Abstract
Susceptibility testing results for Streptococcus pneumoniae isolates (n = 2,279) from eight European countries, examined in the PneumoWorld Study from 2001 to 2003, are presented. Overall, 24.6% of S. pneumoniae isolates were nonsusceptible to penicillin G and 28.0% were resistant to macrolides. The prevalence of resistance varied widely between European countries, with the highest rates of penicillin G and macrolide resistance reported from Spain and France. Serotype 14 was the leading serotype among penicillin G- and macrolide-resistant S. pneumoniae isolates. One strain (PW 158) showed a combination of an efflux type of resistance with a 23S rRNA mutation (A2061G, pneumococcal numbering; A2059G, Escherichia coli numbering). Six strains which showed negative results for mef(A) and erm(B) in repeated PCR assays had mutations in 23S rRNA or alterations in the L4 ribosomal protein (two strains). Fluoroquinolone resistance rates (levofloxacin MIC ≥ 4 μg/ml) were low (Austria, 0%; Belgium, 0.7%; France, 0.9%; Germany, 0.4%; Italy, 1.3%; Portugal, 1.2%; Spain, 1.0%; and Switzerland, 0%). Analysis of quinolone resistance-determining regions showed eight strains with a Ser81 alteration in gyrA; 13 of 18 strains showed a Ser79 alteration in parC. The clonal profile, as analyzed by multilocus sequence typing (MLST), showed that the 18 fluoroquinolone-resistant strains were genetically heterogeneous. Seven of the 18 strains belonged to new sequence types not hitherto described in the MLST database. Europe-wide surveillance for monitoring of the further spread of these antibiotic-resistant S. pneumoniae clones is warranted.
Streptococcus pneumoniae continues to be a significant cause of morbidity and mortality in humans (35). The worldwide increase in the rates of antibiotic resistance in this species has become a serious problem within the last 20 years (1). Macrolide resistance in S. pneumoniae is usually caused by the presence of the erm(B) or the mefA resistance determinant. The erm(B) protein encodes a 23S rRNA methylase, and most pneumococcal strains that harbor this gene are resistant to 14-, 15-, and 16-membered-ring macrolides, lincosamides, and streptogramin B (MLSB phenotype). The mef(A) protein encodes an efflux pump that leads to resistance only to 14- and 15-membered-ring macrolides (45, 52). Other mechanisms of macrolide resistance include changes clustered in a highly conserved region of domain V of 23S rRNA, which plays a key role in macrolide binding (6, 10, 49, 55), and in ribosomal proteins L4 and L22. In addition, erm(TR) mutations (53) have also been described in a few clinical pneumococcal isolates (6, 10, 49, 55).
Newer fluoroquinolones with greater potencies against S. pneumoniae licensed in Europe include levofloxacin and moxifloxacin. Because of the emergence of antimicrobial resistance in pneumococci (1), newer fluoroquinolones are now recommended for the empirical treatment of pneumonia in adults when antimicrobial resistance is suspected (28). Fluoroquinolones are also recommended for initial empirical therapy of selected outpatients with community-acquired respiratory tract infections (e.g., patients with acute exacerbations of chronic bronchitis) in several countries (28, 59). Other therapeutic options (macrolides and doxycycline) are generally preferred for the treatment of uncomplicated infections in outpatients, but there is increasing concern about the misuse and overuse of fluoroquinolones, and it is believed that if abuse of this class of drugs continues unabated, we may see the demise of fluoroquinolones as useful antibiotics within the next 5 to 10 years (28).
Pneumococcal resistance to quinolones is usually due to mutations in either parC or gyrA, or both (40). Strains usually become fully fluoroquinolone resistant with the addition of a mutation in the other target gene (either gyrA or parC). Mutations in parE and gyrB may also contribute to resistance (37). In addition, an efflux mechanism has been described (16).
The PneumoWorld surveillance study was established in 2001 to study the susceptibilities of Streptococcus pneumoniae isolates from patients with noninvasive and invasive pneumococcal disease. This paper presents the results of the 2001 to 2003 PneumoWorld surveillance study of isolates submitted by 31 participating centers in eight European countries and focuses on the prevalence of macrolide and fluoroquinolone resistance geno- or phenotypes in these countries.
(This study was presented in part at the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, 27 to 30 September 2002, San Diego, Calif. [R. R. Reinert, P. Appelbaum, and the PneumoWorld European Study Group, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1623, 2002].)
MATERIALS AND METHODS
Study design.
The PneumoWorld Study was established to study the antimicrobial susceptibilities of S. pneumoniae isolates from Latin America and Europe. In the European part of the study, 31 centers in eight European countries took part in the study: Austria (three centers), Belgium (two centers), France (5 centers), Germany (6 centers), Italy (6 centers), Portugal (3 centers), Spain (4 centers), and Switzerland (2 centers).
Bacterial isolates.
Only pneumococcal isolates of probable clinical significance from adults ≥16 years of age were included. Strains were isolated and preliminarily identified (optochin sensitivity and bile solubility testing) in each center and were stored frozen at −70°C in porous beads (MICROBANK; Mast Diagnostica GmbH, Rheinfeld, Germany) for up to 3 months. Strains were then sent by courier in batches of up to 100 strains in transport medium (Port-A-Cul; Difco, Germany) to the German National Reference Centre for Streptococci for confirmation of species identification, susceptibility testing, and further investigation. Confirmation of the identities of the S. pneumoniae strains was performed by optochin sensitivity and bile solubility testing (2, 15). Demographic data collected during the study included the age and sex of the patient, infection type, culture source, inpatient or outpatient status, and the date of sample collection.
Susceptibility testing.
MIC testing was performed by the broth microdilution method recommended by CLSI (formerly the National Committee for Clinical Laboratory Standards) (38). Microtiter plates (Sensititre susceptibility plates; TREK Diagnostic Systems Ltd., East Grinstead, England) containing penicillin G, amoxicillin, cefotaxime, cefuroxime, cefpodoxime, clarithromycin, clindamycin, gatifloxacin, levofloxacin, trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol with cation-adjusted Mueller-Hinton broth (Oxoid, Wesel, Germany) plus 5% lysed horse blood (Oxoid) were used. The final inoculum was 5 × 105 CFU/ml. MICs were determined following incubation at 35°C for 20 to 24 h in ambient air. S. pneumoniae ATCC 49619 was included as a control strain. Current CLSI interpretive criteria were used to define antimicrobial resistance (38). The isolates were stored at −70°C as described above (MICROBANK). For determination of macrolide-resistant phenotypes, disks (Oxoid Ltd., Basingstoke, United Kingdom) of erythromycin (15 μg) and clindamycin (2 μg) were placed 15 to 20 mm apart on Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, MD) with 5% sheep blood (Oxoid, Wesel, Germany). The plates had previously been inoculated with a swab dipped into a 0.5 McFarland standard bacterial suspension (32). MICs of fluoroquinolone-resistant S. pneumoniae were additionally determined for clinafloxacin (Pfizer, Karlsruhe, Germany), grepafloxacin (Glaxo Wellcome, Hamburg, Germany), sparfloxacin (Aventis, Bad Soden, Germany), moxifloxacin (Bayer, Leverkusen, Germany), and ciprofloxacin (Bayer).
Determination of resistance genes.
For the detection of erm(B) and mef(A), the primers described by Trieu-Cuot et al. (57) and by Tait-Kamradt et al. were chosen (54). Preparation of DNA and reverse transcription-PCR were performed as described previously (48). For seven isolates (six mef(A)- and erm(B)-negative isolates and one mef(A)-positive isolate with a constitutive MLSB [cMLSB] resistance phenotype), sequencing of the 23S rRNA genes and the ribosomal protein L4 and L22 genes was performed with an ABI Prism Big Dye terminator kit (Applied Biosystems, Foster City, CA). The sequencing primers were as reported earlier by Canu et al. (6) and Tait-Kamradt et al. (55). The nucleotide sequences of the 23S rRNA and the L4 and L22 ribosomal proteins in Escherichia coli, S. pneumoniae R6, and S. pneumoniae serotype 4 (56) were obtained from the Institute for Genomic Research website (http://www.tigr.org). For fluoroquinolone-resistant strains, prepared chromosomal DNA was used as a template for PCR amplification of the genes encoding the quinolone resistance-determining region (QRDR). The primers and PCR conditions were those defined previously (24, 47). Some strains with unusual resistance phenotypes (strains PW 555, PW 169, PW 1905, PW 2216, and PW 1571) were screened for resistance determinants ermA, ermC, msrA, msrB, and msrD as described by Shortridge et al. (51) and Daly et al. (7).
Serotyping.
Pneumococcal strains with clarithromycin resistance, penicillin G resistance, or reduced susceptibility to fluoroquinolones were serotyped by Neufeld's Quellung reaction (39) by using type- and factor-specific sera provided by the Statens Serum Institut, Copenhagen, Denmark.
MLST.
Multilocus sequence typing (MLST) of seven strains [six mef(A)- and erm(B)-negative isolates and one mef(A)-positive isolate with a cMLSB phenotype] was carried out as described previously (11). Briefly, internal fragments of the aroE, gdh, gki, recP, spi, xpt, and ddl genes were amplified by PCR from chromosomal DNA with the primer pairs described by Enright and Spratt (11). The alleles at each of the seven loci provide the allelic profile of each isolate and also define their sequence type (ST). The allelic profiles are shown as the alleles at each of the seven loci, in the order aroE, gdh, gki, recP, spi, xpt, and ddl.
Data on antibiotic consumption was obtained from the European Surveillance of Antimicrobial Consumption, University of Antwerp (http://www.ESAC.UA.AC.be). The correlation between antibiotic consumption and fluoroquinolone resistance was analyzed by linear regression by using the Winstat statistical module (R. Fitch Software, Staufen, Germany).
RESULTS
A total of 2,279 isolates of S. pneumoniae from 31 European centers in eight countries were examined. Strains were isolated from the following sources: sputum (n = 1,055 [46.3%]), blood cultures (n = 421 [18.5%]), bronchoalveolar lavage fluid (n = 416 [18.2%]), sinus punctures (n = 150 [6.6%]), cerebrospinal fluid (CSF; n = 57 [2.5%]), other sterile body sites (n = 81 [3.6%]), pus from the middle ear (n = 54 [2.4%]), fluid obtained by tympanocentesis (n = 29 [1.2%]), and other sources (n = 16 [0.7%]). Pneumococci were isolated from individuals in the following age groups: ≥16 to 30 years (n = 261 [11.5% of cases]), >30 to 40 years (n = 275 [12.0% of cases]), >40 to 50 years (n = 276 [12.1% of cases]), >50 to 60 years (n = 421 [18.5% of cases]), >60 to 70 years (n = 494 [21.7% of cases]), and > 70 (n = 506 [22.2% of cases]). Among all patients, 63.6% were males and 35.1% were females (no data on the sex of the patients were available for 1.3% of the patients). The following clinical diagnoses were recorded: pneumonia (n = 1,056 [46.3%]), acute bacterial exacerbation of chronic bronchitis (n = 392 [17.2%]), chronic obstructive lung disease (n = 194 [8.5%]), acute sinusitis (n = 150 [6.6%]), acute otitis media (n = 85 [3.7%]), and other or unknown diagnosis (n = 402 [17.7%]).
Overall, 12.5% of the isolates were penicillin G intermediate (MIC = 0.12 to 1 μg/ml) and 12.1% were penicillin G resistant (MIC ≥ 2 μg/ml). However, there was considerable intercountry variation in these overall resistance rates (Table 1).
TABLE 1.
MIC50s, MIC90s, MIC ranges, and antibiotic resistance of 2,279 isolates of S. pneumoniae in eight European countriesa
| Country (no. of strains) | Antibiotic | MIC (μg/ml)
|
% Resistance (I + R) | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Austria (n = 160) | Penicillin G | 0.016 | 0.03 | 0.008-2 | 4.4 |
| Amoxicillin | 0.016 | 0.03 | 0.008-2 | 0 | |
| Cefotaxime | 0.03 | 0.03 | 0.03-1 | 0 | |
| Cefuroxime | 0.03 | 0.06 | 0.03-4 | 0.6 | |
| Cefpodoxime | 0.03 | 0.06 | 0.03-2 | 0.6 | |
| Clarithromycin | 0.125 | 0.25 | 0.125-≥32 | 10.0 | |
| Clindamycin | 0.125 | 0.125 | 0.125-≥32 | 4.4 | |
| Gatifloxacin | 0.25 | 0.25 | 0.125-0.5 | 0 | |
| Levofloxacin | 0.5 | 1 | 0.25-1 | 0 | |
| COT | 0.25/4.75 | 1/19 | 0.125/2.37-8/152 | 11.3 | |
| Tetracycline | 0.5 | 4 | 0.125-≥32 | 10.6 | |
| Chloramphenicol | 2 | 2 | 2-8 | 1.9 | |
| Belgium (n = 148) | Penicillin G | 0.016 | 0.125 | 0.008-2 | 11.5 |
| Amoxicillin | 0.016 | 0.03 | 0.008-2 | 0 | |
| Cefotaxime | 0.03 | 0.06 | 0.03-2 | 3.4 | |
| Cefuroxime | 0.03 | 0.25 | 0.03-8 | 8.8 | |
| Cefpodoxime | 0.03 | 0.06 | 0.03-4 | 6.8 | |
| Clarithromycin | 0.125 | ≥32 | 0.125-≥32 | 23.7 | |
| Clindamycin | 0.125 | ≥32 | 0.125-≥32 | 18.2 | |
| Gatifloxacin | 0.25 | 0.25 | 0.06-4 | 0.7 | |
| Levofloxacin | 0.5 | 1 | 0.125-≥32 | 0.7 | |
| COT | 0.25/4.75 | 4/76 | 0.125/2.37-16/304 | 18.9 | |
| Tetracycline | 0.5 | ≥32 | 0.125-≥32 | 23.7 | |
| Chloramphenicol | 2 | 2 | 2-16 | 2.7 | |
| France (n = 443) | Penicillin G | 0.06 | 2 | 0.008-4 | 47.6 |
| Amoxicillin | 0.03 | 2 | 0.008-8 | 3.6 | |
| Cefotaximeb | 0.06 | 2 | 0.016-4 | 11.1 | |
| Cefuroxime | 0.25 | 8 | 0.03-8 | 39.1 | |
| Cefpodoxime | 0.125 | 4 | 0.03-4 | 38.4 | |
| Clarithromycin | 0.125 | ≥32 | 0.125-≥32 | 46.1 | |
| Clindamycin | 0.125 | ≥32 | 0.125-≥32 | 44.2 | |
| Gatifloxacin | 0.25 | 0.25 | 0.125-4 | 0.9 | |
| Levofloxacin | 1 | 1 | 0.25-≥32 | 0.9 | |
| COT | 0.5/9.5 | 8/152 | 0.125/2.37-16/304 | 42.0 | |
| Tetracycline | 0.5 | ≥32 | 0.125-≥32 | 40.4 | |
| Chloramphenicol | 2 | 8 | 2-16 | 14.7 | |
| Germany (n = 530) | Penicillin G | 0.016 | 0.03 | 0.008-2 | 6.0 |
| Amoxicillin | 0.016 | 0.03 | 0.008-4 | 0.2 | |
| Cefotaxime | 0.03 | 0.03 | 0.016-2 | 0.6 | |
| Cefuroxime | 0.03 | 0.06 | 0.03-8 | 1.5 | |
| Cefpodoxime | 0.03 | 0.06 | 0.03-4 | 1.3 | |
| Clarithromycin | 0.125 | 1 | 0.125-≥32 | 10.6 | |
| Clindamycin | 0.125 | 0.125 | 0.06-≥32 | 5.1 | |
| Gatifloxacin | 0.25 | 0.25 | 0.06-4 | 0.2 | |
| Levofloxacin | 1 | 1 | 0.06-≥32 | 0.4 | |
| COT | 0.25/4.75 | 1/19 | 0.125/2.37-≥32/≥1,216 | 14.5 | |
| Tetracycline | 0.25 | 4 | 0.125-≥32 | 11.3 | |
| Chloramphenicol | 2 | 2 | 2-≥32 | 1.9 | |
| Italy (n = 462) | Penicillin | 0.016 | 0.25 | 0.008-4 | 13.0 |
| Amoxicillin | 0.016 | 0.06 | 0.008-4 | 0.2 | |
| Cefotaxime | 0.03 | 0.125 | 0.016-2 | 2.8 | |
| Cefuroxime | 0.03 | 0.5 | 0.03-8 | 7.4 | |
| Cefpodoxime | 0.03 | 0.25 | 0.03-4 | 8.0 | |
| Clarithromycin | 0.125 | ≥32 | 0.06-≥32 | 35.5 | |
| Clindamycin | 0.125 | ≥32 | 0.06-≥32 | 24.7 | |
| Gatifloxacin | 0.25 | 0.5 | 0.125-4 | 1.1 | |
| Levofloxacin | 1 | 1 | 0.125-64 | 1.3 | |
| COT | 0.5/9.5 | 4/76 | 0.125/2.37-≥32/≥1,216 | 41.1 | |
| Tetracycline | 0.5 | ≥32 | 0.125-≥32 | 23.8 | |
| Chloramphenicol | 2 | 2 | 2-≥32 | 6.7 | |
| Portugal (n = 174) | Penicillin G | 0.016 | 2 | 0.008-4 | 19.0 |
| Amoxicillin | 0.016 | 2 | 0.008-8 | 2.3 | |
| Cefotaxime | 0.03 | 1 | 0.016-4 | 6.9 | |
| Cefuroxime | 0.06 | 8 | 0.03-8 | 14.4 | |
| Cefpodoxime | 0.06 | 2 | 0.03-8 | 13.8 | |
| Clarithromycin | 0.125 | 1 | 0.125-≥32 | 10.3 | |
| Clindamycin | 0.125 | 0.125 | 0.125-≥32 | 8.6 | |
| Gatifloxacin | 0.25 | 0.25 | 0.06-8 | 1.1 | |
| Levofloxacin | 1 | 1 | 0.06-≥32 | 1.2 | |
| COT | 0.25/4.75 | 4/76 | 0.125/2.37-8/152 | 24.1 | |
| Tetracycline | 0.5 | 8 | 0.125-≥32 | 12.1 | |
| Chloramphenicol | 2 | 2 | 2-16 | 2.3 | |
| Spain (n = 310) | Penicillin G | 0.25 | 2 | 0.008-4 | 61.9 |
| Amoxicillin | 0.25 | 2 | 0.008-8 | 9.0 | |
| Cefotaximec | 0.25 | 2 | 0.016-4 | 11.0 | |
| Cefuroxime | 1 | 8 | 0.03-8 | 43.9 | |
| Cefpodoxime | 0.5 | 4 | 0.03-8 | 44.8 | |
| Clarithromycin | 0.125 | ≥32 | 0.125-≥32 | 43.6 | |
| Clindamycin | 0.125 | ≥32 | 0.125-≥32 | 39.4 | |
| Gatifloxacin | 0.25 | 0.5 | 0.125-4 | 0.7 | |
| Levofloxacin | 1 | 1 | 0.25-≥32 | 1 | |
| COT | 2/38 | 8/152 | 0.125/2.37-8/152 | 66.5 | |
| Tetracycline | 1 | ≥32 | 0.125-≥32 | 47.1 | |
| Chloramphenicol | 2 | 16 | 2-≥32 | 25.8 | |
| Switzerland (n = 52) | Penicillin G | 0.016 | 0.5 | 0.008-2 | 17.3 |
| Amoxicillin | 0.016 | 0.25 | 0.008-2 | 0 | |
| Cefotaxime | 0.03 | 0.5 | 0.03-2 | 3.9 | |
| Cefuroxime | 0.03 | 2 | 0.03-8 | 11.5 | |
| Cefpodoxime | 0.03 | 0.5 | 0.03-4 | 9.6 | |
| Clarithromycin | 0.125 | 0.25 | 0.125-≥32 | 17.3 | |
| Clindamycin | 0.125 | 0.125 | 0.125-≥32 | 11.5 | |
| Gatifloxacin | 0.25 | 0.25 | 0.125-0.5 | 0 | |
| Levofloxacin | 1 | 1 | 0.125-1 | 0 | |
| COT | 0.25/4.75 | 1/19 | 0.125/2.37-8/152 | 28.9 | |
| Tetracycline | 0.5 | 4 | 0.125-≥32 | 13.5 | |
| Chloramphenicol | 2 | 2 | 2 | 0 | |
| All strains (n = 2,279) | Penicillin G | 0.016 | 2 | 0.008-4 | 24.6 |
| Amoxicillin | 0.016 | 1 | 0.008-8 | 2.2 | |
| Cefotaximed | 0.03 | 1 | 0.016-4 | 5.1 | |
| Cefuroxime | 0.06 | 4 | 0.03-8 | 17.7 | |
| Cefpodoxime | 0.03 | 2 | 0.03-8 | 17.5 | |
| Clarithromycin | 0.125 | ≥32 | 0.125-≥32 | 28.0 | |
| Clindamycin | 0.125 | ≥32 | 0.06-≥32 | 22.6 | |
| Gatifloxacin | 0.25 | 0.25 | 0.06-8 | 0.7 | |
| Levofloxacin | 1 | 1 | 0.06-≥32 | 0.8 | |
| COT | 0.25/4.75 | 4/76 | 0.125/2.37-≥32/≥1,216 | 33.4 | |
| Tetracycline | 0.5 | ≥32 | 0.125-≥32 | 25.2 | |
| Chloramphenicol | 2 | 2 | 2-≥32 | 8.6 | |
Abbreviations: I, intermediate; R, resistant; COT, trimethoprim-sulfamethoxazole.
Ninety-seven strains showed an MIC of 1 μg/ml. Of the 97 strains, 1 was a CSF isolate and was classified as cefotaxime intermediate. Of 46 isolates with an MIC of 2 μg/ml, all were nonmeningitis strains and were classified as cefotaxime intermediate. Two strains exhibited an MIC of 4 μg/ml and were classified as cefotaxime resistant, so that in total, 49 strains (11.1%) were cefotaxime intermediate or cefotaxime resistant.
Seventy-six strains (all nonmeningitis isolates) showed an MIC of 1 μg/ml and were classified as cefotaxime susceptible. Of 28 isolates with an MIC of 2 μg/ml, all were nonmeningitis strains and were classified as cefotaxime intermediate. Six strains exhibited an MIC of 4 μg/ml and were classified as cefotaxime resistant, so that in total, 34 strains (11.0%) were cefotaxime intermediate or cefotaxime resistant.
Two hundred seven strains showed an MIC of 1 μg/ml. Of the 207 strains, 1 was a CSF isolate and was classified as cefotaxime intermediate. Of the 108 isolates with an MIC of 2 μg/ml, all were nonmeningitis strains and were classified as cefotaxime intermediate. Nine strains exhibited an MIC of 4 μg/ml and were classified as cefotaxime resistant, so that in total, 118 strains (5.1%) were cefotaxime intermediate or cefotaxime resistant.
A total of 853 pneumococcal strains showed resistance to either macrolides or penicillin G, or both (n = 27), and were chosen for pneumococcal serotyping. Serotypes 14 (18.4%), 23F (13.7%), 6B (13.7%), and 19F (12.5%) were the leading serotypes among the antibiotic-resistant strains (Table 2). The rates of antibiotic resistance varied within countries (Table 3).
TABLE 2.
Serotype distributions of 853 antibiotic-resistanta pneumococcal strains isolated in eight European countries
| Serotype | No. of isolates | % of isolates |
|---|---|---|
| 14 | 157 | 18.4 |
| 23F | 117 | 13.7 |
| 6B | 117 | 13.7 |
| 19F | 107 | 12.5 |
| 19A | 68 | 8.0 |
| 9V | 66 | 7.7 |
| Rough | 36 | 4.2 |
| 6A | 31 | 3.6 |
| 15A | 23 | 2.7 |
| 11A | 20 | 2.3 |
| 3 | 13 | 1.5 |
| 15B | 11 | 1.3 |
| 35B | 10 | 1.2 |
| 9A | 10 | 1.2 |
| 9N | 7 | 0.8 |
| 1 | 5 | 0.6 |
| 10F | 5 | 0.6 |
| 31 | 5 | 0.6 |
| 33F | 5 | 0.6 |
| 20 | 4 | 0.5 |
| 4 | 4 | 0.5 |
| Others | 32 | 3.8 |
| Total | 853 | 100.0 |
The selection of strains included all penicillin G- or macrolide-resistant isolates. Serotype data for strains showing reduced sensitivities to fluoroquinolones are presented in Table 7.
TABLE 3.
Antibiotic resistance of 2,279 isolates of S. pneumoniae in eight European countries, by center
| Country and city | % Resistance
|
||||||
|---|---|---|---|---|---|---|---|
| Penicillin G | Clarithromycin | Clindamycin | Levofloxacin | Trimethoprim- sulfamethoxazole | Tetracycline | Chloramphenicol | |
| Austria | |||||||
| Vienna | 9.6 | 5.8 | 1.9 | 0 | 7.7 | 7.7 | 3.9 |
| Linz | 0 | 12.5 | 0 | 0 | 50.0 | 25 | 12.5 |
| Innsbruck | 2 | 12 | 6 | 0 | 10 | 11 | 0 |
| Belgium | |||||||
| Leuven | 8.1 | 22.2 | 16.2 | 0 | 18.2 | 26.3 | 2.0 |
| Brussels | 18.4 | 26.5 | 22.5 | 2.0 | 20.4 | 18.4 | 4.1 |
| France | |||||||
| Le Chemay | 74.3 | 54.3 | 54.3 | 0 | 65.7 | 54.3 | 25.7 |
| Strasburg | 39.2 | 49.5 | 45.4 | 1.0 | 28.9 | 41.2 | 9.3 |
| Caen | 47.0 | 45.2 | 44.4 | 0 | 43.5 | 35.7 | 13.9 |
| Lyon | 31.6 | 28.6 | 27.6 | 0 | 25.5 | 27.6 | 7.1 |
| Creteil | 63.3 | 58.2 | 56.1 | 3.1 | 61.2 | 53.1 | 24.5 |
| Germany | |||||||
| Aachen | 6.7 | 13.3 | 6.7 | 0 | 8.3 | 13.3 | 3.3 |
| Leipzig | 2.3 | 18.6 | 4.7 | 0 | 23.3 | 4.7 | 0 |
| Kaiserslautern | 6.9 | 10.3 | 6.9 | 0 | 17.2 | 13.8 | 0 |
| Berlin | 8.7 | 11.6 | 8 | 0.7 | 17.4 | 15.2 | 2.9 |
| Jena | 5.7 | 6.7 | 2.9 | 0 | 17.2 | 15.2 | 1.9 |
| Weingarten | 4.5 | 9.0 | 3.2 | 0.7 | 9.7 | 5.2 | 1.3 |
| Italy | |||||||
| Genoa | 9.1 | 32.3 | 18.2 | 0 | 37.4 | 20.2 | 2.0 |
| Rome | 22.5 | 33.8 | 26.8 | 1.4 | 40.9 | 26.8 | 9.9 |
| Bari | 10.6 | 48.5 | 43.9 | 0 | 40.9 | 34.9 | 9.1 |
| Mailand | 13.6 | 28.4 | 24.7 | 1.2 | 29.6 | 27.2 | 9.9 |
| Padua | 16.3 | 20.4 | 18.4 | 4.1 | 28.6 | 20.4 | 4.1 |
| Catania (Sicily) | 9.4 | 44.8 | 19.8 | 2.1 | 61.5 | 16.7 | 6.3 |
| Portugal | |||||||
| Lisbon | 19 | 10.3 | 8.6 | 1.2 | 24.1 | 12.1 | 2.3 |
| Porto | 18.8 | 12.9 | 9.9 | 1.0 | 27.7 | 14.9 | 3.0 |
| Coimbra | 12.8 | 10.3 | 10.3 | 2.6 | 12.8 | 7.7 | 2.6 |
| Spain | |||||||
| Salamanca | 71.4 | 46.2 | 40.7 | 0 | 70.3 | 50.6 | 23.1 |
| Valencia | 41.2 | 21.6 | 17.7 | 2.0 | 41.2 | 27.5 | 9.8 |
| Madrid-1 | 60.6 | 49.5 | 45.5 | 2.0 | 70.7 | 51.5 | 26.3 |
| Madrid-2 | 66.7 | 47.8 | 44.9 | 0 | 73.9 | 50.7 | 40.6 |
| Switzerland | |||||||
| Basel | 18.4 | 18.4 | 13.2 | 0 | 29.0 | 15.8 | 0 |
| La Chaux-de Fonds | 14.3 | 14.3 | 7.1 | 0 | 28.6 | 7.1 | 0 |
In total, 618 pneumococcal strains showed macrolide resistance. The prevalence of macrolide resistance geno- and phenotypes varied greatly between countries. In Spain and France, where among the eight countries in this study the highest rates of macrolide resistance were reported, erm(B) was the leading genotype (Spain, 119 of 129 strains [92.3%]); France, 199 of 202 strains [98.5%]) (Table 4). By contrast, in Germany and Austria, where relatively low rates of macrolide resistance were found, the proportion of macrolide-resistant strains with efflux was relatively high (Germany, 26 of 56 [46.2%]; Austria, 5 of 12 [41.7%]). In addition, in Italy a relatively high rate of mef(A)-positive strains was also observed (25.0% of all macrolide-resistant strains). Six strains from Spain (n = 2), Germany (n = 2), Italy (n = 1), and Belgium (n = 1) which showed no PCR product in repeated assays for mef(A) or erm(B) were further analyzed for mutations in 23S rRNA and alterations in ribosomal proteins L4 and L22 (Table 5). One strain from Spain (strain PW 160), which exhibited a cMLSB phenotype, showed a 23S rRNA mutation at A2060 (pneumococcal numbering; A2058 by E. coli numbering). Interestingly, this strain was ST 156 and serotype 13, indicating a serotype switch from serotype 14 to serotype 13. Two isolates, a serotype 3 strain from Belgium (PW 1571) and a serotype 6A strain from Italy (PW 1905), showed alterations in the L4 ribosomal protein (Ser20Asn). A German strain (PW 555) with an inducible MLSB (iMLSB) resistance phenotype and a clarithromycin MIC of 2 μg/ml showed no alterations in ribosomal proteins L4 and L22 and only A2937G and G2939T mutations located outside the 3′end of 23S rRNA (internal transcribed spacer). Interestingly, strain PW 158 from Spain showed a combination of an efflux mechanism and a 23S rRNA mutation, explaining the discrepancy between the preliminary genotype [mef(A) positive] and phenotype (cMLSB).
TABLE 4.
Overview of macrolide resistance geno- and phenotypes of 618 pneumococcal strains isolated in eight European countries
| Country | No. of strains | % of strains | No. of isolates with the following resistance phenotype:
|
No. of isolates with the following resistance genotype
|
||||
|---|---|---|---|---|---|---|---|---|
| cMLSB | iMLSB | M | erm(B) | mef(A) | erm(B) mef(A) negative | |||
| Spain | 129 | 20.9 | 120 | 1 | 8 | 119 | 8 | 2 |
| Switzerland | 8 | 1.3 | 6 | 0 | 2 | 6 | 2 | 0 |
| Portugal | 18 | 2.9 | 15 | 1 | 2 | 16 | 2 | 0 |
| Austria | 12 | 1.9 | 7 | 0 | 5 | 7 | 5 | 0 |
| Italy | 160 | 25.9 | 112 | 7 | 41 | 119 | 40 | 1 |
| France | 202 | 32.7 | 188 | 11 | 3 | 199 | 3 | 0 |
| Germany | 56 | 9.1 | 27 | 1 | 28 | 28 | 26 | 2 |
| Belgium | 33 | 5.3 | 26 | 2 | 5 | 27 | 5 | 1 |
| Total | 618 | 100 | 501 | 23 | 94 | 521 | 91 | 6 |
TABLE 5.
Mutations in 23S rRNA and changes in ribosomal proteins L4 and L22 in macrolide-resistant pneumococcal strains isolated in eight European countriesg
| PW strain no. | Country | Age of patient (yr) | Source | CLA MIC (μg/ml) | Genotype | Phenotype | 23S rRNA mutation(s)c | L4 mutation | L22 mutation | Serotype | STa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 158 | Spain | 45 | Sputum | ≥32 | mef(A) | cMLSB | A138G, A260G, A2061G,d A1745T | WT | WT | 14 | 156 |
| 160 | Spain | 39 | Blood | ≥32 | Negativeb | cMLSB | A2060Gd | WT | WT | 13 | 156 |
| 169 | Spain | 47 | Sputum | 1 | Negativebe | M | T389C | WT | WT | 35B | 558 |
| 555 | Germany | 37 | Sputum | 2 | Negativebe | iMLSB | A2937G,f G2939Tf | WT | WT | 11A | 62 |
| 1571 | Belgium | 88 | Sputum | 16 | Negativebe | M | T389C, A138G | Ser20Asn | WT | 3 | 180 |
| 1905 | Italy | 70 | RT | 2 | Negativebe | M | T449C, T389C | Ser20Asn | WT | 6A | 1547 |
| 2216 | Germany | 47 | BAL | 8 | Negativebe | M | A138G, A373T, T389C, A1745T | WT | WT | 19F | 1585 |
The strains exhibited the following alleles and sequence types (aroE, gdh, gki, recP, spi, xpt, ddl, ST): PW 158, 7, 11, 10, 1, 6, 8, 1, ST 156; PW 160, 7, 11, 10, 1, 6, 8, 1, ST 156; PW 169, 18, 12, 4, 44, 14, 77, 97, ST 558; PW 555, 2, 5, 29, 12, 16, 3, 14, ST 62; PW 1571, 7, 15, 2, 10, 6, 1, 22, ST 180; PW 1905, 2, 13, 9, 1, 6, 19, 5, ST 1547 ST; PW 2216, 18, 5, 1, 1, 6, 4, 21, ST 1585.
In these strains, neither mef(A) nor erm(B) was detected in repeated PCRs.
Pneumococcal numbering.
Mutations in the erythromycin binding site of the 23S rRNA: A2060 (pneumococcal numbering; A2058, E. coli numbering) and A2061 (pneumococcal numbering; A2059, E. coli numbering).
Strains were negative for ermA, ermC, msrA, msrB, and msrD.
Mutations outside the 23S rRNA (see text).
Abbreviations: WT, wild type; CLA, clarithromycin; RT, respiratory tract; BAL bronchoalveolar lavage fluid.
Eighteen strains showed resistance to fluoroquinolones (defined as a levofloxacin MIC ≥ 4 μg/ml). Fluoroquinolone resistance rates (mean rate, 0.8%; 18 of 2,279 pneumococcal strains) in the eight participating countries were as follows: Austria, 0% (n = 160); Belgium, 0.7% (n = 148); France, 0.9% (n = 443); Germany, 0.4% (n = 530); Italy, 1.3% (n = 462); Portugal, 1.2% (n = 174); Spain, 1.0% (n = 310); and Switzerland, 0% (n = 52). The MICs and the fluoroquinolone resistance genotypes are presented in Table 6. Sixteen of the 18 strains showed high-level ciprofloxacin resistance (MICs ≥ 32 μg/ml). Among the fluoroquinolones tested, clinafloxacin (MIC90, 0.5 μg/ml; MIC range, 0.125 to 0.5 μg/ml) showed the greatest potency against the levofloxacin-nonsusceptible strains. Gatifloxacin (MIC90, 4 μg/ml; MIC range, 0.5 to 8 μg/ml) and moxifloxacin (MIC90, 4 μg/ml; MIC range, 0.25 to 4 μg/ml) were also potent against levofloxacin-nonsusceptible isolates. Infections caused by fluoroquinolone-resistant pneumococci occurred more often in older patients (mean age, 66 years versus a mean age of 55.5 years for patients from whom fluoroquinolone-susceptible isolates were recovered; P = 0.05). Ten of the 18 strains additionally showed reduced sensitivity to penicillin G; and 10 strains also showed MLSB resistance (multiply resistant isolates). Nine of the strains showed additional tetracycline resistance; and five strains had a combination of fluoroquinolone, penicillin G, MLSB, and tetracycline resistance. Two clonally related strains exhibited MLSB resistance in combination with tetracycline resistance (strains PW 2304 and PW 2305) (Table 6). Eight strains showed the classical Ser81 mutations (either Ser81Phe or Ser81Tyr) in gyrA. One strain (PW 904) showed a combination of Ser81Tyr and Ser114Gly alterations. A Glu85 alteration was found in two strains (PW 1698 and PW 1752). With the exception of one strain (PW 1601; Gly486Glu alteration) all isolates showed the wild type for gyrB. Thirteen of 18 strains showed the classical Ser79 alteration (either Ser79Phe or Ser79Tyr) in parC. The second most widespread parC alteration was Lys137Asn, found in 8 of the 18 isolates. This alteration was also seen in combination with Ser79Phe (n = 5). Nine of the 18 strains showed an Ile460Val parE alteration.
TABLE 6.
Characteristics of 18 pneumococcal strains with reduced susceptibility to fluoroquinolones isolated in Europe, 2001 and 2002b
| Strain no. | ST | Fluoroquinolone resistance genotype by alteration in:
|
MIC (μg/ml)
|
Antibiotic(s) to which additional resistance was detecteda | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | gyrB | parC | parE | GAT | CLI | GRE | SPA | MOX | CIP | LEV | |||
| PW 735 | 1558 | Ser81Phe | WT | Ser79Phe, Lys137Asn | Ile460Val | 4 | 0.5 | ≥32 | 32 | 2 | ≥32 | ≥32 | PEN, MLSB, T |
| PW 1698 | 315 | Glu85Lys | WT | Ser79Phe | Ile460Val | 4 | 0.5 | ≥32 | >32 | 4 | ≥32 | ≥32 | PEN, MLSB, T |
| PW 2304 | 66 | WT | WT | Ser79Phe, Lys137Asn | Ile460Val | 4 | 0.5 | ≥32 | >32 | 4 | >32 | ≥32 | MLSB, T |
| PW 2305 | 66 | WT | WT | Ser79Phe, Lys137Asn | Ile460Val | 4 | 0.5 | ≥32 | ≥32 | 4 | ≥32 | ≥32 | MLSB, T |
| PW 1443 | 513 | WT | WT | Ser79Phe | WT | 0.5 | 0.125 | 1 | 1 | 0.25 | 8 | 4 | |
| PW 239 | 156 | Ser81Phe | WT | Asp83Tyr, Lys137Asn | Ile460Val | 2 | 0.25 | 16 | 32 | 1 | ≥32 | 8 | PEN |
| PW 1752 | 156 | Glu85Gly | WT | Lys137Asn | Asp435Asn | 2 | 0.25 | 16 | 32 | 2 | ≥32 | ≥32 | PEN, MLSB |
| PW 603 | 416 | WT | WT | Ser79Phe | WT | 4 | 0.5 | >32 | >32 | 4 | ≥32 | ≥32 | |
| PW 802 | 1596 | WT | WT | Asp83Gly | Ile460Val | 1 | 0.25 | 4 | 8 | 0.5 | ≥32 | 4 | |
| PW 836 | 1596 | Ser81Tyr | WT | Asp83Gly | Ile460Val | 2 | 0.25 | 8 | 32 | 1 | ≥32 | 8 | |
| PW 1601 | 81 | WT | Gly486Glu | Ser79Phe, Lys137Asn | WT | 1 | 0.25 | 2 | 2 | 0.25 | ≥32 | 4 | PEN, T |
| PW 1891 | 1574 | Ser81Phe | WT | Ser79Tyr | WT | 4 | 0.5 | ≥32 | ≥32 | 4 | ≥32 | ≥32 | |
| PW 786 | 1570 | Ser81Phe | WT | Ser79Phe, Lys137Asn | Ile460Val | 4 | 0.5 | ≥32 | ≥32 | 4 | ≥32 | ≥32 | PEN, MLSB, T |
| PW 904 | 90 | Ser81Tyr | WT | Ser79Phe | Ile460Val | 4 | 0.25 | ≥32 | ≥32 | 2 | ≥32 | 16 | PEN, MLSB |
| Ser114Gly | Asn91Gly | ||||||||||||
| PW 931 | 1559 | WT | WT | Ser79Tyr | Ile460Val | 4 | 0.5 | ≥32 | ≥32 | 2 | ≥32 | ≥32 | PEN, MLSB, T |
| PW 1026 | 81 | WT | WT | Ser79Phe | WT | 8 | 0.5 | ≥32 | ≥32 | 4 | ≥32 | ≥32 | PEN, MLSB, T |
| PW 1872 | 1598 | Ser81Tyr | WT | Lys137Asn | Asp435Asn | 2 | 0.25 | 1 | 2 | 1 | 16 | 8 | PEN, MLSB, T |
| PW 2243 | 355 | Ser81Tyr | WT | Ser79Phe | WT | 4 | 0.5 | ≥32 | ≥32 | 2 | ≥32 | ≥32 | |
Resistance to the following additional antibiotics was observed: PEN, penicillin G nonsusceptibility (MIC ≥ 0.1 μg/ml); MLSB, resistance to macrolides, lincosamides, and streptogramin B; T, tetracycline resistance.
Abbreviations: GAT, gatifloxacin; CLI, clinafloxacin; GRE, grepafloxacin; SPA, sparfloxacin; MOX, moxifloxacin; CIP, ciprofloxacin; LEV, levofloxacin; WT, wild type.
Serotype, MLST, and demographic data are presented together in Table 7. Fluoroquinolone resistance was associated with pneumococcal serogroup 19 (19A, n = 5; 19F, n = 2). The clonal profile of fluoroquinolone-resistant S. pneumoniae strains in Europe was heterogeneous, and only a very few clones belonged to identical sequence types (ST 66, ST 156, and a new ST, 1-5variant-6-5-6-20-1). Of note, 7 of the 18 strains belonged to new sequence types not described in the MLST database before.
TABLE 7.
MLSTs and demographic data for 18 pneumococcal strains with reduced susceptibility to fluoroquinolones isolated in Europe, 2001 and 2002b
| Strain | Yra | Country | Specimen | Serotype | Age (yr) | Sex | MLST allele
|
ST | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | ||||||||
| PW 735 | 2001 | Spain | Sputum | 19A | 75 | M | 7 | 13 | 8 | 12 | 9 | 1 | 1 | 1558 |
| PW 1698 | 2002 | Spain | Sputum | 6B | 72 | M | 20 | 28 | 1 | 1 | 15 | 14 | 14 | 315 |
| PW 2304 | 2002 | Italy | Sputum | 9N | 62 | M | 2 | 8 | 2 | 4 | 6 | 1 | 1 | 66 |
| PW 2305 | 2002 | Italy | Sputum | 9N | 67 | M | 2 | 8 | 2 | 4 | 6 | 1 | 1 | 66 |
| PW 1443 | 2002 | Germany | Sputum | 11A | 73 | F | 2 | 5 | 29 | 12 | 6 | 3 | 14 | 513 |
| PW 239 | 2001 | Portugal | Blood | 14 | 68 | M | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 156 |
| PW 1752 | 2001 | France | Sputum | 14 | 67 | M | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 156 |
| PW 603 | 2001 | Italy | Blood | 19A | 65 | M | 1 | 13 | 14 | 4 | 17 | 51 | 14 | 416 |
| PW 802 | 2001 | Italy | BAL | 19A | 35 | F | 1 | 100 | 6 | 5 | 6 | 20 | 1 | 1596 |
| PW 836 | 2001 | Italy | Sputum | 19A | 37 | F | 1 | 100 | 6 | 5 | 6 | 20 | 1 | 1596 |
| PW 1601 | 2002 | Spain | Others | 19F | 95 | M | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 81 |
| PW 1891 | 2002 | Italy | Sputum | 19F | 42 | F | 8 | 14 | 4 | 12 | 9 | 1 | 14 | 1574 |
| PW 786 | 2001 | Belgium | Sputum | 1 | 70 | M | 8 | 11 | 10 | 1 | 9 | 8 | 1 | 1570 |
| PW 904 | 2001 | France | Sputum | 23F | 70 | M | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 90 |
| PW 931 | 2001 | France | Sputum | 19A | 62 | M | 7 | 19 | 2 | 17 | 9 | 22 | 14 | 1559 |
| PW 1026 | 2001 | Portugal | Sputum | 23F | 72 | F | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 81 |
| PW 1872 | 2002 | France | SA | 23F | 55 | F | 11 | 4 | 2 | 4 | 4 | 1 | 169 | 1598 |
| PW 2243 | 2003 | Germany | Sputum | 23F | 91 | F | 1 | 8 | 4 | 2 | 6 | 4 | 6 | 355 |
Year of isolation.
Abbreviations: SA, sinus aspirate; V, variant; BAL, bronchoalveolar lavage fluid; M, male; F, female.
Fluoroquinolone resistance has now reached some of the multiply antibiotic resistant pneumococcal clones described by the pneumococcal epidemiology network (31), such as the Poland6B-20, the serotype 14 variant of the penicillin resistant Spain9V-3, the multiply resistant Spain23F-1 clone, the 19F variant, and the multiply resistant Spain23F-1. Interestingly, S. pneumoniae PW 904 (sequence type 90) showed a serotype exchange from serotype 6B (Spain6B-2) to serotype 23F, and the present study is the first to describe a serotype 23F variant of this clone (Table 8).
TABLE 8.
Genetic relatedness of fluoroquinolone-resistant S. pneumoniae strains isolated in Europe
| Strain | ST | Countries where strains were found | Serotype(s) describeda | Other countries where strain was also reportedd | Time | Resistancef | Clonec |
|---|---|---|---|---|---|---|---|
| PW 1698 | 315 | Spain | 6B | Poland, Bulgaria, Italy, Sri Lanka | 1992-2001 | P, E, T | Poland6B-20 |
| PW 2304, PW 2305 | 66 | Italy | 9N | Sweden, United Kingdom, Australia, Brazil | 1993-2001 | E, T | |
| PW 1443 | 513 | Germany | 11A | Finland | 1995 | NDg | |
| PW 239, PW 1752 | 156 | Portugal, France | 14, 9V, 19F, 11A | Spain, United Kingdom, Denmark, Uruguay, Poland, France, The Netherlands, Czech Republic, Brazil, Israel | 1993-2003 | P, E | Serotype 14 variant of Spain9V-3 |
| PW 603 | 416 | Italy | 19A | England | 1999-2000 | T | |
| PW 1601 | 81 | Spain | 19F | Spain, Poland, throughout Asia | 1990-2001 | P, E, T | Multiresistant Spanish serotype 23F clone (Spain23F-1), serotype 19F variant |
| PW 904 | 90 | Portugal | 6B, 23Fb | Spain, The Netherlands, Iceland, USA, Australia | 1988-1999 | P, E, T | Multiresistant Spanish serotype 6B clone (Spain6B-2) |
| PW 1026 | 81 | Italy | 23F | Worldwide | 1984-2001 | P, E, T | Multiresistant Spanish serotype 23F clone (Spain23F-1) |
| PW 2243 | 355 | Germany | 23F | Germanye | 1999 | P |
The serotype observed in the present study is given in boldface; other information was obtained from the MLST database (www.mlst.net).
The present study is the first to describe the serotype 23F variant of the multiply resistant Spanish serotype 6B clone (Spain6B-2).
Clones as assigned by the pneumococcal molecular epidemiology network (31).
As available in the MLST database by December 2004.
Isolated from a 4-year-old child in Germany as part of a nationwide study (48).
P, penicillin G; E, erythromycin; T, tetracycline.
ND, not determined.
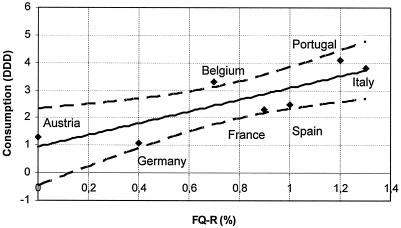
Analysis of a correlation of antibiotic consumption and fluoroquinolone resistance by linear regression showed that, with the exception of Belgium, all data were in the 95% confidence interval (r2 = 0.76) (Fig. 1). These findings confirm that the consumption of fluoroquinolones may drive the development of fluoroquinolone resistance in Europe.
FIG. 1.
Regression analysis of fluoroquinolone consumption and fluoroquinolone resistance in eight European countries. The analysis demonstrates a correlation between the resistance rate and the rate of consumption of fluoroquinolones (r2 = 0.76). The correlation is within the 95% confidence interval (dashed lines) for all countries except Belgium. FQ-R, level of fluoroquinolone resistance (in percent); consumption is defined as the DDD per 1,000 inhabitants; data were obtained from the European Study of Antibiotic Consumption (see the text).
DISCUSSION
Large-scale surveillance programs are required to look for trends in antimicrobial resistance and are essential in the establishment of evidence-based guidelines for the treatment of infections, such as community-acquired respiratory tract infections, where pneumococci are regularly found as the key pathogen. Overall, 24.6% of the isolates of S. pneumoniae from European centers examined in the present study exhibited reduced sensitivity to penicillin G, with a generally low prevalence in countries of Central and Western Europe (Germany, 6.0%; Austria, 4.4%) and particularly high rates in France (47.6%) and Spain (61.9%). Moderate levels of resistance were found in Belgium (11.5%), Portugal (19.0%), Switzerland (17.2%), and Italy (13.0%). This intercountry variability was also found in the PROTEKT 1999 to 2000 study (12), the Alexander Project (22, 50), and the SENTRY Antimicrobial Surveillance Program (13, 18). The prevalence of strains with reduced sensitivity to penicillin G in Italy and Switzerland has significantly increased compared to the prevalence in previous studies from these countries (29, 34, 60). It remains to be seen whether this accurately represents evolution to higher rates of resistance in these countries.
Macrolides are important alternatives to β-lactams for the treatment of lower respiratory tract infections involving S. pneumoniae. The level of macrolide resistance in S. pneumoniae is increasing worldwide but varies widely between countries, as confirmed by the present study. In S. pneumoniae macrolide resistance rates are highest in Asian countries, such as Korea and Japan (>80%). In Europe, Italy, and France, >40% of pneumococcal isolates are reported to be macrolide resistant (12). These large differences in resistance profiles may be explained in part by the strong association between increasing rates of macrolide usage, especially the longer-acting compounds, such as clarithromycin and azithromycin, and the development of resistance to this class of antimicrobials, as reported previously from studies in Spain and Germany (19, 46). The importance of ribosomal mutations in the development of macrolide resistance has only recently been recognized in pneumococci (6, 9, 10). As observed in the present study, the incidence of these strains is low (6 of 2,279) in Europe. Most information available on these resistance mechanisms today is based on in vitro selection studies showing that certain structures participate in the binding of macrolides involving domains V and II of 23S rRNA (6, 9, 10). In clinical isolates, most point mutations were identical to those found in in vitro selection studies, but new mutations were also observed. Point mutations at positions A2058, A2059, and A2062 (E. coli numbering; by pneumococcal numbering, positions A2060, A2061, and A2063) were associated with the development of macrolide resistance (27). The A2058G and A2058U (E. coli numbering; A2060 by pneumococcal numbering) substitutions confer the highest level of MLSB resistance, with MICs of erythromycin and related macrolides of between 32 and >200 μg/ml (6, 26, 27, 36, 43, 55, 58). The relevance of some mutations in the 23S rRNA for macrolide resistance development in S. pneumoniae, such as A138G, A373T, A260G, T389C, A449C, and A1745T, as described in the present study, needs further investigation. Alterations in the L22 and L4 proteins also play an increasing role in macrolide resistance in pneumococci. Mutations in the L4 protein occur in a region of 32 amino acids and interfere with the binding of the protein to rRNA. Nagai and coworkers (36) studied the rates of macrolide resistance among 992 isolates of S. pneumoniae from clinical specimens collected in 1999 and 2000 in Central and Eastern European countries. Among 180 erythromycin-resistant S. pneumoniae isolates, L4 protein mutations were seen in 28 strains (15.6%). Similar pulsed-field gel electrophoresis patterns suggested that some strains from the Slovak Republic, Bulgaria, and Latvia that contained L4 mutations were clonally related (36). Identical amino acid alterations were seen in one isolate from Finland and two strains from Russia (26, 43).
Fluoroquinolone-resistant S. pneumoniae strains are currently relatively rare in the eight European countries participating in the study, and the present surveillance reports resistance levels between ranging from 0% in Austria to 1.2% in Portugal. By contrast, in Hong Kong, Ireland, and some areas of Spain, fluoroquinolone resistance has reached levels of 17.8%, 15.2%, and 5%, respectively (14, 17, 21). The newer fluoroquinolones are now recommended for the treatment of community-acquired respiratory tract infections in many European countries (59), with a corresponding increase in drug use. In 2001, the highest levels of consumption of quinolones were observed in Portugal (4.1 defined daily doses [DDD]/1,000 inhabitants/day, Italy (3.8 DDD/1,000 inhabitants/day), Belgium (3.3 DDD/1,000 inhabitants/day), Spain (2.5 DDD/1,000 inhabitants/day), and France (2.3 DDD/1,000 inhabitants/day). Significantly lower levels of consumption were reported from Austria (1.3 DDD/1,000 inhabitants/day) and Germany (1.1 DDD/1,000 inhabitants/day) (data for Switzerland were not available). Despite generally low resistance levels, linear regression analysis showed a strong correlation between the rates of fluoroquinolone consumption and the rates of resistance (r2 = 0.76) (Fig. 1).
The clinical relevance of fluoroquinolone resistance has been proven by numerous studies (8, 42). Clearly, the spread of fluoroquinolone resistance determinants to international clones possessing the potential for worldwide spread is worrisome, and the present study documented that fluoroquinolone resistance was associated with these international clones (Poland6B-20, serotype 14 variant of Spain9V-3, serotype 19F variant of Spain23F-1, the newly described 23F variant of the Spain6B-2, and the Spain23F-1 clone) in one-third of cases. Other investigators have shown this association of fluoroquinolone resistance with international clones, such as the Spain23F-1 (20), Spain9V-3 (30), Tennessee23F-4 (5), Spain14-5 (41), and England14-9, Taiwan19F-14, Taiwan23F-15, and Maryland6B-17 (5) clones. Furthermore, 6 of the 18 fluoroquinolone-resistant strains showed multiple resistances (additional reduced sensitivity to penicillin G, MLSB resistance, and resistance to tetracycline). Therefore, further spread of these clones may be driven not only by an increase in the rate of fluoroquinolone consumption but also by an increased rate of use of β-lactams, macrolides, and tetracycline. In contrast to the close genetic relatedness observed in macrolide- and penicillin G-resistant pneumococcal clones throughout Europe (44), the fluoroquinolone-resistant clones observed in the present study were characterized by a relatively high genetic diversity.
In the present investigations, QRDR analysis showed that a combination of gyrA (Ser81) and parC (Ser81 and Ser79) mutations was the most widespread and responsible for high-level levofloxacin resistance. In total, 14 different alterations could be detected in gyrA (Ser81Phe, Ser81Tyr, Glu85Lys, Glu85Gly, Ser114Gly), gyrB (Gly486Glu), parC (Ser79Phe, Lys137Asn, Asp83Tyr, Asp83Gly, Ser79Tyr, Asn91Gly), and parE (Ile460Val, Asp435Asn). The parC Asp83 alteration has only recently been described among six isolates from the United States (4) and two strains from Italy (33) and may contribute to a slight increase in the levofloxacin MIC. When this alteration was seen in combination with a gyrA alteration (Ser81), the levofloxacin MIC increases to 8 μg/ml (strains PW 239 and PW 836). The parC Lys137Asn alteration, which was found to be widespread among European isolates, has also been observed among multidrug-resistant S. pneumoniae isolates in the international SENTRY study (23); but as it was also observed among levofloxacin-susceptible strains in Korea, the effect on fluoroquinolone resistance may be low (25). The parC Asn91Gly change has previously been found in S. pneumoniae and may be a consequence of the uptake of DNA from viridans group streptococci (3, 4). This change was detected in one isolate that showed five alterations in the QRDR and a serotype exchange from serotype 6B to serotype 23F, underscoring the possibility that this strain may be highly competent and capable of gene exchange with other streptococci. A single or double alteration in parC leads to a lower level of levofloxacin resistance, and some strains remained levofloxacin intermediate (levofloxacin MIC = 4 μg/ml) but susceptible to moxifloxacin (strains PW 1443, PW 802, and PW 1601). In contrast, three strains (strains PW 603, PW 931, and PW 1026) showed high-level levofloxacin and ciprofloxacin resistance in combination with a relatively high moxifloxacin MIC, even though they had only one alteration in parC and remained the wild type for gyrA. The latter may indicate the existence of other unknown mechanisms of fluoroquinolone resistance which require further investigation.
In summary, the present study showed that antibiotic resistance rates are highly variable in eight different Western European countries. Furthermore, the distribution of macrolide resistance phenotypes and genotypes varied between these countries. Presently, strains with 23S rRNA mutation or alterations in the ribosomal protein L4 do not play an important role in the spread of macrolide resistance in Western Europe. The spread of fluoroquinolone resistance to multiple-antibiotic-resistant clones is alarming, and further dissemination of these clones throughout Europe may be expected. Europe-wide surveillance to monitor the further spread of these antibiotic resistant S. pneumoniae strains is warranted.
Acknowledgments
We thank the PneumoWorld Study Group for cooperation and for providing the isolates. We thank Nelli Neuberger, Maria Lemperle, and Claudia Cremer for excellent technical assistance.
The study was supported in part by grant RKI-415/1369235 from the German Ministry of Health (Bundesminister für Gesundheit) and by a grant of Grünenthal GmbH, Aachen, Germany.
The members of the PneumoWorld Study Group are H. Adler, Kanonsspital Bakteriologielabor, Basel, Switzerland; F. Allerberger, Bundesstatliche Bakteriologische Serologische Untersuchungsanstalt, Innsbruck, Austria; P. Allouch, C. H. de Versailles, Le Chesmay, France; J. M. Amorium, Hospital Geral Santo António, Porto, Portugal; M. L. A. Boaventura, Servico de Patologia Clinica Microbiologia Hospital da Universidade de Coimbra, Coimbra, Portugal, J. J. P. de la Garza, Hospital Clinico San Carlos, Madrid, Spain; J. Etienne, Hóspital Edouard Herriot, Lyon Cedex, France; G. Fadda, Laboratorio di Microbiologia, Policlinico Gemelli, Rom, Italy; G. Funke, Department of Medical Microbiology, Gärtner and Colleagues Laboratories, Weingarten, Germany; J. Garau, Hospital Mútua de Terrassa, Terrassa, Spain; A. Georgopoulus, Microbiology Research Laboratory, Vienna, Austria; G. Gesu, Laboratorio di Microbiologia, Ospendale S. Raffaele, Milan, Italy; M. Gobernado, Hospital la Fe, Valencia, Spain; R. Leclercq, CHU-Hóspital Cóte de Nacre, France; H. Mauch; Institut für Mikro-, Immuno-, und Laboratoriumsmedizin, Berlin, Germany; J. A. G. Melo Christino, Facultad de Medicina Universidade de Lisboa, Lisbon, Portugal; G. Miragliotta, Instituto di Microbiologia, Universitá degli Studi, Bari, Italy; H. Mittermayer, Institut für Hygiene und Mikrobiologie, Linz, Austria; H. Monteil, Ulp-Université Louis Pasteur Strasbourg, Strasbourg, France; G. Nicoletti Dipartimento Scienze Microbiologiche e Ginecologiche, Universitá degli Studi di Catania, Catania, Italy; G. Palu, Laboratorio di Microbiologia, Azienda Ospedaliera, Padua, Italy; W. Pfister, Institut für Med. Mikrobiologie, Jena, Germany; J. P. Prieto, Servicio de Microbiologia, Universidad Complutense, Madrid, Spain; A. C. Rodloff, Institut für Medizinische Mikrobiologie, Leipzig, Germany; J. A. G. Rodrigues, Hospital Clinico de Salamanca, Salamanca, Spain; G. C. Schito, Institute of Microbiology, University of Genoa Medical School, Genova, Italy; S. Schmitt, Medical Laboratory, Kaiserslautern, Germany; H. H. Siegrist, Institut Neuchatelois de Microbiologie, La Chaux-de-Fonds, Switzerland; C.-J. Soussy, Service de Bactériologie-Virologie-Hygiéne CHU-Henri Mondor, Créteil Cedex, France; M. Streulens, Hópital Erasme, Brussels, Belgium; and J. Verhaegen, Universitaire Ziekenhuizen K. U. Leuven, Leuven, Belgium.
REFERENCES
- 1.Appelbaum, P. C. 2002. Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34:1613-1620. [DOI] [PubMed] [Google Scholar]
- 2.Arbique, J. C., C. Poyart, P. Trieu-Cuot, G. Quesne, G. Carvalho Mda, A. G. Steigerwalt, R. E. Morey, D. Jackson, R. J. Davidson, and R. R. Facklam. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42:4686-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, L., M. J. Ferrandiz, J. Linares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, S. D., D. J. Farrell, and I. Morrissey. 2004. Prevalence and molecular analysis of macrolide and fluoroquinolone resistance among isolates of Streptococcus pneumoniae collected during the 2000-2001 PROTEKT US Study. J. Clin. Microbiol. 42:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 6.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daly, M. M., S. Doktor, R. Flamm, and D. Shortridge. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J. Clin. Microbiol. 42:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, R., R. Cavalcanti, J. L. Brunton, D. J. Bast, J. C. de Azavedo, P. Kibsey, C. Fleming, and D. E. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 9.Davies, T. A., B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro development of resistance to telithromycin (HMR 3647), four macrolides, clindamycin, and pristinamycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50 (Suppl. S1):25-37. [DOI] [PubMed] [Google Scholar]
- 13.Fluit, A. C., M. E. Jones, F. J. Schmitz, J. Acar, R. Gupta, and J. Verhoef. 2000. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin. Infect. Dis. 30:454-460. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Rey, C., L. Aquilar, and F. Baquero. 2000. Influences of different factors on prevalence of ciprofloxacin resistance in Streptococcus pneumoniae in Spain. Antimicrob. Agents Chemother. 44:3481-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardam, M. A., and M. A. Miller. 1998. Optochin revisited: defining the optimal type of blood agar for presumptive identification of Streptococcus pneumoniae. J. Clin. Microbiol. 36:833-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill, M. J., N. P. Brenwald, and R. Wise. 1999. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldsmith, C. E., J. E. Moore, P. G. Murphy, and J. E. Ambler. 1998. Increased incidence of ciprofloxacin resistance in penicillin-resistant pneumococci in Northern Ireland. J. Antimicrob. Chemother. 41:420-421. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, K. A., D. J. Biedenbach, and R. N. Jones. 2003. Comparison of Streptococcus pneumoniae and Haemophilus influenzae susceptibilities from community-acquired respiratory tract infections and hospitalized patients with pneumonia: five-year results for the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 46:285-289. [DOI] [PubMed] [Google Scholar]
- 19.Granizo, J. J., L. Aguilar, J. Casal, C. Garcia-Rey, R. Dal-Re, and F. Baquero. 2000. Streptococcus pneumoniae resistance to erythromycin and penicillin in relation to macrolide and beta-lactam consumption in Spain (1979-1997). J. Antimicrob. Chemother. 46:767-773. [DOI] [PubMed] [Google Scholar]
- 20.Ho, P. L., W. C. Yam, T. K. Cheung, W. W. Ng, T. L. Que, D. N. Tsang, T. K. Ng, and W. H. Seto. 2001. Fluoroquinolone resistance among Streptococcus pneumoniae in Hong Kong linked to the Spanish 23F clone. Emerg. Infect. Dis. 7:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, R. N. Gruneberg, and The Alexander Project. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D., M. Stilwell, M. Toleman, and R. Jones. 2004. Emergence and epidemic clonality of multidrug resistant (MDR) S. pneumoniae (SPN; 1997-2003): report from the SENTRY antimicrobial surveillance program. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-830, p. 100. American Society for Microbiology, Washington, D.C.
- 24.Jones, M. E., D. F. Sahm, N. Martin, S. Scheuring, P. Heisig, C. Thornsberry, K. Kohrer, and F. J. Schmitz. 2000. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997-1998 respiratory season. Antimicrob. Agents Chemother. 44:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, S., Y. Kwak, M. Lee, N. Kim, J. Jeong, and Y. Kim. 2004. Prevalence of parC mutation and efflux phenotypes among levofloxacin-susceptible clinical strains of S. pneumoniae isolated in Korea. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1729. American Society for Microbiology, Washington, D.C.
- 26.Kozlov, R. S., T. M. Bogdanovitch, P. C. Appelbaum, L. Ednie, L. S. Stratchounski, M. R. Jacobs, and B. Bozdogan. 2002. Antistreptococcal activity of telithromycin compared with seven other drugs in relation to macrolide resistance mechanisms in Russia. Antimicrob. Agents Chemother. 46:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchese, A., E. A. Debbia, A. Arvigo, A. Pesce, and G. C. Schito. 1995. Susceptibility of Streptococcus pneumoniae strains isolated in Italy to penicillin and ten other antibiotics. J. Antimicrob. Chemother. 36:833-837. [DOI] [PubMed] [Google Scholar]
- 30.McGee, L., C. E. Goldsmith, and K. P. Klugman. 2002. Fluoroquinolone resistance among clinical isolates of Streptococcus pneumoniae belonging to international multiresistant clones. J. Antimicrob. Chemother. 49:173-176. [DOI] [PubMed] [Google Scholar]
- 31.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montanari, M. P., M. Mingoia, E. Giovanetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montanari, M. P., E. Tili, I. Cochetti, M. Mingoia, A. Manzin, and P. E. Varaldo. 2004. Molecular characterization of clinical Streptococcus pneumoniae isolates with reduced susceptibility to fluoroquinolones emerging in Italy. Microb. Drug Resist. 10:209-217. [DOI] [PubMed] [Google Scholar]
- 34.Mühlemann, K., H. C. Matter, M. G. Tauber, and T. Bodmer. 2003. Nationwide surveillance of nasopharyngeal Streptococcus pneumoniae isolates from children with respiratory infection, Switzerland, 1998-1999. J. Infect. Dis. 187:589-596. [DOI] [PubMed] [Google Scholar]
- 35.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801-807. [DOI] [PubMed] [Google Scholar]
- 36.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai, K., T. A. Davies, G. A. Pankuch, B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro selection of resistance to clinafloxacin, ciprofloxacin, and trovafloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2740-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. Supplement M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 39.Neufeld, F. 1902. Über die Agglutination der Pneumokokken und über die Theorie der Agglutination. Z. Hyg. Infektionskrankh. 34:54-72. [Google Scholar]
- 40.Pan, X. S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Trallero, E., J. M. Marimon, A. Gonzalez, and L. Iglesias. 2003. Spain14-5 international multiresistant Streptococcus pneumoniae clone resistant to fluoroquinolones and other families of antibiotics. J. Antimicrob. Chemother. 51:715-719. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Trallero, E., J. M. Marimon, L. Iglesias, and J. Larruskain. 2003. Fluoroquinolone and macrolide treatment failure in pneumococcal pneumonia and selection of multidrug-resistant isolates. Emerg. Infect. Dis. 9:1159-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pihlajamaki, M., J. Kataja, H. Seppala, J. Elliot, M. Leinonen, P. Huovinen, and J. Jalava. 2002. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 46:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinert, R., A. Ringelstein, M. van der Linden, M. Cil, A. Al-Lahham, and F. Schmitz. 2005. Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 43:1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinert, R. R. 2004. Clinical efficacy of ketolides in the treatment of respiratory tract infections. J. Antimicrob. Chemother. 53:918-927. [DOI] [PubMed] [Google Scholar]
- 46.Reinert, R. R., A. Al-Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lütticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 49:61-68. [DOI] [PubMed] [Google Scholar]
- 47.Reinert, R. R., A. Al-Lahham, R. Lütticken, M. Boos, and F. J. Schmitz. 2002. Characterization of clinical Streptococcus pneumoniae strains from Germany with decreased susceptibility to fluoroquinolones. J. Antimicrob. Chemother. 49:1015-1018. [DOI] [PubMed] [Google Scholar]
- 48.Reinert, R. R., S. Simic, A. Al-Lahham, S. Reinert, M. Lemperle, and R. Lütticken. 2001. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients with respiratory tract infections in Germany from 1998 to 1999: results of a national surveillance study. J. Clin. Microbiol. 39:1187-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinert, R. R., A. Wild, P. Appelbaum, R. Lütticken, M. Y. Cil, and A. Al-Lahham. 2003. Ribosomal mutations conferring resistance to macrolides in Streptococcus pneumoniae clinical strains isolated in Germany. Antimicrob. Agents Chemother. 47:2319-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schito, G. C., E. A. Debbia, and A. Marchese. 2000. The evolving threat of antibiotic resistance in Europe: new data from the Alexander Project. J. Antimicrob. Chemother. 46(Suppl. T. 1):3-9. [DOI] [PubMed] [Google Scholar]
- 51.Shortridge, V. D., R. K. Flamm, N. Ramer, J. Beyer, and S. K. Tanaka. 1996. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 26:73-78. [DOI] [PubMed] [Google Scholar]
- 52.Sutcliffe, J., A. Tait Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. MefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 57.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel, F., and H. Scholz. 2002. PEG-Empfehlungen: Rationaler Einsatz oraler Antibiotika bei Erwachsenen. Chemother. J. 2:47-58. [Google Scholar]
- 60.Wüst, J., E. Huf, and F. H. Kayser. 1995. Antimicrobial susceptibilities and serotypes of invasive Streptococcus pneumoniae strains in Switzerland. J. Clin. Microbiol. 33:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]