Abstract
The Haemophilus influenzae P4 lipoprotein (hel) is a potential component of a nontypeable H. influenzae otitis media vaccine. Since P4 is known to be an enzyme, nonenzymatically active forms of recombinant P4 are required. After site-directed mutagenesis of the hel gene, three of the mutated proteins were shown to be vaccine candidates.
The P4 lipoprotein of nontypeable Haemophilus influenzae (NTHi) is a leading vaccine candidate against nontypeable H. influenzae infections. Previous research in this laboratory has demonstrated that the P4 protein is highly conserved among all 103 H. influenzae strains tested at the antigenic level (3) and among 10 strains at the DNA sequence level (>95% identity) (5). Purified P4 elicits antibodies that are broadly cross-reactive and have bactericidal (BC) activity against both NTHi and H. influenzae type b (Hib), and a recombinant form of lipidated P4 protein (rP4) has recently been shown to reduce intranasal colonization in a mouse model after intranasal immunization (6, 9).
Reilly et al. (12) showed that the P4 protein is a highly specific acid phosphomonoesterase. Since the recombinant P4 protein is considered to be a vaccine candidate for humans, use of enzymatically active rP4 may be problematic, especially in infants. Thus, nonenzymatically active recombinant P4 proteins may be crucial to further development of rP4. Reilly et al. demonstrated that mutation of critical aspartate residues common to P4 and other class C bacterial acid phosphatases could eliminate enzymatic activity of rP4 (13), but it was not determined if these mutations had any effect on immunogenicity. These studies describe the production and characterization of additional rP4 mutant proteins.
Mutant proteins D64A, D66A, and F48C were derived from plasmid pHel3 as previously described (12). All other site-directed mutants were derived from plasmid pLP339 (6), a vector containing the wild-type (WT) hel gene from H. influenzae strain Rd KW20 (3). Most of the site-directed mutant proteins were constructed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's directions. Primers used to construct the P4 mutations are listed in Table 1. BsmBI seamless cloning was used to generate several of the P4 mutants. Mutagenized P4 genes were subcloned into pBAD18Cam (Invitrogen) in Escherichia coli strain BLR (Invitrogen) for expression of the mutant P4 protein.
TABLE 1.
Primer pairs used for site-directed mutagenesis of hel gene
| Mutation(s) | Sequence (5′ to 3′) |
|---|---|
| F48C | GCAAAAGTTGCATGCGATCACGCAAAAG |
| CTTTTGCGTGATCGCATGCAACTTTTGC | |
| D64A & D66A | GCGGTTGTGGCTGCTTTAGCTGAAACTATGTTAG |
| CTAACATAGTTTCAGCTAAAGCAGCCACAACCGC | |
| K161R | GCGCGTCTCTTGCATTTTATTTGAAAAAAGACAAATCAGCTAGAGCGGCTC |
| GCGCGTCTCGTGCAGATTCTTCCACGCCATTGAAACC | |
| N218Q | GCGCGTCTCTGCTGGGAAGGCGGTTTAGCTGAAG |
| GCGCGTCTCGCAGCCACCGTAGTTTGCTTGAGGTAACATGATGAAAG | |
| D64N | GCGCGTCTCTGGTAAGAAAAAAGCGGTTGTGGCTAATTTAGATG |
| GCGCGTCTCGTACCTTTTGCCACTTTTGCGTG | |
| D64E | GCGCGTCTCGTACCTTTTGCCACTTTTGCGTG |
| GCGCGTCTCTGGTAAGAAAAAAGCGGTTGTGGCTGAATTAGATG | |
| D184A, D185A | CGCCGTCTCGCTGCCTTCGGTAATACCGTATATGGC |
| CGCCGTCTCGGCAGCTAAGTTATCACCTACATAAAGTACG | |
| A35E, A37E | CGCCGTCTCTTAGAATATCAAGCGTACAATGCGGC |
| CCGCGTCTCATCTAATTCTTTATATTCGCCAGAATCTTGC | |
| A35N, A37N | GCCCGTCTCCCTTAAACTATCAAGCGTACAATGCGGC |
| GCCCGTCTCCTAAGTTTTTATATTCGCCAGAATCTTGC | |
| A35Q, A37Q | CCGCGTCTCATTACAATATCAAGCGTACAATGCGGC |
| GCCCGTCTCGGTAATTGTTTATATTCGCCAGAATCTTGC | |
| A35T, A37T | CGGCGTCTCCATTAACTTATCAAGCGTACAATGCGGC |
| CGGCGTCTCCTAATGTTTTATATTCGCCAGAATCTTGC | |
| Q39E | GCATTAGCTTATGAAGCGTACAATGCGG |
| CCGCATTGTACGCTTCATAAGCTAATGC | |
| Y122F | CGGTAAAGTGTTCTTTGTAACAAACCGC |
| GCGGTTTGTTACAAAGAACACTTTACCG |
Native P4 from NTHi strain P860295 was purified as described previously (3). Mutated rP4 proteins were purified from E. coli BLR cultures containing the appropriate plasmid grown in Hy-Soy medium and induced when the optical density at 600 nm reached approximately 2.0. The P4 mutant proteins were purified by a modification of the method previously described for recombinant WT P4 (9). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on denatured samples was performed using the method of Laemmli (8). Each lane of a 12% acrylamide gel was loaded with 10 μg of protein, and the gels were stained with Coomassie.
The purified P4 mutant proteins were then examined for enzymatic activity in a sensitive fluid phase assay. The phosphomonoesterase activity of rP4 was measured in comparison to WT rP4 by an essentially colorimetric assay as described by Reilly et al. (11), and enzymatic activity was expressed as a percentage of WT rP4.
Antisera against the P4 proteins were produced in Swiss Webster mice (6 to 8 weeks old) subcutaneously immunized at weeks 0 and 4 with 5 μg of the appropriate P4 protein mixed with 25 μg MPL adjuvant (Corixa, Seattle, WA) in phosphate-buffered saline (9). Preimmune and week 6 sera were analyzed for anti-P4 antibodies using a P4 enzyme-linked immunosorbent assay (ELISA) (9) with WT rP4 coating the plate. Whole-cell ELISAs (16) to determine the reactivity of the sera against surface-exposed epitopes of P4 were performed using NTHi P860295 (2) as the coating antigen. Geometric mean titers (GMT) were determined using ELISA titers of sera from individual mice.
Serum BC assays were used to examine the biological activity of anti-rP4 sera and were performed as previously described (3), with slight modifications. NTHi strain P860295 was used as the target strain for all BC assays. Human sera adsorbed against NTHi P860295 (4) was used as a complement source.
A total of 13 different rP4 mutants were made (Table 2). Sites for mutation were selected based on sequence homologies to other known bacterial acid phosphatases in which two aspartic acid residues were known to be important for enzymatic activity (14, 15). Additionally, alanine residues at positions 35 and 37 were changed in pairs to examine the effects of changes in residues not conserved among acid phosphatases. The phenylalanine residue at position 48 was changed to a cysteine residue since it is in a putative hemin-binding domain, KVA(F)DH (10), and this affects the enzymatic activity of rP4 (13). All site-directed mutants were confirmed by DNA sequencing of the P4 gene.
TABLE 2.
Immunogenicity and enzymatic activity of rP4 mutants
| Vaccinea | IgGb ELISA titer vs WT | BC titer vs P860295c | Whole-cell ELISA titer vs P860295d | % of WT rP4 enzymatic activity |
|---|---|---|---|---|
| D64N | 1,921,553f | 20 | 208,236 | 0.2 |
| N218Q | 1,475,833g | 200 | 70,854 | 0.1 |
| Y122F | 1,106,822f | 40 | 89,397 | 94 |
| D64E | 988,016f | 20 | 25,426 | 0.07 |
| Q39E | 770,483f | 320 | 7,857 | 3.3 |
| F48C | 687,742g | 240 | 36,208 | <2e |
| rP4 | 197,160g | 160 | 27,505 | 100 |
| K161R | 186,394g | 60 | 10,730 | 0.4 |
| D64A, D66A | 60,358g | 30 | 5,350 | 0 |
| D184A, D185 | 33,185g | 30 | 2,701 | 0 |
| A35Q, A37Q | 14,331g | 50 | 613 | 0 |
| A35N, A37N | 7,121g | 200 | 580 | 0 |
| A35E, A37E | 4,322g | 30 | 7,523 | 0 |
Mice were immunized two times as described in Materials and Methods.
IgG, immunoglobulin G.
Titer is average of two assays. All preimmune sera had BC titers of <20 in these assays.
Whole-cell ELISA is from pool of sera from 10 mice. Preimmune sera all had titers of <50.
As described by Reilly et al. (13).
Titer of pooled sera of 10 mice.
GMT from individual sera of 10 mice. Preimmune sera had titers of <100.
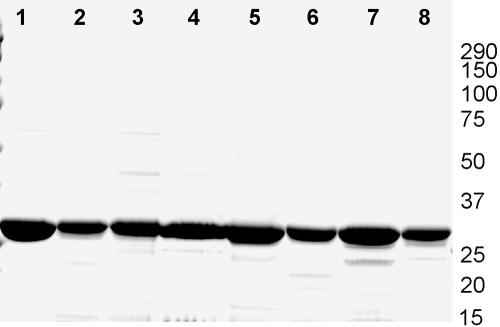
The rP4 mutant proteins were purified to >95% homogeneity (Fig. 1). Silver stain analysis of the purified proteins showed very low levels of E. coli lipooligosaccharide (<0.01 EU/μg protein). The physical properties of the mutant proteins closely resemble native P4 from H. influenzae and WT rP4, although some minor charge differences were detected. All of the proteins were properly processed, resulting in lipidated proteins inserted into the outer membrane of E. coli. Enzymatic activity of most of the mutant rP4s ranged from 0 (nondetectable) to 3.3% of WT rP4 (Table 2). Of all of the mutants examined, only the Y122F mutant had significant levels of enzymatic activity remaining, with essentially WT levels (∼94% in this assay).
FIG. 1.
SDS-PAGE analysis of purified WT and mutant rP4 proteins. Approximately 10 μg of individual purified rP4 protein was run in each lane of a 10 to 20% SDS-PAGE gel that was stained with Coomassie. Seven of the purified rP4 mutant proteins are shown in this analysis. Lanes: 1, WT rP4; 2, rP4 Q39E; 3, rP4 F48C; 4, rP4 D64E; 5, rP4 D64N; 6, rP4 Y122F; 7, rP4 K161R; 8, rP4 N218Q. Positions of molecular weight standards (masses are in kilodaltons) are shown at the right.
Initial immunologic analysis of the rP4 mutants focused on the immune response to WT rP4 protein (Table 2). The rP4 immunogens with double amino acid substitutions had greatly reduced immune responses to the WT rP4 protein. Since the immune responses to the homologous protein were also greatly reduced and the P4 proteins with double amino acid substitutions are readily recognized by the antisera against either native NTHi P4 or rP4 (data not shown), these responses appear to represent reduced immunogenicity in mice and not an immune response to rP4 in a different conformation. The recombinant proteins containing single amino acid substitutions were all more immunogenic than the double mutants (Table 2), and some (D64N, N21Q, D64E, and Y122F) were as immunogenic as WT rP4. The low GMT of WT rP4 in Table 2 resulted from the failure of one animal to respond to the immunogen, giving a very low titer and the low GMT.
When the immune sera were further analyzed using the whole-cell ELISA and BC assays against NTHi P860295, it was apparent that no correlation existed among the protein ELISA, whole-cell ELISA, and BC assays (Table 2). It is also obvious that enzymatic activity of rP4 is not required to elicit high-titer functional antibodies. Taken as a whole, the rP4 proteins containing single amino acid substitutions were more immunogenic and more likely to have higher whole-cell ELISA titers and high BC titers.
No proven correlates of protection against NTHi-mediated diseases such as otitis media exist for humans. BC antibody titers in the blood of children have been associated with reduced rates of NTHi infection (1), but it is not known if BC antibodies are in fact responsible or are just a result of natural infection and some other functional response is required. For a mucosal surface disease such as otitis media, it is possible that large amounts of surface reactive antibodies are required to block adherence or promote clumping and removal of the bacteria from the nasopharynx (2, 7). Thus, the choice of vaccine candidates from the rP4 mutants should rely not upon results from any one assay but on all three aspects of the immune response that were measured. While most of the ELISA titers against purified WT rP4 are GMT, some of the titers shown are pools of 10 animals due to serum limitations in some of the groups. For this reason, direct comparisons of protein ELISA titers of the groups cannot be made, but it is evident from the pool and GMT that the rP4 mutants at the top of the table appear to be more immunogenic than those at the bottom. This is another reason that, in choosing mutants that most closely resemble WT rP4 in immunogenic properties, all three assays are considered, not just ELISA titers. Since all of the mutant proteins except the Y122F mutant had essentially no enzymatic activity, this was not a differentiating criterion. The rP4 mutants with the best combination of greatly reduced enzymatic activity, immunogenicity against WT rP4, whole-cell ELISA titers, and the ability to induce BC activity are the N218Q, Q39E, and F48C mutants and are the preferred vaccine candidates.
Acknowledgments
We acknowledge Leah Fletcher and Rob Hazelo for site-directed mutagenesis, Nan Silliman for work on the rP4 and whole-cell ELISAs, and Alan Howell for work on the NTHi BC assays.
This work was supported in part by grants AI44002 and DC008533 (to A.L.S.) from the National Institutes of Health.
Editor: J. N. Weiser
REFERENCES
- 1.Bernstein, J. M., H. S. Faden, and P. L. Ogra. 1991. Nasopharyngeal colonization by nontypeable Haemophilus influenzae in children: the effect of serum bactericidal antibody. Otolaryngol. Head Neck Surg. 105:406-410. [DOI] [PubMed] [Google Scholar]
- 2.Brinton, C. C., Jr., M. J. Carter, D. B. Derber, S. Kar, J. A. Kramarik, A. C. C. To, and S. W. Wood. 1989. Design and development of pilus vaccines for Haemophilus influenzae diseases. Pediatr. Infect. Dis. J. 8(Suppl.):54-61. [PubMed] [Google Scholar]
- 3.Green, B. A., J. E. Farley, T. Quinn-Dey, R. A. Deich, and G. W. Zlotnick. 1991. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect. Immun. 59:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green, B. A., B. J. Metcalf, T. Quinn-Dey, D. H. Kirkley, S. A. Quataert, and R. A. Deich. 1990. A recombinant non-fatty acylated form of the Hi-PAL (P6) protein of Haemophilus influenzae elicits biologically active antibody against both nontypeable and type b H. influenzae. Infect. Immun. 58:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, B. A., S. B. Olmsted, M. Vazquez, G. Quigley-Reape, and G. W. Zlotnick. 1998. A clinical vaccine formulation for Haemophilus influenzae otitis media containing recombinant P4 and P6 outer membrane proteins, p. 355-362. In M. Tos, J. Thomsen, and V. Balle (ed.), Recent advances in otitis media: proceedings of the fourth extraordinary international symposium. Kugler Publications, Amsterdam, The Netherlands.
- 6.Hotomi, M., Y. Ikeda, M. Suzumoto, K. Yamauchi, B. A. Green, G. Zlotnick, D. S. Billal, J. Shimada, K. Fujihara, and N. Yamanaka. 2005. A recombinant P4 protein of Haemophilus influenzae induces responses biologically active against nasopharyngeal colonization after intranasal immunization. Vaccine 23:1294-1300. [DOI] [PubMed] [Google Scholar]
- 7.Karasic, R., D. J. Beste, S. C.-M. To, W. J. Doyle, S. J. Wood, M. J. Carter, A. C. C. To, K. Tanpowpong, C. D. Bluestone, and C. C. Brinton, Jr. 1988. Evaluation of pilus vaccines for prevention of experimental otitis media caused by nontypable Haemophilus influenzae. Pediatr. Infect. Dis. J. 8(Suppl.):S62-S65. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Mason, K. W., D. Zhu, C. A. Scheuer, J. C. McMichael, G. W. Zlotnick, and B. A. Green. 2004. Reduction of nasal colonization of nontypeable Haemophilus influenzae following intranasal immunization with rLP4/rLP6/UspA2 proteins combined with aqueous formulation of RC529. Vaccine 22:3449-3456. [DOI] [PubMed] [Google Scholar]
- 10.Reidl, J., and J. J. Mekalanos. 1996. Lipoprotein e (P4) is essential for hemin uptake by Haemophilus influenzae. J. Exp. Med. 183:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reilly, T., and A. Smith. 1999. Purification and characterization of a recombinant Haemophilus influenzae outer membrane phosphomonoesterase e. Protein Expr. Purif. 17:401-409. [DOI] [PubMed] [Google Scholar]
- 12.Reilly, T. J., D. L. Chance, and A. L. Smith. 1999. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J. Bacteriol. 181:6797-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reilly, T. J., B. A. Green, G. W. Zlotnick, and A. L. Smith. 2001. Contribution of the DDDD motif of H. influenzae e (P4) to phosphomonoesterase activity and heme transport. FEBS Lett. 494:19-23. [DOI] [PubMed] [Google Scholar]
- 14.Rossolini, G. M., S. Schippa, M. L. Riccio, F. Berlutti, L. E. Macaskie, and M. C. Thaller. 1998. Bacterial nonspecific acid phosphohydrolases: physiology, evolution and use as tools in microbial biotechnology. Cell. Mol. Life Sci. 54:833-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thaller, M. C., S. Schippa, and G. M. Rossolini. 1998. Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 7:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagursky, R. J., P. Ooi, K. F. Jones, M. J. Fiske, R. P. Smith, and B. A. Green. 2000. Identification of a Haemophilus influenzae 5′-nucleotidase protein: cloning of the nucA gene and immunogenicity and characterization of the NucA protein. Infect. Immun. 68:2525-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]