS1-BP KO cells demonstrated increased SG size and decreased SG numbers, akin to observation in p62 KO cells or GABARAP triple KO (TKO) cells, suggesting impaired autophagic clearance. Additionally, NS1-BP KO cells showed increased p62 ubiquitination, leading to p62 autophagic degradation, while NS1-BP overexpression reduced p62 ubiquitination. Additionally, wild-type (WT) p62 expression, not p62 mutants (K420R or K435R), rescued p62 ubiquitination and the cellular phenotype in NS1-BP KO cells. NS1-BP KO cells showed reduced contact between ubiquitinated p62 bodies/aggregates and SGs. Neurons from derived ALS-induced pluripotent stem cells (iPSCs) showed reduced NS1-BP levels, resulting in SG dysregulation. These findings underscore the important role of NS1-BP in p62- and GABARAP-mediated SG regulation, shedding light on the molecular mechanisms of selective autophagic degradation of SGs and the cellular pathogenesis of ALS associated with SGs dysregulation.
Results
NS1-BP regulates the number, size, and dynamics of SGs
Identifying and studying proteins that bind to the LC3 and GABARAP subfamily are crucial for understanding selective autophagy, as many receptors and adaptors involved possess LIR motifs. To accomplish this, a search for proteins containing LIR motifs using the iLIR database was performed (A web resource for LIR motif-containing proteins in eukaryotes) (Supplementary Table 1)34. Several LIR motif-containing proteins were found to potentially bind to the GABARAP subfamily. Among these, NS1-BP, a Kelch family protein, was identified as having a potential LIR motif within its second Kelch motif (Supplementary Fig. 1a). NS1-BP is recognized for its interactions with the influenza virus non-structural protein 1 and regulates viral or host RNA processing/export35, cancer progression36, and regulation of neurite/dendritic spines37.
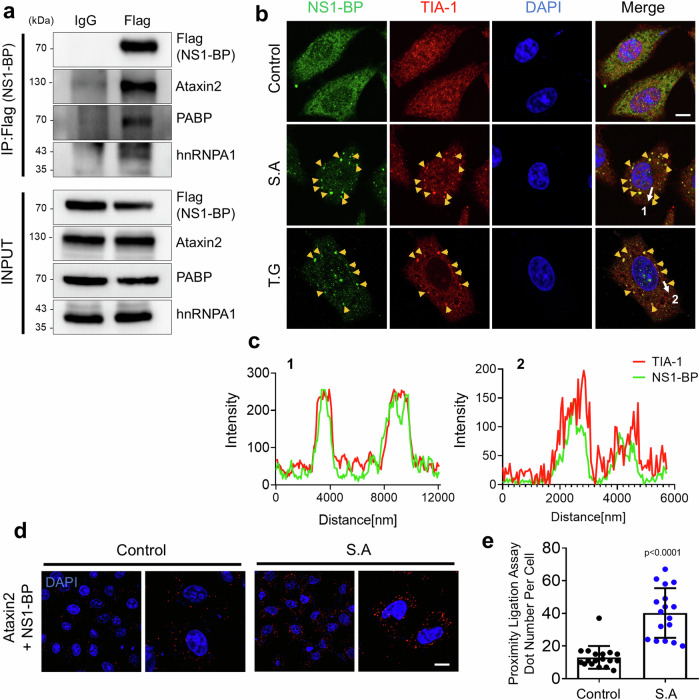
To investigate the binding properties of the LIR motifs from NS1-BP for each mATG8 protein, a previously developed in vitro assay was utilized33. Since NS1-BP functions as a dimer through its BTB/POZ domain38, duplicated LIR motif of NS1-BP (2xLIR[NS1-BP]) fused with mRFP-3x nuclear localization signal (2xLIR[NS1-BP]-mRFP-3xNLS), along with EGFP-tagged mATG8 mutants (EGFP-mATG8[GA]) in HeLa cells were co-expressed. In these mutants, the C-terminal glycine residue was replaced with alanine to impair phosphatidylethanolamine (PE) conjugation (lipidation), inhibiting cellular localization to autophagosomes. The assay revealed that when 2xLIR(NS1-BP)-mRFP-3xNLS interacted with specific EGFP-mATG8(GA), cytosolic EGFP-mATG8(GA) was sequestered into the nucleus based on its binding preference in live cells (Supplementary Fig. 1b, c). EGFP-GABARAP(GA), GABARAPL1(GA), and GABARAPL2(GA) were localized to the nucleus, indicating that the 2xLIR motifs from NS1-BP preferentially bind to GABARAP subfamily proteins rather than LC3 subfamily proteins. In contrast, the 2xLIR mutant (NS1-BP [W443A,V446A]) was diffusely localized to the nucleus and cytoplasm, suggesting a LIR-dependent binding pattern (Supplementary Fig. 1b, c). Further, GST affinity isolation assay using HeLa cell lysates supported the interaction between endogenous NS1-BP and GABARAP subfamily proteins (Supplementary Fig. 1d), suggesting its possible role in stress-induced responses such as autophagy. Furthermore, to confirm that the binding of NS1-BP to GABARAP subfamily proteins is indeed LIR motif-dependent, we have generated an NS1-BP mutant with the WIPV sequence mutated to AIPA (NS1-BP[W443A,V446A]) and performed GST-pulldown assays to assess its interaction with mATG8 proteins. Our results show that the NS1-BP(W443A, V446A) mutant has significantly reduced binding to mATG8s compared to the WT NS1-BP, indicating that the WIPV motif contributes to this interaction (Supplementary Fig. 1e, f). Based on the observation of the residual reduced binding to mATG8s, the LIR motif within NS1-BP may be functional. However, given that approximately 40% of the interaction between the NS1-BP(W443A,V446A) mutant and GST-GABARAP remains, there may be other binding mechanisms for LC3/GABARAP family proteins besides the LIR-LDS interaction. These mechanisms could involve different binding sites within NS1-BP or indirect binding through other mATG8 proteins within the cell.To elucidate NS1-BP functional roles in the stress response pathway associated with autophagy, co-IP and LC-MS/MS analyses were performed using the HEK293T cell lysates expressing FLAG-NS1-BP under oxidative stress induced by sodium arsenite. The silver stained SDS-PAGE analysis of elutes showed 35, 60, and 70 kDa in NS1-BP samples but not in control IgG samples (Supplementary Fig. 2). The co-IP and LC-MS/MS analysis results showed major binding partners, including heat shock protein A12B (HSPA12B) from the HSP70 family, mitochondrial heat shock protein 75 (MTHSP75) belonging to the HSP90 family, and heterogeneous nuclear ribonucleoprotein K (hnRNPK) and hnRNPA2B1 from the heterogeneous nuclear ribonucleoproteins family (Supplementary Fig. 2b, Supplementary Table 2 and Supplementary Data 1). Interestingly, the interaction between NS1-BP and hnRNPK has been previously reported even under untreated conditions39. To validate our findings, a co-immunoprecipitation (co-IP) assay was performed. The co-IP experiments confirmed that FLAG-NS1-BP interacts with MYC-hnRNPA2B1, MTHSP75, or HSPA12B in HEK293T cells under oxidative stress conditions (Supplementary Fig. 2c–e). These results are significant because all of the identified proteins are involved in the stress response pathway. Among them, hnRNPA2B1 or hnRNPK is known to localize to SGs, and mutations in these proteins have been related to neurodegenerative diseases including ALS and membrane scaffold protein (MSP)40 or Au–Kline syndrome with multiple malformation syndrome41. Furthermore, additional co-IP experiments using HEK293T cell lysates expressing FLAG-NS1-BP revealed interactions with core SGs components, including Ataxin2, PABP, and hnRNPA1, upon oxidative stress (Fig. 1a). NS1-BP predominantly colocalizes with TIA-1-containing SGs under oxidative stress or endoplasmic reticulum stress conditions (Fig. 1b, c). Proximity ligation assays (PLA) further demonstrated an increased association of NS1-BP with Ataxin2 under oxidative stress, suggesting its potential role in the stress response pathway associated with SGs regulation (Fig. 1d, e).
Fig. 1. Interaction of NS1-BP with stress granule (SG) components.
a Immunoprecipitation and western blot analysis using HEK293T cell lysates expressing Flag-NS1-BP with anti-Flag (or IgG), anti-Ataxin2, anti-PABP, and anti-hnRNPA1 antibodies. The experiment was repeated three times independently. b Representative images illustrating the cellular localization of NS1-BP (green) with TIA-1 (red) in HeLa cells under sodium arsenite (S.A) (0.5 mM, 1 h) or thapsigargin (T.G) (50 μM, 1 h) treatment conditions. Yellow and white arrows indicate each SGs and the region and direction for line scan, respectively. Scale bar, 10 μm. c Line scan graphs depicting the co-localization of TIA-1 (red) with NS1-BP (green). d Representative images displaying the results of the proximity ligation assay (PLA) for Ataxin2 with NS1-BP under sodium arsenite (0.5 mM, 1 h) condition. Scale bar, 10 μm. e Bar graph showing the proximity ligation assay dots. Data were quantified with a two-tailed unpaired Student’s t-test and presented as mean ± SEM; Control, n = 18; S.A, n = 17. Source data are provided as a Source Data file.
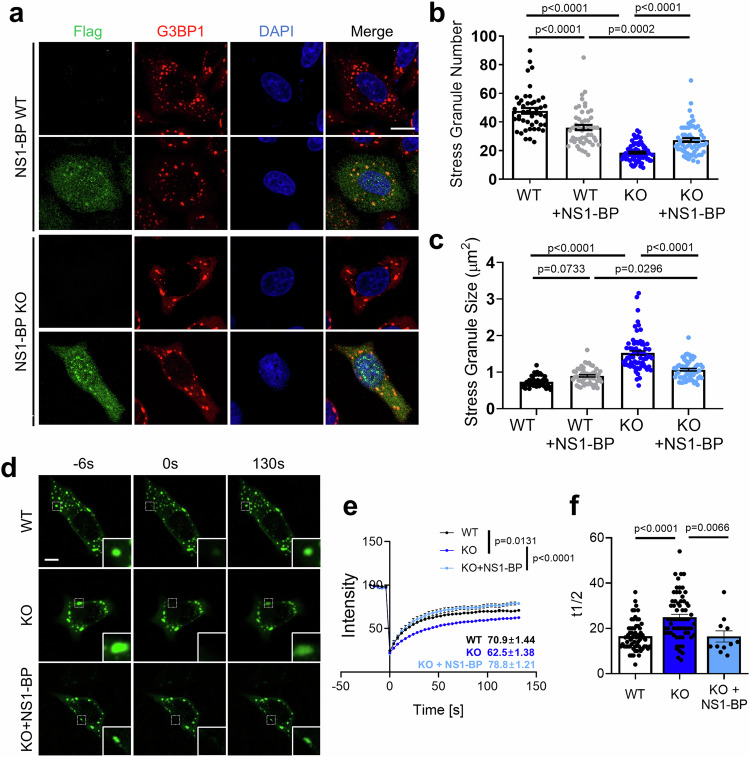
To investigate the physiological roles of NS1-BP in regulating SGs, NS1-BP KO HeLa cells were generated using clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 technology, confirmed by genomic polymerase chain reaction (PCR) (Supplementary Fig. 3b). Immunoblot and immunocytochemistry analyses confirmed the absence of NS1-BP protein in the NS1-BP KO cells than in WT cells (Supplementary Fig. 3d, e). Subsequently, SG number and size in WT and NS1-BP KO cells were assessed. A significant reduction in SGs number in NS1-BP KO cells compared to WT HeLa cells was found, accompanied by increased size under oxidative stress (Fig. 2a–c). Expression of FLAG-NS1-BP rescued the size increase in NS1-BP KO cells (Fig. 2a–c).
Fig. 2. Regulation of stress granule (SG) dynamics by NS1-BP.
a Representative images depicting G3BP1-positive SGs (red) in NS1-BP WT and KO HeLa cells expressing either Flag-vector or Flag-NS1-BP (green). Scale bar, 10 μm. b, c Bar graph illustrating the number(b) and size(c) of SGs. Each dots represent the average number or size of SGs per cell. Data were quantified by one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 44; WT + NS1-BP, n = 50; KO, n = 71; KO + NS1-BP, n = 61. d Representative images showing the GFP-G3BP1-containing SGs before and after photobleaching in NS1-BP WT, NS1-BP KO, or Flag-NS1-BP expressing NS1-BP KO HeLa cells. Scale bar, 5 μm. e The graph displaying normalized GFP-G3BP1 intensity after photobleaching. Data were quantified with a two-tailed unpaired Student’s t-test and presented as mean ± SEM; WT, n = 65; KO, n = 66; KO + NS1-BP, n = 11. f The graph showing the half-time of recovery (t1/2) of GFP-G3BP1 positive stress granules. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 65; KO, n = 66; KO + NS1-BP, n = 11. Source data are provided as a Source Data file.
Furthermore, fluorescence recovery after photobleaching (FRAP) analysis showed significantly reduced mobile fractions and increased recovery half-time of G3BP1-positive SGs in NS1-BP KO cells compared to WT cells, which was restored upon NS1-BP reintroduction (Fig. 2d–f). Approximately 60% of the fractions were recovered in NS1-BP WT cells, whereas only about 50% were recovered in NS1-BP KO cells. These data suggest a regulatory role of NS1-BP in SG dynamics, highlighting its importance in maintaining the proper mobility and recovery of SGs.
Overall, these results show that NS1-BP, characterized as a GABARAP subfamily-binding protein, localizes to SGs and modulates their size, number, or dynamics during oxidative stress.
NS1-BP is associated with p62 under oxidative stress conditions
To understand how NS1-BP regulates the size, number, or dynamics of SGs during oxidative stress, it is essential to consider SG disassembly mechanisms. Typically, after stress cessation, SGs disassemble through various kinds of chaperons, VCP, and RNA helicase42. However, under prolonged stress, SGs can also be selectively degraded by autophagy. Recent studies indicate that autophagy components, including mATG8s or p62, localize to SGs, with p62 playing a role in SG clearance19,43.
To investigate the interaction between NS1-BP and GABARAP subfamily proteins, their association in HEK293T cells expressing GFP-GABARAP subfamily proteins under oxidative stress conditions was examined. Supplementary Fig. 4a shows that the association between NS1-BP and GABARAP subfamily proteins was oxidative stress-dependent. Additionally, under oxidative stress conditions, NS1-BP predominantly co-localized with GABARAP subfamily-containing SGs, suggesting its potential role in SGs regulation (Supplementary Fig. 4b, c). In selective autophagy, mATG8s binds to p62, a selective autophagy adaptor, via the LIR motif to recruit ubiquitinated substrates into the autophagosomal membrane44. Therefore, NS1-BP was investigated to determine if it could associate with p62 under oxidative stress using HEK293T cell lysates expressing FLAG-NS1-BP or FLAG-p62, as well as non-expressing HEK293T cell lysates to assess their endogenous interaction. Figure 3a–c show that NS1-BP is strongly associated with p62 in an oxidative stress-dependent manner. NS1-BP only interacts with p62 under oxidative stress conditions and not under other stressors such as thapsigargin, sorbitol, or heat shock (Supplementary Fig. 5a).
Fig. 3. Interaction of NS1-BP with p62 via its kelch motif and UBA domain in p62 in an oxidative stress-dependent manner.
a Immunoprecipitation and western blot using HEK293T cell lysates expressing Flag-NS1-BP or Flag-p62, treated with sodium arsenite (0.5 mM, 1 h) with anti-Flag (or IgG), anti-p62, and anti-NS1-BP antibodies. The experiments were repeated five times independently. b Immunoprecipitation and western blot using HeLa cell lysates with anti-p62 (or IgG) antibodies. Cells were treated with sodium arsenite (0.5 mM, 1 h). c Bar graph showing NS1-BP normalization. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; Control, n = 5; S.A, n = 5. d Schematic model illustrating the constructs of 3xFlag-NS1-BP, 3xFlag-p62, and their deletion mutants (3xFlag-NS1-BP ΔKelch, 3xFlag-NS1-BP ΔBTB/POZ, 3xFlag-p62 ΔPB1, 3xFlag-p62 ΔUBA). e Immunoprecipitation and western blot using HEK293T cell lysates expressing Flag-NS1-BP WT, Flag-NS1-BP ΔKelch, or Flag-NS1-BP ΔBTB/POZ with anti-Flag (or IgG) and anti-p62 antibodies. f Bar graph showing the normalization of p62. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 6; ΔKelch, n = 6; ΔBTB/POZ, n = 6. g Immunoprecipitation and western blot using HEK293T cell lysates expressing Flag-p62 WT, Flag-p62 ΔUBA, or Flag-p62 ΔPB1 with anti-Flag (or IgG) and anti-NS1-BP antibodies. h Bar graph showing the normalization of NS1-BP. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 3; ΔUBA, n = 3; ΔPB1, n = 3. Source data are provided as a Source Data file.
To further investigate the cellular localization of NS1-BP with p62 under oxidative stress conditions, GFP-G3BP1-expressing HeLa cells were immunostained for NS1-BP and p62. Interestingly, NS1-BP interacts with p62 in the cytoplasm near GFP-G3BP1-positive SGs (Supplementary Fig. 6). These data suggest that p62 associates with SGs through its interaction with NS1-BP under oxidative stress conditions.
The domain involved in the association between NS1-BP and p62 during oxidative stress was identified. NS1-BP contains a BTB/POZ domain and Kelch motif, which forms a β-propeller structure38. Conversely, p62 contains multiple binding domains, including the N-terminal Phox-Bem1p (PB1) and C-terminal UBA domains25. PB1 is known to facilitate p62 oligomerization, while UBA is crucial for p62 multimerization and ubiquitination, essential for its function in sequestering ubiquitinated substrates. Therefore, co-IP experiments with anti-FLAG antibodies were performed using deletion mutants of NS1-BP or p62 (FLAG-NS1-BP, FLAG-NS1-BP ΔKelch, FLAG-NS1-BP ΔBTB/POZ, FLAG-p62, FLAG-p62 ΔUBA, or FLAG-p62 ΔPB1) (Fig. 3d). Figure 3e–h shows that the interaction between endogenous p62 and FLAG-NS1-BP ΔKelch or between endogenous NS1-BP and FLAG-p62 ΔUBA was significantly reduced, indicating that the Kelch domain in NS1-BP and UBA domain in p62 are necessary for their association. Additionally, the association between FLAG-p62 ΔPB1 and NS1-BP was slightly reduced, suggesting that PB1 oligomerization might partially contribute to their association.
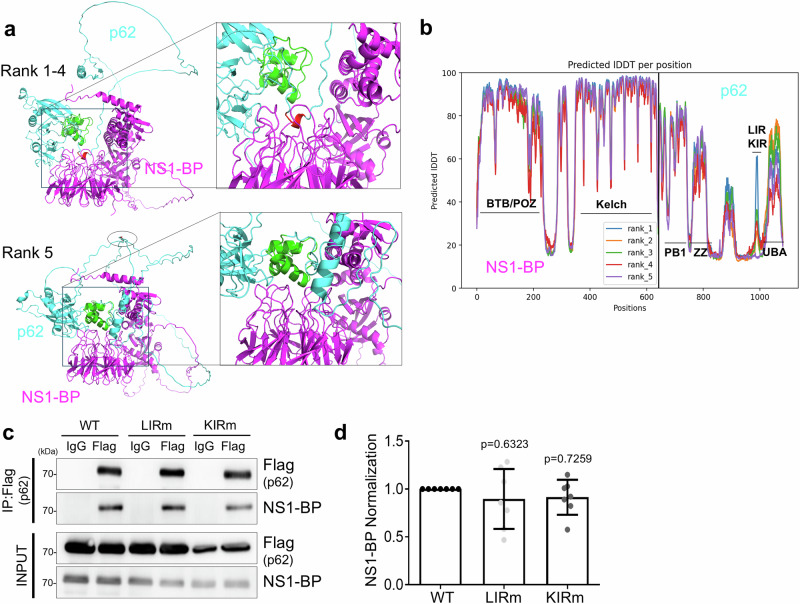
The binding of NS1-BP to p62 may not occur in their monomeric state under naïve conditions; however, to get the structural insight of the association of NS1-BP with p62, their binding regions were predicted using ColabFold modeling45. The prediction results showed two possibilities (Fig. 4a, b). The kelch motif in NS1-BP was predicted to directly interact with the Kelch-like ECH-associated protein 1 (Keap1)-interacting region (KIR) in p62 as shown in the Keap1-p62 structure46. Additionally, the UBA domain in p62 was also predicted to be in close proximity to the kelch motif in NS1-BP as a different model, suggesting a possible association of p62 with NS1-BP, probably facilitated by post-translational modification such as phosphorylation or ubiquitination.
Fig. 4. Interaction between p62 and NS1-BP mediated through its UBA domain and KIR motif.
a ColabFold prediction models of the complex depicting NS1-BP (magenta) and p62/SQSTM1 (cyan). Close-up view of the interacting boxed regions in both models is on the right. Rank 1-4 models are shown in the upper model. The NS1-BP interacts with the KIR of p62 (red) whereas the UBA domain (green) in p62 does not interact with NS1-BP in this complex model. Another predicted model of NS1-BP (magenta) and p62 (cyan) ranked fifth in ColabFold prediction (lower in this panel). The UBA domain (green) in p62/SQSTM1 involved in the interaction with NS1-BP whereas the KIR motif (black oval) locate at far from the NS1-BP. b Predicted local distance difference test (pLDDT) of the ColabFold prediction top5 ranked models. The predicted model of NS1-BP is more accurate than that of p62. c Immunoprecipitation and western blot using HEK293T cell lysates expressing Flag-p62 WT, Flag-p62 LIR mutant, or Flag-p62 KIR mutant, treated with S.A (0.5 mM, 1 h) with anti-Flag (or IgG) and anti-NS1-BP antibodies. d Bar graph showing the normalization of NS1-BP. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 7; LIRm, n = 7; KIRm, n = 7. Source data are provided as a Source Data file.
Based on the predicted association sites identified through the ColabFold modeling, it was clarified whether the KIR motif in p62 could interact with NS1-BP upon oxidative stress condition. However, the co-IP results using a FLAG-p62 KIR mutant showed that the interaction between the KIR mutant and NS1-BP was not significantly affected (Fig. 4c, d). This suggests that the kelch domain of NS1-BP strongly associates with the UBA domain in p62 under oxidative stress rather than the Keap1-KIR type interaction, suggesting that NS1-BP may modulate p62 function via the UBA domain in p62 under oxidative stress.
NS1-BP negatively regulates p62 ubiquitination
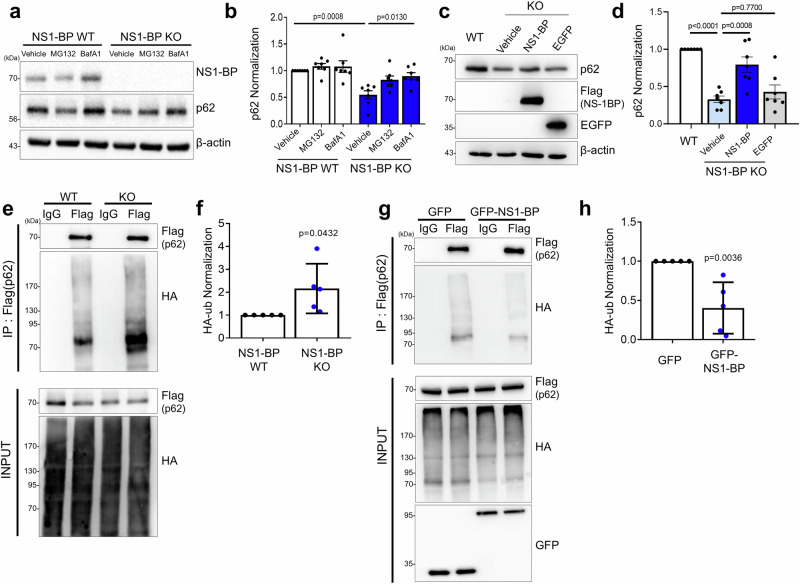
Subsequently, the role of NS1-BP on the function of p62 during oxidative stress was queried. Accumulating evidence showed that during oxidative stress, p62 interacts with Keap1-Cul3, activating nuclear factor erythroid 2-related factor 2 (Nrf2), which in turn induces the expression of antioxidant-response genes47,48. Lysine 420 within the UBA domain of p62 is responsible for its ubiquitination by Keap1/Cul3, and ubiquitination of the UBA domain increases p62 degradation by facilitating its sequestration into autophagic vacuoles49. Additionally, the report shows that NS1-BP (called KLHL39) inhibits KLHL20-mediated substrate ubiquitination by Cul3 and substrate degradation36. Therefore, it was hypothesized that NS1-BP might regulate p62 ubiquitination by Keap1/Cul3 and its degradation. To test this hypothesis, the protein level of p62 was initially assessed in WT and NS1-BP KO cells. Figure 5a–d shows that the protein level of p62 was significantly reduced in NS1-BP KO cells, which was rescued by overexpression of NS1-BP and inhibiting autophagy with bafilomycin A1, suggesting that NS1-BP negatively regulates the autophagic degradation of p62. Subsequently, the effect of NS1-BP on the ubiquitination of p62 was examined. NS1-BP KO cells showed increased p62 ubiquitination than the WT cells, while its overexpression reduced p62 ubiquitination, further supporting the hypothesis that NS1-BP inhibits p62 ubiquitination and degradation during oxidative stress (Fig. 5e–h and Supplementary Fig. 8a).
Fig. 5. Negative regulation of p62 ubiquitination by NS1-BP.
a NS1-BP WT and KO HeLa cells were treated with MG132 (10 μM, 6 h) or BafilomycinA1 (100 nM, 6 h) and assayed for western blot using anti-NS1-BP, anti-p62, and anti-β-actin antibodies. b The bar graph represents the normalization of p62 protein levels. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT vehicle, n = 7; WT MG132, n = 7; WT BafA1, n = 7; KO vehicle, n = 7; KO MG132, n = 7; KO BafA1, n = 7. c Immunoblotting showing the p62 protein levels in NS1-BP WT and Flag-NS1-BP or EGFP lentivirus infected NS1-BP KO HeLa cells. d Bar graph showing p62 Normalization. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 7; KO vehicle, n = 7; KO NS1-BP, n = 7; KO EGFP, n = 7. e Ubiquitination immunoprecipitation and western blot using NS1-BP WT and KO HeLa cell lysates expressing 3xFlag-p62 and HA-ub, treated with sodium arsenite (0.5 mM, 1 h) with anti-Flag (or IgG) and anti-HA antibodies. f Bar graph showing the normalization of HA-ub. Data were quantified with a two-tailed unpaired Student’s t-test and presented as mean ± SEM; NS1-BP WT, n = 5; NS1-BP KO, n = 5. g Ubiquitination immunoprecipitation and western blot using HEK293T cell lysates expressing 3xFlag-p62, HA-ub, and GFP or GFP-NS1-BP, treated with sodium arsenite (0.5 mM, 1 h) with anti-Flag (or IgG) and anti-HA antibodies. h Bar graph showing the normalization of HA-ub. Data were quantified with a two-tailed unpaired Student’s t-test and presented as mean ± SEM; GFP, n = 5; GFP-NS1-BP, n = 5. Source data are provided as a Source Data file.
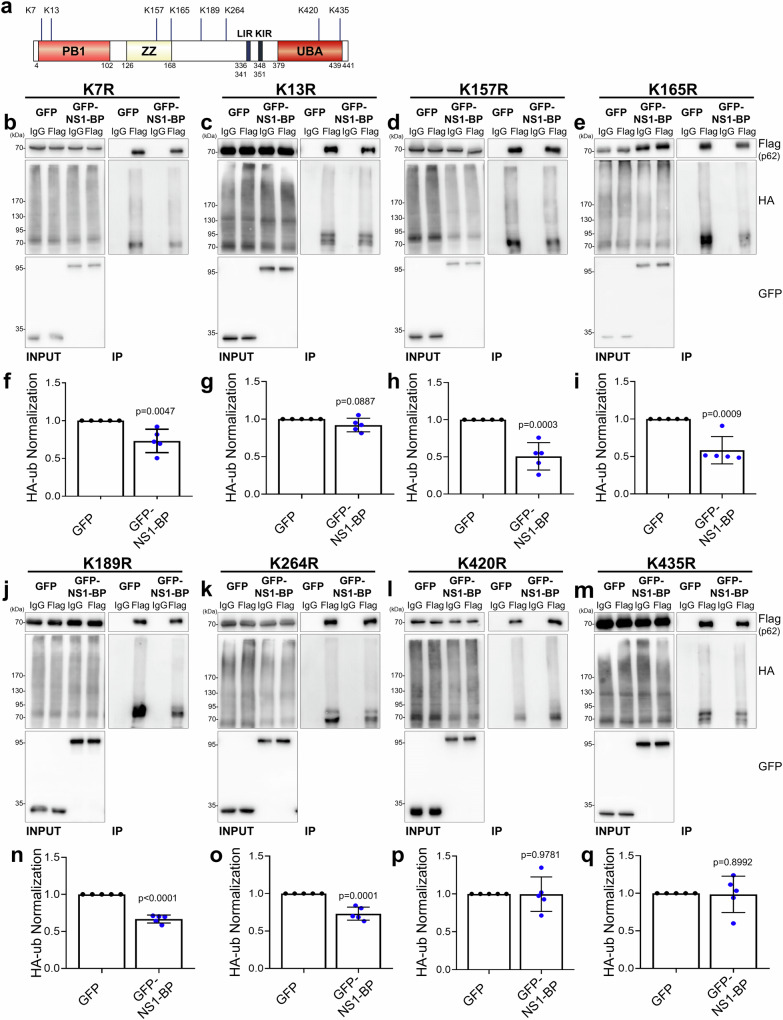
To pinpoint the critical residues within p62 responsible for NS1-BP-mediated inhibition of p62 ubiquitination among the lysine residues essential for p62 ubiquitination, site-specific mutations were introduced by substituting lysine with arginine at lysine 7, 13, 157, 165, 189, 264, 420, and 435 in p6244,49,50. Ubiquitination IP using HA-Ubiquitin was performed in FLAG-p62 WT, K7R, K13R, K157R, K165R, K189R, K264R, K420R, or K435R expressing HEK293T cells. Figure 6 shows that while the expression of p62 lysine mutants K7R, K157R, K165R, K189R, or K264R significantly reduced p62 ubiquitination, similar to what was observed with p62 WT, the K13R mutant (within the PB1 domain) and K420R or K435R mutants (within the UBA domain) did not show a reduction in p62 ubiquitination as observed with p62 WT. Subsequently, the effect of these lysine mutations on the association of NS1-BP with p62 was investigated. Co-immunoprecipitation revealed that compared to p62 WT, the p62 lysine mutant K420R exhibited reduced association with NS1-BP, underscoring the importance of lysine 420 in the UBA domain for this interaction. However, the association of NS1-BP with other p62 lysine mutants did not differ significantly from that with p62 WT (Supplementary Fig. 5b, c). Collectively, these findings suggest that NS1-BP inhibits overall ubiquitination of p62 and autophagic degradation, and lysine residues 13, 420, and 435 in p62 may be crucial for its inhibitory effect on overall p62 ubiquitination under oxidative stress conditions.
Fig. 6. Regulation of p62 ubiquitination by NS1-BP on Lysine 13, Lysine 420, and Lysine 435.
a Schematic model illustrating p62 ubiquitination sites. b, c, d, e, j, k, l, m Ubiquitination immunoprecipitation (Ub-IP) and western blot using HEK293T cells lysates expressing EGFP or EGFP-NS1-BP and Flag-p62 K7R(b), Flag-p62 K13R(c), Flag-p62 K157R(d), Flag-p62 K165R(e), Flag-p62 K189R(j), Flag-p62 K264R(k), Flag-p62 K420R(l), or Flag-p62 K435R(m) treated with sodium arsenite (0.5 mM, 1 h) with anti-Flag (or IgG), anti-GFP, and anti-HA antibodies. f, g, h, i, n, o, p, q. Graphs showing the normalization of HA-ub. Data were quantified with a two-tailed unpaired Student’s t-test and presented as mean ± SEM; GFP, n = 5; GFP-NS1-BP, n = 5. Source data are provided as a Source Data file.
Reduced levels of p62 in NS1-BP KO cells impair SG clearance
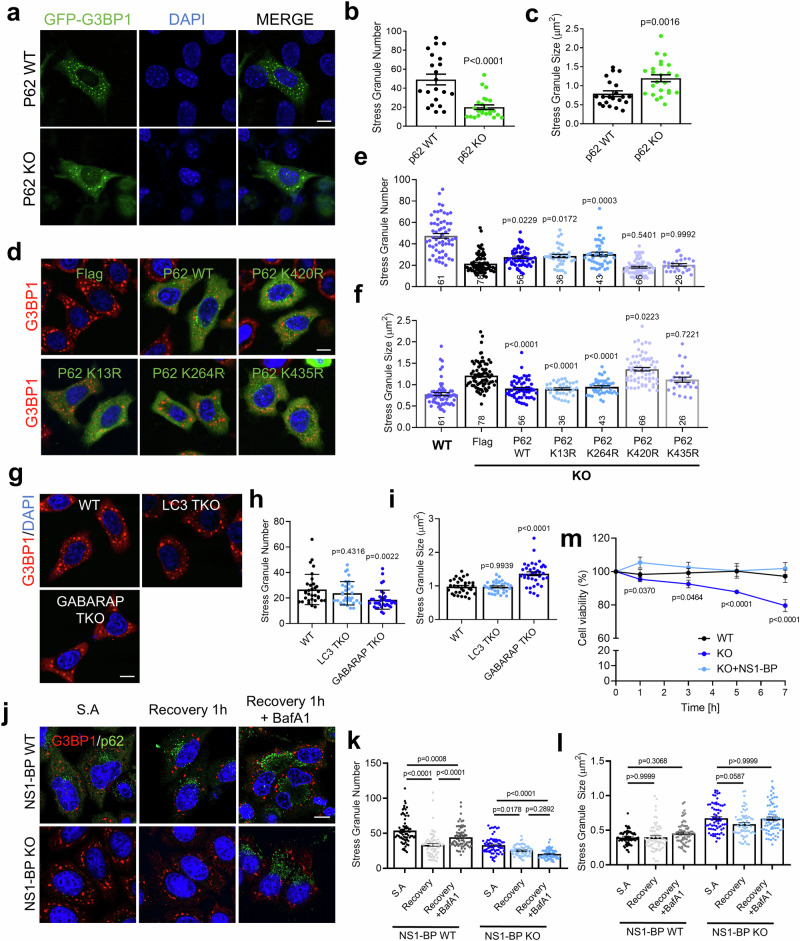
To further examine whether the reduced protein level of p62 in NS1-BP KO cells is responsible for the dysregulation of SGs, the number and size of SGs in p62 KO mouse embryonic fibroblasts (MEFs) were assessed. Figure 7a–c shows the observed increase in SGs size and a decrease in their number in p62 KO MEFs, resembling the cellular phenotype observed in NS1-BP KO cells. To ascertain whether the decreased level of p62 contributes to the altered dynamics of SGs in NS1-BP KO cells, p62 WT or p62 lysine mutants were reintroduced into NS1-BP KO cells. The expression of p62 WT, K13R, or K264R in NS1-BP KO cells significantly restored the number and size of SGs, while p62 K420R or K435R did not show any change (Fig. 7d–f). These results show that p62 K420 or K435 play key roles in ubiquitination and subsequent degradation of p62 in NS1-BP KO cells, thereby contributing to SGs dysregulation.
Fig. 7. Regulation of stress granule (SG) dynamics by p62 and GABARAP family proteins.
a Representative images showing the cellular localization of EGFP-G3BP1 in p62 WT and KO MEFs. Cells were transfected with EGFP-G3PB1 and treated with sodium arsenite (0.5 mM, 1 h). Scale bar, 10 μm. b, c Graphs showing the number(b) and size(c) of SGs per cell. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; p62 WT, n = 21; p62 KO, n = 24. d Representative images showing G3BP1 positive SGs in NS1-BP KO HeLa cells expressing Flag vector, Flag-p62 WT, Flag-p62 K13R, Flag-p62 K264R, Flag-p62 K420R, or Flag-p62 K435R treated with sodium arsenite (0.5 mM, 1 h). Scale bar, 10 μm. e, f Bar graphs showing the number(e) and size(f) SGs per cell. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 61; KO, n = 78; KO+p62 WT, n = 56; KO+p62 K13R, n = 36; KO+p62 K264R, n = 43; KO+p62 K420R, n = 66; KO+p62 K435R, n = 26. g Representative images show G3BP1-positive SGs in LC3 TKO and GABARAP TKO HeLa cells under sodium arsenite (0.5 mM, 1 h) treatment condition. Scale bar, 10 μm. h, i Bar graphs showing the number(h) and size(i) of SGs per cell. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; WT, n = 32; LC3 TKO, n = 32; GABARAP TKO, n = 37. j Representative images show G3BP1-positive SGs in NS1-BP WT and KO HeLa cells upon sodium arsenite treatment and recovery 1 h with or without BafA1 (10 nM, 24 h). Scale bar, 10 μm. k, l Bar graphs showing the number(k) and size(l) of SGs per cell. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; WT S.A, n = 70; WT recovery, n = 67; WT BafA1, n = 70; KO S.A, n = 69; KO recovery, n = 66; KO BafA1, n = 70. m Cell viability of NS1-BP WT, NS1-BP KO, and NS1-BP overexpressed KO cells was assessed after 1, 3, 5, 7 h of sodium arsenite treatment (0.5 mM). The viability was compared with that of untreated cells. Data were quantified using one-way ANOVA in conjunction with the Tukey post hoc test and presented as mean ± SEM; WT, n = 5; KO, n = 5; KO + NS1-BP, n = 5. Source data are provided as a Source Data file.
Given that NS1-BP interacts not only with p62 but also with GABARAP family proteins, it was further investigated whether NS1-BP modulates GABARAP-mediated autophagic SGs degradation. Therefore, the number and size of SGs were examined in GABARAP TKO HeLa or HEK293T cells expressing a selective deconjugase for GABARAP/L1/L2-PE33. Figure 7g–i and Supplementary Fig. 7a–d show that GABARAP/L1/L2-deficient cells exhibited an increase in SG size and a decrease in the number of SGs compared to WT or other LC3A/B-PE deconjugase expressing cells, resembling the phenotype observed in NS1-BP KO cells. Additionally, HeLa cells expressing siRNA targeting ATG16L1 showed a similar increase in SG size and decrease in SG number, akin to observations in NS1-BP KO cells, p62 KO MEFs, and GABARAP TKO HeLa cells (Supplementary Fig. 7e–h). More intriguingly, ubiquitinated G3BP1-positive SGs were significantly decreased in NS1-BP KO cells compared to WT cells and this reduction was rescued by reintroduction of p62 (Supplementary Fig 8b, c), suggesting that stabilized p62 promotes SGs clearance. Moreover, NS1-BP KO cells showed a significant reduction in the localization of GABARAPL1 into G3BP1-positive SGs (Supplementary Fig. 8d, e), suggesting that NS1-BP is crucial for the proper recruitment of GABARAPL1 to SGs, thereby facilitating their autophagic clearance. This suggests that autophagic degradation of SGs may be impaired in NS1-BP KO cells due to reduced p62 levels, potentially caused by increased p62 ubiquitination leading to its degradation.
To confirm whether SG degradation is indeed impaired in NS1-BP KO cells, autophagic degradation of SGs was assessed using the autophagy blocker bafilomycin A1 (BafA1). In NS1-BP WT cells, treatment with BafA1 led to a sustained number of SGs after 1 hour of recovery, suggesting that autophagic degradation may play a role in SG clearance during this early phase (Fig. 7j–l). However, given the dynamic nature of SG disassembly, it is possible that an extended recovery period may reveal additional disassembly of SGs even in the presence of BafA1. This observation suggests that further investigation at later time points could provide a clearer understanding of SG dynamics under these conditions.
Next, to determine whether the accumulation of SGs observed in NS1-BP KO cells plays a beneficial or detrimental role, a cell viability assay was performed following oxidative stress treatment. Interestingly, the results showed that cell viability was significantly reduced in NS1-BP KO cells treated with sodium arsenite compared to WT cells (Fig. 7m). However, the introduction of NS1-BP into NS1-BP KO cells restored cell viability, suggesting that the presence of NS1-BP is crucial for maintaining cell survival under oxidative stress conditions.
Dysregulation of SG dynamics in ALS patient motor neurons
The dysregulation of SGs is increasingly recognized as being associated with various neurodegenerative diseases. In the case of ALS, sporadic and familial cases often involve motor neurons experiencing oxidative stress and exhibiting abnormal accumulation of protein aggregates, including TDP-43. Therefore, it was initially investigated whether deficiency of NS1-BP also causes SG dysregulation in post-mitotic neurons. Supplementary Fig. 9 shows that cultured cortical neurons expressing NS1-BP shRNA demonstrated increased size and decreased number of SGs. These findings suggest a potential involvement of NS1-BP in the cellular processes related to SG dysregulation in ALS, highlighting a possible connection to ALS pathogenesis.
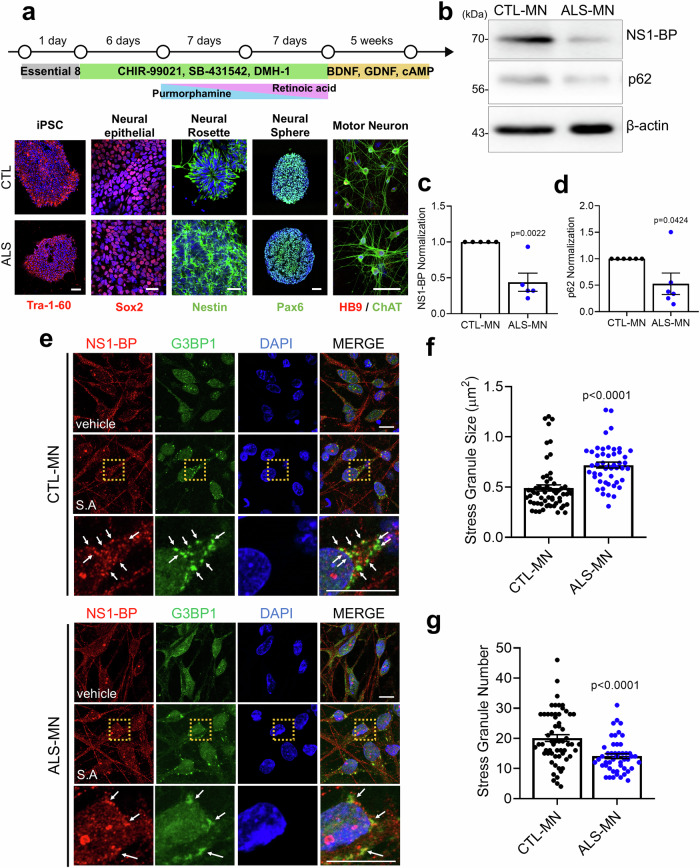
To investigate the protein level of NS1-BP, p62 and its relationship with SGs dysregulation in ALS, patients with ALS-derived motor neurons generated from induced pluripotent stem cells (iPSCs) were examined (Fig. 8a). The findings revealed a significant reduction in NS1-BP and p62 protein levels in patients with ALS derived motor neurons compared to control motor neurons (Fig. 8b–d). Furthermore, an increase in SG size and a decrease in SG number was observed, indicating SG dysregulation in patients with ALS-derived motor neurons (Fig. 8e–g). These results were consistent with the results obtained from NS1-BP KO cells and NS1-BP deficient post-mitotic cortical neurons. Overall, this study offers preliminary insights into the cellular mechanisms that may contribute to SG dysregulation, with potential implications in diseases like ALS and ALS-FTD.
Fig. 8. Dysregulation of stress granules (SGs) due to reduced NS1-BP levels in ALS-patient iPSC-derived motor neurons.
a Schematic model illustrating iPSC differentiation to motor neurons. b Analysis of NS1-BP and p62 protein levels through western blot using anti-NS1-BP, anti-p62, anti-beta-actin in CTL-MN and ALS-MN. c Bar graph indicating the normalization of NS1-BP protein levels. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; CTL-MN, n = 5; ALS-MN, n = 5. d Bar graph indicating the normalization of p62 protein levels. Data werequantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; CTL-MN, n = 6; ALS-MN, n = 6. e Representative images showing NS1-BP (red) and G3BP1-positive SGs (green) in CTL-MN and ALS-MN upon sodium arsenite (0.5 mM, 90 min) treatment or not. Scale bar, 10 μm. f, g Bar graphs showing the SG size(f) and number(g). White arrows indicate NS1-BP and G3BP1-positive SGs. Data were quantified using a two-tailed unpaired Student’s t-test and presented as mean ± SEM; CTL-MN, n = 60; ALS-MN, n = 48. Source data are provided as a Source Data file.
Discussion
SGs are dynamic cytoplasmic structures that form in response to various stressors such as oxidative stress, heat shock, or viral infection, temporarily sequestering mRNAs and RNA-binding proteins to halt translation4. They play a critical role in cellular stress responses and are implicated in the pathogenesis of neurodegenerative diseases, aging, and cancer18,51–53. Recent studies have highlighted the link between SG dynamics and autophagy8,24,43; however, the precise mechanisms, particularly those involving post-translational modifications, remain incompletely understood.
This study identifies NS1-BP as a key regulator of SG dynamics by interacting with autophagy components, GABARAP and p62, under oxidative stress, influencing p62 ubiquitination and facilitating GABARAP localization to SGs for autophagic clearance. Reduced NS1-BP levels in ALS patient-derived neurons, coupled with SG dysregulation, suggest a role in ALS pathogenesis, highlighting the link between impaired autophagy and neurodegenerative disease progression.
NS1-BP plays a critical role in regulating SG dynamics through its interactions with p62 and GABARAP, influencing both the assembly and disassembly of SGs. Our findings reveal that NS1-BP deficiency leads to increased SG size and decreased SG numbers, closely resembling the effects observed when p62 or GABARAP levels are altered. The observed increase in SG size and reduction in SG number in NS1-BP knockout (KO) cells indicate a disruption in SG dynamics and impaired clearance. This failure to efficiently disassemble or degrade SGs can lead to prolonged sequestration of key cellular components, disrupting cellular stress responses and reducing cell viability. The accumulation of larger SGs suggests a bottleneck in the degradation process, preventing the proper recycling of proteins and RNAs necessary for cellular recovery, ultimately contributing to cellular dysfunction and increased susceptibility to stress. This dysfunction may contribute to cellular dysfunction and has been linked to neurodegenerative disease like ALS.
The interplay between SG formation, autophagy, and cellular stress response is complex and involves intricate regulatory mechanisms. Although our study provides evidence of NS1-BP’s involvement in these processes, the precise mechanisms by which NS1-BP regulates SG dynamics remain not fully understood. Beyond its interactions with p62 and GABARAP, NS1-BP might influence SGs through associations with other SG-related proteins or cellular signaling pathways. Future studies exploring the broader interactome of NS1-BP, especially its connections with other autophagy and SG components, will be crucial in uncovering additional roles and regulatory mechanisms, shedding light on its contributions to cellular homeostasis and disease pathogenesis.
Ubiquitination is essential in regulating the dynamics and function of SGs under stress, with several SG proteins like G3BP1, PABP, and FUS undergoing ubiquitination, which influences SG stability and their roles in stress response21,24,54,55. The autophagic clearance of SGs, particularly granulophagy, depends on the ubiquitination of SG components21, including p62, which serves as a selective cargo adaptor for autophagic degradation. Specific lysine residues in p62, such as K7 and K420, are critical for its recognition and subsequent sequestration into autophagosomes44,49. Ubiquitination of p62 itself also targets it for autophagic degradation, contributing to SG clearance and maintaining cellular homeostasis. However, the precise regulatory mechanisms governing p62 ubiquitintion remain incompletely understood.
Our study identifies a role of NS1-BP in negatively regulating p62 ubiquitination, which is crucial for autophagic degradation. By modulating p62 ubiquitination, NS1-BP affects the protein stability and availability of p62 for autophagic processes and potentially for the ubiquitin-proteasome system (UPS). This modulation impacts the levels of p62, a key player in autophagy and selective cargo degradation, which in turn influences SG dynamics. K420 in p62 plays a crucial role in its interaction with NS1-BP and is essential for the regulation of p62 ubiquitination. This lysine residue, located within the UBA domain of p62, is not only pivotal for binding to NS1-BP but also significantly influences the ubiquitination process, impacting p62’s stability and its function in autophagic degradation. K13 is located in the PB1 domain, while K420 and K435 are within the UBA domain of p62, indicating spatial separation. This suggests that NS1-BP may alter p62’s overall structure or accessibility, indirectly affecting these residues’ susceptibility to ubiquitination. ColabFold modeling predicts a strong interaction between NS1-BP’s kelch motif and p62’s UBA domain, potentially inducing a conformational change that impacts ubiquitination at distant sites like K13. Previous studies have shown that such interactions can influence the ubiquitination landscape across structurally distant lysine residues46,49. The significant phenotypic effects observed with single mutations (K13R, K420R, or K435R) suggest that these lysine residues individually play pivotal roles in maintaining p62’s ubiquitination pattern, with lysine 420 being particularly essential for interaction with NS1-BP. Given the modular nature of p62 domains, disruptions at these key lysines can lead to substantial alterations in p62’s regulatory function and its role in SG dynamics.
p62 comprises several domains, including a PB1 domain for oligomerization, the zinc finger (ZZ) domain for N-degron recognition56, TRAF6-binding domain, a LIR motif for interaction with autophagy-related proteins, the KIR, and a C-terminal UBA domain essential for binding polyubiquitin chains25. NS1-BP, a Kelch family protein, features a BTB/POZ domain that facilitates dimerization and Kelch repeats forming a β-propeller structure38. The β-propeller of Kelch repeats in the Keap1 is known to recognize the KIR segments in the p6246 and similar interactions were predicted for the NS1-BP and p62 complex in ColabFold models (Fig. 4b).
Interestingly, NS1-BP’s interaction with p62 was restricted to oxidative stress condition, coinciding with p62’s oligomeric state57. This suggests an alternative model where the UBA domain of p62 contributes to its interaction with NS1-BP, supported by IP analysis showing the kelch motifs of NS1-BP and the UBA domain of p62 as crucial for this binding (Fig. 3e-h). These findings imply that NS1-BP may alter p62 behavior specifically under oxidative stress. Given that small molecule inhibitors targeting the Kelch domain of Keap1 have been developed to address oxidative stress-related conditions58, detailed structural analysis of NS1-BP interactions with p62 and GABARAP could reveal critical molecular interfaces and post-translational modifications. This knowledge could pave the way for targeted therapeutic strategies involving NS1-BP’s modulation of autophagic processes.
GABARAP proteins play a key role in SG dynamics and autophagy, interacting with SG components such as NUFIP2 and G3BP1. NS1-BP enhances the recruitment of GABARAP to SGs, facilitating their incorporation into autophagosomes and subsequent degradation. In NS1-BP KO cells, reduced localization of GABARAP to SGs highlights NS1-BP’s role in promoting autophagic SG clearance, crucial for cellular stress recovery. Additionally, p62, a key autophagy adaptor, also interacts with SG components and plays a significant role in their regulation, further supporting the concept that NS1-BP’s interaction with GABARAP and p62 is essential for the efficient clearance of SGs.
These findings suggest that NS1-BP modulates the recruitment of autophagic cargo, such as damaged organelles or protein aggregates, by influencing GABARAP localization and function. This process is vital during cellular stress recovery, where efficient SG clearance is necessary to maintain cellular homeostasis. By regulating SG size, number, and dynamics, NS1-BP helps coordinate the autophagic degradation of SGs, linking autophagy to stress granule regulation.
Our results indicate that BafA1 treatment inhibits the decrease in SG numbers during the early phase of recovery, suggesting a role for autophagic degradation in SG clearance. However, we recognize the limitation of the 1 hour recovery time point in fully capturing SG disassembly dynamics. SGs typically disassemble over a 1-2 hour period after stress removal, and thus, additional studies examining SG persistence at extended recovery intervals (e.g., 2 h) would be beneficial to determine whether a higher proportion of SGs would disassemble even in the presence of BafA1. This would provide a more comprehensive view of autophagy’s role in SG turnover, aligning our conclusions with the complex temporal nature of SG degradation. Furthermore, we cannot entirely exclude the possibility that altered fusion/fission dynamics may also contribute to the observed effects. Future studies will be needed to specifically investigate SG fusion and fission processes in NS1-BP KO cells to fully delineate their contribution.
This study identifies NS1-BP as a critical regulator of stress granule (SG) dynamics through its interactions with key autophagy components, GABARAP and p62, under oxidative stress conditions. NS1-BP modulates p62 ubiquitination and stability, enhances GABARAP recruitment to SGs, and facilitates their autophagic degradation. In ALS patient-derived motor neurons, reduced NS1-BP levels correlate with altered SG dynamics, suggesting its involvement in ALS pathogenesis by influencing autophagic processes. The findings highlight the complex interplay between SG regulation and autophagy, revealing NS1-BP’s role in maintaining cellular homeostasis under stress. Future research into the detailed mechanisms of NS1-BP’s interactions with autophagy-related proteins could uncover potential therapeutic strategies for neurodegenerative diseases associated with SG dysregulation and autophagy impairment.
Methods
Cell lines and growth conditions
HEK293T(CRL-3216) and HeLa(CCL-2) cells were obtained from American Type Culture Collection(ATCC). To generate NS1-BP KO HeLa cells, cluster regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing was utilized to disrupt the genes encoding human NS1-BP in HeLa cells. The CRISPR/Cas9 system, consisting of CMV-Cas9-EF1a-puromycin/GFP-U6 optimized for cell transfection, was sourced from YSY Biotech Company Ltd (Nanjing, China). To develop the double-nicking NS1-BP-sgRNA-guided CRISPR/Cas9 plasmids, a pair of oligos were designed and subcloned into AflII-digested gRNA_Cloning Vector (Addgene 41824). These plasmids were co-transfected with pCas9-GFP (Addgene 44719) into HeLa cells using lipofectamine 2000 (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA). HeLa cells transfected with pCas9-GFP/NS1-BP two gRNAs were selected with puromycin, and single colonies were isolated. To generate cells stably expressing either 3xFlag-p62 WT or K420R, NS1-BP KO HeLa cells were infected with lentiviral particles and grown in a medium containing 10 μg/ml polybrene (Sigma Aldrich H9268) for 48 h. Stably expressing cells were selected with puromycin, and single colonies were isolated.
All cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin in a humidified atmosphere containing 5% (v/v) of CO2 at 37 °C.
DNA constructs
NS1-BP was PCR-amplified from a human cDNA library and inserted into the pEGFP-C1 vector using BglII and BamHI restriction enzymes or into the 3xFlag-CMV7.1 vector through EcoRI and BamHI restriction enzymes. GST-LC3A, GST-LC3B, GST-LC3C, GST-GABARAP, GST-GABARAPL1, and GST-GABARAPL2 were obtained from Addgene (Cambridge, USA). Additionally, p62 was PCR-amplified from an HA-p62 construct from Addgene and inserted into the 3xFlag-CMV7.1 vector through EcoRI and KpnI restriction enzymes.
To generate domain deletion or mutant constructs, p62 (ΔPB1), p62 (ΔUBA), p62 (KIR mutant), p62 (LIR mutant), p62 K7R, p62 K13R, p62 K157R, p62 K165R, p62 K189R, p62 K264R, p62 K420R, p62 K435R, p62 K420Q, p62 K435Q, p62 K420/435Q, NS1-BP (ΔKelch), and NS1-BP (ΔBTB/POZ) were PCR-amplified with specific primers (Supplementary Table 3) and inserted into the 3xFlag-CMV7.1 vector through EcoRI or BglII (for 5’ region) and KpnI or BamHI for (3’ region) restriction enzymes. Additionally, p62 K13R, p62 K165R, and p62 K435R were amplified by PCR with specific primers and inserted into the 3xFlag-CMV7.1 vector using the infusion cloning method (Takara Bio 639648). To further generate p62 lentiviral constructs, 3xFlag-p62 WT and 3xFlag-p62 K420R were amplified by PCR using specific primers and inserted into the pLVX-EF1a-IRES-Puro (Takara Bio 631988).
Lentivirus production
To produce lentiviruses for infection, Lenti-X 293 T (Takara Bio 632180) cells were co-transfected with pLVX-EF1a-3xFlag-NS1-BP, p62 WT or K420R, psPAX2, and pMD2.G using the CalPhos Mammalian Transfection Kit (Takara Bio 631312). Culture supernatant was collected at 48 h and 72 h post-transfection and passed through a 0.45-μm filter. The viral particles were concentrated by ultracentrifugation (22,600 x g for 3 h) and resuspended in Dulbecco’s phosphate-buffered saline (DPBS).
Transfection, gene silencing, and drug treatment
To transiently express DNA constructs, cells were transfected with plasmid DNA using either the calcium phosphate method or FuGENE HD (Promega E2311) following the instruction of the manufacturer. Knockdown experiments in mouse cortical neurons and HeLa cells were performed using Lipofectamine 2000 (Thermo Fisher Scientific 11668-019) and the shRNA construct or using Lipofectamine RNAiMax (Thermo Fisher Scientific 13778-075) and the siRNA following the instructions of the manufacturer. To induce SG formation, cells were treated with sodium arsenite (S.A 0.5 mM, 1 h), thapsigargin (T.G 20 uM, 1 h), sorbitol (0.4 M, 2 h) or heat shock (42 °C, 2 h).
Immunoblot analysis
Protein samples were separated through SDS-PAGE, transferred to PVDF membranes (Millipore IPVH00010) and incubated with primary antibodies overnight at 4 °C. After washing with TBST (150 mM NaCl, 20 mM Tris-HCl (pH 7.4), 0.05% Tween 20), membranes were exposed to secondary antibodies conjugated with horseradish peroxidase for 1 h. Signals were detected with ECL solution (Millipore, WBKLS0500, USA). Antibodies used included NS1-BP (Santa Cruz sc-373909, 1:100), Flag (Sigma-Aldrich F1804, 1:10,000), MTHSP75 (Proteintech 14887-1-AP), HSPA12B (Novus Biologicals NBP1-88293), p62 (Abnova H00008878-M01, 1:10,000), GABARAPL1 (Cell Signaling 26632, 1:1,000), G3BP1 (Proteintech 13057-2-AP, 1:1,000), Ataxin2 (Proteintech 21776-1-AP, 1:5,000), hnRNPA1(Abcam ab137780, 1:2000), PABP (Abcam ab21060, 1:1,000), HA (Cell Signaling 3724, 1:1000), GFP (Neuromab 75-131, 1:10,000), and Beta-actin (Sigma A5441, 1:10,000). Quantification was performed using ImageJ (NIH) software.
Immunocytochemistry and confocal microscopy
For immunostaining, transfected cells were washed with PBS, then fixed and permeabilized with methanol (Samchun) for 30 min at -20 °C. After blocking with 3% bovine serum albumin for 1 h at room temperature, cells were incubated overnight at 4 °C with TIA-1 (Santa Cruz sc-1751), G3BP1 (Proteintech 13057-2-AP), Flag (Sigma-Aldrich F1804), FK2 (Sigma-Aldrich ST1200), and GABARAPL1 (Genetex GTX132664). Subsequently, they were treated with anti-mouse or anti-rabbit secondary antibodies (Jackson Laboratory) for 2 h at room temperature. Following three washes with 1X PBS, the cells were mounted on glass slides and analysed using an LSM 880 microscope (Carl Zeiss, Germany).
Silver staining and LC-MS/MS analysis
For the silver staining assay, 3xFlag-NS1-BP expressing HEK293T cells were performed IP using Flag antibody according to the co-immunoprecipiration instructions in methods. Silver staining was performed using PageSilver™ Silver staining Kit (Thermo Fisher Scientific K0681) following the instruction of the manufacturer. LC-MS/MS analysis was performed at the Yonsei Proteomic Research Center (Seoul, Korea).
In-gel tryptic digestion
1DE band of interest for analysis were excised from the preparative gel, and the spots were transferred into each 1.5 mL tube. The band was washed with 100 μL of distilled water; then, 100 μL of 100 mM Na2S2O3 and 30 mM K3Fe(CN)6 (1:1) were added to the band and shook for in 10 min. This process was repeated at least three times until the Silver Stain dye disappeared. The supernatant was decanted, and the band was dried in speed vacuum concentrator (LaBoGeneAps, Lynge, Denmark) for 10 min. Reduction of the sample was performed in 10 mM dithiothreitol/25 mM NH4HCO3 (pH 8.0) for over 30 min at 56 °C. The solution was decanted and the band was stored at room temperature in 55 mM iodoacetamide in the dark for 20 min. The solution was decanted and then digested with Pierce trypsin protease, MS grade (Thermo fisher Scientific) (enzyme to substrate ratio = 1:50) at 37 °C with shaking for 16 h. The digested peptide mixture was desalted using ziptip with POROS R2 resin.
LC-MS/MS for peptides analysis_Q-TOF
Nano LC–MS/MS analysis was performed with a nano HPLC system (Agilent, Wilmington, DE). The nano chip column (Agilent, Wilmington, DE, 150 mm × 0.075 mm) was used for peptide separation. The mobile phase A for LC separation was 0.1% formic acid in deionized water and the mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was designed for a linear increase from 3% B to 45% B in 30 min, 45% B to 95% B in 1 min, 95% B in 4 min, and 3% B in 10 min. The flow rate was maintained at 300 nL/min. Product ion spectra were collected in the information-dependent acquisition (IDA) mode and were analyzed by Agilent 6530 Accurate-Mass Q-TOF using continuous cycles of one full scan TOF MS from 300-2000 m/z (1.0 s) plus three product ion scans from 150-2000 m/z (1.5 s each). Precursor m/z values were selected starting with the most intense ion, using a selection quadrupole resolution of 3 Da. The rolling collision energy feature was used, which determines collision energy based on the precursor value and charge state. The dynamic exclusion time for precursor ion m/z values was 60 s.
Database searching
The mascot algorithm (Matrixscience, USA) was used to identify peptide sequences against NCBInr human protein sequence database (2018; 319,713 entries). Database search criteria were, taxonomy; Homo sapiens, fixed modification; carbamidomethylated at cysteine residues, variable modification; oxidized at methionine residues, maximum allowed missed cleavage; 2, MS tolerance; 100 ppm, MS/MS tolerance; 0.1 Da. Only peptides resulting from trypsin digests were considered. The peptides were filtered with a significance threshold of P < 0.05.
GST affinity isolation assay
For the GST affinity isolation assay, HeLa cells or NS1-BP KO HeLa cells were washed with PBS, harvested, and lysed in GST affinity isolation buffer solution (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, supplemented with protease inhibitor cocktail). After centrifugation at 13,000 x g for 20 min, the cleared cell lysates were incubated overnight with purified GST-mATG8 proteins and glutathione-conjugated agarose beads (Sigma Aldrich G4510) at 4 °C. The following day, the samples were washed three to five times with the same GST affinity isolation buffer solution at 4 °C, and the remaining supernatant was removed. The samples were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled immediately, and subjected to SDS-PAGE with Coomassie Brilliant Blue (Sigma Aldrich 1154440025) staining.
Co-Immunoprecipitation
Cells were collected and lysed with 1% NP40 lysis buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% NP40), supplemented with protease inhibitor (Sigma P8340) and phosphatase inhibitor (Thermo Fisher Scientific 78426) on ice for 30 min. After centrifugation at 13,000 rpm at 4 °C for 15 min, the supernatant fractions were collected and incubated with protein A/G-agarose beads (Santa Cruz sc-2002) for 2 h at 4 °C. This step served as a pre-clearing step, with the supernatants containing 1 mg of protein incubated overnight at 4 °C with appropriate antibodies. To precipitate the target protein, the supernatants were incubated with protein A/G agarose bead for 7 h at 4 °C. Afterwards, the beads were washed with lysis buffer three times by centrifugation at 3,000 rpm at 4 °C. The immunoprecipitated proteins were separated via SDS-PAGE, and western blotting was performed as previously described.
Proximity ligation assay (PLA)
The in situ PLA was performed using a Duolink in situ kit (Sigma DUO92101) following the instruction of the manufacturer. Initially, cells were cultured on Matrigel-coated glass. After sodium arsenite treatment, cells were fixed and permeabilized using 100% methanol and then blocked with a blocking solution. Subsequently, cells were incubated with primary antibodies (NS1-BP, Santa Cruz sc-373909, 1:50; Ataxin-2, Proteintech 21776-1-AP, 1:100; p62, Abnova H00008878-M01, 1:100; Flag, Sigma-Aldrich F1804, 1:100), diluted in antibody diluent overnight at 4 °C. Following this, secondary antibodies (PLA probe anti-mouse MINUS and PLA probe anti-rabbit PLUS) were applied for 1 h at 37 °C. Cells were treated with ligase for 30 min at 37 °C and amplified with polymerase diluted in amplification buffer for 100 min at 37 °C.
In vivo p62 ubiquitination IP
Cells were transfected with either 3xFlag-p62 WT or ubiquitination mutant (K7R, K13R, K157R, K165R, K189R, K264R, K420R, or K435R) along with HA-ub constructs, and either EGFP or EGFP-NS1-BP. After 24 h, cells were treated with MG132 (1 uM, 16 h) to inhibit proteasome. Cells were lysed with 1% SDS denaturing buffer (1% SDS, 50 mM Tris-HCl pH7.6, 150 mM NaCl) and supplemented with protease inhibitors. Lysates were boiled for 10 min and disrupted with gentle sonication. Additionally, these samples were diluted 1:20 with 1% Triton X-100 lysis buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100). Subsequent steps were consistent with the co-IP method described above.
Cell viability assay
NS1-BP WT and KO HeLa cells were plated into 24 well plates and cultured in DMEM medium supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin for 2 days. The medium was then replated with fresh medium and 0.5 mM sodium arsenite was treated for 0, 1, 3, 5, and 7 h, respectively. After treatment, MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega) labeling reagent was added to each well, and the plates were incubated at 37 °C for an additional 30 min. Following MTS incubation, the spectrophotometric absorbance of the samples was measured at 490 nm.
FRAP analysis
HeLa cells expressing EGFP-G3BP1 were initially activated by imaging every 4 s for 2.5 min using a 100% 488-nm laser power and 100-ms exposure time to SGs. Subsequently, the EGFP-G3BP1-positive SGs were photobleached, and the EGFP signal intensity was measured before and after photobleaching.
Structure prediction
The predicted complex model between NS1-BP and p62/SQSTM1 was obtained by using ColabFold45. Five ranked models were obtained for the NS1-BP and p62 complex, and the binding modes of these complexes were classified into two categories. PyMOL software was utilized to visualize the models (https://pymol.org/).
iPSC generation from control and patients with ALS-derived primary fibroblast
Control and ALS iPSC lines were reprogrammed from fibroblasts (Control fibroblast, ND29178, age 66, Male; ALS fibroblast, ND41003, age 62, Female) obtained from the Coriell Institute for Medical Research (NINDS collection) using a non-integrating induction method with OSKM factors. The fibroblasts were cultured at 37 °C in Dulbecco’s modified Eagle’s medium (Corning) supplemented with 15% FBS (Corning) and 1% penicillin-streptomycin (Gibco).
For gene delivery, electroporation (Invitrogen™ Neon™ Transfection System) was performed. Seven days after electroporation, the cells were transferred onto MEF feeder cells with iPSC maintenance medium containing 20% KO serum (Gibco 10828028), β-mercaptoethanol (Gibco 21985023), Gluta-MAX (Gibco 35050-061), MEM-Non Essential Amino Acids solution (Gibco 11140050), penicillin/streptomycin (Hyclone SV30010), and DMEM-F12 (Gibco 11320033). After 4 weeks, iPSC-like colonies were picked and cultured on new feeder cells. Furthermore, at an iPSC passage number of 5–7, the culture system was transitioned to a feeder-free system using a truncated recombinant human vitronectin (Gibco A14700) coated plate with Essential 8 Medium (Gibco A1517001).
Motor neuron differentiation
To generate highly pure motor neurons, a previous method was slightly modified59. To induce differentiation into neuronal precursor cells (NPC), iPSCs were dissociated using Accutase (Innovative Cell Technologies AT104) and seeded at a 1:5 ratio on Matrigel-coated plates. On the following day, the iPSC medium was replaced with a chemically defined neural medium (NEP media), including KO DMEM-F12 (Gibco 12660-012) and Neurobasal medium (Gibco 21103-049) at 1:1, 0.5x N2 (Gibco 17502-048), 0.5x B-27 (Gibco 17504-044), 1x GlutaMax supplements (Gibco 35050-061), 1x penicillin–streptomycin (Hyclone SV30010), 100 µM Ascorbic acid (Sigma A4403), 3 µM CHIR99021 (Stem Cell 72054), 2 µM DMH-1 (Stem Cell 73634), and 2 µM SB4315429 (Stem cell 72234). The culture medium was altered every other day. To induce motor neuron patterning (MNP), NPC cells were dissociated with Accutase and seeded at a 1:3 on Matrigel-coated plates with NEP medium supplemented with 0.1 µM Retionic acid (Sigma R2625) and 0.5 µM Puromorphamine (Sigma 540220) 7 days after seeding. The culture medium was altered every other day. For motor neuron differentiation, MNP cells were dissociated with Accutase and cultured in suspension in the NEP medium supplemented with 0.5 µM Retionic acid and 0.1 µM Puromorphamine 7 days after MNP induction. The culture medium was altered every other day. To mature the motor neuron, MND cells were dissociated into single cells with Accutase and plated on mouse glial cells in a motor neuron mature medium. The mature medium include Neurobasal medium, 1x B-27 supplement, 1x GlutaMax, 1x penicillin-streptomycin (Gibco), 100 µM Ascorbic acid (Sigma), 2.6 µg/ml Adenosine 3’,5’-cyclic monophoaphate (Sigma A9501), 0.5 µM Compound E (Stem cell 73954), 10 µg/ml Laminin (Sigma L2020), 10 µg/ml BDNF (Thermo Fisher Scientific 450-02-50UG), and 10 µg/ml GDNF (Thermo Fisher Scientific PHC7041). Medium refreshment with motor neuron mature medium occurred weekly for a total maturation period of 5 weeks.
Statistics and reproducibility
Statistical analyses were performed using a two-tailed unpaired Student’s t-test or one-way ANOVA in conjunction with the Tukey post hoc test using GraphPad Prism version 8.0 software. All data were presented as mean ± SEM. A p-value of <0.05 was considered statistically significant. All experiments were repeated independently at least three times. Cell images were selected and analyzed randomly. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The work was supported by the Science Research Center Program of the National Research Foundation NRF (Grant No. 2020R1A5A1019023, J.-A L.); Neurological Disorder Research Program of the NRF (Grant No. 2020M3E5D9079911, J.-A L.); Basic research program of the NRF (Grant No. 2023R1A2C2007082, J.-A L.); the NRF grant funded by the Korea government (MSIT) (Grant No. RS-2023-00218515, D.-J J.). This work was also supported by JSPS KAKENHI (Grant No. JP19H05706; JP21H004771; 23K20044; 24H00060, M.K.); AMED (Grant No. JP22gm1410004h0003, M.K.) and by the Takeda Science Foundation to M.K.
Author contributions
P.J. conducted the majority of the experiments, analyzed the data, and contributed to manuscript drafting. H-J.H., H.C., S.P., J-W.J., and S-W.P. conducted and assisted with IP experiments. D-H.C. contributed to the interpretation of results and edited the manuscript. H.-J.L. analyzed protein interactions using ColabFold modeling. H.K.S. designed the experiment and wrote the manuscript. K.M. contributed to the interpretation of results and edited the manuscript. D.H. contributed to the interpretation of LC-MS/MS results and edited the manuscript. D-J.J. & J-A.L. conceptualized the study, designed the experiments, and wrote the manuscript. All authors reviewed the manuscript, made necessary corrections, and approved the final version for publication. Consent for publication was obtained from each author.
Peer review
Peer review information
Nature Communications thanks Nobuo Noda, Udai Pandey and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. Source data are provided as a Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pureum Jeon, Hyun-Ji Ham, Haneul Choi.
Change history
2/11/2025
A Correction to this paper has been published: 10.1038/s41467-025-56909-4
Contributor Information
Deok-Jin Jang, Email: jangdj@knu.ac.kr.
Jin-A Lee, Email: leeja@hnu.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55446-w.
References
- 1.Glick, D., Barth, S. & Macleod, K. F. Autophagy: cellular and molecular mechanisms. J Pathol.221, 3–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murrow, L. & Debnath, J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol.8, 105–137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan, J. R., Yoon, J. H. & Parker, R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci.124, 228–239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, P. & Kedersha, N. Stress granules. Curr Biol19, R397–R398 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Kedersha, N. et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol212, 845–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders, D. W. et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell181, 306–324 e328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler J. R., Matheny T., Jain S., Abrisch R., Parker R. Distinct stages in stress granule assembly and disassembly. Elife5, (2016). [DOI] [PMC free article] [PubMed]
- 8.Buchan, J. R., Kolaitis, R. M., Taylor, J. P. & Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell153, 1461–1474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markmiller, S. et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell172, 590–604.e513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isabelle, M., Gagne, J. P., Gallouzi, I. E. & Poirier, G. G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci.125, 4555–4566 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Amen, T. & Kaganovich, D. Stress granules inhibit fatty acid oxidation by modulating mitochondrial permeability. Cell Rep35, 109237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedersha, N., Ivanov, P. & Anderson, P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci.38, 494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portz, B., Lee, B. L. & Shorter, J. FUS and TDP-43 phases in health and disease. Trends Biochem Sci.46, 550–563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramaswami, M., Taylor, J. P. & Parker, R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell154, 727–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J Cell Biol201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron, D. M. et al. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener8, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon, P. & Lee, J. A. Dr. Jekyll and Mr. Hyde? Physiology and pathology of neuronal stress granules. Front Cell Dev Biol.9, 609698 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitiprolu, M. et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun.9, 2794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu, H. H. et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging35, 2822–2831 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Gwon, Y. et al. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science (New York, NY)372, eabf6548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, B. et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell74, 742–757.e748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seguin, S. J. et al. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ21, 1838–1851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C., et al. Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules. Autophagy, 1-18 (2023). [DOI] [PMC free article] [PubMed]
- 25.Berkamp, S., Mostafavi, S. & Sachse, C. Structure and function of p62/SQSTM1 in the emerging framework of phase separation. FEBS J288, 6927–6941 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Bjorkoy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol171, 603–614 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu, M. et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kageyama, S. et al. p62/SQSTM1-droplet serves as a platform for autophagosome formation and anti-oxidative stress response. Nat Commun.12, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippai, M. & Low, P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int.2014, 832704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, W. J. et al. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol Biol Lett.21, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, S. W. et al. Monitoring LC3- or GABARAP-positive autophagic membranes using modified RavZ-based probes. Sci Rep.9, 16593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S. W. et al. Development of new tools to study membrane-anchored mammalian Atg8 proteins. Autophagy, 12010.1080/15548627.2022.2132040 (2022). [DOI] [PMC free article] [PubMed]
- 34.Jacomin, A.-C., Samavedam, S., Promponas, V. & Nezis, I. P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy12, 1945–1953 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff, T., O’Neill, R. E. & Palese, P. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol72, 7170–7180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, H. Y. et al. KLHL39 suppresses colon cancer metastasis by blocking KLHL20-mediated PML and DAPK ubiquitination. Oncogene34, 5141–5151 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Fujii, R. & Takumi, T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci118, 5755–5765 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Sasagawa, K. et al. Identification of Nd1, a novel murine kelch family protein, involved in stabilization of actin filaments. J Biol Chem277, 44140–44146 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Tsai, P. L. et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog9, e1003460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, H. J. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Au, P. Y. B. et al. Phenotypic spectrum of Au-Kline syndrome: a report of six new cases and review of the literature. Eur J Hum Genet26, 1272–1281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Protter, D. S. W. & Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, Y. et al. HDAC6, a novel cargo for autophagic clearance of stress granules, mediates the repression of the type i interferon response during coxsackievirus a16 infection. Front Microbiol11, 78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng, H. et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res.27, 657–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat Methods19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komatsu, M. et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology12, 213–223 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Jain, A. et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem285, 22576–22591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichimura, Y. et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell51, 618–631 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Lee, Y. et al. Keap1/cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep.19, 188–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng, L. et al. Sirt1 deacetylates and stabilizes p62 to promote hepato-carcinogenesis. Cell Death Dis.12, 405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao, B. et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathog.17, e1008690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayabalan, A. K. et al. Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc. Natl Acad. Sci. USA118, e2021719118 (2021). [DOI] [PMC free article] [PubMed]
- 53.Bentmann, E., Haass, C. & Dormann, D. Stress granules in neurodegeneration-lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. Febs j280, 4348–4370 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Yang, Y. et al. TRIM25-mediated ubiquitination of G3BP1 regulates the proliferation and migration of human neuroblastoma cells. Biochimica et biophysica acta Gene regulatory mechanisms1866, 194954 (2023). [DOI] [PubMed] [Google Scholar]
- 55.Maxwell, B. A. et al. Ubiquitination is essential for recovery of cellular activities after heat shock. Science (New York, NY)372, eabc3593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwon, D. H. et al. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat Commun.9, 3291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carroll, B. et al. Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat Commun9, 256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narayanan, D. et al. Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery. Journal of medicinal chemistry65, 14481–14526 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Du, Z. W. et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun6, 6626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. Source data are provided as a Source Data file. Source data are provided with this paper.