Abstract
Legionella pneumophila uses a type IV secretion system to deliver effector molecules into the host cell and establish its replication vacuole. In this study, we investigated the role of LidA, a translocated substrate associated with the surface of the L. pneumophila-containing vacuole. LidA is secreted into the host cell throughout the replication cycle of the bacteria and associates with compartments of the early secretory pathway. When overexpressed in mammalian cells or yeast, LidA interferes with the early secretory pathway, probably via a domain predicted to be rich in coiled-coil structure. Finally, during intracellular replication, the replication vacuoles are in close contact with the endoplasmic reticulum-Golgi intermediate compartment and the Golgi apparatus, suggesting a positive correlation between intracellular growth and association of the vacuole with compartments of the early secretory pathway. We propose that LidA is involved in the recruitment of early secretory vesicles to the L. pneumophila-containing vacuole and that the vacuole associates with the secretory pathway to facilitate this process.
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular gram-negative pathogen (10). In environmental reservoirs, the bacterium is found associated with freshwater amoebae, while human disease is caused by inhalation of contaminated aerosols, followed by multiplication of the bacteria within alveolar macrophages (26, 44).
Upon uptake into macrophages, the bacterium is internalized into a compartment that escapes the endocytic pathway and establishes a replication vacuole (16, 17). Fifteen minutes after internalization, the vacuolar membrane lacks plasma membrane or endocytic markers and is surrounded by mitochondria and small vesicles (15). Based on the thickness of their membranes (40) and on the presence of calnexin around the nascent L. pneumophila vacuole (8), these vesicles are thought to be derived from the endoplasmic reticulum (ER). In addition, the L. pneumophila vacuole intercepts vesicular traffic from ER exit sites and vesicle budding from the ER (18), as well as proteins of the early secretory pathway (8, 19), which appear to be required for establishment of the replication vacuole. Six hours after internalization, the bacterium starts to multiply in an ER-like compartment (38, 40). As replication proceeds, >50% of the L. pneumophila vacuoles acquire markers of endocytic organelles (37). At the end of the replication cycle, the vacuoles contain dozens of bacteria, leading to host cell lysis and infection of the neighboring cells.
A type IV secretion system, known as the Dot/Icm complex, is essential for intracellular growth and for translocation of bacterial effector molecules into the host cell (5, 34, 43). These effectors proteins are thought to promote uptake and to allow the establishment of the replication vacuole. Several of them have been identified in the last 2 years, but their exact functions remain mostly unknown. Chen et al. have recently reported that translocation of LepB and LepC, two proteins with weak homology to SNARE proteins, are important for the specific dissemination of L. pneumophila from amoebae, perhaps by routing the vacuole containing bacteria into the protozoan exocytic pathway (4). Luo et al. have identified nine families of Sid (for substrate of Icm/Dot) proteins that are transferred between bacterial cells by the Dot/Icm system (23). Some of the Sid proteins are translocated to the surface of the L. pneumophila vacuole in infected cells, and some families of Sid proteins contain up to five paralogs. The functional redundancy among the Dot/Icm substrates may explain why many of the sid mutants remain competent for intracellular replication.
Two other Dot/Icm substrates with no paralogs in the L. pneumophila genome have been identified. RalF, a guanidine nucleotide exchange factor, activates Arf family members on the L. pneumophila vacuole (24), and LidA, a protein of unknown function, is required for high-efficiency formation of replication vacuoles within mouse macrophages (6). After infection of macrophages with a lidA mutant that expresses limiting amounts of DotA protein, a fraction of the vacuoles fail to escape fusion with late endocytic compartments. Bacteria in vacuoles that avoid the endocytic pathway, however, grow with wild-type (WT) kinetics, producing, overall, a marginal defect in intracellular growth. The LidA protein has no apparent transmembrane domain, but it contains a central region that is rich in predicted coiled-coil structures. Within 20 min after internalization of the bacteria, LidA is found on the surfaces of the L. pneumophila-containing vacuoles in a Dot/Icm-dependent fashion, with approximately 50% of the vacuoles displaying polar localization of the protein (6). At later times of infection, LidA is exposed uniformly on the membranes of vacuoles in which the bacteria were replicating, as well as dispersed throughout the infected cells.
Although it is well accepted that there must be a connection between L. pneumophila Dot/Icm substrates and the early secretory pathway, very little information is available. In this study, we have investigated the role of LidA, focusing on the localization of the protein within the host cell and its effect on cellular morphology and physiology. We report that LidA is secreted into the host cell throughout the replication cycle of the bacteria, where it associates with membrane compartments. In addition, LidA interacts with and interferes with compartments of the early secretory pathway and therefore appears to be involved in the recruitment of ER-derived vesicles to the vacuole containing bacteria. Moreover, our results suggest that the mature replication vacuole is integrated into the early secretory pathway to facilitate this process.
MATERIALS AND METHODS
Bacterial strains, mammalian cells, yeast, and media.
L. pneumophila Lp02 (thyA hsdR rpsL) and Lp03 (Lp02 dotA) strains are derivatives of Philadelphia-1 strain Lp01 (hsdR rspL) (2). L. pneumophila Lp02-GFP and Lp03-GFP have been described previously (36). The lidA mutant that expresses limiting amounts of DotA protein has been described previously (6). L. pneumophila strains were maintained on charcoal yeast extract solid medium and ACES yeast extract broth culture media (9, 11, 38). U937 cells and COS-1 (African green monkey kidney cells transformed by an origin-defective mutant of simian virus 40) cells were grown as described previously (1, 2). Saccharomyces cerevisiae cells (W303 [ade2 can1 leu2 his3 trp1 ura3 ho::LYS2]) were maintained on YPD or SC medium (35). DNA transformation into yeast was performed as described previously (29).
Detergent fractionation of L. pneumophila.
Sixty microliters of postexponential L. pneumophila expressing green fluorescent protein (GFP) was washed once and resuspended in 5 ml of phosphate-buffered saline (PBS) containing either 1% Triton X-100 (Bio-Rad), 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate; Sigma}, 1% octylglucoside (Calbiochem), or 1% purified digitonin (Calbiochem) and incubated for 20 min at room temperature with intermittent vortexing. The cell lysates were centrifuged for 15 min at 10,000 rpm, and the top 3 ml of the supernatant was collected. The extracted proteins present in the supernatant were precipitated with methanol-chloroform (46). The detergent-soluble and -insoluble materials were suspended in polyacrylamide gel electrophoresis (PAGE) sample buffer containing 5.5 M urea and analyzed by sodium dodecyl sulfate (SDS)-PAGE and immunoblotting.
Antibodies.
The following antibodies (dilution and origin) were used in this study. Anti-LidA (1:10,000 [6]), anti-GFP (1:2,000; Molecular Probes), anti-calnexin (1:10,000; BD Bioscience), anti-RhoGDI (1:500; Santa Cruz Biotechnology), anti-ERGIC53 (1:1,000; gift of H. P. Hauri, Biozentrum, Basel, Switzerland), anti-GM130 (1:100; BD Transduction Laboratories), anti-p115 (1:100; gift of G. Watters, Merck, Inc.), anti-TGN46 (1:100; Serotec), goat anti-rabbit horseradish peroxidase conjugate (1:10,000; Zymed), and donkey anti-rabbit or -mouse Texas Red or fluorescein isothiocyanate conjugate (1:500; Jackson Immunoresearch).
Digitonin and mechanical fractionation of U937 cells infected with L. pneumophila.
Differentiated U937 cells (107) were incubated with postexponential L. pneumophila (multiplicity of infection, 5) for 1, 6, or 12 h in 10-cm2 dishes. The digitonin fractionation assay was performed as previously described (22). Briefly, adherent infected cells were resuspended in 5 ml of PBS containing 1% purified digitonin and fractionated by incubation for 20 min at room temperature with intermittent vortexing. The cell lysates were processed as described above for the fractionation of L. pneumophila except that the digitonin-insoluble material was further solubilized in 1% SDS. For mechanical fractionation, the infected cells were resuspended in PBS and homogenized using a Dounce homogenizer until 90% of the cells were broken as assessed visually. After centrifugation (1 h; 35,000 rpm; 4°C), the cytosolic fraction was collected, and the membrane fraction was subjected to digitonin solubilization. The samples were then processed as described above.
Plasmid construction.
Vectors allowing expression of GFP-LidA fusion derivatives in mammalian cells were constructed by insertion of an EcoRI/BamHI PCR product corresponding to the lidA gene (full open reading frame or truncated versions) into the EcoRI and BamHI restriction sites of pEGFP-C1 (Clontech).
The vector used to integrate the lidA gene under the control of a galactose-inducible promoter at the URA3 locus of Saccharomyces cerevisiae was constructed by insertion of an EcoRI/BamHI PCR product corresponding to the lidA open reading frame into the EcoRI and BamHI restriction sites of pDK20 (a gift from A. Murray, Harvard University, Cambridge, MA). The plasmid was digested with StuI prior to transformation and integrated at the URA3 locus. The transformants were selected on SC medium containing 2% glucose and lacking uracil.
Transfection and immunofluorescence.
COS1 cells (3 × 104) were seeded onto glass coverslips in 24-well dishes and were transfected the next day with 1 μg of plasmid DNA using the calcium phosphate transfection method (Invitrogen). The cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 for 5 min, and stained with the appropriate antibody. The coverslips were mounted using Fluoguard Antifade reagent (Bio-Rad).
Quantification of the expression level of GFP-LidA.
Based on visual inspection, pictures of cells expressing the GFP-LidA fusion protein at a low or high level were captured using a Hammatsu Orca 100 cooled charge-coupled device camera. The saved images were analyzed with IPLab Spectrum on a Macintosh computer, and the total pixel intensity in the region of interest corresponding to the perinuclear area in which LidA accumulated was measured. A low or a high level of expression was represented by the mean of the values of the total pixel intensity obtained from 10 different transfected cells.
L. pneumophila infection and infectious-center assay.
Postexponential-phase L. pneumophila strains were introduced onto monolayers of 105 COS1 cells in 24-well dishes (multiplicity of infection, 5) by centrifugation for 5 min at 1,000 rpm. After incubation for 2 h at 37°C in the presence of 5% CO2, the monolayers were washed to synchronize the infection and further incubated for 12 h at 37°C, 5% CO2 prior to fixation. The samples were stained for extracellular and total bacteria, and the number of intracellular bacteria in each vacuole was recorded by visual inspection.
Yeast labeling and immunoprecipitation of CPY.
S. cerevisiae was grown with aeration at 30°C in SC medium containing 2% galactose and lacking methionine and cysteine from an A600 of 0.3 to an A600 of 1 (4 to 5 h), followed by transfer to SC medium containing 2% glucose and lacking methionine and cysteine for the duration of the procedure. The cells were pulse-labeled with [35S]Met/Cys (50 μCi/A600 unit) (Pro-Mix; Amersham) for 5 min, and a chase was initiated by the addition of unlabeled methionine and cysteine (2 mM final concentration). At the indicated time points, 3 A600 units were collected, and the reaction was stopped by the addition of an equal volume of ice-cold H2O containing 20 mM sodium azide and 10 μg/ml of cycloheximide. Cell lysis and carboxypeptidase Y (CPY) immunoprecipitation were performed as previously described (32). The anti-CPY antibody was obtained from U.S. Biological and used at 1:1,000. Immunoprecipitates were visualized using a phosphorimager. The intensities of the bands corresponding to CPY precursors and mature forms were determined for each time point by using IQMac software, and the percentage of CPY precursors was calculated for each time point.
Electron microscopy.
S. cerevisiae was grown at 30°C in SC medium containing 2% galactose and lacking methionine and cysteine to an A600 of 1 as described for the CPY assay and processed for electron microscopy as described previously (12). Samples were sectioned and analyzed using a Philips CM-10 transmission electron microscope.
RESULTS
LidA is membrane associated after its translocation into the host cell.
In a previous study (6), we observed continued association of LidA with the phagosomal membrane during L. pneumophila intracellular growth. This result indicated that LidA was translocated early after infection and that its presence in the cell was maintained throughout the growth cycle of the bacteria. This is in contrast to a previous proposal that substrates of the Dot/Icm system are involved only in the earliest steps of intracellular growth (30). To investigate translocation further, we used a biochemical assay to determine if LidA accumulates during intracellular growth and to distinguish whether it is present in a cytosolic or membrane-associated compartment. An assay was used based on the selective detergent extraction of proteins from the cytoplasm and the membranes of infected cells without releasing proteins associated with the bacteria (22). To identify the proper detergent, the resistance of L. pneumophila to a variety of agents was tested.
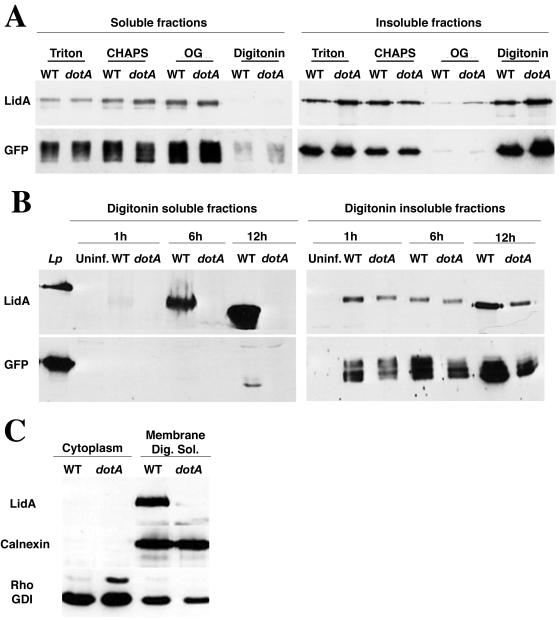
L. pneumophila or a strain harboring a disrupted Dot/Icm complex (dotA mutant) expressing GFP was incubated for 20 min in PBS containing either Triton X-100, CHAPS, octylglucoside (OG), or 1% digitonin (see Materials and Methods). The detergent-soluble and -insoluble fractions were analyzed by immunoblotting using antibodies directed against LidA or GFP (Fig. 1A). Incubation of the bacteria in Triton, CHAPS, or OG led to the extraction of both LidA and GFP (Fig. 1A, compare lanes 1 to 6 from right and left blots). LidA and GFP, however, remained mainly in the detergent-insoluble fractions after incubation of the bacteria in digitonin (Fig. 1A, compare lanes 7 to lanes 8), indicating that L. pneumophila is much more resistant to extraction with this detergent than the others tested. Therefore, incubation of infected macrophages with 1% digitonin will selectively solubilize bacterial proteins that have been exported into the mammalian cell. These results also indicate that GFP, a protein present in large amounts in the cytosol of the bacteria and which is not secreted by the type IV secretion system, can be used as a negative control for translocation.
FIG. 1.
Membrane association of LidA after translocation into host cells. (A) L. pneumophila (WT) and an isogenic dotA mutant (dotA), both expressing GFP, were grown to postexponential phase and resuspended in PBS containing either 1% Triton X-100, 1% CHAPS, 1% OG, or 1% digitonin (see Materials and Methods). The detergent-soluble and -insoluble fractions were analyzed by immunoblotting using antibodies directed against LidA or GFP. (B) U937 cells were incubated for 1, 6, or 12 h with either L. pneumophila (WT) or the isogenic dotA mutant (dotA), both expressing GFP, prior to incubation for 20 min in PBS containing 1% digitonin. The detergent-soluble and -insoluble fractions were analyzed by immunoblotting using antibodies directed against LidA or GFP. Lp, wild-type L. pneumophila grown in bacteriological medium and directly resuspended in sample buffer; Uninf., uninfected. (C) U937 cells were incubated for 6 h with either L. pneumophila (WT) or the isogenic dotA mutant (dotA), both expressing GFP, prior to mechanical fractionation (see Materials and Methods). The cytosolic fraction was separated from the membrane fraction by ultracentrifugation, and the membrane fraction was subjected to digitonin extraction (see Materials and Methods). The cytosolic fraction (Cytoplasm) and the digitonin-soluble fraction (Membrane Dig. Sol.) were analyzed by immunoblotting using antibodies directed against LidA, calnexin, or RhoGDI.
To perform a time course of LidA export into the host cell, U937 macrophage-like cells were incubated for 1, 6, or 12 h with either wild-type L. pneumophila or a dotA mutant and then subjected to digitonin fractionation with subsequent SDS extraction of the digitonin-insoluble fraction (see Materials and Methods). The digitonin-soluble and -insoluble fractions were analyzed by immunoblotting using antibodies directed against LidA and GFP (see Materials and Methods). As shown in Fig. 1B, LidA, but not GFP, was found in the digitonin-soluble fraction (Fig. 1B, left, WT). Moreover, LidA appeared to accumulate in the host cells, as the amount found in the digitonin-soluble fraction increased over time (Fig. 1B, left, compare WT at 1, 6, and 12 h). In addition, we observed a difference in the apparent molecular weight of LidA that was dependent on incubation of the bacteria with eukaryotic cells and digitonin (Fig. 1B, left, compare Lp to digitonin soluble). A similar result was observed with GFP, arguing against LidA processing once it is translocated in the host cell. The presence of LidA in the digitonin-soluble fraction was dependent on an intact Dot/Icm complex (Fig. 1B, left, dotA), and a lidA mutant showed no immunoreactive band corresponding to LidA (data not shown). The immunoreactive signal was also dependent on the presence of L. pneumophila in the cell, since LidA and GFP were not detected in the digitonin-soluble and -insoluble fractions of uninfected U937 cells (Fig. 1B, Uninf.).
To determine whether translocated LidA was free in the cytoplasm or membrane associated, U937 cells were incubated for 6 h with L. pneumophila (WT or dotA mutant) and mechanically disrupted. The membranes were separated from the cytoplasm by high-speed centrifugation and then subjected to digitonin solubilization (see Materials and Methods). As shown in Fig. 1C, analysis of the cellular localization of LidA showed that the protein was not present in the soluble fraction of cells infected with either wild-type L. pneumophila or the dotA mutant (Fig. 1C, top). However, LidA was present in the digitonin-solubilized membrane fraction when the cells were infected with wild-type L. pneumophila (Fig. 1C, top). Calnexin, an ER membrane protein, was found only in the digitonin-soluble membrane fraction (Fig. 1C, middle), while the cytosolic marker, RhoGDI, was enriched in the soluble supernatant fraction relative to the membrane, in contrast to the fractionation of LidA (Fig. 1C, bottom). Therefore, translocated LidA appears to accumulate in host cell membranes during the replication cycle of L. pneumophila.
LidA localizes to early secretory compartments and causes their disruption when overexpressed.
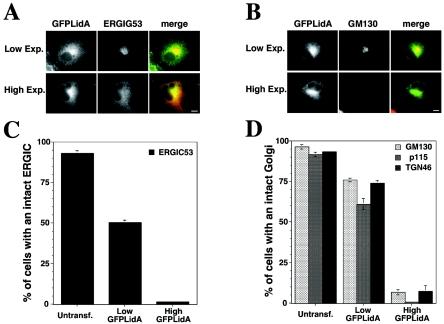
In order to investigate the function of LidA, we analyzed the cellular localization of a GFP-LidA fusion protein and the effect of its overexpression on transiently transfected COS1 cells. Mechanical fractionation of the transfected cells expressing GFP-LidA indicated that the fusion protein was membrane associated (data not shown). Immunofluorescence analysis indicated that the GFP-LidA fusion protein was concentrated in a perinuclear area of the host cell (Fig. 2A and B, left). Immunostaining using antibodies directed against proteins of the ERGIC (endoplasmic reticulum-Golgi intermediate compartment) (ERGIC53 [39]) (Fig. 2A, top) or the Golgi (GM130 [25]) (Fig. 2B, top) revealed that this region corresponded to the ERGIC and the Golgi. These cellular compartments were intact in cells that expressed GFP-LidA at a low level (∼598 arbitrary units/perinuclear region of interest) (Fig. 2A and B, top). However, in cells expressing large amounts of GFP-LidA (∼2,392 arbitrary units/perinuclear region of interest), the ERGIC was redistributed throughout the cells (Fig. 2A, bottom) and the Golgi was undetectable (Fig. 2B, bottom). Quantification of ERGIC disruption indicated that in the absence of GFP-LidA (Fig. 2C, Untransf.), the ERGIC was intact in 95% of the cells. However, in cells expressing low levels of GFP-LidA, only 75% of the cells showed an intact ERGIC (Fig. 2C, Low GFPLidA; P ≤ 5 × 10−5), while less than 5% of the cells expressing large amounts of GFP-LidA had an intact ERGIC (Fig. 2C, High GFPLidA). A similar result was obtained when the disruption of the Golgi apparatus was analyzed. In the absence of GFP-LidA, the Golgi was intact in 90% of the cells (Fig. 2D, Untransf.). In contrast, only 60% of the cells expressing GFP-LidA at a low level (Fig. 2D, Low GFPLidA; P ≤ 0.0007) and 10% of the cells expressing large amounts of GFP-LidA (Fig. 2D, High GFPLidA) had an intact Golgi apparatus based on staining with anti-GM130. Both the cis- and the trans-Golgi appeared disrupted, because we observed similar results when the cells were probed with antibody directed against either p115 (cis-Golgi marker) (45) or TGN46 (trans-Golgi marker) (28) (Fig. 2D).
FIG. 2.
The GFP-LidA fusion protein localizes to early secretory compartments and causes their disruption when overexpressed. (A and B) Immunofluorescence micrographs of COS1 cells expressing GFP-LidA at low (top row) or high (bottom row) levels and stained with antibodies directed against either the ERGIC marker, ERGIC53 (A), or the Golgi marker, GM130 (B). Left, GFPLidA; middle, ERGIC53/GM130; right, merge. Bars, 10 μm. (C and D) Quantification of ERGIC (C) or Golgi (D) disruption in either untransfected COS1 (Untransf.) or transfectants expressing either low levels (Low GFPLidA) or high levels (High GFPLidA) of GFPLidA. The cells were stained with anti-ERGIC53 to assay for ERGIC disruption and with anti-GM130, p115, or TGN46 to assay for disruption of the Golgi, the cis-Golgi, and the trans-Golgi, respectively. The values represent the mean ± standard deviation of three independent experiments.
The coiled-coil-rich region of LidA targets the protein to compartments of the secretory pathway and causes ERGIC and Golgi disruption.
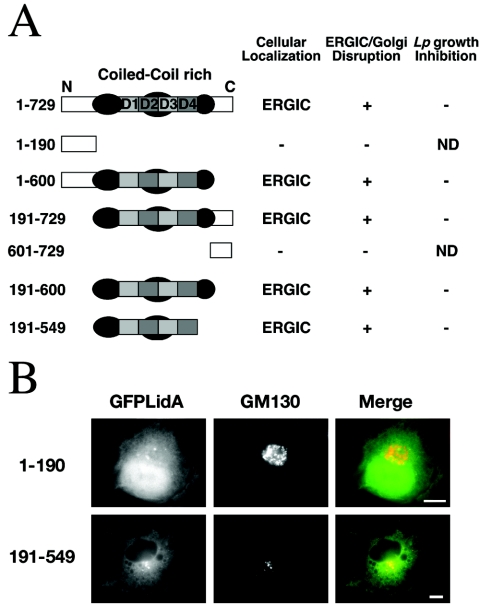
We next investigated which region of LidA was responsible for its localization to compartments of the secretory pathway and its ability to disrupt the ERGIC and the Golgi apparatus. LidA contains a central region predicted to be rich in coiled-coil structures (amino acids [aa] 191 to 600) that includes four subdomains, called here D1 (aa 303 to 358), D2 (aa 344 to 401), D3 (aa 434 to 490), and D4 (aa 491 to 548) (Fig. 3A). D1 and D3 share 47% amino acid identity, while D2 and D4 are 30% identical. Based on domain analysis, a series of truncated GFP-tagged versions of LidA were constructed and expressed in COS1 cells. Immunofluorescence of the transfected cells indicated that the amino- and carboxy-terminal regions of LidA were dispensable for the localization of the protein to the ERGIC (Fig. 3A, constructs 1 to 600 and 191 to 729). In addition, neither of these regions alone localized to the ERGIC (Fig. 3A and B, constructs 1 to 190 and 601 to 729). However, the region predicted to be rich in coiled-coil structure is sufficient to target LidA to the ERGIC of the cell (Fig. 3A and B, construct 191 to 600), although the most carboxy-terminal coiled-coil region was dispensable for this localization (Fig. 3A, construct 191 to 541). Smaller derivatives that further cut this region into subdomains failed to target to the ERGIC (data not shown). We observed a positive correlation between the ability of the truncated versions of LidA to localize to the ERGIC of the host cell and their ability to disrupt the ERGIC and the Golgi apparatus when expressed in large amounts (Fig. 3A). We have previously shown that L. pneumophila replication within COS1 cells was similar to that observed in macrophage-like cell types (8), so we next tested if expression of a truncated version of LidA would affect L. pneumophila intracellular growth. For this purpose, transiently transfected COS1 cells expressing the constructs described above were incubated with L. pneumophila, and the number of bacteria per vacuole was determined 14 h postinfection. Even though the ERGIC and the Golgi apparatus were clearly disrupted upon expression of large amounts of these constructs, none of them had an effect on the intracellular replication of L. pneumophila (Fig. 3A), as in each transfectant, at least 70% of the vacuoles showed more than 10 bacteria at 14 h postinfection. This indicates that in COS1 cells LidA-mediated disruption of compartments of the early secretory pathway did not affect intracellular growth.
FIG. 3.
The predicted coiled-coil domain of LidA targets the protein to early secretory compartments and causes ERGIC/Golgi disruption. (A) Schematic representation of truncated LidA polypeptides that were fused to GFP. The extent of each deletion is shown on the left. The coiled-coil-rich domain is indicated in grey. Dark-grey circles indicate the predicted coiled-coil structures, and dark- and light-grey squares indicate the repeats (D1 to D4), with the square color indicating regions of highest sequence similarity (see the text). The cellular localization of each construct and its ability to disrupt the ERGIC or the Golgi apparatus or to affect L. pneumophila intracellular growth is indicated on the right. +, present; −, absent; ND, not determined. (B) Immunofluorescence micrographs of COS1 cells expressing truncated versions of LidA: amino acids 1 to 190 (top) or amino acids 191 to 549 (bottom) fused to the GFP and stained with antibodies directed against the Golgi marker, GM130. Left, GFP-LidA; middle, GM130; right, merge. Bars, 10 μm.
LidA interferes with the secretory pathway of Saccharomyces cerevisiae.
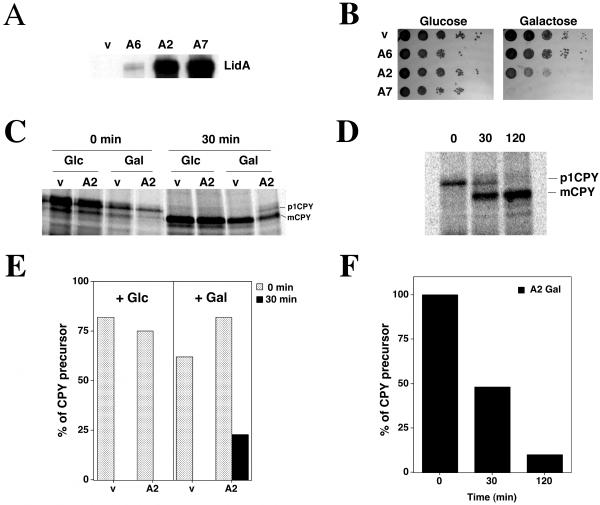
The disruption of the Golgi apparatus observed in mammalian cells may be due to the perturbation of the early secretory pathway. To directly demonstrate that LidA affects secretion, the protein was expressed in the yeast Saccharomyces cerevisiae, which has many elements of its secretory pathway conserved in higher eukaryotes and is an attractive model to study bacterial pathogenesis (41). When the lidA gene, under the control of a galactose-inducible promoter, was integrated at the URA3 locus, three categories of clones (illustrated by clones A6, A2, and A7) were obtained based on their survival on solid media (Fig. 4A). The growth defects observed were positively correlated with the amount of LidA produced (Fig. 4A and B).
FIG. 4.
Delay in CPY maturation in yeast cells expressing LidA. (A) Yeast strains harboring chromosomal copies of lidA under the control of the galactose-inducible promoter (A6, A2, and A7) or containing a vector control (v) grown in YPGal were analyzed by immunoblotting using antibody directed against LidA. The strains were adjusted to equivalent numbers of 0.1 A600 units. (B) Tenfold serial dilutions of different yeast strains containing lidA under the control of a galactose-inducible promoter (A6, A2, and A7) or containing a vector control (v) were spotted onto YPD (Glucose) or YPGal (Galactose) plates and incubated at 30°C for 3 days. (C) Yeast cells containing a vector control (v) or a chromosomal copy of lidA under the control of a galactose-inducible promoter (A2) were grown at 30°C in glucose (Glc) or galactose (Gal) to an A600 of 1. The cells were pulse-labeled with [35S]Met/Cys for 5 min and chased for 0 min (0 min) or 30 min (30 min) (see Materials and Methods). Immunoprecipitations of CPY were performed, immunoprecipitates were subjected to SDS-PAGE, and relative abundance was determined as described in Materials and Methods. (D) Yeast cells containing a chromosomal copy of lidA under the control of a galactose-inducible promoter (A2) were grown at 30°C in galactose to an A600 of 1 and were processed as described for panel C, except that samples were collected 0, 30, and 120 min after the beginning of the chase. (E) Quantification of CPY maturation. The percentages of CPY precursor, corresponding to the experiment shown in panel C, were calculated (see Materials and Methods) for both strains (v and A2) grown in glucose (+ Glc) or galactose (+ Gal) after 0 min or 30 min of chase. (F) Quantification of CPY maturation corresponding to the experiment shown in panel D.
When spotted on solid media using noninducing conditions (glucose), the growth of all three clones was comparable to that of a strain containing a vector control (v) (Fig. 4B, left). Under conditions in which LidA was expressed (galactose), however, only the growth of the clone A6 was comparable to that of a clone containing a vector control. A2 showed an intermediate growth defect, and A7 was completely defective for growth (Fig. 4B, right), suggesting that LidA expression interfered with a critical cellular function.
The transit of the CPY enzyme (42) from the ER to the vacuole was investigated in these clones to determine whether transport through the secretory pathway was affected by LidA. CPY is synthesized in the ER as a precursor (p1CPY; 67 kDa) and travels through the Golgi, where the protein is highly glycosylated (p2CPY; 69 kDa), prior to targeting the vacuole, where it is processed and accumulates as a mature form (mCPY; 61kD). This maturation was followed in a pulse-chase experiment, analyzing labeled CPY by immunoprecipitation (see Materials and Methods). When yeast cells containing a vector control were grown in glucose (noninducing conditions) or galactose (inducing conditions), the precursor form of CPY (p1CPY) was processed into its mature form during a 30-min chase after 5 min of pulse-labeling (Fig. 4C, compare v Glc/Gal, 0 min, to v Glc/Gal, 30 min). The same result was observed when yeast cells containing an integrated copy of the lidA gene (clone A2) expressing intermediate amounts of the protein were grown under noninducing conditions (Fig. 4C, A2, Glc, 30 min). However, when LidA expression was induced, p1CPY accumulated (Fig. 4C, compare A2, Glc, 30 min, to A2, Gal, 30 min). Under conditions in which 100% of CPY precursor was processed in a strain harboring a vector control, 25 to 50% of CPY remained in precursor form in the LidA-expressing A2 clone (Fig. 4E and F). Moreover, the precursor of CPY was maintained for an extended period of time in the LidA-expressing strain, as 10% of the total CPY was still present even after 2 h of chase (Fig. 4D and F). No defect in CPY secretion was observed in yeast expressing LidA at a lower level (clone A6) (data not shown), while the highest LidA-expressing clone (A7) was not analyzed further because it was completely defective for growth shortly after transfer into galactose-containing medium (data not shown).
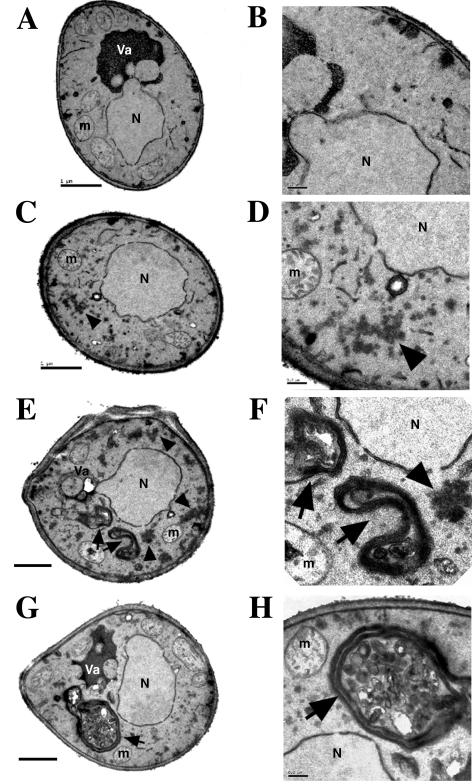
The observed secretion defect caused by LidA expression in S. cerevisiae was associated with intracellular morphological alterations, as determined by electron microscopy of yeast cells grown under inducing conditions. Cells expressing LidA at a low level (clone A6) showed accumulation of dense material that appeared to be clusters of vesicles approximately 50 nm in diameter (Fig. 5C and D) that was not present in the vector-containing control (Fig. 5A and B). In addition, cells expressing LidA at a higher level (clone A2) showed accumulation of multimembranous compartments containing vesicular structures of various sizes and shapes (Fig. 5E, F, G, and H). The changes are reminiscent of morphological abnormalities observed in yeast secretory mutants (7, 20). Therefore, high expression levels of LidA disrupt the growth of S. cerevisiae, and this appears to be the consequence of dysfunction of the secretory pathway.
FIG. 5.
Yeast cells expressing LidA accumulate vesicles and multimembranous compartments. Yeast cells containing a vector control (A and B) or a chromosomal copy of lidA under the control of a galactose-inducible promoter (C and D, A6; E, F, G, and H, A2) were grown at 30°C in galactose to an A600 of 1 and processed for electron microscopy. The panels on the right represent enlargements of a section of the pictures presented in the panels on the left. Arrowheads and arrows indicate accumulation of vesicles and multimembranous compartments, respectively. N, nucleus; Va, vacuole; m, mitochondria. Bars, 1 μm (right panels) and 0.2 μm (left panels).
L. pneumophila replication vacuole is in close contact with the ERGIC and the Golgi apparatus.
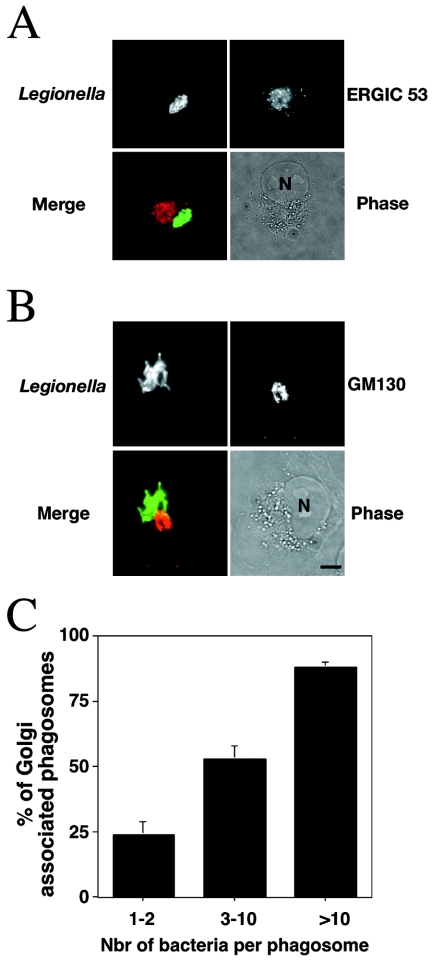
Artificial expression of large amounts of LidA led to disruption of early secretory compartments in both mammalian cells and yeast, suggesting that the protein interacts with those compartments. LidA-mediated disruption of the ERGIC and the Golgi, however, did not affect L. pneumophila intracellular replication (Fig. 3A), raising the possibility that the ERGIC and the Golgi may also be disrupted when LidA is exported into the host cell under infection conditions. Therefore, the integrity of the ERGIC and Golgi was analyzed in mammalian cells 14 h postinfection with wild-type L. pneumophila. We observed that in COS1 cells, both the ERGIC and the Golgi apparatus were intact in cells harboring replicating L. pneumophila. In addition, replicative vacuoles harboring dozens of bacteria were found localized in a perinuclear region that corresponded to the ERGIC (Fig. 6A), and they were also found in close contact with the Golgi apparatus, based on staining with anti-GM130 (Fig. 6B). For the most part, the Golgi was located between the vacuoles and the nucleus, and it was not unusual to observe hand-like extensions of the replicative vacuole that were surrounding part of the Golgi (data not shown). We performed a quantitative analysis of the association of L. pneumophila vacuoles with the Golgi, depending on the size of the replication vacuole. A vacuole was considered to be in association with the Golgi if at least one bacterium was in close contact with it. The results indicated a positive correlation between the size of the vacuole containing bacteria and its association with the Golgi (Fig. 6C). Therefore, it seems likely that the L. pneumophila-containing vacuole integrates into the secretory pathway in order to facilitate the recruitment of early secretory vesicles necessary for the establishment and maintenance of the replication vacuole.
FIG. 6.
The L. pneumophila replication vacuole is in close contact with early secretory compartments. (A and B) Immunofluorescence of COS1 cells incubated for 14 h with wild-type L. pneumophila and stained for the ERGIC (A; anti-ERGIC53) or the Golgi apparatus (B; anti-GM130). (Top rows) L. pneumophila is shown in green, and the ERGIC or the Golgi apparatus in red. (Bottom rows) Merged and phase images. N, nucleus. Bar, 10 μm. (C) Quantification of the association of the replication vacuoles with the Golgi apparatus. Displayed are the numbers (Nbr) of cells for which the vacuoles are in association with the Golgi apparatus and that have the indicated numbers of bacteria per vacuole. The values represent the means plus standard deviations of three independent experiments.
DISCUSSION
LidA in the host cell.
We have previously shown by immunofluorescence analysis that LidA is exposed on the surfaces of L. pneumophila vacuoles in a Dot/Icm-dependent manner (6). To further investigate LidA secretion into the host cell, we used a biochemical assay previously reported by Lee et al. to study the secretion of Yersinia Yop proteins in HeLa cells (22) and based on the selective solubilization of eukaryotic membranes of infected cells. We showed that LidA was translocated and accumulated within the host cell throughout the intracellular replication cycle of L. pneumophila. The apparent molecular weight of LidA in the digitonin-soluble fraction was smaller than that observed in bacterial extracts, but the same phenomenon was observed with GFP, so we suspect that the difference in apparent size was due to effects of digitonin on the electrophoretic mobility of the samples. Substrates of type III or type IV secretion systems, such as the EPEC receptor, Tir (21), or CagA from Helicobacter pylori (33), are phosphorylated after translocation into the host cell, so we cannot exclude the possibility that LidA may be processed in the host cell.
After its translocation, LidA was found associated with the membranes of the host cell. We have previously shown by immunofluorescence on infected macrophages that, at late times of infection, LidA was detected throughout the cell in addition to decorating the phagosomal membrane (6). Therefore, once translocated into the host cell, LidA localization appears to extend beyond the vacuolar membrane into the cell. The GFP-LidA fusion protein was membrane associated and concentrated in the ERGIC/Golgi area when overexpressed in mammalian cells, so it seems likely that LidA is targeted to membranes of early secretory compartments. This hypothesis is supported by the fact that overexpression of large amounts of GFP-LidA in mammalian cells appeared to disrupt the ER-to-Golgi traffic, causing the redistribution of the ERGIC throughout the cell and dissipation of the Golgi. This phenomenon is reminiscent of the consequences of overexpression of a GDP-bound form of Rab1 (Rab1S25N), a small GTPase involved in regulating the ER-to-Golgi traffic, which causes apparent loss of the Golgi apparatus (27). Consistent with our results for LidA overexpression in mammalian cells, yeast expressing LidA exhibits characteristics of secretory mutants, including delays in CPY maturation and accumulation of small vesicles (20). In addition, yeast expressing the greatest amount of LidA also accumulated multimembranous compartments that resembled mammalian autophagic compartments. The degree of defective CPY secretion appeared to correlate with LidA expression levels. A small amount of LidA led to accumulation of vesicles but left the ER-to-Golgi traffic functional, based on CPY processing. Higher expression levels of LidA, however, led to accumulation of more vesicles, exacerbating perturbation of the secretory pathway, leading to engulfment of excess material in multimembranous compartments. Similar structures had been previously observed in yeast cells expressing a GTP-bound form of Ypt1 (Rab1) in a background deficient for its dedicated GTPase-activating protein (GAP) (7). We believe that LidA overexpression in eukaryotic cells or yeast reflects an exaggeration of what is happening under infection conditions that leads to a very drastic effect on the secretory pathway. Since the ERGIC and the Golgi were intact in infected cells, it is more likely that under infection conditions the amount of LidA that is delivered into the host cell is sufficient to interfere with the ER-to-Golgi traffic without disrupting it, as observed in COS1 cells or yeast expressing very little of the protein.
Association of the replicative vacuole with the Golgi apparatus.
We observed that during L. pneumophila intracellular multiplication, the replication vacuole was in close contact with the ERGIC and the Golgi apparatus. Moreover, the association of the vacuole with the Golgi was positively correlated with the size of the replication vacuole. One may argue that as the bacteria multiply, the replication vacuole becomes larger and therefore appears closer to the ERGIC or the Golgi apparatus, due to space limitations. This does not appear to be the case, because even after extensive intracellular replication, the vacuole occupies only a small portion of the cytoplasm. Association of bacterial pathogens with the Golgi apparatus has been observed. In epithelial cells, the vacuole containing Salmonella enterica serovar Typhimurium is surrounded by the Golgi membrane (31), and the vacuole containing Chlamydia trachomatis also migrates to the peri-Golgi region of the host cell, where it fuses with a subset of Golgi-derived exocytic vesicles (13, 14). It is proposed that Salmonella and Chlamydia interact with the Golgi apparatus to satisfy nutritional requirements. Despite the close contact with the Golgi apparatus, L. pneumophila does not appear to exchange material with this compartment. Indeed, analysis of isolated vacuoles containing bacteria showed that although proteins of the early secretory pathway were associated with this compartment, it was devoid of Golgi markers (8). Moreover, unlike those of Salmonella and Chlamydia, the L. pneumophila vacuole is surrounded by the ER and located in the peri-ERGIC region of the host cell. Therefore, it seems likely that the L. pneumophila replication vacuole integrates into the secretory pathway, perhaps, as previously proposed (18), to optimize the recruitment of vesicles that traffic between the ER and the Golgi apparatus. The purpose of this localization could be to sequester nutrients for the bacterium or to allow the vacuolar membrane to enlarge around the bacteria.
The model.
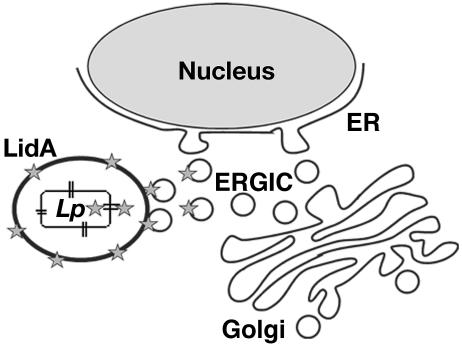
Our results are consistent with the following model (Fig. 7). Upon L. pneumophila uptake by the host cell, LidA is translocated and is associated with the vacuolar membrane and with compartments derived from the early secretory apparatus via its region that is rich in coiled-coil structure. Such structures have been found in many tethering factors, as well as SNARE-like proteins involved in vesicle docking and fusion (3). Therefore, it is possible that LidA interacts with a host protein(s) of the secretory pathway in order to mediate the recruitment of early secretory vesicles to the L. pneumophila-containing vacuole. This process is probably facilitated by the close association of the replication vacuole with the ERGIC and the Golgi apparatus. We believe that the vacuole integrates into the early secretory pathway, where it mimics the Golgi apparatus and intercepts material that would normally head to the Golgi. However, since the ERGIC and the Golgi are left intact as L. pneumophila replicates, the amount of LidA injected into the host cell is probably highly regulated, in order to intercept ER-to-Golgi traffic without disrupting the secretory pathway and to prevent dissipation of preexisting organelles.
FIG. 7.
Schematic representation of LidA-mediated ER-derived vesicle recruitment to the L. pneumophila-containing vacuole. Upon L. pneumophila uptake by the host cell, LidA (stars) is injected into the host cell cytoplasm via the type IV secretion system (double lines). The protein then associates with the phagosomal membrane and with compartments of the secretory pathway, such as the ERGIC. In this model, LidA interacts with a host protein(s) of the secretory pathway to facilitate the recruitment of ER-derived vesicles to the L. pneumophila (Lp)-containing vacuole. This may be facilitated by the migration of the phagosome in the vicinity of the ERGIC and the Golgi apparatus.
Acknowledgments
We thank Susan Van Rheenen, Molly Bergman, Marion Shonn-Dorer, Matthias Machner, and Zhao-Qing Luo for reviewing the manuscript and H. P. Hauri and G. Watters for gifts of reagents.
R.R.I. is an Investigator of the Howard Hughes Medical Institute (HHMI). I.D. is a Human Frontier Science Program Fellow. This work was supported by the HHMI.
Editor: D. L. Burns
REFERENCES
- 1.Alrutz, M. A., and R. R. Isberg. 1998. Involvement of focal adhesion kinase in invasin-mediated uptake. Proc. Natl. Acad. Sci. USA 95:13658-13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S., and B. S. Glick. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153-166. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 5.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conover, G. M., I. Derré, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 7.De Antoni, A., J. Schmitzova, H. H. Trepte, D. Gallwitz, and S. Albert. 2002. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277:41023-41031. [DOI] [PubMed] [Google Scholar]
- 8.Derré, I., and R. R. Isberg. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72:3048-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 11.Gabay, J. E., M. Blake, W. D. Niles, and M. A. Horwitz. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J. Bacteriol. 162:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammie, A. E., V. Brizzio, and M. D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 14.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 92:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4:945-954. [DOI] [PubMed] [Google Scholar]
- 19.Kagan, J. C., M. Stein, M. Pypaert, and C. Roy. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser, C. A., and R. Schekman. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61:723-733. [DOI] [PubMed] [Google Scholar]
- 21.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 22.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 23.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1984. Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J. Clin. Investig. 74:771-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuoffer, C., H. W. Davidson, J. Matteson, J. Meinkoth, and W. E. Balch. 1994. A GDP-bound rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125:225-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prescott, A. R., J. M. Lucocq, J. James, J. M. Lister, and S. Ponnambalam. 1997. Distinct compartmentalization of TGN46 and beta 1,4-galactosyltransferase in HeLa cells. Eur. J. Cell Biol. 72:238-246. [PubMed] [Google Scholar]
- 29.Rose, M., F. Winston, and P. Heiter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 31.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sapperstein, S. K., V. V. Lupashin, H. D. Schmitt, and M. G. Waters. 1996. Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol. 132:755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman, F., G. Fink, and J. Hicks. 1986. Methods in yeast genetics: a laboratory manual, p. 163-169. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, B. L., S. H. Low, H. P. Hauri, and W. Hong. 1995. Segregation of ERGIC53 and the mammalian KDEL receptor upon exit from the 15 degrees C compartment. Eur. J. Cell Biol. 68:398-410. [PubMed] [Google Scholar]
- 40.Tilney, L. G., O. S. Harb, P. S. Connelly, C. G. Robinson, and C. R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637-4650. [DOI] [PubMed] [Google Scholar]
- 41.Valdivia, R. H. 2004. Modeling the function of bacterial virulence factors in Saccharomyces cerevisiae. Eukaryot. Cell 3:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vida, T. A., T. R. Graham, and S. D. Emr. 1990. In vitro reconstitution of intercompartmental protein transport to the yeast vacuole. J. Cell Biol. 111:2871-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 44.Wadowsky, R. M., L. J. Butler, M. K. Cook, S. M. Verma, M. A. Paul, B. S. Fields, G. Keleti, J. L. Sykora, and R. B. Yee. 1988. Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl. Environ. Microbiol. 54:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters, M. G., D. O. Clary, and J. E. Rothman. 1992. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 118:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]