Abstract
A novel spectinomycin/streptomycin resistance gene, designated aadA14, was detected on the mobilizable 5,198-bp plasmid pCCK647 from Pasteurella multocida. The aadA14 gene encodes an aminoglycoside adenylyltransferase of 261 amino acids. Sequence comparisons revealed that the AadA14 protein showed less than 60% identity to the AadA proteins known so far.
Spectinomycin is an aminocyclitol antibiotic which inhibits bacterial protein biosynthesis by reversibly binding to the 30S ribosomal subunit. Resistance to spectinomycin is commonly due to enzymes which inactivate the drug by adenylylation. At least two major groups of adenylyltransferases (AAD)—also known as nucleotidyltransferases (ANT)—involved in spectinomycin resistance can be differentiated. One group consists of enzymes [referred to as AAD(3")(9) or ANT(3")(9)] which adenylylate spectinomycin at the 9-OH position of the spectinomycin actinamine ring but also adenylylate the aminoglycoside antibiotic streptomycin at the 3"-OH position of the streptomycin glucosamine ring and thereby mediate combined resistance to spectinomycin and streptomycin (39). Such enzymes, of which a considerable number of variants have been described, are known to occur in a wide variety of gram-negative bacteria and occasionally also in gram-positive bacteria, such as Enterococcus faecalis (4). The corresponding genes, which are commonly referred to as aadA or ant(3")-I, have been detected on plasmids and in the chromosomal DNA, with many of them being located on gene cassettes in class 1 integrons (1, 5, 15, 17, 20, 22, 23, 25, 27-30, 32, 35, 36, 40). A second group of adenylylating enzymes, including those encoded by the genes spc from transposon Tn554 (19) and aad9 from the E. faecalis plasmid pDL55 (14), exhibits only AAD(9) [or ANT(9)] activity and hence confers only resistance to spectinomycin.
In veterinary medicine, spectinomycin is commonly used to control bovine respiratory tract infections due to Pasteurella multocida, Mannheimia haemolytica, or Histophilus somni. Although P. multocida and M. haemolytica isolates which exhibit high-level resistance to spectinomycin, with MICs of ≥256 μg/ml, have recently been reported from Germany, attempts to identify aadA, spc, or aad9 genes in these isolates failed, as did experimental approaches to horizontally transfer the potential spectinomycin resistance genes (31). In the present study, we identified a first aadA gene on a small plasmid from a bovine P. multocida isolate from Belgium.
The ca. 5.2-kb plasmid pCCK647 was identified in a previously reported P. multocida capsular type F strain which was obtained from a case of fatal peritonitis in calves (3). The plasmid was transferred by electrotransformation into the recipient strains P. multocida P4000 (18) and Escherichia coli JM109 (Stratagene, Amsterdam, The Netherlands), where it mediated resistance to spectinomycin (MIC ≥ 512 μg/ml) and streptomycin (MIC = 256 μg/ml). Since PCR detection for the known spectinomycin/streptomycin or spectinomycin resistance genes (31) yielded negative results, it was assumed that plasmid pCCK647 harbored a so-far-undescribed type of spectinomycin/streptomycin resistance gene. To identify the resistance gene located on this plasmid, pCCK647 was subjected to restriction mapping (Fig. 1), and ClaI-EcoRI fragments of ca. 0.8 and 4.4 kb were cloned into pBluescript II SK+ (Stratagene). Both fragments were sequenced completely on both strands by primer walking starting with the M13 forward and reverse primers (MWG, Ebersberg, Germany).
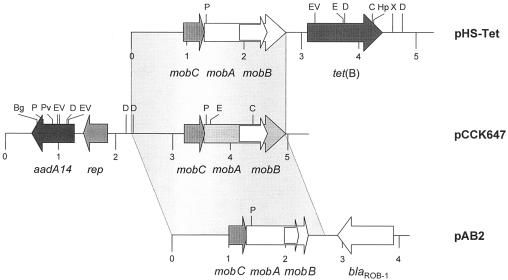
FIG. 1.
Comparison of the maps of plasmid pCCK647 from P. multocida with the maps of plasmid pHS-Tet from H. parasuis (13) and pAB2 from M. haemolytica (37). The arrows indicate the extents of the genes tet(B) (tetracycline resistance), aadA14 (spectinomycin/streptomycin resistance), rep (plasmid replication), blaROB-1 (ampicillin resistance), mobA, mobB, and mobC (plasmid mobilization), with the arrowheads showing the directions of transcription. The regions of similarity between pCCK647, pHS-Tet, and pAB2 are marked by grey shading. A distance scale in kilobases is given below each map. Restriction endonuclease cleavage sites are abbreviated as follows: Bg, BglII; C, ClaI; D, DraI; E, EcoRI; EV, EcoRV; Hp, HpaI; P, PstI; Pv, PvuII; and X, XbaI.
Sequence analysis identified five open reading frames, with one reading frame exhibiting similarity to a plasmid replication gene, three reading frames resembling plasmid mobilization genes, and the remaining reading frame coding for an adenylyltransferase (Fig. 1). The putative rep gene of plasmid pCCK647 coded for a protein of 108 amino acids which showed 57% identity to a 61-amino-acid segment of the 94-amino-acid replication protein RepB from Rhodococcus erythropolis (accession no. AAG29855). A 2,680-bp region of pCCK647 comprising the three reading frames for mobilization proteins showed 86.6% and 86.4% similarity to the corresponding regions of the recently described tetracycline resistance plasmid pHS-Tet from Haemophilus parasuis (13) and the β-lactamase-encoding plasmid pAB2 from Mannheimia haemolytica (37), respectively (Fig. 1). The smallest of the three reading frames, coding for a 102-amino-acid MobC protein, overlapped the mobA reading frame by 3 bp. MobC from pCCK647 exhibited 88% identity to the 101-amino-acid MobC proteins from plasmids pHS-Tet and pAB2. The 160-amino-acid MobB protein showed 89% identity to the 160-amino-acid MobB protein from pHS-Tet and 91% identity to the N-terminal 84 amino acids of the 90-amino-acid MobB protein from pAB2. The largest reading frame in pCCK647 coded for the 474-amino-acid MobA protein. This protein exhibited 79% identity to the 468-amino-acid MobA protein from pHS-Tet and 86% identity to the N-terminal 313 amino acids of the 376-amino-acid MobA protein from pAB2. Since the mob genes of pCCK647 differed from the ones previously described, mobilization of plasmid pCCK647 was experimentally confirmed. The conjugative tet(A)-carrying tetracycline resistance plasmid pEC1591 originally isolated from E. coli and obtained from the strain collection of our institute was chosen to provide the transfer apparatus for the mobilization of plasmid pCCK647. For this, plasmid pCCK647 was first transformed into E. coli JM109 cells which carried the conjugative plasmid pEC1591. Conjugation experiments into the rifampin-resistant E. coli strain HK225 (21) by filter mating followed a previously described protocol (8). Transconjugants were selected on triple-selective Luria-Bertani agar plates supplemented with rifampin (100 μg/ml), tetracycline (15 μg/ml), and spectinomycin (50 μg/ml). Plasmid analysis and determination of the resistance phenotype of the transconjugants confirmed that the transconjugants carried both plasmids, pEC1591 and pCCK647, and were resistant to rifampin, tetracycline, streptomycin, and spectinomycin. This observation suggests that the mobilization system of plasmid pCCK647 is functionally active.
The fifth reading frame in pCCK647 coded for a (3")(9) adenylyltransferase of 261 amino acids, designated AadA14. Comparisons with other AadA proteins on the basis of a multisequence alignment revealed an overall low degree of 51.4% to 56.5% identity to the currently known AadA proteins, with the best matches to the AadA23 protein from Salmonella enterica serovar Agona (17) and its close relative Aad23b from E. coli (accession no. BAD38865). The corresponding homology tree shown in Fig. 2 confirms that AadA14 is only distantly related to the other AadA proteins and clusters with them at 57% identity. In the sequences flanking the aadA14 gene, neither relics of integron sequences nor sequences resembling a 59-base element or parts of the 3′ conserved segments of class 1 or class 2 integrons (27) were detectable. Thus, it is unlikely that the aadA14 gene is a cassette-borne aadA gene.
FIG. 2.
Homology tree of selected AadA proteins involved in combined resistance to spectinomycin and streptomycin based on a multisequence alignment produced with the DNAMAN software (Lynnon-BioSoft, Ontario, Canada). The bacterial source and the database accession number are given for each AadA protein. For a number of AadA proteins, e.g., AadA1 or AadA2, a large number of identical or closely related sequences from different bacterial sources are deposited in the databases. To reduce the complexity of this homology tree, only one representative for each type of AadA protein was chosen. The designations of the different AadA proteins are used as they are deposited in the databases, although these designations do not always reflect the real structural similarities between the different AadA proteins (9). Abbreviations (including reference to the corresponding AadA proteins, if published) are as follows: A. baumannii, Acinetobacter baumannii AadA (20) and AadA4 (32); A. genomosp. 3, Acinetobacter genomospecies 3 AadA1 (40); A. hydrophila, Aeromonas hydrophila; C. jejuni, Campylobacter jejuni AadA2 (22); C. freundii, Citrobacter freundii AadA2 (24); C. glutamicum, Corynebacterium glutamicum AadA2 (34) and AadA9 (33); E. faecalis, Enterococcus faecalis AadA (4); E. coli, Escherichia coli AadA1 (16), AadA2 (28), AadA4 (1), and AadA5 (36); K. oxytoca, Klebsiella oxytoca AadA1 (26); K. pneumoniae, Klebsiella pneumoniae AadA8 (25); P. multocida, Pasteurella multocida AadA1 (38); P. aeruginosa, Pseudomonas aeruginosa AadA1 (15), AadA6 (2), and AadA10 (23); S. Agona, Salmonella enterica serovar Agona AadA23 (17); S. Choleraesuis, Salmonella enterica serovar Choleraesuis; S. Newport, Salmonella enterica serovar Newport AadA7 (6); S. Typhimurium, Salmonella enterica serovar Typhimurium AadA1 (35) and AadA21 (7); S. flexneri, Shigella flexneri; uncult. bacterium, uncultured bacterium; V. cholerae, Vibrio cholerae AadA2 (5).
To determine whether the gene aadA14 also occurs in other epidemiologically unrelated high-level spectinomycin/streptomycin-resistant Pasteurella and Mannheimia isolates, an aadA14-specific PCR assay was developed. The primers aadA14-fw (5′-TCACTTGTTTGGTTCCGCAGT-3′) and aadA14-rev (5′-TCTTTCGGATAAGCTGCCAGA-3′) (annealing temperature, 60°C) were used to amplify an internal 642-bp fragment of the aadA14 gene. Moreover, this amplicon was cloned into pCR-Blunt II Topo (Invitrogen, Groningen, The Netherlands), cut off from the vector by EcoRI digestion, labeled with the Dig-High Prime DNA labeling and detection starter kit I (Boehringer, Mannheim, Germany), and used as a gene probe for Southern blot hybridization of HindIII-digested whole cell DNA (10, 11). Three P. multocida and two M. haemolytica isolates from Germany (31), all exhibiting MICs of spectinomycin of ≥256 μg/ml and MICs of streptomycin of ≥128 μg/ml, were investigated for the presence of the gene aadA14. Another 11 bovine P. multocida isolates which exhibited only spectinomycin resistance, 7 from Germany and 4 from Belgium, were also included. However, negative results were obtained with both methods for all 16 isolates tested. The PCR-based observation that the four Belgian isolates also did not carry so-far-known aadA genes or the genes spc and aad9 is in agreement with previously published findings on the German isolates (31). Attempts to detect the spectinomycin adenylyltransferase gene aadA from Legionella longbeachae (accession no. AF288536) and the aminocyclitol/aminoglycoside phosphotransferase gene aph(9)-Ia from Legionella pneumophila (accession no. U94857) also yielded negative results for all 16 isolates. These results strongly suggest that so-far-undescribed genes are responsible for spectinomycin and spectinomycin/streptomycin resistance in Pasteurella and Mannheimia organisms. Moreover, the results of this study and another recently published study (12) show that Pasteurella isolates carry certain resistance genes that are distantly related to genes from other bacteria which mediate the same resistance phenotype.
Nucleotide sequence accession number.
The sequence of the 5,198-bp plasmid pCCK641 has been deposited in the EMBL database under accession number AJ884726.
Acknowledgments
We thank Vera Nöding and Roswitha Becker for excellent technical assistance.
REFERENCES
- 1.Adrian, P. V., C. J. Thomson, K. P. Klugman, and S. G. B. Amyes. 2000. New gene cassettes for trimethoprim resistance, dfr13, and streptomycin-spectinomycin resistance, aadA4, inserted on a class 1 integron. Antimicrob. Agents Chemother. 44:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended-spectrum β-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catry, B., K. Chiers, S. Schwarz, C. Kehrenberg, A. Decostere, and A. de Kruif. 2005. A case of fatal peritonitis in calves caused by Pasteurella multocida capsular type F. J. Clin. Microbiol. 43:1480-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, N. C., Ø. Olsvik, J. M. Swenson, C. A. Spiegel, and F. C. Tenover. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faldynova, M., M. Pravcova, H. Havlickova, I. Kolackova, A. Cizek, R. Karpiskova, and I. Rychlik. 2003. Evolution of antibiotic resistance in Salmonella enterica serovar Typhimurium strains isolated in the Czech Republic between 1984 and 2002. Antimicrob. Agents Chemother. 47:2002-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frech, G., and S. Schwarz. 1998. Tetracycline resistance in Salmonella enterica subsp. enterica serovar Dublin. Antimicrob. Agents Chemother. 42:1288-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R., and S. Partridge. 2003. Unambiguous numbering of antibiotic resistance genes. Antimicrob. Agents Chemother. 47:3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehrenberg, C., S. A. Salmon, J. L. Watts, and S. Schwarz. 2001. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida, and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of plasmid pMHT1. J. Antimicrob. Chemother. 48:631-640. [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg, C., and S. Schwarz. 2001. Molecular analysis of tetracycline resistance in Pasteurella aerogenes. Antimicrob. Agents Chemother. 45:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehrenberg, C., and S. Schwarz. 2005. dfrA20, a novel trimethoprim resistance gene from Pasteurella multocida. Antimicrob. Agents Chemother. 49:414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lancashire, J. F., T. D. Terry, P. J. Blackall, and M. P. Jennings. 2005. Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 49:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michael, G. B., M. Cardoso, and S. Schwarz. 2005. Class 1 integron-associated gene cassettes in Salmonella enterica subsp. enterica serovar Agona isolated from pig carcasses in Brazil. J. Antimicrob. Chemother. 55:776-779. [DOI] [PubMed] [Google Scholar]
- 18.Miranda, C. D., C. Kehrenberg, C. Ulep, S. Schwarz, and M. C. Roberts. 2003. Diversity of tetracycline resistance genes from bacteria isolated from Chilean salmon farms. Antimicrob. Agents Chemother. 47:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 20.Nemec, A., L. Dolzani, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233-1240. [DOI] [PubMed] [Google Scholar]
- 21.Nüesch-Inderbinen, M. T., F. H. Kayser, and H. Hächler. 1997. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Halloran, F., B. Lucey, B. Cryan, T. Buckley, and S. Fanning. 2004. Molecular characterization of class 1 integrons from Irish thermophilic Campylobacter spp. J. Antimicrob. Chemother. 53:952-957. [DOI] [PubMed] [Google Scholar]
- 23.Partridge, S. R., C. M. Collis, and R. M. Hall. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pepperell, C., J. V. Kus, M. A. Gardam, A. Humar, and L. L. Burrows. 2002. Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 46:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, E. D., M. A. Leverstein-van Hall, A. T. Box, J. Verhoef, and A. C. Fluit. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preston, K. E., R. A. Venezia, and K. A. Stellrecht. 2004. The SHV-5 extended spectrum beta-lactamase gene of pACM1 is located on the remnant of a compound transposon. Plasmid 51:48-53. [DOI] [PubMed] [Google Scholar]
- 27.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 28.Sabate, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbe, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandvang, D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43:3036-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandvang, D. 2001. Aminoglycoside resistance genes and their mobility in gramnegative bacteria from production animals. Ph.D. thesis. The Royal Veterinary and Agricultural University, Copenhagen, Denmark.
- 31.Schwarz, S., C. Kehrenberg, S. A. Salmon, and J. L. Watts. 2004. In vitro activities of spectinomycin and comparator agents against Pasteurella multocida and Mannheimia haemolytica from respiratory tract infections of cattle. J. Antimicrob. Chemother. 53:379-382. [DOI] [PubMed] [Google Scholar]
- 32.Segal, H., R. Thomas, and B. G. Elisha. 2003. Characterisation of class 1 integron resistance gene cassettes and the identification of a novel IS-like element in Acinetobacter baumannii. Plasmid 49:169-178. [DOI] [PubMed] [Google Scholar]
- 33.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48:117-129. [DOI] [PubMed] [Google Scholar]
- 34.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2003. Plasmids in Corynebacterium glutamicum and their classification by comparative genomics. J. Biotechnol. 104:27-40. [DOI] [PubMed] [Google Scholar]
- 35.Tosini, F., P. Visca, I. Luzzi, A. M. Dionisi, C. Pezzella, A. Petrucca, and A. Carattoli. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, P. A., C. J. McIver, Y. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 37.Wood, A. R., F. A. Lainson, F. Wright, G. D. Baird, and W. Donachie. 1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res. Vet. Sci. 58:163-168. [DOI] [PubMed] [Google Scholar]
- 38.Wu, J.-R., H. K. Shieh, J.-H. Shien, S.-R. Gong, and P.-C. Chang. 2003. Molecular characterization of plasmids with antimicrobial resistant genes in avian isolates of Pasteurella multocida. Avian Dis. 47:1384-1392. [DOI] [PubMed] [Google Scholar]
- 39.Yamada, D., D. Tipper, and J. Davies. 1968. Enzymatic inactivation of streptomycin by R-factor resistant Escherichia coli. Nature (London) 219:288-291. [DOI] [PubMed] [Google Scholar]
- 40.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the blaVIM-2 gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]