Abstract
Sixteen homologs of multidrug resistance efflux pump operons of the resistance-nodulation-cell division (RND) family were found in the Bacteroides fragilis genome sequence by homology searches. Disruption mutants were made to the mexB homologs of the four genes most similar to Pseudomonas aeruginosa mexB. Reverse transcription-PCR was conducted and indicated that the genes were transcribed in a polycistronic fashion and that the promoter was upstream of bmeA (the mexA homolog). One of these disruption mutants (in bmeB, the mexB homolog) was more susceptible than the parental strain to certain cephems, polypeptide antibiotics, fusidic acid, novobiocin, and puromycin. The gene for this homolog and the adjacent upstream gene, bmeA, were cloned in a hypersensitive Escherichia coli host. The resultant transformants carrying B. fragilis bmeAB were more resistant to certain agents; these agents also had lower MICs for the B. fragilis bmeB disruption mutants than for the parental strain. The putative efflux pump operon is composed of bmeA, bmeB, and bmeC (a putative outer membrane channel protein homologous with OprM). Addition of the efflux pump inhibitors, carbonyl cyanide m-chlorophenylhydrazone (a proton conductor that eliminates the energy source) and Phe-Arg β-naphthylamide (MC-207,110) (the first specific inhibitor described for RND pumps in P. aeruginosa), resulted in lowered MICs in the parental strain but not in the bmeB disruption mutant, indicating that the bmeB pump is affected by these inhibitors. This is the first description of RND type pumps in the genus Bacteroides.
Bacteroides fragilis, an anaerobic gram-negative rod, is an opportunistic pathogen that can cause significant mortality in infections resulting from abdominal trauma or surgery (7, 19). Although it accounts for only 0.5% of the enteric flora, it is the Bacteroides species most frequently isolated from patients with intra-abdominal infections and/or bacteremia (in which mortality reached 45% if inactive therapy was given). It often presents a serious problem for therapy, since it is resistant to many antibiotics, including most of the penicillins, cephalosporins, and the quinolones (1, 3, 19, 20, 25).
Gram-negative bacteria including B. fragilis are usually more resistant to a large number of antibiotics and other noxious agents than are gram-positive bacteria. Clinically significant levels of antibiotic resistance are caused by interplay between the efficient outer membrane (OM) permeability barrier, ubiquitous periplasmic β-lactamases, and recently recognized multidrug resistance (MDR) efflux pumps (17). These pumps have broad substrate specificity and may act synergistically with the permeability barrier to result in significant intrinsic resistance to many antimicrobials. These pumps expel the antimicrobial from the cell into the surrounding space, and the antimicrobials then have to pass through the OM permeability barrier to regain entry to the cell (18). Thus, the MDR pumps can effect significant resistance even when their transporter activity is quite low, as long as the OM functions as an effective barrier. Antimicrobials which are expelled by these pumps include fluoroquinolones, chloramphenicol, and β-lactam antibiotics that have lipophilic side chains (34). For many β-lactams and carbapenems that are not hydrolyzed by periplasmic enzymes, synergy between efflux and the permeability barrier is necessary for effective drug resistance (18). Pumps belonging to the resistance-nodulation-division (RND) family of transporters are the major multidrug efflux pumps in both Escherichia coli and Pseudomonas aeruginosa. The pumps in this family consist of three components: the inner membrane transporter, a periplasmic lipoprotein, and an OM channel (31).
The first multidrug efflux operon described was mexAB-oprM in P. aeruginosa; acrAB-tolC comprises the corresponding system in E. coli. Both systems have broad substrate specificities. In Pseudomonas, the products MexA, MexB, and OprM (in this order in the operon) are the linker protein, transporter, and OM channel, respectively (9, 21, 22, 34). This system is the major efflux pump associated with intrinsic resistance among 17 possible RND efflux pumps in P. aeruginosa. P. aeruginosa is intrinsically more resistant than E. coli due to both a highly impermeable OM and the presence of multiple efflux systems (5, 23). Inactivation of this pump renders P. aeruginosa even more susceptible to most antibiotics than the average E. coli strain. Therefore, we decided to investigate the presence and function of RND type systems in B. fragilis, a gram-negative anaerobic bacterium that is the major cause of anaerobic infections. Like P. aeruginosa, B. fragilis has a relatively large chromosome (6.3 Mb and 5.2 to 5.4 Mb, respectively).
Little is known about efflux pumps in obligate anaerobic bacteria. The BexA pump, a member of the multidrug and toxic compound extrusion class (MATE), was described in Bacteroides thetaiotaomicron and XepCAB, a member of RND pump family, was described in Porphyromonas gingivalis (6, 13). The aim of this study was to determine whether genes for RND efflux pumps are present in the B. fragilis genome and to determine their involvement in resistance to antimicrobials. Since the B. fragilis ATCC 25285 genome has been completed, we used genomic analysis to locate RND homologs in B. fragilis. This report describes the identification of 16 homologs of the MDR pump system in B. fragilis, construction and characterization of disruption mutants of four of these homologs, and the cloning of a portion of one of the homologs, bmeAB, in a hypersusceptible strain of E. coli. This is the first description of the RND pump system in the genus Bacteroides.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. E. coli strains were grown on Luria-Bertani (LB) agar or broth at 37°C containing appropriate antibiotics when necessary. B. fragilis ATCC 25285 and its mutants were grown anaerobically in GAM broth (Nissui, Tokyo, Japan) or Brucella HK agar (Kyokuto Pharmaceutical Ind., Tokyo, Japan) in an anaerobic chamber (5% CO2, 10% H2, 85% N2) as described previously (32).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Plasmid | Relevant descriptiona | Reference and/or source |
|---|---|---|---|
| Escherichia coli | |||
| DH5α | Host strain for cloning | 4 | |
| S17-1 | Mobilizing strain for Bacteroides spp.; TMPr | 30 | |
| KAM3 | Deletion in the chromosomal acrAB genes from E. coli DH5α | 14 | |
| DH5α | pGERM | Suicide vector in Bacteroides; AMPrb ERYr | 27; N. B. Shoemaker |
| S17-1 | pFMB2744 | pGERM containing a 1,580-bp internal fragment from B. fragilis bmeB | This study |
| S17-1 | pBMA1 | pGERM containing a 828-bp internal fragment from B. fragilis bmeA | This study |
| S17-1 | pBOP3 | pGERM containing a 1,142-bp internal fragment from B. fragilis bmeC | This study |
| TOP10 | pCR-XL TOPO | Cloning vector; KANr | Invitrogenc |
| KAM3 | pTPOUT25 | pCR-XL-TOPO containing a 4,891-bp fragment from bmeAB in B. fragilis | This study |
| KAM3 | pTPIN5 | pCR-XL-TOPO containing a 2,622-bp fragment from bmeAB in B. fragilis | This study |
| Bacteroides fragilis | |||
| ATCC 25285 | Type strain | ATCC | |
| FMB271 | bmeB disruption mutant; ERYr | This study | |
| FMAB1 | bmeAB disruption mutant; ERYr | This study | |
| FMA1 | bmeA disruption mutant; ERYr | This study | |
| FOM1 | bmeC disruption mutant; ERYr | This study | |
| FMB1 | Homolog 1 disruption mutant; ERYr | This study |
Abbreviations: TMP, trimethoprim; AMP, ampicillin; ERY, erythromycin; KAN, kanamycin.
AMPr, ampicillin resistance expressed in E. coli only under aerobic conditions; ERYr, erythromycin resistance in Bacteroides spp.
Invitrogen Corp., Carlsbad, CA.
Search for mexB homologs in the B. fragilis genome sequence.
The sequence data were produced by the Bacteroides fragilis Sequencing Group at the Sanger Institute and can be obtained from http://www.sanger.ac.uk/Projects/B_fragilis/. A basic local alignment search tool (BLAST) homology search was carried out with the B. fragilis NCTC9343 (ATCC 25285) genome sequence using the whole amino acid sequence of MexB (P. aeruginosa PAO1 MexB; accession number AAG03815) as a query.
Construction of disruption mutants.
DNA manipulations were carried out by standard procedures as described by Sambrook and Russell (28). The primers used in PCR are shown in Table 2. The pGERM suicide vector (gift of Abigail Salyers, University of Illinois, Urbana) was used in insertion-mediated mutagenesis to construct gene disruption mutants (27). pGERM cannot replicate in Bacteroides spp. but confers erythromycin resistance for Bacteroides spp. when inserted into the chromosome. Thus, a single homologous recombination event occurring in the internal homologous locus of a potential gene for an efflux pump (e.g., bmeB) will result in the insertion of pGERM::‘bmeB' into the chromosome.
TABLE 2.
Primers used for PCR
| Primer | Associated gene | Starta | 5′-3′ Sequence | End |
|---|---|---|---|---|
| MDU-1 | bmeB | 163288 | TCTTCATTCGGTACCAAACCGGb | 163309 |
| MDU-2 | bmeB | 164868 | TCCTACTGTACGTGTCAACACGb | 164847 |
| MDU-3 | bmeA | 165910 | TCGATCCGGTGCAGTATGAA | 165891 |
| MDU-4 | bmeAB | 166579 | AACAGTTGGGGAACACTTC | 166561 |
| MDU-5 | bmeAB | 161689 | TAGGTCTGTCATATGATTTG | 161708 |
| MDU-6 | bmeA | 165082 | TTGATAGTAGCACCGTCTTTC | 165102 |
| MDU-7 | bmeC | 161629 | TCCGATACTGTCAACTTCGG | 161610 |
| MDU-8 | bmeC | 160488 | GCCGACAATACCTCCAGATA | 160507 |
| MDU-9 | bmeARTS | 165279 | CCCTCAAAAAGCAACTTACGA | 165259 |
| MDU-10 | bmeARTAS | 165019 | TGATTTTTGCGTTCTTCCAA | 165038 |
| MDU-11 | bmeBRTS | 163235 | CGCACTCAGGAAGTGATGTG | 163216 |
| MDU-12 | bmeBRTAS | 162916 | TCCAGACTACCGCCTGTCTT | 162935 |
| MDU-13 | bmeCRTS | 160651 | TTGAATGCCGGAAGTGAAGT | 160632 |
| MDU-14 | bmeCRTAS | 160397 | GCCCAATGCCTGATAGAGG | 160415 |
| MDU-15 | Bf.recBRTS | 2551202 | TATCAGCCTGGGAATGGAAG | 2551183 |
| MDU-16 | Bf.recBRTAS | 2550893 | TTTATGGAGGAGCAGCGAAT | 2550912 |
The number is from the B. fragilis ATCC 25285 genome sequence.
Restriction sites of KpnI and HincII are underlined in MDU-1 and MDU-2, respectively.
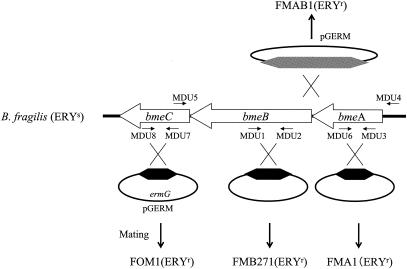
The bmeB disruption mutant was constructed by amplifying a 1,580-bp internal fragment of a mexB homolog open reading frame of B. fragilis ATCC 25285 with high-fidelity, blunt-end-producing Pyrobest DNA polymerase (Takara, Kyoto, Japan) using primers MDU-1 and MDU-2 that included the KpnI and HincII restriction sites, respectively (underlined in Table 2). The amplified DNA was digested with KpnI and HincII, and the fragments were inserted into pGERM that had been digested with KpnI and SmaI, resulting in pFMB2744. E. coli S17-1 (30) was transformed with pFMB2744, and transformants were selected by ampicillin and trimethoprim. Matings with E. coli S17-1/pFMB2744 and B. fragilis ATCC 25285 were conducted (13, 32). Transconjugants were selected on Brucella HK plates containing gentamicin (at 200 μg/ml to select against the donor) and erythromycin (at 10 μg/ml, to select for recombinants). The disruption mutants were maintained by erythromycin selection. Verification of the disruption was obtained by genomic Southern blot analysis. A mutant was obtained in which the bmeB gene was disrupted and was designated B. fragilis FMB271 (Fig. 1).
FIG. 1.
Schematic map of bmeABC in B. fragilis. The main figure consists of a hypothetical operon composed of bmeA (a mexA homolog), bmeB (a mexB homolog), and bmeC (an oprM homolog). A schematic diagram of the insertion-mediated mutagenesis is included. For example, the pGERM recombinant carrying an internal DNA fragment of bmeB (indicated by the black hexagon) is integrated into the recipient by a single homologous recombination event (crossed black lines) to result in FMB271, a disruption mutant of bmeB. Small arrows indicate positions and directions of primers. Abbreviations: ERYs, erythromycin sensitive; ERYr, erythromycin resistant; ermG, erythromycin-resistant marker for Bacteroides spp.
Similarly, another insertion mutant, FMA1, was constructed using primers MDU-3 and MDU-6, in which the insertion was made at an adjacent upstream open reading frame encoding a mexA homolog, which we named bmeA. A third mutant, FOM1, was constructed as an insertion mutant of an adjacent downstream gene, an oprM homolog, which we named bmeC, using a fragment amplified with primers MDU-7 and MDU-8. The insertion mutant FMAB1 was constructed as an insertion mutant of the region between bmeA and bmeB using primers MDU-1 and MDU-3; this fragment included the first half of bmeB and a slightly shortened bmeA, extending almost to the start of bmeA (Table 3). Schematic indications of primers and mutants are shown in Fig. 1 and Table 3.
TABLE 3.
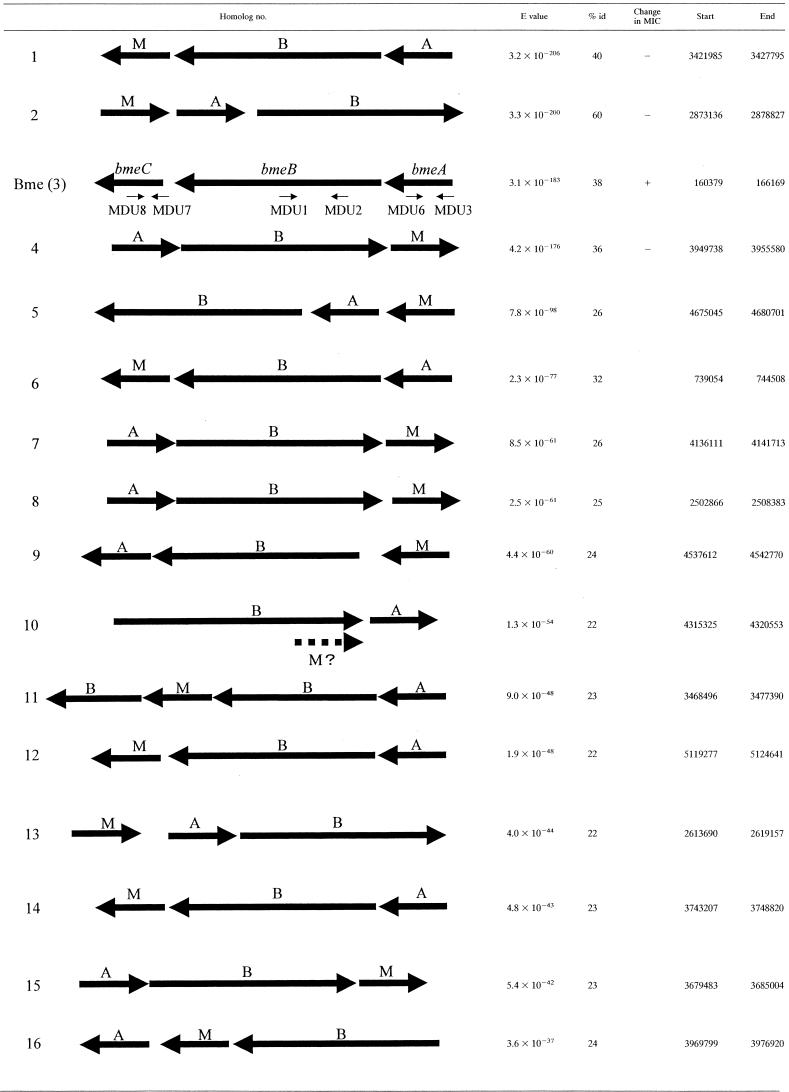
Sixteen homologs of B. fragilis mexAB-oprMa
Orientation of genes with the operons and positions of the operons in the B. fragilis ATCC 25285 genome and E values compared to those of P. aeruginosa MexB are shown. Disruption mutants of homologs 1 to 4 (homolog 3 = bme) were constructed and tested for susceptibility to a variety of antimicrobial agents. A plus sign indicates that the mutant was more susceptible than the parent, and a minus sign indicates no change in susceptibility. % id, percent identification.
Reverse transcription-PCR (RT-PCR) analysis of the disruption mutants.
Bacteria were grown as described above, and RNA was extracted with the PURESCRIPT RNA isolation kit (Gentra Systems, Minneapolis, MN), followed by DNase treatment (Takara). The absence of any contaminating chromosomal DNA was confirmed by conventional PCR. RNA (3.0 μg) was used in the first cycle with random primers (Takara) and reverse transcriptase (Toyobo, Osaka, Japan). cDNA (300 ng) was then used as template with the gene-specific primers (Table 2) and Taq polymerase (2 min at 95°C followed by 25 cycles, each cycle consisting of 1 min at 95°C, 30 s at 58°C, and 1 min at 72°C). PCR products were visualized by gel analysis and quantitated by measurements of optical density at 260 nm. Transcription of recB was used as a positive control (32).
Cloning of bmeAB in E. coli.
The bmeAB region (4,891 bp) of B. fragilis was amplified by PCR using primers MDU-4 and MDU-5 (Table 2) and cloned into an E. coli ΔacrAB mutant KAM3 as a host ((14), a kind gift of T. Tsuchiya, Okayama University). The MDU-4 primer was designed about 350 bp upstream from the start codon of bmeA to include the promoter sequence. The PCR product (4,891 bp) carrying bmeAB was ligated into the TOPO-XL cloning vector (Invitrogen Corp., Carlsbad, CA), resulting in pTPOUT25; E. coli KAM3 was transformed with pTPOUT25, and transformants were selected with kanamycin. The insertion fragment, bmeAB, of pTPOUT25 was sequenced to verify that no misamplification occurred. A truncated bmeAB was also cloned as a control, using primers MDU-1 and MDU-3 (Fig. 1). The plasmid containing the truncated bmeAB was named pTPIN5.
Susceptibility testing.
The MICs of a variety of antimicrobial agents for E. coli and B. fragilis strains were determined by broth microdilution assays in LB broth (overnight assay) and GAM broth (48-h assay) (15), respectively. MICs were originally measured in a twofold dilution series and then redone with smaller dilution intervals (for example, 1, 1.5, 2, 3, 4, 6, 8, 12, 16, and 24 μg/ml) to more accurately measure changes in MIC between the parental and mutant strains.
RESULTS
MDR pump homologs in B. fragilis.
At least 16 possible operons homologous to Pseudomonas mex operons were found in the B. fragilis genome by searching first with the P. aeruginosa MexB amino acid sequence and then with B. fragilis Mex sequences. Upstream and downstream analyses of the sequences surrounding the mexB homologs revealed the associated genes (mexA and oprM homologs). The order of the genes within the operon, E value, percent identity compared to Pseudomonas mexB, and positions within the B. fragilis genome (ATCC 25285) are indicated in Table 3. Analysis of the operon structure illuminated some unusual genetic arrangements for pump operons. All 16 mexB homologs were contained in an operon that also included a mexA homolog and an oprM homolog. Fourteen of the 15 operons had these three genes in the order indicated in Table 3; each gene had its own start and stop codons. In the Mex10 homolog, the OprM homolog was found as a fusion protein at the end of the MexB homolog. This same pattern was seen in both B. fragilis ATCC 25285 and the laboratory strain B. fragilis 638R. Homolog 11 had an additional mexB-like gene after the oprM gene.
We chose the four genes most homologous to Pseudomonas mexB and constructed insertion mutants using the pGERM suicide vector in an insertion-mediated mutagenesis scheme as described above. We isolated mutants FMB1, FMB241, FMB271, and FMB771, corresponding to homolog number 1, 2, 3 (BmeB), and 4, respectively (Table 3).
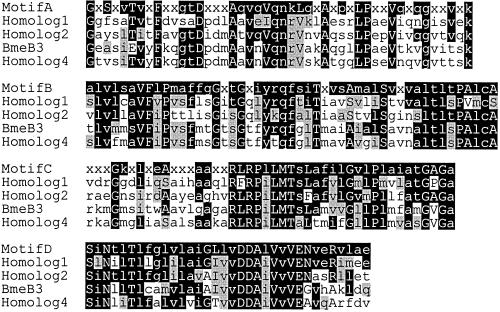
Susceptibilities of the four individual disruption mutants to various antimicrobial agents were determined. Changes in MIC were seen only with FMB271 (Table 4). MICs of FMB1, FMB241, and FMB771 were unchanged (data not shown). The mexB3 homolog that was disrupted in FMB271 (renamed bmeB) showed 38% identity to P. aeruginosa mexB (E value, 3.1 × 10−183) (Table 3). The bmeB gene hypothetically encoded a protein with 1,067 amino acids; hydropathy analysis, using GENETYX-MAC v10.1 software (Genetyx Co., Tokyo, Japan), indicated that it contained 12 transmembrane segments, as do other RND pumps. In addition, BmeB had a high degree of similarity to consensus sequences of conserved motifs in the RND family (Fig. 2) (24). Further, a mexA homolog, named bmeA (encoding 384 amino acids; E value of 1.3 × 10−35 compared to P. aeruginosa MexA), and an oprM homolog, named bmeC (E value of 3.2 × 10−46 compared to P. aeruginosa OprM), were found at the adjacent upstream and downstream positions of bmeB, respectively, corresponding to the order of the mexAB-oprM operon in P. aeruginosa (22). Thus, bmeABC appeared to be a member of the mexB-type RND family of efflux pumps. Based on our finding that bmeB was important in antimicrobial efflux, we constructed disruption mutants of the other components of the bme operon as described above and listed in Table 1.
TABLE 4.
Antibiotic susceptibilities of various strainsa
| MIC (μg/ml)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CEXc | CFZ | ZOX | LEX | CLS | PLB | BCT | VAN | FDA | NVB | PUR | |
| B. fragilis ATCC 25285 | 128 | 256 | 64 | 256 | 512 | 256 | 2,048 | 48 | 2 | 128 | 32 |
| FMB271 (ΩbmeB::pGERM) | 64 | 128 | 16 | 64 | 256 | 64 | 512 | 16 | 1 | 64 | 8 |
| FMAB1 (ΩbmeAB::pGERM) | 64 | 128 | 32 | 64 | 256 | 64 | 1,024 | 32 | 1 | 96 | 24 |
| FMA1 (ΩbmeA::pGERM) | 64 | 128 | 16 | 64 | 256 | 64 | 512 | 16 | 1 | 64 | 8 |
| FOM1 (ΩbmeC::pGERM) | 128 | 256 | 64 | 256 | 512 | 256 | 2,048 | 48 | 2 | 128 | 32 |
| E. coli KAM3 | 8 | 1 | 0.0625 | 16 | 0.5 | 2 | >2,048 | 512 | 4 | 4 | 4 |
| KAM3-pTPOUT25 | 16 | 2 | 0.125 | 32 | 1 | 4 | >2,048 | 512 | 4 | 4 | 16 |
| KAM3-pTPIN5 | 8 | 1 | 0.0625 | 16 | 0.5 | 2 | >2,048 | 512 | 4 | 4 | 4 |
MIC determination by the broth microdilution assay was repeated at least three times in each case, and the consistencies of the MICs were confirmed. MICs were originally measured in a twofold dilution series and then redone with smaller dilution intervals (for example, 1, 1.5, 2, 3, 4, 6, 8, 12, 16, and 24 μg/ml) to more accurately measure changes in MIC between the parental and mutant strains.
No changes in the MICs of the following agents for the B. fragilis strains were seen (MICs are given in parentheses): norfloxacin (32 μg/ml), chloramphenicol (2 μg/ml), tetracycline (0.25 μg/ml), ethidium bromide (32 μg/ml), deoxycholic acid (256 μg/ml), and protamine (>512 μg/ml).
Abbreviations: CEX, cephalothin; CFZ, cefazolin; ZOX, ceftizoxime; LEX, cephalexin; CLS, colistin; PLB, polymyxin-B; BCT, bacitracin; VAN, vancomycin; FDA, fusidic acid; NVB, novobiocin; PUR, puromycin.
FIG. 2.
Multiple sequence alignments of B. fragilis MexB1, MexB2, BmeB (MexB3), and MexB4. Conserved motifs were aligned using ClustalX 1.8 and visualized with MacBoxshade 2.15. The motifs were identified by Putman et al. (24). Black and gray shaded residues are identical and similar to each other, respectively. The consensus sequences of the motifs are displayed as follows: x, any amino acid; uppercase letters, amino acid occurring in >70% of the examined sequences; lowercase letters, amino acid occurs in >40% of sequences examined.
RT-PCR analysis of disruption mutants of bmeA, bmeB, and bmeC.
Results of the RT-PCR analysis are shown in Table 5. FMB1 is a disruption mutant of the mexB-like gene of homolog 1 and was included as an additional positive control. The results indicate that the genes were transcribed in a polycistronic manner, consistent with nearly all characterized tripartite efflux pumps. Thus, if bmeA was disrupted, neither bmeB nor bmeC was transcribed. If bmeB was disrupted, bmeA was transcribed, but not bmeB and bmeC. In the FOM1 disruptant, both bmeA and bmeB were transcribed. The bmeAB disruptant would be predicted to have a full-length bmeA (with its promoter) followed by (in order) a truncated bmeB, pGERM plasmid insert, foreshortened plasmid bmeA (missing the beginning of bmeA and its promoter), and foreshortened bmeB (plasmid chromosomal hybrid). Thus, there were intact bmeA and bmeB genes in FMAB1, but they were not contiguous. As predicted, only the bmeA gene is transcribed.
TABLE 5.
RT-PCR analysis of disruption mutants
| Primers | Analysis of strain (gene disrupted)a
|
|||||
|---|---|---|---|---|---|---|
| FMA1 (ΩbmeA) | FMB271 (ΩbmeB) | FOM1 (ΩbmeC) | FMAB1 (ΩbmeAB) | FMB1 (ΩmexB1) | ATCC 25285 (type strain) | |
| bmeA primers (MDU 9/10) | − | + | + | + | + | + |
| bmeB primers (MDU 11/12) | − | − | + | − | + | + |
| bmeC primers (MDU 13/14) | − | − | − | − | + | + |
| recB primers (MDU 15/16) | + | + | + | + | + | + |
+, gene transcribed; −, gene not transcribed.
Determination of MICs for the disruption mutants of bmeA, bmeB, and bmeC.
The disruption mutant FMB271 carrying the pGERM::‘bmeB' insertion was two- to fourfold-more susceptible to cephems (cephalothin, cefazolin, ceftizoxime, and cephalexin), polypeptide antibiotics (polymyxin-B, colistin, bacitracin, and vancomycin), fusidic acid, novobiocin, and puromycin than the parent strain, B. fragilis ATCC 25285 (Table 4). There was no change in the MICs to quinolones (nalidixic acid, norfloxacin, and ofloxacin), tetracycline, chloramphenicol, and ethidium bromide. FMA1, a disruption mutant of bmeA, had the same pattern as FMB271. The pattern of FOM1 (the OM channel disruptant), in which both bmeA and bmeB are transcribed, was the same as that of the parental strain, suggesting that BmeC, a potential pump channel, might be interchangeable with other homologous OM protein channels. Also, this demonstrates that the phenotype of the bmeB insertion (i.e., lowered MICs) was due to the bmeB disruption and not to a downstream effect on bmeC. As predicted by the results of the transcription analysis, the susceptibility pattern of FMAB1 was the same as those of FMA1 and FMB271, since only the bmeA gene was transcribed.
Cloning of bmeAB in E. coli.
To further investigate the function of bmeAB, we cloned the entire bmeAB locus into the hypersusceptible E. coli KAM3 as a host (KAM3/pTPOUT25). A truncated portion of the gene including the C-terminal portion of bmeA and the N-terminal part of bmeB was also cloned as a negative control (KAM3/pTPIN5). MICs of various antimicrobials for E. coli transformants carrying the genes were determined (Table 4). KAM3/pTPOUT25 (carrying the intact bmeAB) showed moderately increased resistance to some of the drugs that showed lower MICs in the disruption mutant, suggesting that the genes were expressed and functional as a multidrug efflux pump in E. coli, especially for cephems, colistin, polymyxin B, and puromycin.
Effects of efflux inhibitors on the B. fragilis multidrug efflux pump.
Carbonyl cyanide m-chlorophenylhydrazone (CCCP), a proton conductor, and Phe-Arg β-naphthylamide (MC-207,110), the first specific inhibitor described for RND pumps in P. aeruginosa, were used to characterize the BmeAB pump (10, 26). As shown in Table 6, addition of either CCCP or MC-207,110 resulted in two- to fourfold-lower MICs of cephalexin, polymyxin-B, fusidic acid, and novobiocin for the parental strain. These effects occurred at concentrations well below the inhibitory concentrations of either CCCP or MC-207,110. When the BmeB pump was disrupted (FMB 271), the effects of CCCP and MC-207,110 were not seen. As CCCP is a proton pump inhibitor, this suggests that the BmeB pump is driven by a proton gradient, consistent with other RND pumps. The finding that FMB271 is still fairly resistant to certain agents suggests that there are other pumps that are active in efflux of these agents. However, it appears that bmeB is the only pump affected by CCCP and MC-207,110 that is active in efflux of these agents. Apparently, the other pumps were not affected by these two efflux inhibitors. Further study will more fully resolve these issues.
TABLE 6.
Susceptibility of the BmeB pump to potential inhibitorsa
| Inhibitor | LEX
|
PLB
|
FDA
|
NVB
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − | +MCb | +CC | − | +MC | +CC | − | +MC | +CC | − | +MC | +CC | |
| ATCC 25285 | 256 | 64 | 64 | 256 | 64 | 64 | 2 | 1 | 1 | 128 | 64 | 64 |
| FMB271 | 64 | 64 | 64 | 64 | 64 | 64 | 1 | 1 | 1 | 64 | 64 | 64 |
Abbreviations: LEX, cephalexin; PLB, polymyxin-B; FDA, fusidic acid; NVB, novobiocin; MC, MC-207,110 (Phe-Arg β-naphthylamide); CC, CCCP (carbonyl cyanide m-chlorophenylhydrazone). Values are MICs (in micrograms per milliliter). A minus sign indicates that no inhibitor was added.
MC-207,110 and CCCP were added at 20 μg/ml and 200 μM, respectively. The MICs of MC-207,110 and CCCP for B. fragilis ATCC 25285 and FMB271 were >128 μg/ml and 200 μM, respectively.
DISCUSSION
Our original searches of the B. fragilis genome were conducted using the Pseudomonas mexB sequence and revealed nine homologs. Analysis of the upstream and downstream regions of these homologs were conducted and confirmed that each of the homologs was associated with both a MexA homolog and an OprM homolog, in a variety of arrangements. Since the genome sequencing of B. fragilis ATCC 25285 has been completed and the genome sequence of B. thetaiotaomicron has been published (33), we searched them for homologs of the BmeB protein by using TBLASTN. We have not yet similarly searched the B. fragilis YCH46 genome sequence, since it was only very recently published (8). With this search in B. fragilis ATCC 25285, 7 additional homologs (i.e., 16 total) of the mex operon were identified (E value, <9 × 10−15). There were no significant differences in homolog analysis using either the latest or the past raw unfinished sequences at the early stage of this study. Interestingly, we also found 17 MDR pump homologs (E value, <2 × 10−36) in the B. thetaiotaomicron genome. Both B. fragilis and B. thetaiotaomicron are predominantly colonic anaerobes and both have multiple homologs of a variety of genes. Perhaps the multiplicity of pumps gives the organisms an advantage in specifically regulating the efflux of a wide range of toxic substances that they may encounter in the gut.
We constructed disruptions of four B. fragilis mexB homologs to evaluate their contribution to antimicrobial resistance. We chose the genes to be disrupted by ranking in order of degree of homology with P. aeruginosa mexB. Of these mexB-like disruption mutants, FMB271 was the only mutant that was more susceptible than the parental strain for the drugs tested. There are several possible explanations for these results. First, only 4 of the 16 mex-like homologs were tested. Although these were the four homologs most like Pseudomonas mexB, other homologs may be important. Second, the drugs tested might not be the appropriate substrates for the pumps investigated. Third, potential mexB homologs may not be expressed constitutively and may be “silent” or unexpressed under the conditions tested. Also, any efflux pump impaired by mutation may be compensated for by increased action of the other pumps.
Analysis of the disruption mutants of the bme homolog (homolog 3) indicated that the genes are transcribed in a polycistronic manner. Analysis of the susceptibility patterns of the various disruptants is consistent with the results of the transcription analysis. Thus, if bmeA is disrupted, none of the downstream genes is transcribed. The patterns of FMA1, FMB271, and FMAB1 (in which transcription of both bmeA and bmeB [FMA1] or bmeB [FMB271 and FMAB1] are disrupted) are similar. The pattern of FOM1 (the outer membrane channel disruptant), in which both bmeA and bmeB are transcribed, is similar to that of the parental strain, suggesting that another outer membrane channel may be utilized when bmeC is unavailable. This pattern is also seen with other MDR pumps (31).
Gene arrangement of the MDR pumps in B. fragilis differs from other well-characterized systems. In P. aeruginosa, 17 possible RND family secondary transporters have now been identified (http://www.membranetransport.org/). Among them, at least six RND efflux pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY-OprM, MexHI-OpmD, and MexJK-OprM) have been characterized (2, 11, 22, 29, 31). In Pseudomonas MDR pump operons, genes encoding the RND transporter (MexB homolog) and periplasmic membrane fusion protein (MexA homolog) are always present, while the gene encoding the OM channel (OprM homolog) is not always present (34). In E. coli RND pumps, the genes for transporter and membrane fusion proteins (acrAB) occur together as an operon, and the gene for the TolC OM channel occurs elsewhere on the genome (34). In contrast, each of the 16 MDR pump operons in B. fragilis contains all three of the components of the tripartite pump complex. In addition, to our knowledge, there are no other homologs in which the OprM portion is transcribed as a fusion protein at the end of MexB (Mex10).
Pseudomonas MexAB-OprM is expressed constitutively in cells under the standard conditions and contributes to intrinsic resistance to many antimicrobials; its disruption mutant becomes hypersusceptible to various antimicrobial agents, indicating a significant level of expression of this operon in the wild-type strain (9, 17). To date, none of the pumps we identified in B. fragilis appeared to have such a significant effect on the susceptibility of the organism. However, it is possible that insertion mutagenesis of one pump gene may be compensated for by increased expression of another pump gene. Investigations are under way with multiple deletion mutants and assays of pump transcription and expression to further investigate this possibility. Also, we are investigating the role of potential regulators in pump expression. While our early searches with E. coli AcrR and P. aeruginosa MexR (efflux pump regulators) (23) did not reveal any homologs in B. fragilis, subsequent analysis using a MexR conserved domain sequence revealed at least two potential regulators (one adjacent to Mex homolog 10 and one adjacent to Mex homolog 15). Subsequently, the AcrR consensus sequence was used to BLAST search the NCBI Protein database; the resulting hits above an E value threshold of e−8 (247 sequences) were then used to BLAST search the B. fragilis ATCC 25285 genome at the Sanger Center, resulting in three additional putative homologs. One of these potential regulators is immediately upstream of the mex5 operon. Mutations within the repressor have been shown to lead to overexpression of the corresponding efflux pumps in other organisms (17). Experiments to determine the effect of disrupting these selected regulator genes in B. fragilis are under way.
Phenograms indicating the phylogenetic relatedness of these proteins to the E. coli and P. aeruginosa efflux pumps were constructed (data not shown). The 16 pumps can be roughly separated into three groups, and the groups are generally, but not always, consistent among the MexB, MexA, and OprM families. Mex 1, Mex 2, Bme, and Mex 4 are the most homologous to P. aeruginosa MexAB-OprM, and thus were initially chosen for the disruption experiments. Among the OprM homologs, nine have the typical conserved domain structure (two adjacent OEP [outer membrane efflux protein] motifs) and seven have some combination of an OEP and/or TolC motif (but not necessarily a double OEP). The MexA and MexB homologs all have the expected motifs.
To date, the only multidrug efflux pump described in Bacteroides has been BexA (12, 13) (a MATE family pump) in B. thetaiotaomicron. XepCAB (6), an RND family pump, has been described in P. gingivalis (16), a related gram-negative anaerobic rod. XepCAB was the first example of an RND pump in an anaerobic organism, and the order of genes in the operon was xepC, xepA, and xepB. With B. fragilis, we have identified at least 16 RND-type pumps in the genome. We have constructed disruptions in four of these pumps. Thus far, we have demonstrated efflux activity only with the BmeABC homolog. B. fragilis and the related anaerobe, B. thetaiotaomicron, carry numerous homologs of a variety of pump genes; both genera also carry many MATE family efflux pumps, and B. thetaiotaomicron contains 60 proteins predicted to be components of drug efflux systems (13, 33). The sheer number of pump genes carried by both B. fragilis and B. thetaiotaomicron suggests that active efflux is a major mechanism underlying antimicrobial resistance in these anaerobes. Future studies will involve systematic investigation on the function and expression of each of the efflux pump homologs of the various families in the genome.
Acknowledgments
We thank Abigail Salyers and Nadja D. Shoemaker (University of Illinois, Urbana, Ill.) for sending bacterial strains and plasmids for genetics of Bacteroides spp., Tamami Saito for helpful advice and technical assistance, and the Sanger Center for making the B. fragilis ATCC 25285 genomic sequence freely available to the public throughout the sequencing stages.
This study was supported by a Scientific Research Special Grant from Matsumoto Dental University (to O.U.), by a Grant-in-Aid for Scientific Research (C) (15591957 to F.Y.) from the Japan Society for the Promotion of Science (JSPS) and the AGU High-Tech Research Center Project for Private Universities, by a matching fund subsidy from MEXT (Ministry of Education, Culture, Sports, Science and Technology), Japan (1001 to O.U. and F.Y.), and by Merit Review Funds from the U.S. Department of Veterans Affairs (to H.M.W.).
REFERENCES
- 1.Aldridge, K. E., D. Ashcraft, M. O'Brien, and C. V. Sanders. 2003. Bacteremia due to Bacteroides fragilis group: distribution of species, β-lactamase production, and antimicrobial susceptibility patterns. Antimicrob. Agents Chemother. 47:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falagas, M. E., and E. Siakavellas. 2000. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int. J. Antimicrob. Agents 15:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Gardner, R. G., J. B. Russell, D. B. Wilson, G.-R. Wang, and N. B. Shoemaker. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed β-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl. Environ. Microbiol. 62:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock, R. E. W., and F. S. L. Brinkman. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda, T., and F. Yoshimura. 2002. A resistance-nodulation-cell division family xenobiotic efflux pump in an obligate anaerobe, Porphyromonas gingivalis. Antimicrob. Agents Chemother. 46:3257-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jousimies-Somer, H. R., P. H. Summanen, H. Wexler, S. M. Finegold, S. E. Gharbia, and H. N. Shah. 2003. Bacteroides, Porphyromonas, Prevotella, Fusobacterium, and other anaerobic gram-negative bacteria, p. 880-901. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 8.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamae, S., H. Nikaido, Y. Tanaka, and F. Yoshimura. 1998. Active efflux of norfloxacin by Bacteroides fragilis. Antimicrob. Agents Chemother. 42:2119-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 1997. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 4th ed. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 16.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido, H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 12:215-223. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, I., C. O. Solberg, and S. M. Finegold. 1999. A primer on anaerobic bacteria and anaerobic infections for the uninitiated. Infection 27:159-165. [DOI] [PubMed] [Google Scholar]
- 20.Oteo-Iglesias, J., J.-I. Alós, and J.-L. Gómez-Garcés. 2002. Increase in resistance to new fluoroquinolones from 1998 to 2001 in the Bacteroides fragilis group. J. Antimicrob. Chemother. 50:1055-1057. [DOI] [PubMed] [Google Scholar]
- 21.Poole, K., K. Krebes, C. Mcnally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 23.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 24.Putman, M., H. W. Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen, B. A., K. Bush, and F. P. Tally. 1997. Antimicrobial resistance in anaerobes. Clin. Infect. Dis. 24(Suppl. 1):S110-S120. [DOI] [PubMed] [Google Scholar]
- 26.Renau, T. E., R. Léger, E. M. Flamme, J. Sangalang, M. W. She, R. Yen, C. L. Gannon, D. Griffith, S. Chamberland, O. Lomovskaya, S. J. Hecker, V. J. Lee, T. Ohta, and K. Nakayama. 1999. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42:4928-4931. [DOI] [PubMed] [Google Scholar]
- 27.Salyers, A. A., G. Bonheyo, and N. B. Shoemaker. 2000. Starting a new genetic system: lessons from Bacteroides. Methods 20:35-46. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sekiya, H., T. Mima, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and characterization of a multidrug efflux pump, MexHI-OpmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47:2990-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology 1:784-791. [Google Scholar]
- 31.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda, O., and F. Yoshimura. 2003. Transposon-induced norfloxacin-sensitive mutants of Bacteroides thetaiotaomicron. Microbiol. Immunol. 47:17-25. [DOI] [PubMed] [Google Scholar]
- 33.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 34.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]