Abstract
Streptococcus agalactiae UCN36 was resistant to lincomycin (MIC = 16 μg/ml) but susceptible to clindamycin (MIC = 0.12 μg/ml) and erythromycin (MIC = 0.06 μg/ml). A 4-kb HindIII fragment was cloned from S. agalactiae UCN36 total DNA on plasmid pUC18 and introduced into Escherichia coli AG100A, where it conferred resistance to lincomycin. The sequence analysis of the fragment showed the presence of a 1,724-bp element delineated by imperfect inverted repeats (22 of 25 bp) and inserted in the operon for capsular synthesis of S. agalactiae UCN36. This element carried two open reading frames (ORF). The deduced amino acid sequence of the upstream ORF displayed similarity with transposases from anaerobes and IS1. The downstream ORF, lnu(C), encoded a 164-amino-acid protein with 26% to 27% identity with the LnuAN2, LnuA, and LnuA′ lincosamide nucleotidyltransferases reported for Bacteroides and Staphylococcus, respectively. Crude lysates of E. coli AG100A containing the cloned lnu(C) gene inactivated lincomycin and clindamycin in the presence of ATP and MgCl2. Mass spectrometry experiments demonstrated that the LnuC enzyme catalyzed adenylylation of lincomycin.
Streptococcus agalactiae (group B streptococcus) is a major cause of invasive infection in neonates and pregnant women. It has also been increasingly recognized as a significant pathogen in nonpregnant adults, especially among patients with underlying conditions (13, 26). S. agalactiae is responsible for acute and chronic diseases, such as respiratory tract infections, endocarditis, sepsis, meningitis, pyelonephritis, and neurological problems (12). Penicillin G and ampicillin, which are always active against this pathogen, are the therapy of choice for S. agalactiae infections. However, in the case of intolerance to penicillins or lack of clinical response, clindamycin and macrolides are major alternatives (2).
Lincosamide antibiotics include lincomycin and clindamycin. Their structure consists of a hygric acid alkylated in position 4 and linked to a 6 amino-thio-octopyranoside residue. Clindamycin is a semisynthetic derivative obtained by chlorination of lincomycin (4). The spectrum of activity of lincosamides include gram-positive cocci, with some exceptions, such as Enterococcus faecalis (25). They prevent the protein synthesis by inhibition of the peptidyltransferase in binding mainly the A2058 of the 23S rRNA in the 50S subunit of the bacterial ribosome (9).
The most common mechanism of resistance to lincosamides involves N6 dimethylation of a specific adenine residue (A2058) of the 23S rRNA molecule (17, 28). This alteration of the antibiotic target site is invariably catalyzed by an rRNA methyltransferase encoded by erm genes. This resistance mechanism confers cross-resistance to macrolides, lincosamides, and streptogramin B (MLS phenotype) (17, 28). In contrast to the MLS phenotype, specific resistance to lincosamides is due to enzymatic inactivation of those antibiotics. Phosphorylation and nucleotidylation of the hydroxyl group at position 3 of lincosamides have been detected in several species of Streptomyces (1, 21). Lincosamide nucleotidyltransferases encoded by lnu genes (formerly lin) was observed in both animal and human strains (8, 10, 11, 18). In clinical isolates, six lnu genes have been described: lnu(A), lnu(A′), lnu(B), lnu(B-like), lnu(AN2), and linF (3, 4, 5, 15, 27). The O-nucleotidyltransferases encoded by these genes inactivate lincosamides by adenylylation (3, 5). lnu(A) and lnu(A′) have been reported in Staphylococcus haemolyticus and Staphylococcus aureus, respectively (4, 5). They encode two isoenzymes of 161 amino acids differing by 14 amino acids. An lnu(AN2) gene homologous to lnu(A) and lnu(A′) (55% of identity) was evidenced in Bacteroides spp. (27). This gene would be carried by a mobilizable transposon. The lnu(B) gene from Enterococcus faecium has been described (3). This gene does not display homology with the other lnu genes and is carried by a large conjugative plasmid. More recently, an lnu(B-like) gene [79% identity with lnu(B)] and an linF gene [34.9% identity with lnu(B)] were identified in Eubacterium and Escherichia coli, respectively (15). In E. coli, the linF gene confers cross-resistance to lincomycin and clindamycin, whereas in the other organisms, the lnu genes confer resistance to lincomycin but not to clindamycin. However, the bactericidal activity of clindamycin, which is already weak against susceptible strains, is totally abolished (18). In this study, we characterized the function and genetic support of a new lnu(C) gene that confers resistance to lincomycin in a clinical strain of S. agalactiae UCN36.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. agalactiae UCN36 was isolated in our laboratory from the vaginal tract of a pregnant woman. S. haemolyticus BM4610 [lnu(A) gene], S. aureus BM4611 [lnu(A′) gene], and E. coli(pVMM25) [containing a cloned lnu(B) gene] were used as controls in PCR experiments (3, 18). E. coli AG100A, kindly provided by Hiroshi Nikaido was used in cloning experiments (23). E. coli AG100A is a mutant susceptible to lincosamides resulting from inactivation of the AcrAB pump responsible for active efflux of lincosamides by transposon Tn903 harboring a kanamycin resistance gene. Strains were grown in brain heart infusion broth and agar incubated aerobically at 37°C. S. agalactiae BM132 was used as a negative control in lincosamide inactivation experiments (16).
Antibiotic susceptibility testing.
Susceptibility to antibiotics was determined by the disk diffusion technique (6). MICs of antibiotics were determined by the agar dilution method with Mueller-Hinton medium supplemented with 5% sheep blood and incubated at 37°C under an aerobic atmosphere (6). Erythromycin was from Aventis Pharma (Romainville, France). Lincomycin and clindamycin were from Pfizer (Groton, Conn.).
Lincosamide inactivation.
The kinetics of lincomycin inactivation by resting cells were determined in liquid medium as previously described (19). S. agalactiae UCN36 cells, suspended in 0.01 M phosphate buffer (pH 7) containing 14 μg of lincomycin per ml, were incubated at 37°C for various periods of time. The pH of this suspension remained constant. Inactivation of lincomycin was followed by a bioassay with Micrococcus luteus ATCC 9341 as an indicator organism.
For preparation of modified lincomycin, E. coli AG100A cells containing the cloned lnu(C) gene were lysed by sonication. Cell debris were removed by centrifugation at 40,000 × g for 45 min. Lincomycin (200 μg/ml) was added in the supernatants and then incubated at 37°C for 18 h in the presence of ATP (2.5 mM) and MgCl2 (50 mM). Inactivation of antibiotics was monitored as indicated above. Aliquots of inactivated lincomycin were freeze-dried.
Mass spectrometry.
Samples were analyzed by using an electrospray ion trap mass spectrometer (LCQ Deca XP; Thermofinnigan, San Jose, CA) coupled on line with high-performance liquid chromatography (HPLC) (Surveyor LC). They were separated by reverse-phase HPLC on a C18 capillary column (ThermoHyPurity C18 150 by 0.18). A linear gradient (flow rate, 5 μl/min) from 5 to 95% B was used, where solvent A was a 2 mM ammonium acetate aqueous solution and B was a 2 mM ammonium acetate solution in methanol. The electrospray ionization parameters were as follows: spray voltage, 4.5 kV; spray current, 80 μA; sheath gas flow rate, 35; auxiliary gas flow rate, 10; capillary temperature, 250°C; capillary voltage, 10 V; tube lens offset, −5 V. These parameters were issued from an optimization of the detection of lincomycin. Spectra were acquired in a mode that alternated a full mass spectrometry (MS) scan (mass range from m/z 200 to 1,000; 3 microscans; maximum ion time, 100 ms), followed by a collision-induced dissociation (CID)-MS2 and a CID-MS3 (3 microscans; maximum ion time, 400 ms; collision energy, 35%) of the most abundant ion detected in the previous spectra.
PCR conditions.
The primers used for the amplification of lnu(A), lnu(A′), and lnu(B) genes were previously described (3, 18). The PCR consisted of denaturation (95°C, 30 s), annealing (50°C, 30 s), and extension (72°C, 30 s to 3 min). The products were stored at 4°C until ready for analysis. The Taq DNA polymerase was obtained from Eurobio (Les Ulis, France).
Cloning and sequencing of a DNA fragment conferring resistance to lincosamides.
Chromosomal DNA from S. agalactiae strain UCN36 was digested with various restriction enzymes and ligated at 4°C to plasmid vector pUC18 digested with the corresponding restriction enzymes. Recombinant plasmids were transformed by electroporation (Gene Pulser; Bio-Rad, Ivry-sur-Seine, France) into electrocompetent E. coli AG100A cells. E. coli AG100A transformants were selected on media containing clindamycin (6 μg/ml), ampicillin (100 μg/ml), and kanamycin (20 μg/ml). Both DNA strands were sequenced in an automated ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, CA). Nucleotide and amino acid sequences were analyzed by using the BLAST and FASTA software available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Multiple-sequence alignments were performed with the ClustalX program available at the Centre de Ressources Infobiogen website (http://www.infobiogen.fr/).
Nucleotide sequence accession number.
The nucleotide sequence of the 1,724-bp element from S. agalactiae UCN36 has been deposited in the GenBank database under accession no. AY928180.
RESULTS
Lincomycin resistance in S. agalactiae UCN36.
S. agalactiae UCN36 displayed an unexpected phenotype of resistance to antibiotics. The strain was resistant to lincomycin (MIC = 16 μg/ml) but susceptible to erythromycin (MIC = 0.06 μg/ml), clindamycin (MIC = 0.12 μg/ml), quinupristin (MIC = 16 μg/ml), and dalfopristin (MIC = 2 μg/ml). This phenotype contrasted with the usual phenotype of cross-resistance between macrolides, lincosamides, and streptogramins B and was similar to the phenotype due to nucleotidylation of lincosamides conferred by the lnu class of genes in gram-positive cocci (3, 18). However, PCR experiments failed to detect any DNA sequence related to lnu(A), lnu(A′), lnu(B), or lnu(B-like) genes.
To investigate the mechanism of resistance, a lincomycin inactivation bioassay was implemented. The kinetics of lincomycin inactivation by resting cells of S. agalactiae UCN36 showed that the concentration of native lincomycin (14 μg/ml) decreased within 6 h to 2.5 μg/ml, which is the limit of detection of the method in contrast to the negative-control S. agalactiae BM132. Clindamycin was similarly inactivated despite a low MIC. This result suggested the presence of a lincosamide-inactivating enzyme in S. agalactiae UCN36.
Characterization and localization of the lnu(C) gene.
Restricted DNA fragments from S. agalactiae UCN36 were cloned on plasmid pUC18, and recombinant plasmids were introduced into the lincosamide-sensitive E. coli mutant strain AG100A. The plasmid content of 10 transformants resistant to ampicillin, kanamycin, and clindamycin was analyzed by agarose gel electrophoresis of crude bacterial lysates. The smallest recombinant plasmid, pUV14, with a 4-kb HindIII insert was studied further. Acquisition of the recombinant plasmid by E. coli AG100A led to an increase in the MICs of clindamycin and lincomycin from 2 to 32 μg/ml and 64 to 256 μg/ml, respectively. The transformant was found able to inactivate lincomycin and clindamycin.
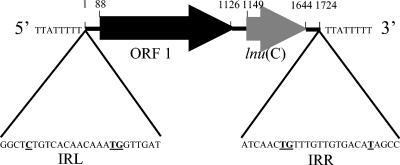
The insert was entirely sequenced. Analysis of the sequence revealed homology with known genes. The 5′ end sequence of the insert was identical to that of a chromosomal fragment of S. agalactiae NEM316 bearing the 3′ end of the cpsD gene and the first 272 nucleotides of the cpsE gene. The cpsD and cpsE genes belong to the capsular synthesis operon and encode two galactosyl transferases (14). The open reading frame of the cpsE gene of S. agalactiae UCN36 was interrupted after nucleotide 272 by the insertion of a 1,724-bp fragment and continued after the insertion. As a consequence, the strain was nontypeable. Two open reading frames (ORFs) in the same orientation, ORF 1 and ORF 2 of 1,038 and 495 bp, respectively, were identified within the inserted fragment (Fig. 1). Both translation ATG start codons were preceded at 9 bp by ribosome binding site-like sequences, 5′-CCGAAGGAGG-3′ and 5′-TTTCTGGAGA-3′, complementary at 7 and 4 bases (underlined) to the 3′-OH-terminal (5′-UCUUUCCUCC-3′) sequence of Bacillus subtilis 16S rRNA, respectively (22).
FIG. 1.
Schematic map of the 1,724-bp genetic element bearing the lnu(C) gene. The element contains ORF 1 (black arrow), putatively encoding a homologue of IS1 transposase, and the lnu(C) gene (grey arrow). The 25-bp imperfect inverted repeats (IRL and IRR) are shown, and the mismatches are in bold and underlined. The element is flanked by 8-bp direct repeats.
The deduced amino acid sequence (164 amino acids) obtained from ORF 2 showed 26 to 27% of identity with lincosamide nucleotidyltransferases, including LnuAN2 from Bacteroides sp. and LnuA and LnuA′ from staphylococci. The lnu-related gene of S. agalactiae UCN36 was thus designated lnu(C) (designation provided by Marilyn Roberts [http://faculty.washington.edu/marilynr/]). ORF 1 was located 23 bp upstream from lnu(C). The deduced protein sequence (345 amino acids) of ORF 1 was distantly related to several transposases described for anaerobic strains and IS1. The protein displayed 35% identity with the IS1 transposase of E. coli (24) and 33 to 42% identity with putative transposases reported for Clostridium sordellii, Clostridium tetani, and Clostridium acetobutylicum (GenBank accession numbers BAC57548.1, NP_782873.1, and NP_348122.1, respectively). A conserved domain shared with transposases belonging to the IS1 family was detected between amino acid 52 and amino acid 126.
The 1,724-bp fragment bearing ORF 1 and lnu(C) was bound at both extremities by 25-bp imperfect inverted repeats. Twenty-two nucleotides of 25 were complementary (Fig. 1). This organization suggested that lnu(C) was borne by a transposon-like structure. The element was flanked by an AT-rich 8-bp sequence which could correspond to the duplication of the target during transposition.
Mechanism of resistance to lincosamides.
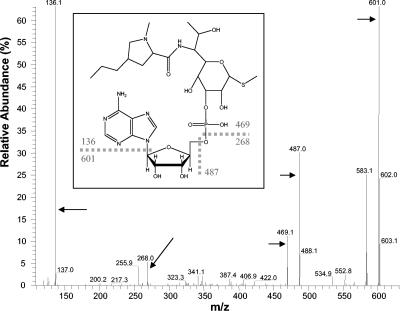
Inactivation of 200 μg of lincomycin per ml was obtained when crude extracts of E. coli AG100A/pUV14 were incubated with ATP and MgCl2 but not when cells were incubated in the absence of ATP. HPLC analysis of the treated samples revealed a single peak eluted at 8.6 min. MS analysis of this fraction revealed three major peaks, displaying m/z of 736.4, 369.4, and 758.3. These 3 peaks corresponded to a unique 736.4-atomic mass unit (amu) compound and to its doubly charged and sodium adduct forms, respectively. The mass observed for this compound was in agreement with adenylylation of lincomycin (+329). The structure of this compound was further characterized by CID-tandem mass spectrometry of both the singly and the doubly charged forms (Fig. 2). Spectra revealed major fragments at 601 and 136 amu, resulting from leakage of the adenine moiety of the compound, and fragments at 469 and 268 amu, resulting from the leakage of the adenosine moiety.
FIG. 2.
CID-MS2 analysis of the 736.4-amu product resulting from lincomycin modification. (Inset) Proposed structure for adenylyl lincomycin, as deduced from fragments observed after CID. Positions of fragments are indicated by arrows. Note that the ester bond was arbitrarily positioned on C-3 of lincomycin on the figure but could also involve the hydroxyl group of C-2 or that of C-4.
DISCUSSION
Isolated resistance to lincosamides which defines the L phenotype, appears to be rare in S. agalactiae. Recently, S. agalactiae isolates intermediate or resistant to clindamycin and lincomycin but susceptible to erythromycin have been reported from New Zealand (20). However, resistance to lincosamides was combined with high MICs of dalfopristin (4 to 32 μg/ml), a streptogramin A antibiotic, defining the so-called LSA phenotype. The biochemical and genetic basis for this resistance remained unknown. In Canada, a single clindamycin-resistant and erythromycin-susceptible strain has been reported which contained an lnu(B) [lin(B)] gene similar to that initially reported for E. faecium and responsible for lincosamide nucleotidylation (3, 7). In this study, we report a new lincosamide resistance gene called lnu(C) distantly related to the other lnu genes. Mass spectrometry experiments showed that lincomycin resistance was due to nucleotidylation of the antibiotic. The precise site of nucleotidylation of lincomycin and clindamycin was not characterized in this study. The LnuA nucleotidyltransferase modifies a hydroxyl group of clindamycin and lincomycin at positions 3 and 4, respectively. By contrast, LnuB modifies a hydroxyl at position 3 in both clindamycin and lincomycin.
In the original gram-positive host and in the E. coli transformant, both lincomycin and clindamycin were inactivated. However, resistance to lincomycin only was detected in S. agalactiae and resistance to both lincomycin and clindamycin was detected in E. coli. The reason for the difference in phenotypic expression of the resistance determinant in the two backgrounds remains unexplained. Hypothetically, the difference between the two lincosamides, which was also reported for the lnu(A) gene, might be related to differences in relative affinities of clindamycin and lincomycin for the ribosomes of gram-positive and gram-negative organisms and for the LnuC enzyme: clindamycin might have better affinity for the gram-positive ribosomes than for LnuC.
The lnu(C) gene was located on a genetic element which bore a homologue of the IS1 transposase gene and which was delineated by imperfect inverted repeats. The structure of the genetic element displayed characteristics similar to those of a transposon. However, it differed from the classical insertion sequences, which are small and compact DNA sequences encoding only functions involved in their translocation, and from the classical transposons, which contain resolvase genes. Further characterization of this putative transposon is currently in progress.
On a practical point of view, lincomycin resistance may be misidentified in strains with the L phenotype if only erythromycin is tested. In addition, the test of clindamycin does not predict for lincomycin resistance. Although the activity of clindamycin against S. agalactiae is only weakly affected by the mechanism of resistance, a 100-fold increase in the bacterial inoculum led to a 3-dilution increase in the MIC of clindamycin for S. agalactiae UCN36 (data not shown). This inoculum effect was greater than that observed for the lincosamide-susceptible strain, and the clinical significance of this modest increase in the MIC of clindamycin remains to be evaluated.
Acknowledgments
We thank Hiroshi Nikaido for the gift of E. coli AG100A, the Fondation pour la Recherche Médicale for financial support, and Marilyn Roberts for providing the designation for the lnu(C) gene.
REFERENCES
- 1.Argoudelis, A. D., J. H. Coats, and S. A. Mizsak. 1997. Microbial transformation of antibiotics. Clindamycin ribonucleotides. J. Antibiot. 30:474-487. [DOI] [PubMed] [Google Scholar]
- 2.Betriu, C., E. Culebras, M. Gomez, I. Rodriguez-Avial, B. A. Sanchez, M. C. Agreda, and J. J. Picazo. 2003. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob. Agents Chemother. 10:128-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozdogan, B., L. Berrezouga, M. Kuo, D. Yurek, K. Farley, B. Stockman, and R. Leclercq. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisson-Noël, A., and P. Courvalin. 1986. Nucleotide sequence of gene linA encoding resistance to lincosamide in Staphylococcus haemolyticus. Gene 43:247-253. [DOI] [PubMed] [Google Scholar]
- 5.Brisson-Noël, A., P. Delrieu, D. Samain, and P. Courvalin. 1988. Inactivation of lincosamide O-nucleotidyltransferases and comparison of the corresponding resistance gene. J. Biol. Chem. 263:15880-15887. [PubMed] [Google Scholar]
- 6.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2004. Communiqué 2004. [Online.] Société Française de Microbiologie, Paris, France. http://www.sfm.asso.fr/.
- 7.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devriese, L. A. 1980. Two new types of resistance to lincomycin in pathogenic staphylococci from animals. Ann. Inst. Pasteur (Paris) 131B:261-266. [PubMed] [Google Scholar]
- 9.Douthwaite, S. 1992. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 20:4717-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta, G. N., and L. A. Devriese. 1981. Degradation of macrolide-lincosamide-streptogramin antibiotics by lactobacillus strains from animals. Ann. Inst. Pasteur (Paris) 132A:51-57. [PubMed] [Google Scholar]
- 11.Dutta, G. N., and L. A. Devriese. 1982. Resistance to macrolide, lincosamide and streptogramin antibiotics and degradation of lincosamide in streptococci from bovine mastitis. J. Antimicrob. Chemother. 10:403-408. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, M. S., and C. J. Baker. 1995. Streptococcus agalactiae (group B Streptococcus), p. 1835-1845. In G. L. Mandell, J. E. Bennett, and R. Dollin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 13.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 14.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 15.Heir, E., B. A. Lindstedt, T. M. Leegaard, E. Gjernes, and G. Kapperud. 2004. Prevalence and characterisation of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class I integron-located lincosamide resistance gene. Ann. Clin. Microbiol. Antimicrob. 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horodniceanu, T., L. Bougueleret, N. El-Solh, D. H. Bouanchaud, and Y. A. Chabbert. 1979. Conjugative R plasmids in Streptococcus agalactiae (group B). Plasmid 2:197-206. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 18.Leclercq, R., A. Brisson-Noël, J. Duval, and P. Courvalin. 1987. Phenotypic expression and genetic heterogeneity of lincosamide inactivation in Staphylococcus spp. Antimicrob. Agents Chemother. 31:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leclercq, R., C. Carlier, J. Duval, and P. Courvalin. 1985. Plasmid-mediated resistance to lincomycin by inactivation in Staphylococcus haemolyticus. Antimicrob. Agent Chemother. 28:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malbruny, B., A. M. Werno, T. P. Anderson, D. R. Murdoch, and R. Leclercq. 2004. A new phenotype of resistance to lincosamide and streptogramin A-type antibiotics in Streptococcus agalactiae in New Zealand. J. Antimicrob. Chemother. 54:1040-1044. [DOI] [PubMed] [Google Scholar]
- 21.Marshall, V. P., W. F. Liggett, and J. I. Cialdella. 1989. Enzymic inactivation of lincosamide and macrolide antibiotics: divalent metal cation and coenzyme specificities. J. Antibiot. 42:826-830. [DOI] [PubMed] [Google Scholar]
- 22.Moran, C. P., Jr., N. Lang, and R. Losick. 1981. Nucleotide sequence of a Bacillus subtilis promoter recognized by Bacillus subtilis RNA polymerase containing sigma 37. Nucleic Acids Res. 9:5979-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saedler, H., and B. Heiss. 1973. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol. Gen. Genet. 122:267-277. [DOI] [PubMed] [Google Scholar]
- 25.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyrrell, G. J., L. D. Senzilet, J. S. Spika, D. A. Kertesz, M. Alagaratnam, M. Lovgren, J. A. Talbot, and the Sentinel Health Unit Surveillance System Site Coordinators. 2000. Invasive disease due to group B streptococcal infection in adults: results from Canadian, population-based, active laboratory surveillance study-1996. J. Infect. Dis. 182:168-173. [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., N. Shoemaker, G. R. Wang, and A. Salyers. 2000. Characterization of a Bacteroides mobilizable transposon of a functional lincomycin resistance gene. J. Bacteriol. 182:3559-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]