Abstract
The in vitro activities of DX-619, des-fluoro(6) quinolone, against 1,208 clinical isolates were examined. DX-619 was particularly potent against staphylococci, including ciprofloxacin- and methicillin-resistant strains; the MIC at which 90% of the strains tested were inhibited was 0.5 μg/ml. In addition, DX-619 was also active against gram-negative bacteria.
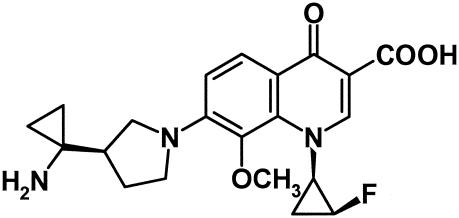
The development of resistance to antimicrobial agents and the emergence of multidrug-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA), methicillin-resistant coagulase-negative staphylococci (MRCNS), penicillin-resistant Streptococcus pneumoniae, vancomycin-resistant enterococci (VRE), extended-spectrum β-lactamase-producing gram-negative rods, and multidrug-resistant Pseudomonas aeruginosa (1, 8, 10, 13, 17, 20), have generated worldwide concern in the medical community. Among these, MRSA and VRE are common gram-positive pathogens of nosocomial infections which account for outbreaks and are increasing in frequency (4, 5, 14). Furthermore, community-acquired MRSA infections have been reported in recent years (6). Vancomycin is still widely used against serious infections caused by MRSA and enterococci, because there are only a few therapeutic options (19). The emergence of vancomycin-resistant strains of MRSA has been reported sporadically since 2002 (2, 9, 18). Recently, linezolid, a new synthetic oxazolidinone active against MRSA and VRE, has been a potential alternative (3, 22). However, linezolid- and vancomycin-resistant enterococci have been reported already (15). These problems reveal an urgent need for new antibacterials that are active against multidrug-resistant gram-positive bacteria. In this context, a novel des-fluoro(6) quinolone, DX-619, has been synthesized, with the chemical structure shown in Fig. 1.
FIG. 1.
Chemical structure of DX-619.
In this study, we compared the antimicrobial activity of DX-619 with those of other quinolones and the other classes of antibacterial agents, including anti-gram-positive bacterial agents, against freshly isolated bacteria.
(This study was presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 14 to 17 September 2003 [H. Inagaki et al., abstr. F-1054].)
DX-619, ciprofloxacin, clinafloxacin, garenoxacin, gatifloxacin, levofloxacin, moxifloxacin, sitafloxacin, and linezolid were synthesized at Daiichi Pharmaceutical Co. Ltd., Tokyo, Japan. Ampicillin, benzylpenicillin, cefaclor, ceftazidime, ceftriaxone, imipenem, oxacillin, arbekacin, clindamycin, metronidazole, minocycline, quinupristin-dalfopristin, teicoplanin, and vancomycin were purchased from the manufacturers or Sigma Aldrich Japan (Tokyo, Japan). Each drug was used as an anhydrous free base.
Bacterial strains were collected by the Levofloxacin Surveillance Group from patients in Japan in 2000 (20), with the exception of the strains mentioned below. Streptococcus agalactiae, Neisseria gonorrhoeae, Stenotrophomonas maltophilia, and anaerobic bacteria isolated in Japan were obtained from BML, Inc. (Saitama, Japan). Five ciprofloxacin-resistant strains of Streptococcus pneumoniae were isolated in Asia and Europe in 1997 and 1998 (16), and nine such strains were collected by the Levofloxacin Surveillance Group in Japan in 2002 (21). VRE were obtained from Creighton University (Omaha, Nebr.) and from Kyoto Pharmaceutical University and Gunma University in Japan.
MICs were determined according to the standard agar dilution method recommended by NCCLS (11) for bacterial species other than Haemophilus influenzae and anaerobes, for which the agar dilution method recommended by the Japanese Society of Chemotherapy was used (7). Mueller-Hinton agar (Becton Dickinson, Sparks, Md.)supplemented with 5% sheep blood (Kohjin Bio Co., Ltd., Saitama, Japan) was used for streptococci and Moraxella catarrhalis, and GC agar (Becton Dickinson) was used for N. gonorrhoeae. Mueller-Hinton agar supplemented with 5% Fildes enrichment (Becton Dickinson) was used for H. influenzae, and modified Gifu anaerobe medium agar (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) was used for anaerobic bacteria. Drug-containing agar plates were incubated with one loopful of inoculum, corresponding to about 104 CFU (about 105 CFU for S. pneumoniae) per spot, and were incubated at 35°C for 20 h (48 h for Peptostreptococcus spp. and Clostridium difficile). N. gonorrhoeae was incubated under 5% CO2, and anaerobic bacteria were incubated under an anaerobic atmosphere. The MIC was defined as the lowest drug concentration that prevented visible growth of bacteria. Staphylococci, S. pneumoniae, enterococci, H. influenzae, P. aeruginosa, and N. gonorrhoeae were classified into three categories, susceptible, intermediate, or resistant, according to the breakpoint of NCCLS standards (12). The quality control strains recommended by NCCLS were included as internal controls throughout the study.
Table 1 shows the antibacterial activity of DX-619 against gram-positive bacteria in comparison with those of reference compounds. The MIC90s (MICs at which 90% of isolates are inhibited) of DX-619 against methicillin-susceptible Staphylococcus aureus and methicillin-susceptible coagulase-negative staphylococci were both 0.015 μg/ml. MIC90s of DX-619 against ciprofloxacin-susceptible MRSA, ciprofloxacin-resistant MRSA, and MRCNS were 0.008, 0.5, and 0.12 μg/ml, respectively. Against staphylococci, DX-619 showed the most potent activity among the compounds tested, including anti-gram-positive agents. DX-619 was especially potent against ciprofloxacin-resistant MRSA, inhibiting the growth of all strains at 1 μg/ml, a MIC 2-fold lower than those of vancomycin and linezolid, 4-fold lower than those of teicoplanin and quinupristin-dalfopristin, and at least 16-fold lower than those of the other compounds tested. MIC90s of DX-619 against penicillin-susceptible S. pneumoniae, penicillin-resistant S. pneumoniae, Streptococcus pyogenes, and S. agalactiae were 0.015, 0.03, 0.015, and 0.12 μg/ml, respectively. Against 19 strains of ciprofloxacin-resistant S. pneumoniae, MICs of DX-619 ranged from 0.015 to 0.12 μg/ml, and the activity was also the highest among the compounds tested. MIC90s of DX-619 against vancomycin-susceptible and -resistant Enterococcus faecalis were 0.25 and 0.5 μg/ml, respectively, and MIC90s against vancomycin-susceptible and -resistant Enterococcus faecium and vancomycin-resistant Enterococcus gallinarum were all 2 μg/ml. The activity against these VRE was also the highest among the reference compounds. DX-619 inhibited 90% of isolates of Peptostreptococcus spp. and C. difficile at 0.5 and 2 μg/ml, respectively.
TABLE 1.
Antibacterial activities of DX-619 and reference compounds against gram-positive bacteria
| Organism (no. of strains) and compound | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Staphylococcus aureus | ||||
| Methicillin susceptible (48) | ||||
| DX-619 | ≦0.004-0.06 | 0.008 | 0.015 | |
| Levofloxacin | 0.12-8 | 0.25 | 0.5 | |
| Sitafloxacin | 0.008-1 | 0.03 | 0.06 | |
| Ciprofloxacin | 0.12-32 | 0.5 | 2 | |
| Moxifloxacin | 0.03-2 | 0.06 | 0.12 | |
| Gatifloxacin | 0.06-4 | 0.12 | 0.25 | |
| Garenoxacin | 0.008-1 | 0.03 | 0.06 | |
| Vancomycin | 1-2 | 1 | 1 | |
| Teicoplanin | 0.25-1 | 0.5 | 0.5 | |
| Quinupristin-dalfopristin | 0.12-0.25 | 0.25 | 0.25 | |
| Linezolid | 2-4 | 2 | 4 | |
| Arbekacin | 4-128 | 8 | 16 | |
| Imipenem | 0.03-0.06 | 0.06 | 0.06 | |
| Ceftriaxone | 2-16 | 4 | 4 | |
| Minocycline | 0.12-16 | 0.12 | 0.12 | |
| Oxacillin | 0.25-1 | 0.5 | 1 | |
| Methicillin resistant, ciprofloxacin susceptible (24) | ||||
| DX-619 | ≦0.004-0.015 | 0.008 | 0.008 | |
| Levofloxacin | 0.12-0.25 | 0.25 | 0.25 | |
| Sitafloxacin | ≦0.004-0.03 | 0.03 | 0.03 | |
| Ciprofloxacin | 0.12-0.5 | 0.5 | 0.5 | |
| Moxifloxacin | 0.015-0.06 | 0.03 | 0.06 | |
| Gatifloxacin | 0.03-0.12 | 0.12 | 0.12 | |
| Garenoxacin | 0.008-0.03 | 0.015 | 0.03 | |
| Vancomycin | 1-2 | 2 | 2 | |
| Teicoplanin | 0.25-2 | 0.5 | 1 | |
| Quinupristin-dalfopristin | 0.12-0.5 | 0.25 | 0.5 | |
| Linezolid | 2 | 2 | 2 | |
| Arbekacin | 4-32 | 16 | 32 | |
| Imipenem | 0.25-64 | 1 | 32 | |
| Ceftriaxone | 32->128 | 128 | >128 | |
| Minocycline | 0.12-16 | 0.12 | 4 | |
| Oxacillin | 16->128 | 64 | >128 | |
| Methicillin resistant, ciprofloxacin resistant (99) | ||||
| DX-619 | 0.03-1 | 0.06 | 0.5 | |
| Levofloxacin | 4->128 | 8 | >128 | |
| Sitafloxacin | 0.25-16 | 0.5 | 8 | |
| Ciprofloxacin | 8->64 | 32 | >64 | |
| Moxifloxacin | 1-64 | 2 | 32 | |
| Gatifloxacin | 1-128 | 4 | 64 | |
| Garenoxacin | 0.25-64 | 1 | 32 | |
| Vancomycin | 0.5-2 | 1 | 2 | |
| Teicoplanin | 0.25-4 | 1 | 1 | |
| Quinupristin-dalfopristin | 0.12-4 | 0.5 | 0.5 | |
| Linezolid | 0.5-2 | 1 | 1 | |
| Arbekacin | 4->128 | 8 | 32 | |
| Imipenem | 1->128 | 32 | 64 | |
| Ceftriaxone | >128 | >128 | >128 | |
| Minocycline | 0.06-16 | 8 | 16 | |
| Oxacillin | 64->128 | >128 | >128 | |
| Coagulase-negative staphylococci | ||||
| Methicillin susceptible (46) | ||||
| DX-619 | ≦0.004-0.12 | 0.008 | 0.015 | |
| Levofloxacin | 0.12-16 | 0.25 | 0.5 | |
| Sitafloxacin | 0.008-0.25 | 0.015 | 0.03 | |
| Ciprofloxacin | 0.12-32 | 0.25 | 0.5 | |
| Moxifloxacin | 0.03-4 | 0.12 | 0.12 | |
| Gatifloxacin | 0.06-4 | 0.12 | 0.25 | |
| Garenoxacin | 0.015-1 | 0.03 | 0.06 | |
| Vancomycin | 0.5-4 | 1 | 2 | |
| Teicoplanin | 0.12-4 | 0.5 | 2 | |
| Quinupristin-dalfopristin | 0.12-0.25 | 0.12 | 0.25 | |
| Linezolid | 0.5-1 | 1 | 1 | |
| Arbekacin | 1-32 | 2 | 8 | |
| Imipenem | 0.015-0.03 | 0.03 | 0.03 | |
| Ceftriaxone | 0.5-8 | 1 | 2 | |
| Minocycline | 0.06-0.5 | 0.06 | 0.12 | |
| Oxacillin | 0.06-0.25 | 0.12 | 0.12 | |
| Methicillin resistant (45) | ||||
| DX-619 | 0.008-0.5 | 0.06 | 0.12 | |
| Levofloxacin | 0.25->128 | 8 | 32 | |
| Sitafloxacin | 0.015-2 | 0.25 | 0.5 | |
| Ciprofloxacin | 0.12->64 | 8 | 64 | |
| Moxifloxacin | 0.06-64 | 1 | 4 | |
| Gatifloxacin | 0.12-64 | 2 | 4 | |
| Garenoxacin | 0.03-32 | 1 | 4 | |
| Vancomycin | 1-4 | 2 | 2 | |
| Teicoplanin | 0.25-8 | 1 | 4 | |
| Quinupristin-dalfopristin | 0.12-0.25 | 0.12 | 0.25 | |
| Linezolid | 1-2 | 1 | 2 | |
| Arbekacin | 2-64 | 16 | 64 | |
| Imipenem | 0.12-128 | 32 | 64 | |
| Ceftriaxone | 8->128 | 32 | >128 | |
| Minocycline | 0.06-16 | 0.25 | 8 | |
| Oxacillin | 2->128 | 32 | >128 | |
| Streptococcus pneumoniae | ||||
| Penicillin susceptible (48) | ||||
| DX-619 | ≦0.004-0.03 | 0.015 | 0.015 | |
| Levofloxacin | 0.5-2 | 1 | 2 | |
| Sitafloxacin | 0.03-0.12 | 0.06 | 0.06 | |
| Ciprofloxacin | 0.5-4 | 1 | 2 | |
| Moxifloxacin | 0.12-0.25 | 0.12 | 0.25 | |
| Gatifloxacin | 0.25-0.5 | 0.25 | 0.5 | |
| Garenoxacin | 0.015-0.25 | 0.06 | 0.12 | |
| Vancomycin | 0.25-1 | 0.5 | 1 | |
| Quinupristin-dalfopristin | 0.5-2 | 1 | 1 | |
| Linezolid | 0.5-2 | 1 | 2 | |
| Imipenem | ≦0.004-0.06 | 0.008 | 0.015 | |
| Ceftriaxone | 0.015-1 | 0.12 | 0.5 | |
| Benzylpenicillin | 0.015-0.06 | 0.03 | 0.06 | |
| Penicillin intermediate and resistant (50) | ||||
| DX-619 | 0.008-0.06 | 0.03 | 0.03 | |
| Levofloxacin | 1-2 | 1 | 2 | |
| Sitafloxacin | 0.03-0.25 | 0.06 | 0.12 | |
| Ciprofloxacin | 1-8 | 2 | 4 | |
| Moxifloxacin | 0.12-0.5 | 0.25 | 0.5 | |
| Gatifloxacin | 0.25-1 | 0.5 | 0.5 | |
| Garenoxacin | 0.03-0.25 | 0.12 | 0.12 | |
| Vancomycin | 0.25-1 | 0.5 | 0.5 | |
| Quinupristin-dalfopristin | 0.5-4 | 1 | 1 | |
| Linezolid | 1-2 | 1 | 2 | |
| Imipenem | 0.12-0.5 | 0.25 | 0.5 | |
| Ceftriaxone | 0.5-2 | 1 | 2 | |
| Benzylpenicillin | 1-4 | 2 | 2 | |
| Ciprofloxacin resistant (19) | ||||
| DX-619 | 0.015-0.12 | 0.03 | 0.12 | |
| Levofloxacin | 8-32 | 16 | 16 | |
| Sitafloxacin | 0.12-0.5 | 0.25 | 0.5 | |
| Ciprofloxacin | 4-32 | 16 | 32 | |
| Moxifloxacin | 1-4 | 2 | 4 | |
| Gatifloxacin | 2-8 | 4 | 8 | |
| arenoxacin | 0.06-2 | 0.5 | 1 | |
| Vancomycin | 0.12-1 | 0.5 | 1 | |
| Quinupristin-dalfopristin | 0.5-2 | 1 | 1 | |
| Linezolid | 0.25-1 | 1 | 1 | |
| Imipenem | ≦0.004-0.5 | 0.06 | 0.25 | |
| Ceftriaxone | 0.008-4 | 0.25 | 1 | |
| Benzylpenicillin | ≦0.004-4 | 0.25 | 4 | |
| Streptococcus pyogenes (49) | ||||
| DX-619 | 0.008-0.015 | 0.008 | 0.015 | |
| Levofloxacin | 0.25-2 | 0.5 | 1 | |
| Sitafloxacin | 0.015-0.06 | 0.03 | 0.06 | |
| Ciprofloxacin | 0.25-2 | 0.5 | 1 | |
| Moxifloxacin | 0.12-0.5 | 0.25 | 0.5 | |
| Gatifloxacin | 0.25-0.5 | 0.25 | 0.5 | |
| Garenoxacin | 0.06-0.25 | 0.12 | 0.25 | |
| Vancomycin | 0.5-1 | 0.5 | 1 | |
| Teicoplanin | 0.12-0.25 | 0.25 | 0.25 | |
| Quinupristin-dalfopristin | 0.25-0.5 | 0.5 | 0.5 | |
| Linezolid | 0.25-1 | 0.5 | 1 | |
| Imipenem | ≦0.004-0.008 | ≦0.004 | 0.008 | |
| Ceftriaxone | ≦0.004-0.03 | 0.03 | 0.03 | |
| Benzylpenicillin | ≦0.004-0.015 | 0.015 | 0.015 | |
| Streptococcus agalactiae (19) | ||||
| DX-619 | 0.008-0.12 | 0.015 | 0.12 | |
| Levofloxacin | 0.5-32 | 1 | 32 | |
| Sitafloxacin | 0.06-1 | 0.06 | 0.5 | |
| Ciprofloxacin | 0.5-32 | 1 | 32 | |
| Moxifloxacin | 0.12-8 | 0.25 | 8 | |
| Gatifloxacin | 0.25-8 | 0.25 | 8 | |
| Garenoxacin | 0.06-4 | 0.12 | 4 | |
| Vancomycin | 0.5-2 | 1 | 1 | |
| Teicoplanin | 0.12-0.5 | 0.5 | 0.5 | |
| Quinupristin-dalfopristin | 0.5 | 0.5 | 0.5 | |
| Linezolid | 1-2 | 1 | 2 | |
| Imipenem | 0.015-0.03 | 0.03 | 0.03 | |
| Ceftriaxone | 0.06-0.25 | 0.12 | 0.12 | |
| Benzylpenicillin | 0.06-0.12 | 0.06 | 0.12 | |
| Enterococcus faecalis | ||||
| Vancomycin susceptible (50) | ||||
| DX-619 | 0.03-0.5 | 0.06 | 0.25 | |
| Levofloxacin | 1-128 | 2 | 32 | |
| Sitafloxacin | 0.06-4 | 0.12 | 2 | |
| Ciprofloxacin | 0.5-64 | 1 | 64 | |
| Moxifloxacin | 0.12-16 | 0.25 | 8 | |
| Gatifloxacin | 0.25-32 | 0.5 | 16 | |
| Garenoxacin | 0.12-8 | 0.25 | 4 | |
| Vancomycin | 1-4 | 1 | 2 | |
| Teicoplanin | 0.12-0.5 | 0.25 | 0.5 | |
| Quinupristin-dalfopristin | 4-32 | 8 | 16 | |
| Linezolid | 1-2 | 2 | 2 | |
| Imipenem | 1-4 | 1 | 4 | |
| Vancomycin resistant (18) | ||||
| DX-619 | 0.015-0.5 | 0.25 | 0.5 | |
| Levofloxacin | 0.5-64 | 32 | 64 | |
| Sitafloxacin | 0.06-4 | 2 | 4 | |
| Ciprofloxacin | 0.25-128 | 32 | 64 | |
| Moxifloxacin | 0.12-32 | 16 | 32 | |
| Gatifloxacin | 0.25-32 | 16 | 32 | |
| Garenoxacin | 0.06-8 | 4 | 8 | |
| Vancomycin | 8->128 | >128 | >128 | |
| Teicoplanin | 0.25-128 | 64 | 128 | |
| Quinupristin-dalfopristin | 0.5-16 | 8 | 16 | |
| Linezolid | 2 | 2 | 2 | |
| Imipenem | 1-128 | 2 | 64 | |
| Enterococcus faecium | ||||
| Vancomycin susceptible (47) | ||||
| DX-619 | 0.03-4 | 1 | 2 | |
| Levofloxacin | 1-128 | 16 | 64 | |
| Sitafloxacin | 0.06-8 | 1 | 4 | |
| Ciprofloxacin | 1->64 | 16 | 64 | |
| Moxifloxacin | 0.25-64 | 8 | 32 | |
| Gatifloxacin | 0.25-32 | 16 | 32 | |
| Garenoxacin | 0.25-64 | 8 | 32 | |
| Vancomycin | 0.5-2 | 1 | 1 | |
| Teicoplanin | 0.25-2 | 0.5 | 1 | |
| Quinupristin-dalfopristin | 0.25-16 | 0.5 | 2 | |
| Linezolid | 2 | 2 | 2 | |
| Imipenem | 2->128 | >128 | >128 | |
| Vancomycin resistant (19) | ||||
| DX-619 | 0.06-4 | 0.25 | 2 | |
| Levofloxacin | 2-64 | 4 | 64 | |
| Sitafloxacin | 0.12-8 | 0.5 | 8 | |
| Ciprofloxacin | 1->128 | 4 | >128 | |
| Moxifloxacin | 0.5-32 | 4 | 32 | |
| Gatifloxacin | 0.5-32 | 4 | 32 | |
| Garenoxacin | 0.25-32 | 8 | 32 | |
| Vancomycin | 32->128 | >128 | >128 | |
| Teicoplanin | 0.5-64 | 32 | 64 | |
| Quinupristin-dalfopristin | 0.5-4 | 0.5 | 2 | |
| Linezolid | 2 | 2 | 2 | |
| Imipenem | 16->128 | >128 | >128 | |
| Enterococcus gallinarum Vancomycin resistant (13) | ||||
| DX-619 | 0.12-2 | 1 | 2 | |
| Levofloxacin | 2-64 | 16 | 64 | |
| Sitafloxacin | 0.25-8 | 2 | 8 | |
| Ciprofloxacin | 2->128 | 128 | >128 | |
| Moxifloxacin | 0.5-32 | 4 | 32 | |
| Gatifloxacin | 0.5-64 | 8 | 32 | |
| Garenoxacin | 0.25-8 | 2 | 8 | |
| Vancomycin | 8->128 | >128 | >128 | |
| Teicoplanin | 0.5->128 | 32 | >128 | |
| Quinupristin-dalfopristin | 0.25-2 | 0.5 | 2 | |
| Linezolid | 2 | 2 | 2 | |
| Imipenem | 1->128 | >128 | >128 | |
| Peptostreptococcus spp. (13) | ||||
| DX-619 | ≦0.004-1 | 0.06 | 0.5 | |
| Levofloxacin | 0.06-32 | 4 | 16 | |
| Sitafloxacin | 0.015-2 | 0.06 | 0.5 | |
| Ciprofloxacin | 0.12-16 | 2 | 8 | |
| Moxifloxacin | 0.03-32 | 0.25 | 2 | |
| Gatifloxacin | 0.03-128 | 0.12 | 32 | |
| Garenoxacin | 0.12-64 | 1 | 8 | |
| Vancomycin | 0.25-4 | 0.25 | 1 | |
| Teicoplanin | 0.06-0.12 | 0.12 | 0.12 | |
| Quinupristin-dalfopristin | 0.25-0.5 | 0.5 | 0.5 | |
| Linezolid | 0.5-1 | 1 | 1 | |
| Imipenem | ≦0.004-1 | 0.03 | 0.12 | |
| Ceftriaxone | 0.06->128 | 0.25 | 4 | |
| Clostridium difficile (17) | ||||
| DX-619 | 0.25-32 | 2 | 2 | |
| Levofloxacin | 4-128 | 64 | 128 | |
| Sitafloxacin | 0.25-32 | 1 | 2 | |
| Ciprofloxacin | 16-64 | 32 | 64 | |
| Moxifloxacin | 2-128 | 32 | 32 | |
| Gatifloxacin | 2-64 | 32 | 32 | |
| Garenoxacin | 1-128 | 32 | 64 | |
| Vancomycin | 1-4 | 2 | 2 | |
| Teicoplanin | 0.06-0.25 | 0.12 | 0.25 | |
| Quinupristin-dalfopristin | 1-2 | 1 | 2 | |
| Linezolid | 0.5-4 | 1 | 4 | |
| Imipenem | 8-32 | 16 | 16 | |
| Ceftriaxone | 16->128 | 128 | >128 | |
The antibacterial activity of DX-619 against gram-negative strains is shown in Table 2. DX-619 showed good antibacterial activity against H. influenzae, including ampicillin-resistant strains, and M. catarrhalis, against which the highest MIC was 0.06 μg/ml. DX-619 inhibited 90% of isolates of Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., Citrobacter spp., Salmonella spp., Proteus mirabilis, indole-positive Proteus, and Serratia marcescens at 1, 0.12, 0.5, 1, 0.06, 2, 0.5, and 2 μg/ml, respectively, and these activities were comparable to those of levofloxacin. DX-619 also showed activity comparable to that of levofloxacin against ciprofloxacin-susceptible P. aeruginosa, with a MIC90 of 1 μg/ml. The MIC90 of DX-619 against ciprofloxacin-resistant P. aeruginosa was 64 μg/ml. Against Acinetobacter spp. and N. gonorrhoeae including ciprofloxacin-resistant strains, DX-619 showed good antibacterial activity; the highest MIC against these species was 2 μg/ml. The MIC90 of DX-619 was 0.5 μg/ml against Bacteroides fragilis. The MICs of the compounds tested against the reference strains for quality control were reproducible throughout the study.
TABLE 2.
Antibacterial activities of DX-619 and reference compounds against gram-negative bacteria
| Organism (no. of strains) and compound | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | ||||
| Haemophilus influenzae | ||||||
| Ampicillin susceptible (20) | ||||||
| DX-619 | ≦0.004-0.008 | ≦0.004 | ≦0.004 | |||
| Levofloxacin | 0.008-0.015 | 0.015 | 0.015 | |||
| Sitafloxacin | ≦0.004 | ≦0.004 | ≦0.004 | |||
| Ciprofloxacin | ≦0.004-0.015 | 0.008 | 0.008 | |||
| Moxifloxacin | ≦0.004-0.015 | 0.008 | 0.015 | |||
| Gatifloxacin | ≦0.004-0.015 | 0.008 | 0.008 | |||
| Garenoxacin | ≦0.004-0.008 | ≦0.004 | 0.008 | |||
| Imipenem | 0.06-4 | 0.5 | 1 | |||
| Ceftazidime | 0.06-0.5 | 0.12 | 0.25 | |||
| Cefaclor | 1-8 | 2 | 4 | |||
| Ampicillin | 0.12-0.5 | 0.25 | 0.5 | |||
| β-Lactamase positive, ampicillin resistant (21) | ||||||
| DX-619 | ≦0.004-0.008 | ≦0.004 | ≦0.004 | |||
| Levofloxacin | ≦0.004-0.015 | 0.015 | 0.015 | |||
| Sitafloxacin | ≦0.004 | ≦0.004 | ≦0.004 | |||
| Ciprofloxacin | ≦0.004-0.015 | 0.008 | 0.008 | |||
| Moxifloxacin | ≦0.004-0.03 | 0.015 | 0.015 | |||
| Gatifloxacin | ≦0.004-0.015 | 0.008 | 0.008 | |||
| Garenoxacin | ≦0.004-0.015 | ≦0.004 | 0.008 | |||
| Imipenem | 0.25-4 | 1 | 2 | |||
| Ceftazidime | 0.06-0.5 | 0.12 | 0.5 | |||
| Cefaclor | 1-32 | 8 | 16 | |||
| Ampicillin | 4-128 | 16 | 32 | |||
| β-Lactamase negative, ampicillin intermediate and resistant (25) | ||||||
| DX-619 | ≦0.004-0.015 | ≦0.004 | 0.015 | |||
| Levofloxacin | 0.008-0.015 | 0.015 | 0.015 | |||
| Sitafloxacin | ≦0.004 | ≦0.004 | ≦0.004 | |||
| Ciprofloxacin | 0.008-0.015 | 0.008 | 0.015 | |||
| Moxifloxacin | 0.008-0.03 | 0.015 | 0.03 | |||
| Gatifloxacin | ≦0.004-0.015 | 0.008 | 0.015 | |||
| Garenoxacin | ≦0.004-0.015 | 0.008 | 0.015 | |||
| Imipenem | 0.25-8 | 4 | 8 | |||
| Ceftazidime | 0.12-1 | 0.5 | 0.5 | |||
| Cefaclor | 8-128 | 64 | 128 | |||
| Ampicillin | 2-8 | 4 | 8 | |||
| Moraxella catarrhalis (48) | ||||||
| DX-619 | 0.008-0.06 | 0.015 | 0.03 | |||
| Levofloxacin | 0.03-0.06 | 0.03 | 0.06 | |||
| Sitafloxacin | ≦0.004-0.015 | 0.008 | 0.015 | |||
| Ciprofloxacin | 0.03-0.06 | 0.03 | 0.06 | |||
| Moxifloxacin | 0.03-0.12 | 0.06 | 0.06 | |||
| Gatifloxacin | 0.03-0.06 | 0.03 | 0.06 | |||
| Garenoxacin | 0.008-0.03 | 0.015 | 0.03 | |||
| Imipenem | 0.12-2 | 1 | 1 | |||
| Ceftazidime | 0.12->32 | 0.5 | 2 | |||
| Ampicillin | 0.25-8 | 4 | 8 | |||
| Escherichia coli (48) | ||||||
| DX-619 | 0.015-8 | 0.03 | 1 | |||
| Levofloxacin | 0.015-32 | 0.06 | 4 | |||
| Sitafloxacin | ≦0.004-2 | 0.015 | 0.5 | |||
| Ciprofloxacin | ≦0.004-64 | 0.015 | 4 | |||
| Moxifloxacin | 0.015-32 | 0.06 | 8 | |||
| Gatifloxacin | 0.008-16 | 0.03 | 4 | |||
| Garenoxacin | 0.015-64 | 0.06 | 8 | |||
| Imipenem | 0.06-0.5 | 0.12 | 0.25 | |||
| Ceftazidime | 0.06-8 | 0.25 | 0.5 | |||
| Ampicillin | 1->128 | 4 | >128 | |||
| Klebsiella pneumoniae (49) | ||||||
| DX-619 | 0.03-1 | 0.06 | 0.12 | |||
| Levofloxacin | 0.015-2 | 0.06 | 0.12 | |||
| Sitafloxacin | ≦0.004-0.5 | 0.03 | 0.03 | |||
| Ciprofloxacin | ≦0.004-0.5 | 0.03 | 0.06 | |||
| Moxifloxacin | 0.06-2 | 0.12 | 0.25 | |||
| Gatifloxacin | 0.015-2 | 0.06 | 0.12 | |||
| Garenoxacin | 0.06-2 | 0.12 | 0.25 | |||
| Imipenem | 0.12-0.5 | 0.12 | 0.25 | |||
| Ceftazidime | 0.06-1 | 0.12 | 0.25 | |||
| Ampicillin | 1->128 | 32 | 32 | |||
| Enterobacter spp. (26) | ||||||
| DX-619 | 0.03-2 | 0.06 | 0.5 | |||
| Levofloxacin | 0.03-2 | 0.06 | 0.5 | |||
| Sitafloxacin | 0.015-0.5 | 0.015 | 0.12 | |||
| Ciprofloxacin | 0.008-2 | 0.015 | 0.25 | |||
| Moxifloxacin | 0.03-4 | 0.06 | 0.5 | |||
| Gatifloxacin | 0.03-2 | 0.03 | 0.5 | |||
| Garenoxacin | 0.03-8 | 0.12 | 1 | |||
| Imipenem | 0.12-1 | 0.25 | 1 | |||
| Ceftazidime | 0.12->128 | 0.25 | 64 | |||
| Citrobacter spp. (26) | ||||||
| DX-619 | 0.03-4 | 0.25 | 1 | |||
| Levofloxacin | 0.015-4 | 0.12 | 0.5 | |||
| Sitafloxacin | 0.008-2 | 0.06 | 0.5 | |||
| Ciprofloxacin | 0.008-2 | 0.03 | 0.25 | |||
| Moxifloxacin | 0.03-8 | 0.25 | 2 | |||
| Gatifloxacin | 0.015-4 | 0.12 | 1 | |||
| Garenoxacin | 0.03-16 | 0.5 | 4 | |||
| Imipenem | 0.25-1 | 0.5 | 1 | |||
| Ceftazidime | 0.12->128 | 1 | 128 | |||
| Salmonella spp. (26) | ||||||
| DX-619 | 0.06-0.25 | 0.06 | 0.06 | |||
| Levofloxacin | 0.03-0.12 | 0.06 | 0.06 | |||
| Sitafloxacin | 0.015-0.12 | 0.015 | 0.015 | |||
| Ciprofloxacin | 0.015-0.12 | 0.015 | 0.015 | |||
| Moxifloxacin | 0.06-0.5 | 0.12 | 0.12 | |||
| Gatifloxacin | 0.03-0.25 | 0.06 | 0.06 | |||
| Garenoxacin | 0.06-0.5 | 0.06 | 0.12 | |||
| Imipenem | 0.12-0.25 | 0.25 | 0.25 | |||
| Ceftazidime | 0.25-0.5 | 0.25 | 0.5 | |||
| Proteus mirabilis (26) | ||||||
| DX-619 | 0.06-4 | 0.12 | 2 | |||
| Levofloxacin | 0.03-8 | 0.06 | 2 | |||
| Sitafloxacin | 0.015-1 | 0.03 | 0.25 | |||
| Ciprofloxacin | 0.015-4 | 0.03 | 1 | |||
| Moxifloxacin | 0.25-32 | 0.5 | 8 | |||
| Gatifloxacin | 0.06-16 | 0.12 | 2 | |||
| Garenoxacin | 0.12-32 | 0.5 | 16 | |||
| Imipenem | 0.25-4 | 1 | 2 | |||
| Ceftazidime | 0.03-0.25 | 0.06 | 0.12 | |||
| Indole-positive Proteus (25) | ||||||
| DX-619 | 0.03-8 | 0.12 | 0.5 | |||
| Levofloxacin | 0.015-4 | 0.06 | 0.25 | |||
| Sitafloxacin | ≦0.004-1 | 0.015 | 0.06 | |||
| Ciprofloxacin | ≦0.004-4 | 0.015 | 0.06 | |||
| Moxifloxacin | 0.06-32 | 0.25 | 0.5 | |||
| Gatifloxacin | 0.03-8 | 0.12 | 0.5 | |||
| Garenoxacin | 0.12-128 | 0.25 | 2 | |||
| Imipenem | 0.25-4 | 2 | 4 | |||
| Ceftazidime | 0.06-16 | 0.12 | 0.25 | |||
| Serratia marcescens (26) | ||||||
| DX-619 | 0.12-8 | 0.5 | 2 | |||
| Levofloxacin | 0.06-8 | 0.25 | 2 | |||
| Sitafloxacin | 0.03-1 | 0.12 | 0.5 | |||
| Ciprofloxacin | 0.03-8 | 0.06 | 2 | |||
| Moxifloxacin | 0.12-16 | 0.5 | 4 ?xpp Tr> Gatifloxacin | 0.12-8 | 0.25 | 2 |
| Garenoxacin | 0.5-64 | 1 | 4 | |||
| Imipenem | 0.5-2 | 1 | 1 | |||
| Ceftazidime | 0.12-16 | 0.5 | 4 | |||
| Pseudomonas aeruginosa | ||||||
| Ciprofloxacin susceptible (51) | ||||||
| DX-619 | 0.25-2 | 1 | 1 | |||
| Levofloxacin | 0.25-2 | 0.5 | 1 | |||
| Sitafloxacin | 0.03-0.25 | 0.12 | 0.25 | |||
| Ciprofloxacin | 0.06-0.5 | 0.12 | 0.25 | |||
| Moxifloxacin | 0.5-4 | 2 | 2 | |||
| Gatifloxacin | 0.25-2 | 1 | 1 | |||
| Garenoxacin | 0.25-4 | 1 | 2 | |||
| Imipenem | 0.5-32 | 2 | 16 | |||
| Ceftazidime | 0.5-128 | 2 | 4 | |||
| Ciprofloxacin intermediate and resistant (52) | ||||||
| DX-619 | 2->128 | 16 | 64 | |||
| Levofloxacin | 4->128 | 32 | >128 | |||
| Sitafloxacin | 0.5-32 | 4 | 16 | |||
| Ciprofloxacin | 2->64 | 16 | 64 | |||
| Moxifloxacin | 8->128 | 64 | >128 | |||
| Gatifloxacin | 4->128 | 32 | 64 | |||
| Garenoxacin | 8->128 | 64 | >128 | |||
| Imipenem | 0.5->128 | 16 | 128 | |||
| Ceftazidime | 1->128 | 16 | >128 | |||
| Stenotrophomonas maltophilia (20) | ||||||
| DX-619 | 0.12-2 | 0.5 | 2 | |||
| Levofloxacin | 0.06-4 | 1 | 4 | |||
| Sitafloxacin | 0.03-0.5 | 0.12 | 0.5 | |||
| Ciprofloxacin | 0.015-8 | 2 | 4 | |||
| Moxifloxacin | 0.12-2 | 0.5 | 2 | |||
| Gatifloxacin | 0.12-4 | 1 | 2 | |||
| Garenoxacin | 0.25-8 | 2 | 8 | |||
| Imipenem | 2->128 | >128 | >128 | |||
| Ceftazidime | 2-128 | 32 | 128 | |||
| Acinetobacter spp. (26) | ||||||
| DX-619 | 0.03-1 | 0.03 | 0.12 | |||
| Levofloxacin | 0.06-4 | 0.12 | ||||
| Sitafloxacin | 0.015-1 | 0.03 | 0.12 | |||
| Ciprofloxacin | 0.06-8 | 0.12 | 0.5 | |||
| Moxifloxacin | 0.03-2 | 0.06 | 0.5 | |||
| Gatifloxacin | 0.03-2 | 0.06 | 0.25 | |||
| Garenoxacin | 0.015-2 | 0.03 | 0.12 | |||
| Imipenem | 0.12-0.5 | 0.25 | 0.5 | |||
| Ceftazidime | 1-16 | 4 | 8 | |||
| Neisseria gonorrhoeae | ||||||
| Ciprofloxacin susceptible (23) | ||||||
| DX-619 | ≦0.004-0.015 | ≦0.004 | 0.015 | |||
| Levofloxacin | ≦0.004-0.12 | 0.008 | 0.06 | |||
| Sitafloxacin | ≦0.004 | ≦0.004 | ≦0.004 | |||
| Ciprofloxacin | ≦0.004-0.06 | 0.008 | 0.06 | |||
| Moxifloxacin | ≦0.004-0.06 | 0.008 | 0.06 | |||
| Gatifloxacin | ≦0.004-0.03 | 0.008 | 0.03 | |||
| Garenoxacin | ≦0.004-0.03 | ≦0.004 | 0.015 | |||
| Imipenem | 0.03-2 | 0.06 | 0.25 | |||
| Ceftazidime | 0.008-0.25 | 0.06 | 0.12 | |||
| Benzylpenicillin | 0.06-1 | 0.12 | 0.25 | |||
| Ciprofloxacin resistant (26) | ||||||
| DX-619 | 0.25-2 | 0.5 | 1 | |||
| Levofloxacin | 4-16 | 8 | 16 | |||
| Sitafloxacin | 0.12-0.5 | 0.25 | 0.5 | |||
| Ciprofloxacin | 4-32 | 16 | 32 | |||
| Moxifloxacin | 2-8 | 4 | 8 | |||
| Gatifloxacin | 1-4 | 2 | 4 | |||
| Garenoxacin | 1-8 | 2 | 4 | |||
| Imipenem | 0.03-2 | 1 | 2 | |||
| Ceftazidime | 0.06-1 | 0.5 | 1 | |||
| Benzylpenicillin | 0.06-4 | 2 | 4 | |||
| Bacteroides fragilis (20) | ||||||
| DX-619 | 0.06-0.5 | 0.06 | 0.5 | |||
| Levofloxacin | 1-32 | 1 | 32 | |||
| Sitafloxacin | 0.03-0.5 | 0.06 | 0.25 | |||
| Ciprofloxacin | 4-64 | 4 | 64 | |||
| Moxifloxacin | 0.25-8 | 0.25 | 4 | |||
| Gatifloxacin | 0.25-8 | 0.5 | 8 | |||
| Garenoxacin | 0.12-2 | 0.12 | 2 | |||
| Imipenem | 0.06-2 | 0.12 | 1 | |||
| Ceftazidime | 4->128 | 32 | >128 | |||
| Metronidazole | 0.25-1 | 0.5 | 1 | |||
| Clindamycin | 0.06->128 | 0.25 | >128 | |||
This study showed that DX-619, a recently discovered des-fluoro(6) quinolone, possesses the most potent antibacterial activity among the compounds tested against gram-positive bacteria, including ciprofloxacin-resistant MRSA, MRCNS, VRE, and ciprofloxacin-resistant S. pneumoniae. The most common resistant pathogen in hospitals is MRSA, which accounts for outbreaks and is increasing in frequency in many facilities (5). The MIC90 of DX-619 for ciprofloxacin-resistant MRSA was 0.5 μg/ml, which was lower than those of linezolid and vancomycin. This finding may be attributable to the high inhibitory activity of DX-619 against altered target enzymes of MRSA (M. Tanaka et al., 43rd ICAAC, abstr. F-1060). The relative potency of DX-619 will be better understood when the human pharmacokinetics are available. Further studies of DX-619 are warranted based on the available data.
Acknowledgments
We thank T. Otani for valuable comments and critical review of the manuscript.
REFERENCES
- 1.Bell, J. M., and J. D. Turnidge. 2002. High prevalence of oxacillin-resistant Staphylococcus aureus isolates from hospitalized patients in Asia-Pacific and South Africa: results from SENTRY Antimicrobial Surveillance Program, 1998-1999. Antimicrob. Agents Chemother. 46:879-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, and the Vancomycin-Resistant Staphylococcus aureus Investigative Team. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 3.Clemett, D., and A. Markham. 2000. Linezolid. Drugs 59:815-827. [DOI] [PubMed] [Google Scholar]
- 4.Diekema, D. J., B. J. BootsMiller, T. E. Vaughn, R. F. Woolson, J. W. Yankey, E. J. Ernst, S. D. Flach, M. M. Ward, C. L. J. Franciscus, M. A. Pfaller, and B. N. Doebbeling. 2004. Antimicrobial resistance trends and outbreak frequency in United States hospitals. Clin. Infect. Dis. 38:78-85. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou, N. H. 2002. Infectious disease 2001: drug resistance, new drugs. Drug Resist. Update 5:181-191. [DOI] [PubMed] [Google Scholar]
- 6.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identifiable predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 7.Japan Society of Chemotherapy. 1981. Method for the determination of minimum inhibitory concentration (MIC) of aerobic bacteria by agar dilution method. Chemotherapy (Tokyo) 29:76-79. (In Japanese.) [Google Scholar]
- 8.Jones, M. E., R. S. Blosser-Middleton, C. Thornsberry, J. A. Karlowsky, and D. F. Sahm. 2003. The activity of levofloxacin and other antimicrobials against clinical isolates of Streptococcus pneumoniae collected worldwide during 1999-2002. Diagn. Microbiol. Infect. Dis. 47:579-586. [DOI] [PubMed] [Google Scholar]
- 9.Kacica, M. 2004. Vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 10.Mathai, D., R. N. Jones, M. A. Pfaller, and the SENTRY Participant Group North America. 2001. Epidemiology and frequency of resistance among pathogens causing urinary tract infections in 1,510 hospitalized patients: a report from the SENTRY Antimicrobial Surveillance Program (North America). Diagn. Microbiol. Infect. Dis. 40:129-136. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2001. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 5th ed. M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing; 13th informational supplements. M100-S13. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Nilgate, S., P. Nunthapisud, and A. Chongthaleong. 2003. Vancomycin-resistant enterococci in King Chulalongkorn Memorial Hospital: a 5-year study. J. Med. Assoc. Thai. 86(Suppl. 2):S224-S229. [PubMed] [Google Scholar]
- 14.NNIS System. 2003. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control 31:481-498. [DOI] [PubMed] [Google Scholar]
- 15.Rahim, S., S. K. Pillai, H. S. Gold, L. Venkataraman, K. Inglima, and R. A. Press. 2003. Linezolid-resistant, vancomycin-resistant Enterococcus faecium infection in patients without prior exposure to linezolid. Clin. Infect. Dis. 36:E146-E148. [DOI] [PubMed] [Google Scholar]
- 16.Sahm, D. F., M. E. Jones, M. L. Hickey, D. R. Diakun, S. V. Mani, and C. Thornsberry. 2000. Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis isolated in Asia and Europe, 1997-1998. J. Antimicrob. Chemother. 45:457-466. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz, F. J., A. C. Fluit, A. Brisse, J. Verhoef, K. Kohrer, and D. Milatovic. 1999. Molecular epidemiology of quinolone resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol. Med. Microbiol. 26:281-287. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanakunakorn, C. 1981. The antibacterial action of vancomycin. Rev. Infect. Dis. 3:S210-S215. [PubMed] [Google Scholar]
- 20.Yamaguchi, K., A. Ohno, F. Kashitani, M. Iwata, and the Levofloxacin Surveillance Group. 2003. Activity of antimicrobial agents against 8,474 clinical isolates obtained from 37 medical institutions during 2000 in Japan. Jpn. J. Antibiot. 56:5-24. [PubMed] [Google Scholar]
- 21.Yamaguchi, K., A. Ohno, F. Kashitani, M. Iwata, and the Levofloxacin Surveillance Group. 2005. In vitro susceptibilities to levofloxacin and various antimicrobial agents of 11,475 clinical isolates obtained from 52 centers in 2002. Jpn. J. Antibiot. 58:17-44. [PubMed] [Google Scholar]
- 22.Zurenko, G. E., B. H. Yagi, R. D. Schaadt, J. W. Allison, J. O. Kilburn, S. E. Glickman, D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner. 1996. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob. Agents Chemother. 40:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]