Abstract
Multidrug-resistant Pseudomonas aeruginosa with combined decreased susceptibility to ceftazidime, ciprofloxacin, imipenem, and piperacillin is increasingly being found as a cause of nosocomial infections. It is important to look for combinations of drugs that might be synergistic. Ciprofloxacin resistance by P. aeruginosa is mediated in part by an efflux pump mechanism. Gatifloxacin, an 8-methoxyfluoroquinolone, inhibits a staphylococcal efflux pump. An earlier in vitro study using an Etest synergy method and time-kill assay suggested synergy of ciprofloxacin and gatifloxacin against P. aeruginosa. Synergy testing was performed by Etest and time-kill assay for 31 clinically unique, plasmid DNA distinct, U.S. P. aeruginosa isolates. Etest MICs for ciprofloxacin were 4 to >32 μg/ml, and for gatifloxacin they were >32 μg/ml. Ciprofloxacin plus gatifloxacin showed synergy by the Etest method for 6 (19%) of the 31 P. aeruginosa isolates using a summation fractional inhibitory concentration of ≤0.5 for synergy. Synergy was demonstrated for 13/31 (42%) of isolates by time-kill assay. No antagonism was detected. The remaining isolates were indifferent to the combination. The Etest method and time-kill assay were 65% (20/31) concordant. The mechanism of the in vitro synergy may include P. aeruginosa ciprofloxacin efflux pump inhibition by gatifloxacin.
It is now accepted that bacterial multidrug antimicrobial resistance is a worldwide problem, in large part related to antimicrobial use in humans and other animals. Serious infections with these resistant bacteria are commonplace. Therapy for Pseudomonas aeruginosa, a major cause of life-threatening nosocomial infection, is problematic because of the propensity for multiple-drug resistance (19). Some P. aeruginosa strains are only susceptible to the polymyxins (18). Mechanisms of resistance of P. aeruginosa are dependent mainly on impermeability and multidrug efflux pumping (9). This raises the MICs of penicillins, cephalosporins, quinolones, tetracyclines, and chloramphenicol. Carbapenem resistance is associated with metallo-beta-lactamases (20). The development of vaccines and new antimicrobial agents has not kept pace with resistance; therefore, the search for other methods of therapy such as synergistic combinations is necessary. Many antimicrobial combinations have been studied for synergy in vitro and in vivo against P. aeruginosa (3-5, 14).
Ciprofloxacin (CIP) is the most in vitro-active anti-P. aeruginosa fluoroquinolone, but increasing resistance (now 25% in most of the United States) frequently precludes its use (7). The mechanism of the resistance includes efflux pumping. Gatifloxacin (GAT) is an 8-methoxyfluoroquinolone with similar substituent configuration to reserpine, a known efflux pump inhibitor. Perhaps related to this, it resists efflux pumping by gram-positive bacteria and has low MICs for these bacteria. Gatifloxacin is inherently not as active in vitro against Pseudomonas aeruginosa. If gatifloxacin were to interfere with the efflux pumping of ciprofloxacin by ciprofloxacin-resistant Pseudomonas aeruginosa, then a potentially active combination might result. We tested the combination of gatifloxacin with ciprofloxacin in vitro to see if synergy against Pseudomonas aeruginosa could be demonstrated. The Etest synergy screening methodology developed by Pankey and Ashcraft to evaluate antimicrobials against gram-positive cocci using combinations of moxifloxacin and linezolid against methicillin-resistant staphylococci (G. Pankey and D. Ashcraft, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C92, 2001) was used to screen for synergy of ciprofloxacin and gatifloxacin against the Pseudomonas aeruginosa isolates. Time-kill assays were performed for comparison.
(Part of these data was presented at the 102nd General Meeting of the American Society for Microbiology, Salt Lake City, Utah, May 2002.)
MATERIALS AND METHODS
Antimicrobial agents.
Standard laboratory powders of CIP (Bayer Corporation, West Haven, CT) and GAT (Bristol-Myers Squibb, Princeton, NJ) were used in this study. Etest strips (AB Biodisk, Solna, Sweden) of CIP and GAT were also used.
Microorganisms and media.
Thirty-one unique clinical, plasmid DNA distinct (by pulsed-field gel electrophoresis) Pseudomonas aeruginosa isolates were collected from October 1999 through June 2003 from five hospitals in the New Orleans, LA, metropolitan area. The isolates were cultured from the lower respiratory tract (11), wound (9), urine (6), blood (2), ear (1), catheter tip (1), and bone (1). All strains were identified by the Vitek system (bioMerieux Inc., Hazelwood, MO). Isolates were stored frozen at −70°C in Columbia broth with 20% glycerol. Each strain was subcultured twice onto a blood agar plate (Trypticase soy agar with 5% sheep blood; Becton-Dickinson Microbiology Systems, Sparks, MD) before use. Pseudomonas aeruginosa ATCC 27853 was included as a quality control strain. Mueller-Hinton II broth (MHB; Becton-Dickinson Microbiology Systems, Sparks, MD) was prepared in the laboratory. Mueller-Hinton II agar (MHA) plates (Becton-Dickinson Microbiology Systems, Sparks, MD) were used for the Etest MIC determinations and the Etest synergy screening method. Trypticase soy agar with 5% sheep blood plates (Becton-Dickinson Microbiology Systems, Sparks, MD) were used for the colony counts in the time-kill assay.
MIC determinations. (i) BMD MICs.
MICs by broth microdilution (BMD) were performed following 2003 NCCLS guidelines (16). The concentration range tested was 0.25 to 32 μg/ml for CIP and GAT.
(ii) Etest MICs.
MICs were determined in triplicate by the Etest method, and testing was performed according to the manufacturer's instructions. MICs in between twofold dilutions were rounded up to the next twofold dilution for purposes of comparison with the BMD MIC.
Synergy studies.
Testing for synergy was determined by both the Etest synergy screen method and a time-kill assay.
Etest synergy screen method.
The Etest synergy method was performed in triplicate, the summation fractional inhibitory concentration (ΣFIC) was calculated for each set of MICs, and the mean ΣFIC was used for comparison to the time-kill assay results. The inoculum and streaked MHA plates for each isolate were prepared the same as for Etest MICs. To check adequate diffusion of the antibiotic from the strip into the agar in 1 h, we performed quality control using P. aeruginosa ATCC 27853 on each drug strip prior to performing the Etest synergy method. Two Etest strips containing the same drug were placed side by side on an inoculated MHA plate. One strip was removed after 1 h (the second strip remained on the agar), and the plate was incubated for 18 h at 35°C. The resulting ellipses and MICs were compared. The ellipse and MIC of the strip removed before overnight incubation equaled the ellipse and MIC of the strip that remained on the agar overnight for both CIP and GAT. Therefore, the 1-h incubation at room temperature was assumed adequate for complete diffusion into the agar.
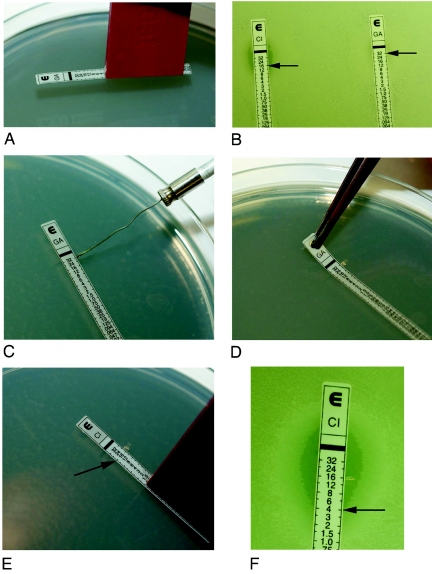
Etest synergy screening was performed by applying CIP and GAT Etest strips to different sections of an MHA plate. The agar was marked adjacent to the previously determined MIC on each Etest strip. The strips were removed after incubating for 1 h at room temperature. Using an Etest applicator, a new GAT strip was placed over the area of the previously removed CIP strip so that the GAT MIC corresponded with the mark of the CIP MIC. CIP strips were applied in reciprocate fashion. This established a concentration ratio of both 1× MICs for the two antimicrobials. The resulting combination ellipses were read after 20 h of incubation at 35°C (Fig. 1).
FIG. 1.
A. An Etest applicator was used to place the Etest strip on the agar surface. B. Etest strips after 18 h of incubation, showing isolate 2598. CIP MIC = 16 μg/ml (left) and GAT MIC = >32 μg/ml (right). C. Agar was marked, using a wire loop, adjacent to 32 on the GAT strip (MIC = >32 μg/ml). D. The GAT strip was removed after 1 h. E. The new CIP strip was placed over the area of the removed GAT strip, matching the MIC of CIP (16 μg/ml) to the MIC agar mark of GAT (32 μg/ml was used). F. The resulting combination ellipse was read after 18 h of incubation. After combination with the GAT strip, the CIP MIC was 4 μg/ml.
To evaluate the effect of the combinations, the FIC was calculated for each antibiotic in each combination. High off-scale MICs (>32 μg/ml) were converted to the next twofold dilution (64 μg/ml). The following formulas were used to calculate the ΣFIC:
FIC of drug A = (MIC of drug A in combination)/(MIC of drug A alone)
FIC of drug B = (MIC of drug B in combination)/(MIC of drug B alone)
ΣFIC = FIC of drug A + FIC of drug B.
Synergy was defined by a ΣFIC of ≤0.5. Antagonism was defined by a ΣFIC of >4. Interactions represented by a ΣFIC of >0.5 but ≤4 were termed indifferent (per 2005 instructions to authors, Antimicrob. Agents Chemother. 49:1-20, 2005).
Time-kill assays.
The time-kill assay was chosen initially to compare it with the Etest method. Time-kill assays were performed on all isolates following guidelines set by the NCCLS (15). Each isolate was tested against CIP and GAT alone and in combination at a concentration equal to 1× MIC. Antimicrobial solutions for the time-kill assays were prepared and diluted the same day of testing. Each isolate was grown to a logarithmic phase, so that the final inoculum was approximately 5 × 105 CFU/ml and was verified after plating in duplicate using a spiral plater (Spiral Biotech, Inc., Bethesda, MD). The spiral plater was used to accurately detect bacterial counts as low as 20 CFU/ml. A bottle containing the organism plus antibiotic-free MHB served as the growth control. A total volume of 30 ml MHB in the bottles was used. Bottles were incubated at 35°C in ambient air for 24 h. Samples (0.5 ml) were removed from each bottle at 0 h and 24 h. Serial 10-fold dilutions (10−1 to 10−5) were performed in 0.85% sterile saline when necessary. Dilutions were plated using the spiral plater, the plates were incubated 18 to 24 h at 35°C in ambient air, and colony counts were determined using a scanner (Spiral Biotech, Inc., Bethesda, MD). The mean colony count (in CFU/ml) from duplicate samples was used in the determination of synergy. Discrepant results were repeated. The possibility of antibiotic carryover was reduced by performing serial dilutions, plating with a spiral plater (which further dilutes and plates the sample), and using the 1× MIC of drug.
Synergy was defined as a ≥2 log10 decrease in colony count at 24 h by the combination compared to the most active single agent. Indifference was defined as a <2 log10 increase or decrease in colony count at 24 h by the combination compared with that by the most active drug alone. Antagonism was defined as a ≥2 log10 increase in colony count at 24 h by the combination compared with that by the most active drug alone (10).
Determination of MBC.
Minimal bactericidal concentration (MBC) testing for CIP and GAT was performed whenever the MIC by Etest and BMD differed by >2 twofold dilutions (15 isolates). All MBC testing was performed according to NCCLS bactericidal testing guidelines (15). The concentration range tested was 0.5 to 128 μg/ml for CIP and GAT. The inoculum was prepared the same as for the broth microdilution procedure.
After 24 h, the inoculum verification plate's colonies were counted and the mean CFU/ml was used. The MIC was determined visually. A 100-μl aliquot from each clear tube was subcultured in duplicate to blood agar plates. Plates were incubated at 35°C for 24 h. Each plate was examined, and colonies were counted (the mean CFU/ml was used). The MBC was defined as the minimal concentration of CIP or GAT needed to kill ≥99.9% of the viable organisms after 24 h of incubation.
RESULTS
In vitro susceptibility testing.
The mean MICs determined by BMD and Etest are presented in Table 1. The BMD MICs were lower than the Etest MICs for 27/31 CIP MICs and 23/31 GAT MICs. All of the P. aeruginosa isolates were Etest resistant to both CIP and GAT, using the NCCLS interpretive standards for ciprofloxacin (17). CIP MICs (in μg/ml) were 4 to >32 by Etest and 1 to >32 by BMD. GAT MICs (in μg/ml) were ≥32 by Etest and ≥4 by BMD.
TABLE 1.
MICs by broth microdilution and Etest, MBCs, Etest synergy method, and time-kill assay
| P. aeruginosa isolate (n = 31) | CIP MIC
|
GAT MIC
|
Synergy testing
|
||||
|---|---|---|---|---|---|---|---|
| Etesta | BMD (MBC) | Etesta | BMD (MBC) | Etesta ΣFICs | Mean ΣFIC | Time-kill assay log10 changeb | |
| B44415 | 8 | 4 | >32 | 8 (64) | 0.6, 0.6, 0.6 | 0.6 | −2.6, SYN |
| 2598 | 16 | 4 | >32 | 16 | 0.6, 0.6, 0.6 | 0.6 | −2.2, SYN |
| 2659 | 16 | 4 | >32 | 4 (64) | 0.5, 0.5, 0.6 | 0.5 | −3.0, SYN |
| F18147 | 8 | 4 | >32 | 16 | 0.4, 0.4, 0.4 | 0.4 | −3.8, SYN |
| 1635 | 8 | 4 | >32 | 8 (64) | 0.8, 0.8, 0.7 | 0.8 | −2.0, SYN |
| 1629 | >32 | 1 (8) | >32 | 4 (32) | 0.1, 0.1, 0.1 | 0.1 | −2.3, SYN |
| 2019 | 8 | 1 (16) | >32 | 4 (64) | 0.8, 0.6, 0.4 | 0.6 | −5.6, SYN |
| 1303 | >32 | 8 (32) | >32 | 32 | 0.8, 0.4, 0.3 | 0.5 | −2.3, SYN |
| 5704-1 | 8 | 4 | 32 | 8 | 0.4, 0.9, 0.8 | 0.7 | −2.2, SYN |
| S12305 | 8 | 2 | >32 | 8 (64) | 0.6, 0.6, 0.8 | 0.7 | −2.2, SYN |
| H21689 | 16 | 4 | 32 | 8 | 0.5, 0.5, 0.6 | 0.5 | −1.9, IND |
| 1659 | 4 | 2 | >32 | 8 (16) | 0.6, 0.8, 0.6 | 0.7 | −1.3, IND |
| 6213 | >32 | 16 | >32 | 32 | 0.9, 0.5, 0.9 | 0.8 | −1.5, IND |
| 2631 | 8 | 2 | >32 | 4 (32) | 0.6, 0.4, 0.6 | 0.5 | +0.01, IND |
| F28200 | 16 | 4 | >32 | 16 | 0.8, 0.8, 0.8 | 0.8 | 0, IND |
| M20078 | 8 | 4 | >32 | 16 | 0.6, 0.8, 0.8 | 0.7 | 0, IND |
| B4467R | >32 | 16 | >32 | 16 | 2.0, 2.0, 2.0 | 2.0 | +0.2, IND |
| B4723R | >32 | >32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | +0.5, IND |
| 2833 | >32 | >32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | +1.0, IND |
| S53290 | >32 | 16 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | +0.2, IND |
| M34690 | >32 | 8 (64) | >32 | 32 | 2.0, 2.0, 2.0 | 2.0 | +0.8, IND |
| T21511 | >32 | >32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | +0.9, IND |
| 2854 | >32 | 32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | −1.5, IND |
| 2033 | >32 | 4 (16) | >32 | 16 | 2.0, 1.1, 1.2 | 1.4 | −0.8, IND |
| 1858 | >32 | 8 (64) | >32 | 8 (128) | 2.0, 2.0, 2.0 | 2.0 | −1.6, IND |
| 64539 | >32 | 32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | −1.3, IND |
| 3963 | >32 | 32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | −0.7, IND |
| T70594 | >32 | 8 (32) | >32 | 32 | 2.0, 2.0, 2.0 | 2.0 | −0.9, IND |
| 1923 | >32 | 8 (64) | >32 | 16 | 2.0, 2.0, 2.0 | 2.0 | −3.5, SYN |
| 6016 | >32 | 8 (64) | >32 | 16 | 1.0, 0.9, 2.0 | 1.3 | −2.9, SYN |
| 298 | >32 | >32 | >32 | >32 | 2.0, 2.0, 2.0 | 2.0 | −2.8, SYN |
Performed in triplicate.
Values represent the log10 change in CFU/ml in the time-kill assay after 24-h exposure to the combination of CIP and GAT compared to the most active drug alone. Negative values indicate a decrease in colony count; positive values indicate an increase in colony count. SYN, synergy; IND, indifference.
The MBCs (Table 1) correlated more closely with the Etest MICs than BMD MICs. MBCs for CIP and GAT were only performed for isolates when the Etest and BMD MICs differed by >2 twofold dilutions (nine CIP and nine GAT). There was a 77.8% essential agreement (±1 twofold dilution) of the CIP and GAT MBCs with the Etest MICs, while MBCs showed only 5.6% essential agreement with the broth microdilution MICs.
Synergy testing.
The Etest synergy method for the P. aeruginosa isolates using CIP plus GAT revealed synergy in 6/31 (19%) (ΣFIC ≤ 0.5). Indifference (ΣFIC, >0.5 to ≤4) occurred in 25/31 (81%) of the isolates. Time-kill studies revealed synergy in 13/31 (42%) and indifference in 18/31 (58%) (Table 1). Isolates with a ciprofloxacin MIC of >32 μg/ml were usually indifferent: 16/18 (89%) by Etest, perhaps relating to the limit of the gradient to 32 μg/ml.
Concordance of the Etest synergy method and the time-kill assay was demonstrated in 20/31 (65%) isolates. All discordant time-kill assay results were repeated. For two isolates, the Etest synergy method results were ΣFICs of 0.5 and 0.5 (synergy), but the time-kill assay showed indifference, −1.9 and +0.01 log10 change in CFU/ml. Nine isolates showed indifference by Etest (ΣFICs of 0.6, 0.6, 0.7, 0.6, 0.7, 0.7, 2.0, 1.3, and 2.0) but were synergistic by the time-kill assay (−2.6, −2.2, −2.0, −5.6, −2.2, −2.2, −3.5, −2.9, and −2.8 log10 change in CFU/ml). No antagonism was detected by either method. The sensitivity and specificity of the Etest method cannot be determined since there is no established reference method for synergy testing.
DISCUSSION
Three methods to detect in vitro synergy have been described: the time-kill assay, checkerboard, and Etest. Synergy testing methods are not standardized for reproducibility and interpretation and, therefore, it is extremely difficult to compare these methods' results from different studies.
In the time-kill assay for synergy, drug concentrations are fixed and do not decrease over time, as they would in vivo. Also, there are no standard concentrations at which antibiotics are tested. The inoculum size and time frame of the time-kill assay add more variability to the test. The time parameter of 24 h can limit or alter results of the experiment if regrowth occurs with one or both antibiotics. Regrowth can be caused by use of subinhibitory concentration of antibiotics. Emergence of resistant subpopulations may account for the regrowth, or regrowth may be due to bacteria that adhere to the surface of the bottle and are subsequently released in the medium. Another factor affecting regrowth is inactivation of the antibiotics in vitro.
In the checkerboard technique, serial dilutions of two drugs are performed in tubes or microtiter wells using drug concentrations equal to, above, and below the MICs of the drugs being tested. The checkerboard method measures inhibitory activity. Only if each microdilution well at the MIC and greater is subcultured for growth would this method predict bactericidal activity. The time-kill assay for synergy testing measures bactericidal activity but is time-consuming and labor-intensive. Because these two methods measure different activities, studies have shown results that have poor agreement (1, 2, 4, 5, 22). There are limitations associated with both methods. In the study by Cappelletty and Rybak (4), methodologies for synergy testing of resistant P. aeruginosa were compared and problems were discussed.
The third method for determining synergy, the Etest synergy method, is relatively new. The use of the Etest strip for synergy has yet to be standardized but has the potential to be a useful screening test for the determination of synergy. Studies by White et al. (22) and Bonapace et al. (2) evaluated the use of the Etest for synergy testing by placing the Etest strips on the agar in a cross formation, with a 90° angle at the intersection between the scales at the respective MICs for the organism. In the study by White et al. (22), the agreement between their Etest method and time-kill assay ranged from 63 to 75% and agreement between the checkerboard method and time-kill assay ranged from 44 to 88%. Correlation was dependent on the bacterium tested (Escherichia coli ATCC 35218, Enterobacter cloacae ATCC 23355, P. aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 29213) and antibiotics tested (cefepime or ceftazidime in combination with tobramycin or ciprofloxacin). Similarly, in the study by Bonapace et al. (2), the agreement between the Etest and time-kill assay ranged from 42 to 97% and 30 to 67% for the checkerboard method and time-kill assay. This study included 10 strains of Acinetobacter baumannii, and antimicrobial combinations evaluated consisted of trovafloxacin or tobramycin in combination with cefepime or piperacillin. Antagonism was difficult to detect with their method. Both studies concluded that additional testing using an Etest method needed to be performed.
An unpublished Etest synergy method has been used in testing for synergy against mycobacteria (A. Bolmstrom and U. Nordstrom, Abstr. 16th Int. Congr. Chemother., abstr. 7001, 1995; A. Wanger and K. Mills, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E7, 1995). An Etest synergy method was also evaluated for Candida spp. using combinations of fluconazole, amphotericin B, and flucytosine (8). Their Etest method was a prediffusion technique, where the first Etest strip was removed after 1 h and the second strip was put directly where the first strip had been removed, rather than matching MIC to MIC. Etest and time-kill assay results were compared. The agreement between Etest and time-kill assay was 83% (15/18) tests.
Checkerboard and three Etest synergy methods were compared for 29 isolates of Xanthomonas maltophilia (J. Poupard, R. Langan, L. Utrup, S. Rittenhouse, and R. Clark, Abstr. 18th Int. Congr. Chemother., abstr. 366, 1993). This study used the combination of timentin plus tobramycin or timentin plus amikacin. Three Etest methods were evaluated: (i) a prediffusion, in which one strip was placed on the agar and removed after 1 h and the second strip was placed directly where the first strip had been removed, rather than matching MIC to MIC; (ii) Etest strips were placed side by side; and (iii) a second Etest strip was placed on top of the first strip. Only the prediffusion method was reproducible, and so it was used for comparison purposes. For both antibiotic combinations, the checkerboard and Etest correlation was low (30% for timentin plus tobramycin and 38% for timentin plus amikacin).
Manno et al. compared the checkerboard and an Etest synergy method using 131 Burkholderia cepacia isolates (13). The Etest synergy method used was the prediffusion technique, as previously described by Poupard et al. The combination of a β-lactam agent plus ciprofloxacin, cotrimoxazole, chloramphenicol, or rifampin was evaluated. The Etest and checkerboard had a 90% agreement.
We have further modified the Etest synergy method to use a concentration equal to 1× MIC for each drug. Depending on the drug, concentration ranges vary on Etest strips. A MIC-to-MIC placement of the strips seems to give a more accurate diffusion of the two drugs and the effects (if any) that each drug has on the other in combination against the organism. Several abstracts have been presented on the technique using gram-positive and gram-negative bacteria (G. Pankey, D. Ashcraft, and O. Prakash, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-1133, 2002; G. Pankey and D. Ashcraft, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. C109, 2002; G. Pankey, M. Wynn, D. Ashcraft, and P. Pankey, Abstr. Natl. Found. Infect. Dis. Conf. Antimicrob. Resist., abstr. 4, 2003; G. Pankey, D. Ashcraft, and P. Pankey, Abstr. 41st Infect. Dis. Soc. Am., abstr. 229, 2003; N. Patel, G. Pankey, and D. Ashcraft, Abstr. 42nd Infect. Dis. Soc. Am., abstr. 306, 2004). Our Etest method was compared to the time-kill assay, but the two methods use totally different test systems, solid medium versus liquid, respectively. However, both methods predict bactericidal activity in vitro. (The Etest MICs had a 77% essential agreement with the MBCs, unlike the BMD MICs, which had 5.6% essential agreement with MBCs.) The Etest was able to detect slight hazes of growth and resistant subpopulations. Since the MICs of both quinolones were read as bactericidal endpoints, these resistant colonies were included when reading the endpoint for the Etest MIC. The BMD method was unable to detect these small amounts of growth in many cases. Lorian showed that bacteria grown on a surface are significantly different from bacteria grown in liquid medium (12). The differences include growth rate, adherence, and susceptibility to antibacterial agents, as well as differences in the biochemical constitutions of the bacteria themselves and their metabolites. One major difference is in the ultrastructure. There is evidence indicating that bacteria in vivo grow and produce disease on surfaces and not in body fluids (6, 11, 12, 21). The identical ultrastructures of bacteria found in vivo and organisms grown in vitro on a surface support the conclusion that in vitro experiments aimed at duplicating in vivo conditions should be done on solid media.
In conclusion, the results of our Etest synergy method show that it may be an alternative to time-kill studies for in vitro synergy testing of P. aeruginosa with ciprofloxacin and gatifloxacin. The technique is simple to use, time-efficient, inexpensive, reproducible, and yielded results comparable to the time-kill assay. Once the MICs were determined, the Etest synergy method results could be obtained in 24 h at a material cost of less than $10. However, the concentration on the Etest strip may limit the use of the Etest synergy method to resistant isolates with an Etest MIC not exceeding the Etest strip concentration. This is a probable explanation for the three isolates that were indifferent by the Etest method but were synergistic by time-kill assay (an exact ΣFIC cannot be calculated without being able to determine the “exact MIC” on an Etest strip). At the present time, we recommend that our Etest synergy method not be used if the MIC is >32 μg/ml for both ciprofloxacin and gatifloxacin.
Although it is interesting that we could demonstrate in vitro synergy of ciprofloxacin and gatifloxacin against some P. aeruginosa isolates, the mechanism of the in vitro synergy is unknown. There is no evidence of in vivo synergy, but we plan to test this combination in an animal model of P. aeruginosa pneumonia. The clinical benefit, if any, of in vitro synergy by ciprofloxacin plus gatifloxacin against any strain of P. aeruginosa remains to be determined. In addition, because of potential toxicity of therapeutic doses of both drugs in combination, treatment of patients with P. aeruginosa infections using ciprofloxacin plus gatifloxacin should be avoided at this time.
Acknowledgments
We thank Chupol Shariffskul, Michael Lacassagne, and Arlene Lefebre, Clinical Laboratory Scientists at the State of Louisiana, Office of Public Health/General Laboratory, New Orleans, LA, for performing the pulsed-field gel electrophoresis analysis on the P. aeruginosa isolates, Pat C. Pankey for laboratory management, and Marion Stafford for editorial support.
REFERENCES
- 1.Bayer, A. S., and J. O. Morrison. 1984. Disparity between timed-kill and checkerboard methods for determination of in vitro bactericidal interactions of vancomycin plus rifampin versus methicillin-susceptible and -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 26:220-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonapace, C. R., R. L. White, L. V. Friedrich, and J. A. Bosso. 2000. Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn. Microbiol. Infect. Dis. 38:43-50. [DOI] [PubMed] [Google Scholar]
- 3.Bustamante, C. L., R. C. Wharton, and J. C. Wade. 1990. In vitro activity of ciprofloxacin in combination with ceftazidime, aztreonam, and azlocillin against multiresistant isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:1814-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappelletty, D. M., and M. J. Rybak. 1996. Comparison of methodologies for synergism testing of drug combinations against resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekar, P. H., L. R. Crane, and E. J. Bailey. 1987. Comparison of the activity of antibiotic combinations in vitro with clinical outcome and resistance emergence in serious infection by Pseudomonas aeruginosa in non-neutropenic patients. J. Antimicrob. Chemother. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 6.Eng, R. H., A. Hsieh, and S. M. Smith. 1995. Antibiotic killing of bacteria: comparison of bacteria on surfaces and in liquid, growing and nongrowing. Chemotherapy 41:113-120. [DOI] [PubMed] [Google Scholar]
- 7.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2001. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32:S146-S155. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, R. E., D. J. Diekema, S. A. Messer, M. A. Pfaller, and M. E. Klepser. 2002. Comparison of Etest, chequerboard dilution and time-kill studies for the detection of synergy or antagonism between antifungal agents tested against Candida species. J. Antimicrob. Chemother. 49:345-351. [DOI] [PubMed] [Google Scholar]
- 9.Livermore, D. M. 2001. Of Pseudomonas, porins, pumps and carbapenems. J. Antimicrob. Chemother. 47:247-250. [DOI] [PubMed] [Google Scholar]
- 10.Lorian, V. (ed.). 1996. Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 11.Lorian, V. 1988. Differences between in vitro and in vivo studies. Antimicrob. Agents Chemother. 32:1600-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorian, V. 1989. In vitro simulation of in vivo conditions: physical state of the culture medium. J. Clin. Microbiol. 27:2403-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manno, G., E. Ugolotti, M. L. Belli, M. L. Fenu, L. Romano, and M. Cruciani. 2003. Use of the E test to assess synergy of antibiotic combinations against isolates of Burkholderia cepacia-complex from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 22:28-34. [DOI] [PubMed] [Google Scholar]
- 14.Mouton, J. W., M. L. van Ogtrop, D. Andes, and W. A. Craig. 1999. Use of pharmacodynamic indices to predict efficacy of combination therapy in vivo. Antimicrob. Agents Chemother. 43:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NCCLS. 1999. Methods for determining bactericidal activity of antimicrobial agents. NCCLS document M26-A. NCCLS, Wayne, Pa.
- 16.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS document M7-A6. NCCLS, Wayne, Pa.
- 17.NCCLS. 2005. Performance standards for antimicrobial susceptibility testing; fourteenth informational supplement. CLSI/NCCLS document M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 18.Ouderkirk, J. P., J. A. Nord, G. S. Turett, and J. W. Kislak. 2003. Polymyxin B nephrotoxicity and efficacy against nosocomial infections caused by multiresistant gram-negative bacteria. Antimicrob. Agents Chemother. 47:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramythiotou, E., J. C. Lucet, J. F. Timsit, D. Vanjak, C. Paugam-Burtz, J. L. Trouillet, S. Belloc, N. Kassis, A. Karabinis, and A. Andremont. 2004. Acquisition of multidrug-resistant Pseudomonas aeruginosa in patients in intensive care units: role of antibiotics with antipseudomonal activity. Clin. Infect. Dis. 38:670-677. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, H. 2000. Questions about the behaviour of bacterial pathogens in vivo. Philos. Trans. R. Soc. London B 355:551-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, R. L., D. S. Burgess, M. Manduru, and J. A. Bosso. 1996. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and Etest. Antimicrob. Agents Chemother. 40:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]