Abstract
Alanine scanning of motif A in the pJHCMW1-encoded aminoglycoside 6′-N-acetyltransferase type Ib identified amino acids important for the ability of the enzyme to confer wild-type levels of resistance to kanamycin and amikacin. The replacement of two amino acids, D117 or L120, with alanine residues resulted in complete loss of the resistance phenotype.
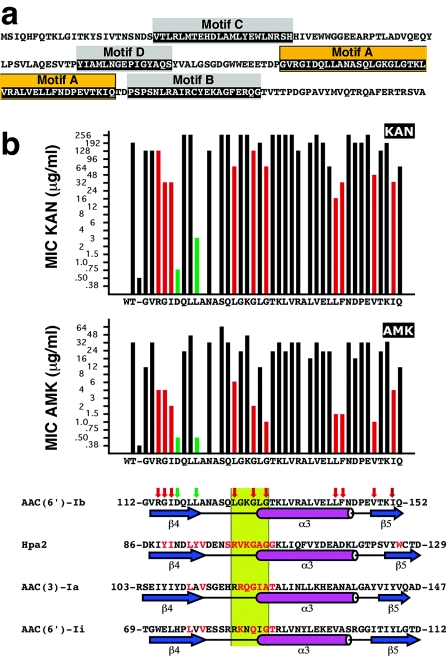
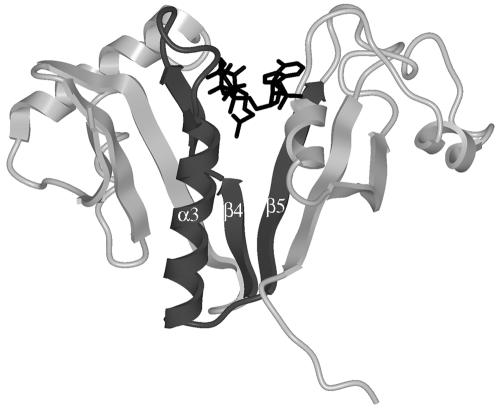
The aminoglycoside N-acetyltransferases belong to the GCN5-related N-acetyltransferase (GNAT) superfamily, a group of enzymes that catalyze the transfer of an acetyl group from acetyl coenzyme A (acetyl-CoA) to a primary amine in a wide variety of acceptor molecules (7, 20). GNAT proteins possess four loosely conserved regions (considering the primary sequence), named motifs A through D, arranged in the order C-D-A-B (11). The aminoglycoside 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] coded for by pJHCMW1 possesses all four conserved motifs (Fig. 1a) (16, 17). The structure of motif A, which plays an important role in acetyl-CoA binding, is the most highly conserved among acetyltransferases (Fig. 1b) (7). Based on primary structure, this motif was defined starting at the strand β4 and extending to the carboxy end of the α3 helix (7, 11). However, on the basis of additional information from structural comparison, other authors proposed that motif A encompasses the whole βαβ unit, i.e., the strand β5 should also be included inside motif A (Fig. 1b) (1, 10, 14). The acetyl-CoA molecule fits into a V-shaped cleft formed between the β4 and β5 strands (18). As an example, Fig. 2 shows the structure of AAC(6′)-Ii bound to acetyl-CoA (23) with the amino acids belonging to motif A identified in dark gray. Figure 1b shows that the predicted secondary structure of the AAC(6′)-Ib motif A includes the βαβ core and is comparable to other GNAT proteins, such as Hpa2, AAC(3)-Ia, and AAC(6′)-Ii. Strand β4 was shown to play an essential role in binding the cofactor acetyl-CoA and includes amino acids important for the catalytic activity in GNAT enzymes (1, 3, 5, 7, 20, 23).
FIG. 1.
(a) Amino acid sequence of the AAC(6′)-Ib protein. The boxes indicate the conserved motifs A through D. Orange-highlighted amino acids indicate motif A. (b) Susceptibilities of E. coli harboring plasmids containing the mutant derivatives of AAC(6′)-Ib and alignment of the motif A with the amino acid sequences of other members of the GNAT superfamily. The MICs of KAN and AMK are plotted on top, the vertical axis shows the antibiotic concentrations as they are in the E-strip, and the bars show the results for the controls (plasmidless E. coli and E. coli harboring the wild-type pJHCMW1) and each substitution with alanine. The bottom part of panel b shows the alignment of the amino acid sequences of the motif A of each of the indicated GNAT proteins. The amino acid sequence of motif A in Hpa2 (accession number AY558056), AAC(3)-Ia (accession number S68049) (9), and AAC(6′)-Ii (accession number L12710) (4), as well as the structures of the regions as determined before (1, 22, 23), are shown aligned and compared to the amino acid sequence and the predicted secondary structure of the motif A of AAC(6′)-Ib (accession number AF479774) (19). Amino acids in red are involved in contacts with the cofactor acetyl-CoA (1, 3, 22). The β strands and α helix are indicated below each sequence; they are named as described by Dyda et al. (7). The green box identifies the conserved region Q/RxxGxG/A. The red vertical arrows on top of the AAC(6′)-Ib amino acid sequence indicate substitutions that reduced the MIC of AMK to 8 μg/ml or less while the MICs of KAN were not significantly reduced or reduced to no less than 16 μg/ml. The green vertical arrows indicate substitutions that resulted in MICs of AMK comparable to that shown by the plasmidless control and in MICs of KAN that were ≤3 μg/ml.
FIG. 2.
Structure of AAC(6′)-Ii (PDB 1B87) in complex with acetyl-CoA (23). The amino acids in motif A have been colored dark gray, and the acetyl-CoA molecule is shown in black. Note that the β4 strand is shown as two strands separated by a short coil (H74-P75) in this model.
Alanine-scanning mutagenesis is useful to perform systematic analysis of a protein region by replacing each amino acid with an alanine residue. This substitution removes the side chain beyond the β carbon without altering the conformation of the main chain or imposing extreme electrostatic or steric effects (21). In this work, we performed a systematic analysis of the predicted motif A βαβ unit of AAC(6′)-Ib (Fig. 1b) by alanine-scanning mutagenesis. Our results indicated that amino acids important for AAC(6′)-Ib to confer wild-type levels of resistance to kanamycin (KAN) and amikacin (AMK) are located in each of the β strands, the α helix, and a region highly conserved among GNATs which encompasses a loop immediately prior to and part of the first turn of the α helix.
Escherichia coli XL1-Blue (Stratagene) and E. coli TOP10 (Invitrogen) were used as hosts for mutagenesis and recombinant cloning, respectively. Substitutions with alanine were generated using the QuikChange mutagenesis kit (Stratagene) and pJHCMW1 DNA as described before (17). All mutants were confirmed by the sequencing of the whole gene. The wild-type aac(6′)-Ib gene and mutant gene derivatives were cloned by using pBAD102 as a cloning vector (Invitrogen). The genes were expressed by adding 0.2% arabinose to log-phase cultures of E. coli TOP10 harboring the recombinant clone, followed by incubation for 4 h at 37°C. MICs were determined by the Etest method with commercial strips (AB Biodisk). The determination of acetyltransferase activity, determination of protein concentration, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were carried out as described before (13, 17). Secondary structure prediction of AAC(6′)-Ib was performed using PROF Secondary Structure Prediction System (http://www.aber.ac.uk/∼phiwww/prof/) (12) and Advanced Protein Secondary Structure Prediction Server (http://imtech.res.in/raghava/apssp/) software. The figure depicting the structure of AAC(6′)-Ii (23) was generated by using ICM-Browser software, version 3.1-07B (MolSoft).
All amino acids in the AAC(6′)-Ib motif A were replaced by alanine residues, and E. coli cells harboring the mutated genes were analyzed by the determination of the MICs of KAN and AMK (Fig. 1b).
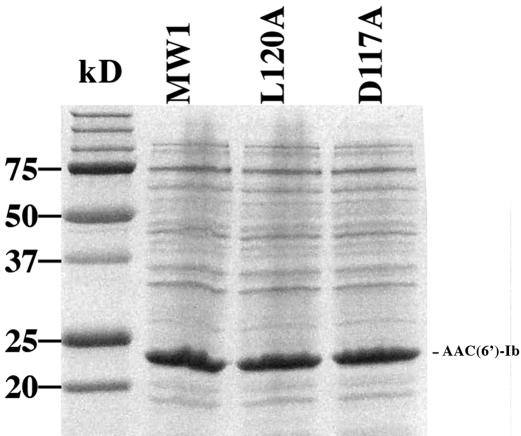
Several substitutions within strand β4 had an effect on the resistance phenotype of the cells. Substitutions R114A, G115A, and I116A reduced MICs of AMK to 4 μg/ml or less (8- to 16-fold reductions) but still conferred relatively high levels of resistance to KAN (1.5- to 6-fold reductions), which indicated that they were able to catalyze acetylation of this aminoglycoside (Fig. 1b, red bars and arrows). Conversely, the substitutions D117A and L120A led to drastic reductions in the MICs of both AMK and KAN (Fig. 1b, green bars and arrows). To test if the reductions in the MICs were due to changes in catalytic activity, the mutant genes were cloned under the control of the araBAD promoter, expressed, and analyzed to determine their acetylating activities in vitro. Figure 3 shows that the levels of expression of the mutant and the wild-type enzymes are similar, and Table 1 shows that the mutant enzymes have poor (L120A) or no (D117A) acetylating activity. Previous results indicated that L residues in the β4 strand are implicated in the acetylation process (5). As shown in Fig. 1b, there is a leucine residue that makes contact with acetyl-CoA at the end of the β4 strands of each of the shown enzymes. Furthermore, it has recently been determined that the AAC(6′)-Ii L76 residue is implicated in transition state or intermediate stabilization besides interacting with the cofactor (5). In the case of AAC(6′)-Ib, there are two leucine residues present at the end of the β4 strand (Fig. 1b). The results obtained by the replacement of L119 and L120 with alanines, which show that the L120A substitution abolished resistance to AMK and KAN and reduced enzymatic activity (Fig. 1b and Table 1), suggest that the residue L120 might play an important role in activity of AAC(6′)-Ib. However, it is interesting that there was a residual activity in the L120A mutant enzyme, and the substitution seems to have a bigger effect on the acetylation of AMK than on the acetylation of KAN. Furthermore, the MIC reductions in E. coli cells harboring the L120A substitution, with respect to the wild type, were 64-fold, while the acetylation activities were reduced to about 10 and 40% for AMK and KAN, respectively. These results could indicate that, in addition to a reduced enzymatic activity, other factors contribute to reduction of MICs in this derivative. Enzymologic and stability studies will permit us to understand the characteristics of this substitution derivative and determine the role played by this amino acid.
FIG. 3.
Expression of AAC(6′)-Ib and mutant derivatives. Total proteins obtained from E. coli TOP10 cells harboring either pBADMW1 (MW1) (wild type), pBADD117A (D117A substitution), or pBADL120A (L120A substitution) induced in the presence of 0.2% arabinose for 4 h were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The sizes of the molecular mass standards are shown to the left, and the position of AAC(6′)-Ib is shown to the right.
TABLE 1.
Acetylating activity
| Plasmid (substitution)a | Mean acetylation ± SD (cpm/μg protein)b
|
|
|---|---|---|
| AMK | KAN | |
| pBADMW1 (no substitution) | 13,632 ± 95 | 12,989 ± 440 |
| pBADD117A (D117A) | 10 ± 3 | 5 ± 2 |
| pBADL120A (L120A) | 1,633 ± 38 | 5,190 ± 357 |
Soluble extracts were obtained from E. coli TOP10 harboring the indicated plasmid.
Values are averages of triplicates.
The substitution D117A is the one that most dramatically affected enzymatic activity. E. coli cells harboring the gene coding for the enzyme derivative with this substitution were as susceptible to AMK and KAN as were plasmidless E. coli cells, demonstrating the critical role played by this amino acid residue for the activity of AAC(6′)-Ib (Fig. 1b and Table 1). Although this amino acid is not highly conserved among GNAT proteins in general, it is conserved among the closely related subgroup of AAC(6′) enzymes formed by AAC(6′)-Ib, AAC(6′)-Ie, AAC(6′)-IIa, AAC(6′)-IIb, AAC(6′)-IIc, and AAC(6′)-IId (8, 15). Experiments carried out by Boehr et al. (2) showed that the D99 residue of AAC(6′)-Ie, which is equivalent to D117 in AAC(6′)-Ib, is essential for activity as well as for the special characteristics of this enzyme, which is capable of catalyzing O acetylation besides the normal N-acetylating activity. Replacement of the AAC(6′)-Ie D99 with alanine or asparagine resulted in a dramatic deleterious effect. Conversely, replacement of D99 with a glutamic acid residue resulted in an enzyme derivative that conserved a small fraction of the activity found in the wild type. These results led the authors to propose that the carboxylate moiety of the amino acid at this position plays a role in enzymatic activity (2). In the case of AAC(6′)-Ib, the MICs of AMK and KAN for derivatives carrying substitution D117A, D117N, or D117E showed a pattern comparable to that found for derivatives with the same replacements of D99 in AAC(6′)-Ie. E. coli cells harboring the D117A or D117N substitution were susceptible to AMK and KAN (Table 2). In the case of the derivatives with the D117E substitution, the levels of resistance to KAN and AMK, while still very low, were higher than those of the cells mediated by the substitutions D117A and D117N (Table 2). In particular, an increase in KAN resistance is clearly observed. This increase is comparable to that found in the case of the experiments performed by mutagenesis of D99 in AAC(6′)-Ie (2). We conclude that, as it is the case of D99 in the AAC(6′)-Ie enzyme, the aspartic acid residue at position 117 is essential for proper enzymatic activity of AAC(6′)-Ib. Furthermore, since the presence of the carboxylate moiety at this position results in a small increment in enzymatic activity with respect to the substitutions D117N and D117A, this group may play a role in the function of the enzyme or contribute to the maintenance of the protein's correct structure. Further research, including enzymologic analysis, will provide information to determine the specific role of D117 in AAC(6′)-Ib.
TABLE 2.
Susceptibilities to AMK and KAN of E. coli harboring the wild-type gene or D117A, D117N, or D117E substitutions
| E. coli cells harboring: | MIC (μg/ml)
|
|
|---|---|---|
| AMK | KAN | |
| No plasmid | 0.25 | 0.25 |
| pJHCMW1 (wild type) | 32 | 192 |
| D117A | 0.25 | 0.38 |
| D117N | 0.75 | 1.5 |
| D117E | 1.5 | 8 |
Other substitution mutants with impaired function are located within the α helix, the 126-LGKGLG-131 segment, which corresponds to the conserved Q/RxxGxG/A segment (1, 6, 11, 22), or the strand β5 (red in Fig. 1b). We do not know if all these mutants are affected in catalytic activity, stability, or both. Enzymology and structural studies are currently being carried out to characterize them. These experiments may lead to a thorough understanding of the structure and function of the AAC(6′)-Ib protein, which may help in future rational drug design.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in the GenBank sequence library and assigned the accession numbers AY648341 through AY648378.
Acknowledgments
This work was supported by National Institutes of Health grant 1R15AI47115-02. M.W., J.N., and D.B. were supported by MSD grant 5R25GM56820-03 from the National Institutes of Health. J.F. was the recipient of an undergraduate research creative award from CalState Fullerton.
We thank B. Dutnall for discussions and suggestions and K. Kantardjieff for help in generating the depiction of the AAC(6′)-Ii structure.
REFERENCES
- 1.Angus-Hill, M. L., R. N. Dutnall, S. T. Tafrov, R. Sternglanz, and V. Ramakrishnan. 1999. Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 294:1311-1325. [DOI] [PubMed] [Google Scholar]
- 2.Boehr, D. D., S. I. Jenkins, and G. D. Wright. 2003. The molecular basis of the expansive substrate specificity of the antibiotic resistance enzyme aminoglycoside acetyltransferase-6′-aminoglycoside phosphotransferase-2". The role of ASP-99 as an active site base important for acetyl transfer. J. Biol. Chem. 278:12873-12880. [DOI] [PubMed] [Google Scholar]
- 3.Burk, D., N. Ghuman, L. Wybenga-Groot, and A. Berghuis. 2003. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12:426-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa, Y., M. Galimand, R. Leclercq, J. Duval, and P. Courvalin. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draker, K., and G. Wright. 2004. Molecular mechanism of the enterococcal aminoglycoside 6′-N-acetyltransferase: role of GNAT-conserved residues in the chemistry of antibiotic inactivation. Biochemistry 43:446-454. [DOI] [PubMed] [Google Scholar]
- 6.Dutnall, R. N., S. T. Tafrov, R. Sternglanz, and V. Ramakrishnan. 1998. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94:427-438. [DOI] [PubMed] [Google Scholar]
- 7.Dyda, F., D. C. Klein, and A. B. Hickman. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29:81-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannecart-Pokorni, E., F. Depuydt, L. de Wit, E. van Bossuyt, J. Content, and R. Vanhoof. 1997. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Il associated with a sulI-type integron. Antimicrob. Agents Chemother. 41:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javier Teran, F., M. Alvarez, J. E. Suarez, and M. C. Mendoza. 1991. Characterization of two aminoglycoside-(3)-N-acetyltransferase genes and assay as epidemiological probes. J. Antimicrob. Chemother. 28:333-346. [DOI] [PubMed] [Google Scholar]
- 10.Magnet, S., T. Lambert, P. Courvalin, and J. Blanchard. 2001. Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC(6′)-Iy aminoglycoside N-acetyltransferase. Biochemistry 40:3700-3709. [DOI] [PubMed] [Google Scholar]
- 11.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 12.Ouali, M., and R. D. King. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9:1162-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panaite, D. M., and M. E. Tolmasky. 1998. Characterization of mutants of the 6′-N-acetyltransferase encoded by the multiresistance transposon Tn1331: effect of Phen171-to-Leu171 and Tyr80-to-Cys80 substitutions. Plasmid 39:123-133. [DOI] [PubMed] [Google Scholar]
- 14.Rao, S. T., and M. G. Rossmann. 1973. Comparison of super-secondary structures in proteins. J. Mol. Biol. 76:241-256. [DOI] [PubMed] [Google Scholar]
- 15.Salipante, S. J., and B. G. Hall. 2003. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol. Biol. Evol. 20:653-659. [DOI] [PubMed] [Google Scholar]
- 16.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shmara, A., N. Weinsetel, K. J. Dery, R. Chavideh, and M. E. Tolmasky. 2001. Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob. Agents Chemother. 45:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sternglanz, R., and H. Schindelin. 1999. Structure and mechanism of action of the histone acetyltransferase GCN5 and similarity to other N-acetyltransferases. Proc. Natl. Acad. Sci. USA 96:8807-8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolmasky, M. E. 1990. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24:218-226. [DOI] [PubMed] [Google Scholar]
- 20.Vetting, M. W., L. P. S. de Carvalho, M. Yu, S. S. Hegde, S. Magnet, S. L. Roderick, and J. S. Blanchard. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212-226. [DOI] [PubMed] [Google Scholar]
- 21.Wells, J. A. 1991. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 202:390-411. [DOI] [PubMed] [Google Scholar]
- 22.Wolf, E., A. Vassilev, Y. Makino, A. Sali, Y. Nakatani, and S. Burley. 1998. Crystal structure of a GCN5-related N-acetyltranferase: Serratia marcescens aminoglycoside 3-N-acetyltransefrase. Cell 94:439-449. [DOI] [PubMed] [Google Scholar]
- 23.Wybenga-Groot, L., K. Draker, G. Wright, and A. Berghuis. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7:497-507. [DOI] [PubMed] [Google Scholar]