Abstract
Purified QnrA blocked ciprofloxacin inhibition of topoisomerase IV, just as QnrA was previously found to prevent quinolone inhibition of DNA gyrase. With a gel displacement assay, tagged QnrA was shown to bind to topoisomerase IV and its subunits in a reaction that did not depend on the presence of DNA, quinolone, or ATP.
The plasmid-encoded quinolone resistance protein QnrA (formerly Qnr) has been shown to protect purified Escherichia coli DNA gyrase from quinolone inhibition (4) and to bind to both the gyrase holoenzyme and its subunits (5). Protection of the second quinolone target, topoisomerase (topo) IV, however, could not be established with an assay involving relaxation of supercoiled plasmid pBR322 DNA (4).
With a more sensitive assay, QnrA protection of purified E. coli topo IV from quinolone inhibition has been demonstrated. The mechanism of this effect has been studied with a gel displacement assay. As with DNA gyrase, QnrA binds to both the topoisomerase holoenzyme and its ParC and ParE subunits.
E. coli strains BL21(DE3) and DH5α (Invitrogen, Carlsbad, CA), plasmid pQE-100 Double Tag (QIAGEN, Valencia, CA)-qnr with IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible QnrA synthesized with N-terminal His6 and C-terminal Tag-100 epitopes, and the media and growth conditions used have been described previously (5).
The parC and parE genes were amplified by PCR from genomic DNA isolated from E. coli KL16 using primers 5′-CATGGATCCAGCGATATGGCAGAG and 5′-CATGGAATTCTTACTCTTCGCTATC for parC and 5′-CATGCTGCAGGACGCAAACTTATAAC and 5′-CATGAAGCTTGATCATCGCGTCAGTAC for parE. The parC PCR product was digested with BamHI and EcoRI restriction enzymes and cloned into expression vector pTrcHisA (Ampr) (Invitrogen), which added N-terminal His6 and Xpress epitopes that were used to facilitate isolation and detection of the translated fusion proteins. The parE PCR product was digested with PstI and HindIII restriction enzymes and cloned into the same pTrcHisA vector. The correct in-frame placement of parC and parE in the pTrcHisA-parC and pTrcHisA-parE recombinant plasmids was confirmed by DNA sequencing (Tufts University Core Facility).
The pTrcHisA-parC and pTrcHisA-parE plasmids were introduced into E. coli BL21(DE3) by electroporation, and the pQE-100 Double Tag-qnrA plasmid was introduced into E. coli DH5α, with selection for transformants in each case on Trypticase soy agar plates containing 100 μg/ml ampicillin. Induction of protein expression with 0.5 mM IPTG (for the topo IV subunits) or 1 mM IPTG (for QnrA) and protein isolation using a HiTrap chelating HP column (Amersham Pharmacia Biotech, Piscataway, NJ) were accomplished as previously described for the gyrase subunits (5).
The topo IV assay for decatenation of kinetoplast DNA (TopoGEN, Inc., Columbus, OH) was performed as described previously (3), except that 1 mM ATP was used and the reactions were incubated for 2 h at 25°C to maximize interactions between topo IV or its subunits and QnrA. The products were analyzed by electrophoresis in 1.2% agarose with staining by ethidium bromide. The topo IV holoenzyme was reconstituted by incubating the ParC and ParE subunits together in reaction buffer containing 300 ng of kinetoplast DNA per 40 μl. Purified QnrA was added, followed by 18 μM ciprofloxacin and 1 mM ATP.
Protein analysis was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Ready Gel precast gels (Bio-Rad, Hercules, CA). After electrophoresis, proteins were electrically transferred onto a nitrocellulose membrane, which was incubated in blocking buffer and then with antibody to the Xpress or Tag-100 epitopes (5). After washing, the bound antibody was localized by incubation with goat anti-mouse IgG1 antibody conjugated to horseradish peroxidase (Southern Biotechnology Associates, Birmingham, AL). The peroxidase was localized by color production in a solution containing 0.4 mg/ml 3-amino-9-ethyl carbazole (Sigma, St. Louis, MO), 50 mM sodium acetate, pH 5.0, and 0.015% hydrogen peroxide.
A standard topoisomerase assay was performed as described above, with the following modifications. The topoisomerase subunit(s) was added to a reaction mixture containing 1.5 μg/80 μl supercoiled pBR322 DNA (New England Biolabs, Beverly, MA), followed by 0.4 μM QnrA, 18 μM ciprofloxacin, and 1 mM 5′-adenylyl-β,γ-imidodiphosphate (ADPNP) (Sigma). ADPNP, a nonhydrolyzable ATP analog, was used to stabilize ParE dimers and to maximize potential interactions between QnrA and the enzyme/DNA/ciprofloxacin complex. The amount of ParC and ParE subunits and QnrA added has been specified for each reaction in the figure legends. The reaction mix was incubated at 25°C for 40 min. Half of the reaction was used for protein localization with Coomassie blue staining, and half was used for Western blotting to detect the Tag-100 epitope. All samples were run under native conditions (without SDS or 2-mercaptoethanol) to preserve any protein interactions. Transfer was carried out overnight at 4°C at a constant voltage of 7 V, using transfer buffer containing 51 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol to enhance transfer of large protein complexes. QnrA detection with antibody to Tag-100 was performed as described above.
The ParC and ParE subunits of E. coli topo IV were purified and demonstrated to be homogeneous. They were combined to reconstitute the holoenzyme. QnrA was also purified to homogeneity as a doubly tagged product with an N-terminal His6 for purification and a C-terminal Tag-100 epitope for detection.
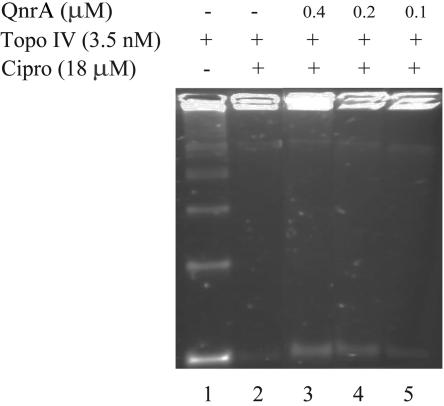
In an assay involving decatenation of kinetoplast DNA, topo IV inhibition by 18 μM ciprofloxacin was blocked by as little as 0.1 μM QnrA (Fig. 1).
FIG. 1.
Tag-100-QnrA-His6 fusion protein protects topo IV from quinolone inhibition of decatenation. Reactions of 40 μl were analyzed by agarose gel electrophoresis. Reaction mixtures contained 0.5 μg catenated kinetoplast DNA (lanes 1 to 5), 3.5 nM topo IV (lanes 1 to 5), 18 μM ciprofloxacin (lanes 3 to 5), and purified QnrA at 0.4 μM (lane 3), 0.2 μM (lane 4), or 0.1 μM (lane 5).
Physical interaction between purified QnrA and topo IV was studied by a nondenaturing gel retardation assay and Western blotting. The reaction mixture from a standard topo IV assay was analyzed by PAGE under nondenaturing conditions that preserved protein-protein interactions. After electrophoresis, proteins were transferred to a membrane that was probed with specific antibody to detect comigration of the His6-QnrA-Tag-100 fusion protein with the larger topoisomerase holoenzyme or its subunits.
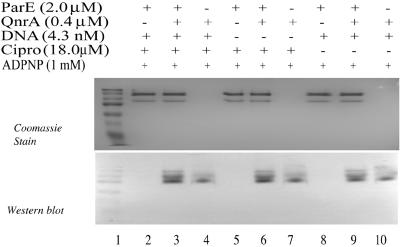
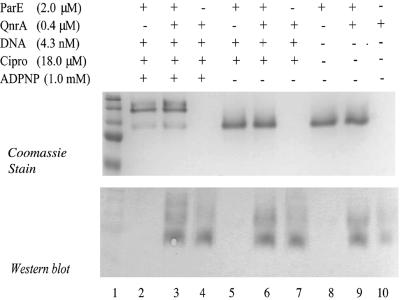
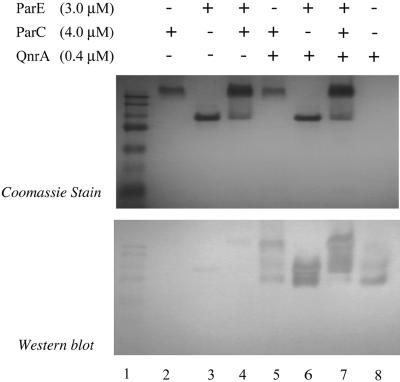
Figure 2 shows the association of QnrA with ParE after incubation with DNA, ciprofloxacin, and ADPNP. Omission of DNA, ciprofloxacin, ADPNP, or all three components (Fig. 2 and 3) did not prevent this association. Similarly, QnrA interacted with the ParC subunit in the absence of the reaction components (Fig. 4) and with the topo IV holoenzyme as well (Fig. 4). We have shown previously that QnrA does not bind to such unrelated proteins as carbonic anhydrase or myosin (5), and it thus appears to have specificity for type 2 topoisomerases.
FIG. 2.
Interaction of QnrA and ParE subunit does not depend on supercoiled plasmid DNA or ciprofloxacin. Reactions of 80 μl (40 μl per gel) were analyzed by native gel electrophoresis (A) and by Western blotting with Tag-100 antibody (QIAGEN) (B). Reaction mixtures contained 2 μM ParE subunit, 0.4 μM QnrA, 4.3 nM supercoiled pBR322 plasmid DNA, 18 μM ciprofloxacin, and 1 mM ADPNP where indicated in the figure. Prestained broad-range molecular weight markers (Bio-Rad) were run in lane 1 to allow comparison of the two gels and not for molecular sizing.
FIG. 3.
Interaction of QnrA and DNA ParE subunit does not depend on relaxed plasmid DNA, ciprofloxacin, and ADPNP. Reactions of 80 μl (40 μl per gel) were analyzed by native gel electrophoresis (A) and by Western blotting with Tag-100 antibody (B). Reaction mixtures contained 2 μM ParE subunit, 4.3 nM supercoiled pBR322 plasmid DNA, 18 μM ciprofloxacin, and 1 mM ADPNP where indicated in the figure. Prestained broad-range molecular weight markers were run in lane 1 to allow comparison of the two gels and not for molecular sizing.
FIG. 4.
Interaction of QnrA and topo IV and its subunits. Reactions of 80 μl (40 μl per gel) were analyzed by native gel electrophoresis (A) and by Western blotting with Tag-100 antibody (B). Reaction mixtures contained 3 μM ParE (lanes 3, 4, 6, and 7), 4 μM ParC (lanes 2, 4, 5, and 7), and 0.4 μM QnrA (lanes 5 to 8). Prestained broad-range molecular weight markers were run in lane 1 to allow comparison of the two gels and not for molecular sizing.
Using a sensitive assay procedure, protection of topo IV by QnrA has been detected. DNA gyrase and topo IV have somewhat different functions in the cell (1), but both are heterodimers, with the GyrA and ParC subunits of E. coli having 29% of their amino acids in common while the GyrB and ParE subunits share a similar percentage identity. Hence, it is not surprising that QnrA can protect both quinolone targets. That this would be the case was suggested by the ability of QnrA to augment the resistance of quinolone-resistant gyrA mutants (2). Also, if QnrA did not protect both topoisomerases, exposure of a cell making QnrA to a quinolone concentration above its MIC would tend to select mutants in the unprotected and hence more susceptible target, something that has not been observed (unpublished observations). Even though E. coli topo IV is less sensitive than gyrase to quinolone inhibition, so that a higher concentration of ciprofloxacin was required to produce inhibition in vitro, a comparable concentration of QnrA was protective with both targets.
By a gel displacement assay, QnrA has been shown to bind to topo IV and its subunits. The binding did not depend on the presence of DNA, ciprofloxacin, or ATP. Hence, like the binding of QnrA to DNA gyrase and its subunits (5), the formation of a QnrA-topo IV complex probably occurs before the formation of the cleavage complex. Whether QnrA binding blocks access of quinolone to the enzyme, reduces the number of targets for quinolone inhibition, or has some other effect on enzyme function is currently under study.
Acknowledgments
This study was supported in part by grants AI43312 (to G.A.J.) and AI57576 (to D.C.H.) from the National Institutes of Health, U.S. Public Health Service.
REFERENCES
- 1.Drlica, K., and D. C. Hooper. 2003. Mechanisms of quinolone action, p. 19-40. In D. C. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents, 3rd ed. ASM Press, Washington, D.C.
- 2.Martínez-Martínez, L., A. Pascual, I. García, J. Tran, and G. A. Jacoby. 2003. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51:1037-1039. [DOI] [PubMed] [Google Scholar]
- 3.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 4.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]