Abstract
Resistance of Mycobacterium tuberculosis to fluoroquinolones (FQ) results mostly from mutations in the gyrA gene. We developed a reverse hybridization-based line probe assay in which oligonucleotide probes carrying the wild-type gyrA sequence, a serine-to-threonine (S95T) polymorphism, and gyrA mutations (A90V, A90V-S95T, S91P, S91P-S95T, D94A, D94N, D94G-S95T, D94H-S95T) were immobilized on nitrocellulose strips and hybridized with digoxigenin-labeled PCR products obtained from M. tuberculosis strains. When a mutated PCR product was used, hybridization occurred to the corresponding mutated probe but not to the wild-type probe. A panel of M. tuberculosis complex strains including 19 ofloxacin-resistant (OFL-R) and 9 ofloxacin-susceptible (OFL-S) M. tuberculosis strains was studied for detection and identification of gyrA mutations by the line probe assay and nucleotide sequencing, in comparison with testing of in vitro susceptibility to FQ. Results were 100% concordant with those of nucleotide sequencing. The S95T polymorphism, which is not related to FQ resistance, was found in 5 OFL-S and 2 OFL-R strains; the other 17 OFL-R strains harbored single mutations associated with serine or threonine at codon 95. No mutations were found in the other OFL-S strains. Overall, on the basis of the MICs on solid medium, the new line probe assay correctly identified all OFL-S and 17 out of 19 (89.5%) OFL-R strains. A nested-PCR protocol was also evaluated for the assay to amplify PCR products from M. tuberculosis-spiked sputa, with a good specificity and a sensitivity of 2 × 103 M. tuberculosis CFU per ml of sputum.
Fluoroquinolones (FQ) are antimicrobial agents with good in vitro and in vivo activities against Mycobacterium tuberculosis (5, 9-11, 29). The use of these drugs as second-line antituberculosis agents is recommended for treating multidrug-resistant (MDR) tuberculosis (TB) (3, 8). As with other antimicrobial agents, the use of FQ can generate resistant mutants (5, 6, 11-13). The principal target of the FQ in M. tuberculosis is DNA gyrase, a type II topoisomerase composed of two A and two B subunits encoded by the gyrA and gyrB genes, respectively (2, 11, 33). Mutations in the so-called quinolone resistance-determining region (QRDR) of gyrA are the primary mechanisms of FQ resistance in M. tuberculosis. Amino acids at positions 88, 90, 91, and 94 of gyrA are those most frequently substituted in the FQ-resistant M. tuberculosis clinical isolates (1, 11, 12, 19, 22, 26, 27, 31-33); mutations of gyrB have rarely been reported (15).
Despite the increasing use of these drugs in TB therapy, standard 3-week culture-based FQ susceptibility testing in solid media is not always performed (11). PCR-based techniques provide new possibilities for the rapid diagnosis of FQ resistance; however, it can be difficult to put them into practice in the mycobacteriology laboratory (7, 24, 33, 34). To this end, we developed a reverse hybridization-based line probe assay for rapid detection of gyrA mutations in M. tuberculosis. The test was based on the same principle as the commercial line probe assay for rapid determination of rifampin (RMP) resistance, the INNO-LiPA Rif. TB (Innogenetics, Ghent, Belgium) (20, 21), which, due to the low frequency of monoresistance to rifampin, serves as a surrogate marker for detection of MDR-TB (30). FQ resistance is rising, especially in MDR strains (5, 11); thus, our test can be useful for evaluating FQ resistance particularly in these organisms when treatment failures and relapses have been documented.
MATERIALS AND METHODS
Microorganisms and drug susceptibility testing.
M. tuberculosis clinical isolates were collected from Italian mycobacteriology laboratories (Rome, Florence, Ancona, Milan, and Siena) (9) and from the TB hospital of Sukhumi (Abkhazia). M. tuberculosis ATCC 27294 (H37Rv) was used as a drug-susceptible reference strain. All strains were grown in Löwenstein-Jensen slants (BioMerieux, Marcy l'Etoile, France). DNA from a Mycobacterium bovis BCG Copenhagen strain harboring the D94H and S95T mutations was kindly provided by Howard E. Takiff, Caracas, Venezuela.
MICs of ofloxacin (OFL), ciprofloxacin (CIP), sparfloxacin (SPX), moxifloxacin (MOX), levofloxacin (LEV), and gatifloxacin (GAT) were determined in Middlebrook 7H11 agar (Difco, Detroit, Mich.) as previously described (9). OFL (Sigma Chemical, St. Louis, Mo.), CIP (Bayer, Milan, Italy), LEV (Glaxo Wellcome, Verona, Italy), and GAT (Grunenthal, Aachen, Germany) were dissolved in distilled water; SPX (Aventis Pharma, Vitry-Alfortville, France) and MOX (Bayer) were dissolved in 0.1 M NaOH. Briefly, plates containing different drug concentrations (0.031 to 32 μg/ml) were inoculated in triplicate with approximately 2 × 103 CFU by a semiautomatic inoculator (Multipoint Inoculator A400; Denley, West Sussex, United Kingdom) and incubated at 37°C under 5% CO2 for 21 days. The MIC was defined as the lowest drug concentration inhibiting more than 99% of the inoculum. A strain was considered resistant to OFL if the MIC was >2 μg/ml of OFL (18).
DNA extraction, PCR, and DNA sequencing.
A loopful of M. tuberculosis colonies was collected from Löwenstein-Jensen slants and suspended in Middlebrook 7H9 broth (Difco). Bacteria were centrifuged at 3,000 × g for 30 min, and the pellet was washed twice in 1 ml of TE (10 mM Tris-HCl, 1 mM EDTA; pH 8). Organisms were suspended in 500 μl of TE and incubated first at 100°C for 30 min and then at −20°C for 60 min. Cellular debris was pelleted at 13,000 × g for 5 min, and the supernatant was used for PCR. Approximately 2 to 5 ng of genomic DNA was used for amplification of a 320-bp fragment of the M. tuberculosis H37Rv gyrA gene with primers gyrA-S (5′-CAGCTACATCGACTATGCGA-3′) and gyrA-AS (5′-GGGCTTCGGTGTACCTCAT-3′) (27) and for amplification of a 346-bp fragment of the gyrB gene with primers gyrB-S (5′-CGCAAGTCCGAACTGTATGTCGTAG-3′) and gyrB-AS (5′-GTTGTGCCAAAAACACAT GC-3′).
After 3 min at 95°C, PCR was performed for 28 cycles with an iCycler thermal cycler (Bio-Rad, Hercules, Calif.) as follows: 40 s at 95°C, 1 min at 55°C, and 1 min at 72°C. Amplicons were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced at MWG Biotech (Ebersberg, Germany). PCR products containing wild-type (WT) and mutated sequences were cloned into pGEM T-Vector (Promega, Madison, Wis.) as a reproducible source of labeled targets for optimization experiments.
Preparation of oligonucleotide probes, strips, and labeled targets.
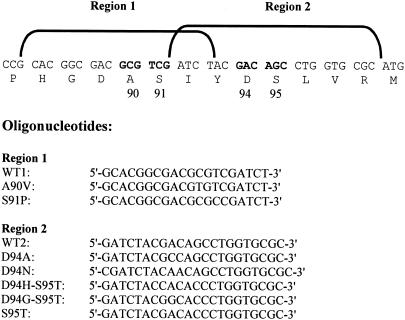
For the development of the line probe assay, oligonucleotide probes were designed to contain (i) the H37Rv wild-type (WT1) and mutated sequences encompassing region 1, covering amino acid substitutions in positions 90 and 91, and (ii) the H37Rv wild-type (WT2) and mutated sequences encompassing region 2, covering amino acid substitutions in positions 94 and 95 (Fig. 1). A poly(dT) tail was added to 20 pmol of each probe in terminal transferase buffer (Roche, Basel, Switzerland), 2 mM CoCl2, 3.2 mM dTTP, and 400 U terminal transferase in a final volume of 25 μl for 2 h at 37°C. Poly(dT) probes (4 pmol per well) were diluted in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), blotted onto Protran BA83 nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) using a 48-well Bio-Dot SF microfiltration apparatus (Bio-Rad), and fixed by baking the membranes at 80°C for 2 h. After fixing, the membranes were cut into 5- by 60-mm strips. For each bacterial strain to be tested, two strips with immobilized oligonucleotide probes were prepared, one containing WT1 and three mutated sequences and the other containing WT2 and four mutated sequences; each strip contained also a digoxigenin (DIG)-labeled 320-bp gyrA fragment of M. tuberculosis H37Rv as a positive-control probe.
FIG. 1.
Nucleotide and amino acid sequences of the QRDR of the gyrA gene. Mutated codons are boldfaced and numbered. Positions of oligonucleotide probes encompassing regions 1 and 2 are shown. Region 1 oligonucleotides are all 20-mers. Region 2 oligonucleotides are 22-mers with the exception of D94N, which is a 23-mer.
Labeled targets (DIG-labeled PCR products) were obtained from genomic DNA or plasmid DNA containing cloned gyrA sequences by using 10 μM DIG-dUTP in addition to standard deoxynucleoside triphosphates, with primers gyrA-S and gyrA-AS and the cycling protocol described above for 32 cycles. PCR products were purified from unincorporated DIG-dUTP using the QIAquick PCR purification kit (QIAGEN).
Line probe assay procedure.
Approximately 100 ng of DIG-labeled PCR products was denatured at 100°C for 5 min, diluted in 1 ml of prewarmed 5× SSC containing 0.5% sodium dodecyl sulfate (SDS), and hybridized to each strip at 63°C for 90 min with gentle shaking, with 1°C variations causing either cross-hybridization or low signals (data not shown). Strips were washed twice with 2× SSC containing 0.1% SDS at room temperature for 15 min and three times in prewarmed 0.5× SSC containing 0.1% SDS at 63°C for 15 min, rinsed in 20 mM Tris (pH 7.6)-285 mM NaCl-0.2% Tween 20 (TBST), and incubated for 30 min in blocking solution (TBST containing 1% of bovine serum albumin).
Strips were incubated for 30 min with an alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche) diluted 1:500 in blocking solution. The antibody was washed out by two 15-min washes at room temperature in washing buffer (0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween 20; pH 7.5), and the strips were equilibrated in AP buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, 5 mM MgCl2) before color development with 8 μl of 5-bromo-4-chloro-3-indolylphosphate (BCIP; 20 μg/μl; Sigma) and 6 μl of nitroblue tetrazolium (50 μg/μl; Sigma) in 1 ml of AP buffer.
Line probe assay for sputa.
Sputa were collected from smear-negative nontuberculous patients and divided into two aliquots, of which one was frozen and one was checked for the absence of M. tuberculosis by cultivation in the MGIT 960 system (Becton Dickinson, Sparks, Md.) (28). Frozen aliquots from M. tuberculosis-negative sputa were pooled and spiked with different numbers of M. tuberculosis H37Rv CFU in a volume of 5 ml; after treatment for decontamination and concentration (16, 17), sediments were used for DNA purification by the guanidinium thiocyanate method (4).
To increase the sensitivity and specificity of the line probe assay for clinical samples, amplification was performed by a nested PCR using external primers gyrA-ex-S (5′-GGATCGAACCGGTTGACAT-3′) and gyrA-ex-AS (5′-GGATATTGGTTGCCATGCC-3′) for the first 25 cycles to amplify a 528-bp fragment, and an additional 25 cycles with primers gyrA-S and gyrA-AS using a 10-fold dilution of the first reaction mixture. For preparation of the 320-bp gyrA DIG-labeled target of M. tuberculosis H37Rv, a 32-cycle nested reaction was preferred due to less efficient amplification in the presence of DIG-dUTP. Target DNA was hybridized to the panel of oligonucleotide probes immobilized on the nitrocellulose strips as described above.
RESULTS
gyrA mutations and FQ susceptibility.
Twenty-nine M. tuberculosis complex strains, including M. tuberculosis strain H37Rv, 27 M. tuberculosis clinical isolates, and 1 BCG strain, were studied for detection and identification of gyrA mutations by DNA sequencing and the line probe assay. No gyrB mutations were found in the 28 M. tuberculosis strains. Mutated gyrA alleles and amino acid changes are shown in Table 1; the patterns of resistance/susceptibility of OFL and the MICs of OFL, CIP, LEV, SPX, GAT, and MOX are also given.
TABLE 1.
gyrA mutations and fluoroquinolone susceptibility of Mycobacterium tuberculosis complex strains
| Strain no. (designation) | Amino acid change | Allele | MIC (μg/ml) of:
|
|||||
|---|---|---|---|---|---|---|---|---|
| OFL (R/S)a | CIP | LEV | SPX | GAT | MOX | |||
| 1 (MTB H37Rv) | None | WT | 1 (S) | 1 | 0.125 | 0.125 | 0.06 | 0.125 |
| 2 (MTB P10) | None | WT | 1 (S) | 0.5 | 0.125 | 0.125 | 0.125 | 0.125 |
| 3 (MTB Ru108) | None | WT | 2 (S) | 1 | 1 | 0.5 | 0.5 | 1 |
| 4 (MTB Ru108-6) | None | WT | 2 (S) | 1 | 1 | 0.5 | 0.5 | 1 |
| 5 (MTB Ru124) | S95T | AGC95ACC | 1 (S) | 0.5 | 0.5 | 0.5 | 0.125 | 0.125 |
| 6 (MTB Ru95-0) | S95T | AGC95ACC | 0.5 (S) | 0.5 | 0.25 | 0.5 | 0.125 | 0.125 |
| 7 (MTB Ru95-3) | S95T | AGC95ACC | 0.5 (S) | 0.5 | 0.25 | 0.5 | 0.06 | 0.125 |
| 8 (MTB F16) | S95T | AGC95ACC | 0.5 (S) | 0.5 | 0.25 | 0.5 | 0.06 | 0.125 |
| 9 (MTB M23) | S95T | AGC95ACC | 2 (S) | 2 | 1 | 2 | 0.25 | 0.5 |
| 10 (MTB R21) | S95T | AGC95ACC | 16 (R) | 8 | 8 | 8 | 2 | 4 |
| 11 (MTB Ru152) | S95T | AGC95ACC | 8 (R) | 8 | 4 | 8 | 1 | 2 |
| 12 (MTB R24) | A90V | GCG90GTG | 8 (R) | 8 | 8 | 4 | 2 | 4 |
| 13 (MTB A1) | A90V | GCG90GTG | 8 (R) | 4 | 4 | 2 | 1 | 1 |
| 14 (MTB A3) | A90V | GCG90GTG | 8 (R) | 4 | 4 | 2 | 1 | 2 |
| 15 (MTB R19) | A90V-S95T | GCG90GTG-AGC95ACC | 4 (R) | 8 | 4 | 2 | 2 | 2 |
| 16 (MTB F9) | A90V-S95T | GCG90GTG-AGC95ACC | 4 (R) | 4 | 4 | 2 | 1 | 1 |
| 17 (MTB F11) | A90V-S95T | GCG90GTG-AGC95ACC | 8 (R) | 4 | 2 | 2 | 1 | 1 |
| 18 (MTB F18) | A90V-S95T | GCG90GTG-AGC95ACC | 8 (R) | 8 | 4 | 4 | 1 | 1 |
| 19 (MTB Ru95-8) | A90V-S95T | GCG90GTG-AGC95ACC | 8 (R) | 4 | 4 | 2 | 0.5 | 1 |
| 20 (MTB Ru95-9) | A90V-S95T | GCG90GTG-AGC95ACC | 8 (R) | 4 | 4 | 2 | 1 | 1 |
| 21 (MTB M6159) | S91P | TCG91CCG | 8 (R) | 4 | 4 | 2 | 1 | 2 |
| 22 (MTB M60) | S91P-S95T | TCG91CCG-AGC95ACC | 4 (R) | 4 | 2 | 1 | 0.5 | 1 |
| 23 (MTB R1) | D94A | GAC94GCC | 4 (R) | 4 | 4 | 2 | 0.5 | 1 |
| 24 (MTB MO) | D94N | GAC94AAC | 8 (R) | 4 | 2 | 2 | 1 | 1 |
| 25 (MTB F7) | D94G-S95T | GAC94GGC-AGC95ACC | 8 (R) | 4 | 4 | 4 | 2 | 4 |
| 26 (MTB F15) | D94G-S95T | GAC94GGC-AGC95ACC | 8 (R) | 4 | 4 | 2 | 1 | 2 |
| 27 (MTB M3) | D94G-S95T | GAC94GGC-AGC95ACC | 8 (R) | 4 | 2 | 2 | 0.5 | 1 |
| 28 (MTB Ru227-15) | D94G-S95T | GAC94GGC-AGC95ACC | 8 (R) | 4 | 4 | 2 | 1 | 1 |
| 29 (BCG HETb) | D94H-S95T | GAC94CAC-AGC95ACC | NDc | ND | ND | ND | ND | ND |
R, resistant; S, susceptible. The strains were considered R or S to OFL if the MIC in Middlebrook 7H11 agar was >2 μg/ml or ≤2 μg/ml of OFL, respectively (18).
BCG HET refers to DNA from a BCG strain received by Howard E. Takiff.
ND, not determined.
Ten different gyrA alleles were found, including the H37Rv wild-type allele, four alleles with one mutation each (A90V, S91P, D94A, D94N), the allele with the S95T polymorphism, and four alleles each containing a single mutation associated with the S95T polymorphism (A90V-S95T, S91P-S95T, D94G-S95T, D94H-S95T). Overall, amino acids at positions 90 (nine strains) and 94 (seven strains) were those most frequently substituted.
Out of 28 M. tuberculosis strains tested, 9 were OFL susceptible (OFL-S) and 19 were OFL resistant (OFL-R). In the resistant strains, the rank order of the median MICs for the various FQ was as follows: OFL (8 μg/ml) > CIP and LEV (4 μg/ml) > SPX (2 μg/ml) > GAT and MOX (1 μg/ml). For the strains harboring A90V and A90V-S95T substitutions, the median MICs of OFL, CIP, LEV, SPX, and GAT were the same while those of MOX differed by 1 dilution. The MIC of CIP was the same, and those of the other FQ were 1 dilution higher, for the strain carrying the S91P mutation compared to the S91P-S95T strain. Among organisms harboring the S95T polymorphism, five OFL-S and two OFL-R strains were observed. All four OFL-S strains harbored the wild-type allele.
Line probe assay.
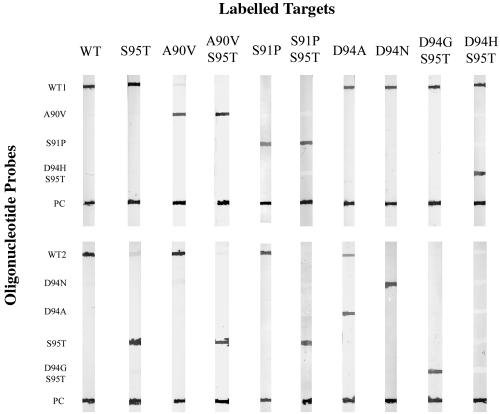
A representative line probe assay pattern obtained by testing the panel of M. tuberculosis complex strains listed in Table 1 is shown in Fig. 2. The PCR products of the four wild-type OFL-S strains hybridized only to the WT1 and WT2 probes, while S95T polymorphism-containing alleles (five OFL-S and two OFL-R strains) hybridized to the WT1 and S95T probes but not to the WT2 probe. Among the 17 M. tuberculosis OFL-R strains, eight hybridization patterns were observed, in agreement with the DNA sequencing results, including the patterns A90V (3 strains), A90V-S95T (6 strains), S91P (1 strain), S91P-S95T (1 strain), D94A (1 strain), D94N (1 strain), D94G-S95T (4 strains), and D94H-S95T (1 BCG strain). Overall, a good specificity in detecting wild-type and mutated alleles, containing the S95T polymorphism or not, was observed using the line probe assay; hybridization signals (visualized as violet bands in the strips) were strong and clear-cut with low background.
FIG. 2.
Representative line probe assay pattern obtained by testing the M. tuberculosis strains listed in Table 1. Oligonucleotide probes, indicated on the left, were blotted onto nitrocellulose strips and hybridized to wild-type and mutated DIG-labeled PCR product targets (top). Each strip was blotted with a wild-type DIG-labeled PCR product as a positive control for the development reaction (PC). PCR products from wild-type strains hybridized only to probes WT1 and WT2, while those containing the S95T polymorphism hybridized to the WT1 and S95T probes. Targets with single mutations in region 1 (A90V or S91P) hybridized to the specific probe and to the WT2 probe in the nonmutated region; when mutations were associated with the S95T polymorphism (A90V-S95T, S91P-S95T), hybridization occurred with the specific probe and with the S95T probe. Targets with a mutation in region 2 (D94A, D94N, D94G-S95T, or D94H-S95T) hybridized to WT1 and to the specific probes in region 2. Some cross-hybridization to the WT2 probe was observed with the D94A target.
In more detail, the A90V PCR product hybridized to the A90V and WT2 probes, but not to the WT1 probe, as expected from the presence of a mutated nucleotide in region 1 (see also Fig. 1). When the A90V-S95T product was used, hybridization occurred to the A90V and S95T probes but not to the WT1 and WT2 probes; a similar pattern was seen for S91P-S95T. Some cross-hybridization was observed only with the PCR products carrying the D94A mutation, which hybridized to the D94A and also the WT2 probe, but still with a lower intensity compared to D94A signal, easily discriminating between the region 2 binding patterns of wild-type and mutated D94A probes. Since in preliminary experiments the PCR products hybridized poorly to the 22-mer D94N probe, a new 23-mer probe was used successfully to improve the signal intensity. When PCR products differed from the wild type in two adjacent amino acids of region 2, as was the case with D94H-S95T and D94G-S95T, hybridization occurred both to the WT1 and the D94H-S95T and D94G-S95T probes, respectively, but not to the WT2 and S95T probes.
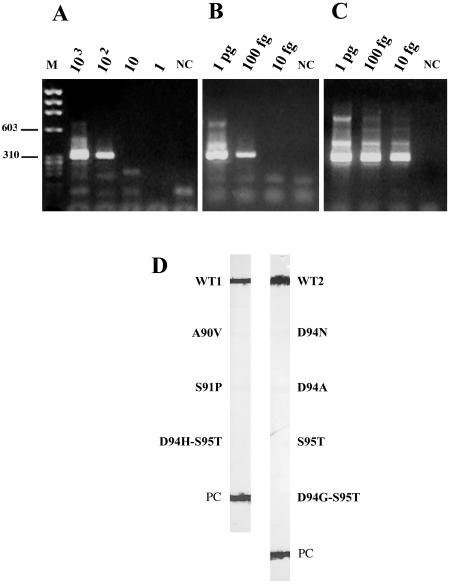
The sensitivity of the new line probe assay for clinical samples was tested by spiking 5 ml of M. tuberculosis-negative sputa with different numbers of M. tuberculosis H37Rv CFU before sample treatment; the nested PCR was performed with 5-μl aliquots of extracted DNA. Figure 3A shows that the PCR detected as many as 102 M. tuberculosis CFU, corresponding to 2 × 103 CFU/ml of untreated sputum. To evaluate DNA loss during purification and the presence of PCR inhibitors, 5-μl portions of treated nonspiked samples and distilled water, respectively, were tested in the presence of different amounts of M. tuberculosis H37Rv DNA. When DNA was added to nonspiked sputum, the sensitivity of the PCR was 100 fg (Fig. 3B), which is the DNA equivalent of approximately 20 mycobacteria (14); when DNA was added to distilled water, the sensitivity was 10 fg (Fig. 3C). The assay was also specific, since the 320-bp fragment of M. tuberculosis H37Rv correctly hybridized to the WT1 and WT2 probes (Fig. 3D). Overall, these observations indicated that despite the presence of PCR inhibitors in the sputum and DNA loss during purification, the assay was able to detect M. tuberculosis DNA in this clinical material.
FIG. 3.
Determination of nested-PCR sensitivity (A-C) and line probe assay specificity (D) using DNA extracted from sputum samples. (A) Sputa spiked with different amounts of mycobacteria. M, size markers; NC, negative control. Bands corresponding to 603 and 310 bp are indicated. PCR products from 103, 102, 10, and 1 M. tuberculosis H37Rv CFU are shown. (B) Different amounts of DNA added to nonspiked sputa processed as for panel A. Shown are PCR products from 1 pg, 100 fg, and 10 fg of M. tuberculosis H37Rv DNA. (C) Different amounts of DNA in distilled water. Shown are PCR products from 1 pg, 100 fg, and 10 fg of M. tuberculosis H37Rv DNA. (D) Results obtained after hybridization of the DIG-labeled PCR products from M. tuberculosis H37Rv-spiked sputa with oligonucleotide probes immobilized on nitrocellulose strips. The 320-bp gyrA fragment of M. tuberculosis H37Rv hybridized only to the WT1 and WT2 probes.
DISCUSSION
In this study, we developed a reverse line blot assay for detecting OFL resistance derived from point mutations in the gyrA gene of M. tuberculosis clinical isolates. Specific oligonucleotide probes spanning the QRDR of the gyrA gene were immobilized on nitrocellulose strips and hybridized with DIG-labeled PCR products obtained from M. tuberculosis DNA; the hybrids formed were detected colorimetrically. If a mutation was present in the PCR product, the mismatch prevented it from hybridizing with the immobilized WT probe, while it hybridized with an immobilized probe carrying a particular mutation.
Results were 100% concordant with those of nucleotide sequencing. The new line probe assay was reliable, sensitive, and unambiguous, due to the use of DIG-labeled PCR products and optimized hybridization conditions. The assay covers all gyrA mutations and polymorphisms reported for M. tuberculosis clinical isolates (1, 7, 11, 12, 19, 22, 26, 27, 31, 32) with the exception of D94Y (7, 26, 27, 32) and the rarely reported C88G mutation (12, 19). However, expansion of the present version of the assay to other mutations is possible by addition of new probes. It has been reported that S95T does not participate in FQ resistance (25, 27, 33). In keeping with these observations, our data showed that the S95T polymorphism did not affect FQ MICs when associated with A90V and S91P; furthermore, MICs for five OFL-S strains were lower than or equal to the critical concentration of OFL in Middlebrook 7H11 agar (2 μg/ml) (18). In our strain collection, 2 out of 19 (10.5%) OFL-R strains carried only an S95T polymorphism and showed that the MICs of OFL were 4 and 8 times higher than the critical concentration. OFL-R strains containing the S95T polymorphism were also found by other investigators (7, 22).
In comparison with in vitro results, the assay correctly identified all OFL-S and 17 out of 19 (89.5%) OFL-R strains. This is in keeping with the knowledge that 75 to 94% of FQ-resistant isolates had gyrA mutations in the QRDR (33). Other possible mechanisms of resistance include mutations in regions of gyrA and gyrB outside the QRDR, decreased cell wall permeability, active drug efflux pump mechanisms, sequestration of drug, and drug inactivation (11). A cross-resistance to the six FQ tested was observed among clinical isolates, with MOX and GAT showing the lowest MICs for both OFL-R and OFL-S strains. Both MOX and GAT are C-8-methoxy-FQ, and acquired resistance is expected to develop less readily than for other drugs due to their lower mutant prevention concentrations (23).
A nested-PCR protocol was also set up for the line probe assay to amplify DNA extracted from sputum samples, with a sensitivity of 2 × 103 M. tuberculosis CFU/ml of sputum. This value is lower than that required for recognition of acid-fast smear positivity by Ziehl-Neelsen staining (0.5 × 104 to 1 × 104 CFU/ml of sputum) (16), thus allowing the assay potentially to be able to detect M. tuberculosis DNA even when the sputum is negative by Ziehl-Neelsen staining.
Overall, the new line probe assay correctly identified OFL susceptibility and resistance in all strains in which a gyrA mutation occurred. To our knowledge, this is the first strip-based test developed for detection of OFL resistance. The assay is a rapid, cost-effective, and technically simple alternative to nucleotide sequencing and may be recommended for laboratories that do not have prompt access to molecular technology facilities and for those processing samples collected in regions in which FQ resistance is rising. Once a stock of the strips has been prepared, it can be stored in the refrigerator and used for detecting gyrA mutations if OFL resistance is suspected. Testing of M. tuberculosis for FQ susceptibility should be routinely considered for patients showing a treatment failure or a relapse following a multidrug anti-TB regimen. The assay can also be performed when RMP resistance, as a surrogate marker of MDR-TB, has been detected by the INNO-LiPA Rif. TB or another rapid test. The knowledge that an M. tuberculosis strain is resistant to both first- and second-line drugs such as RMP and OFL can be relevant to improving the control of MDR TB.
Acknowledgments
This study was supported in part by grants 0AL/F, 1AF/F2, and 4AI/F2 from the Ministero della Salute (1% Projects).
We thank Enrico Tortoli (Careggi Hospital, Florence, Italy), Claudio Piersimoni (Torrette Hospital, Ancona, Italy), Mirella Tronci and Patrizia Chiaradonna (Forlanini Hospital, Rome, Italy), Valeria Penati and Patrizia Vaccarino (Villa Marelli, Milan, Italy), Marco R. Oggioni (University of Siena, Italy), Howard E. Takiff (Lab de Genética Molecular, CMBC, IVIC, Caracas, Venezuela), Francis Varaine (Médecins Sans Frontières, Paris, France), and Hayk Karakozian and Erchanik Arzumanian (TB hospital of Sukhumi, Abkhazia) for providing M. tuberculosis strains and sputum samples.
REFERENCES
- 1.Alangaden, G. J., E. K. Manavathu, S. B. Vakulenko, N. M. Zvonok, and S. A. Lerner. 1995. Characterization of fluoroquinolone-resistant mutant strains of Mycobacterium tuberculosis selected in the laboratory and isolated from patients. Antimicrob. Agents Chemother. 39:1700-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 48:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, A. A. Vernon, et al. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryskier, A., and J. Lowther. 2002. Fluoroquinolones and tuberculosis. Expert Opin. Investig. Drugs 11:233-258. [DOI] [PubMed] [Google Scholar]
- 6.Chan, E. D., V. Laurel, M. J. Strand, J. F. Chan, M. L. Huynh, M. Goble, and M. D. Iseman. 2004. Treatment and outcome analysis of 205 patients with multidrug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 169:1103-1109. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, A. F., W. W. Yew, E. W. Chan, M. L. Chin, M. M. Hui, and R. C. Chan. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crofton, J., P. Chaulet, D. Maher, J. Grosset, W. Harris, N. Horne, M. Iseman, and B. Watt. 1997. Guidelines for the management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland.
- 9.Fattorini, L., E. Iona, M. L. Ricci, O. F. Thoresen, G. Orru, M. R. Oggioni, E. Tortoli, C. Piersimoni, P. Chiaradonna, M. Tronci, G. Pozzi, and G. Orefici. 1999. Activity of 16 antimicrobial agents against drug-resistant strains of Mycobacterium tuberculosis. Microb. Drug Resist. 5:265-270. [DOI] [PubMed] [Google Scholar]
- 10.Fattorini, L., D. Tan, E. Iona, M. Mattei, F. Giannoni, L. Brunori, S. Recchia, and G. Orefici. 2003. Activities of moxifloxacin alone and in combination with other antimicrobial agents against multidrug-resistant Mycobacterium tuberculosis infection in BALB/c mice. Antimicrob. Agents Chemother. 47:360-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginsburg, A. S., J. H. Grosset, and W. R. Bishai. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 3:432-442. [DOI] [PubMed] [Google Scholar]
- 12.Ginsburg, A. S., S. C. Woolwine, N. Hooper, W. H. Benjamin, Jr., W. R. Bishai, S. E. Dorman, T. R. Sterling, and S. R. Timothy. 2003. The rapid development of fluoroquinolone resistance in M. tuberculosis. N. Engl. J. Med. 349:1977-1978. [DOI] [PubMed] [Google Scholar]
- 13.Grimaldo, E. R., T. E. Tupasi, A. B. Rivera, M. I. Quelapio, R. C. Cardano, J. O. Derilo, and V. A. Belen. 2001. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant Mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int. J. Tuberc. Lung Dis. 5:546-550. [PubMed] [Google Scholar]
- 14.Kox, L. F., J. van Leeuwen, S. Knijper, H. M. Jansen, and A. H. Kolk. 1995. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J. Clin. Microbiol. 33:3225-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, A. S., L. L. Tang, I. H. Lim, and S. Y. Wong. 2002. Characterization of pyrazinamide and ofloxacin resistance among drug resistant Mycobacterium tuberculosis isolates from Singapore. Int. J. Infect. Dis. 6:48-51. [DOI] [PubMed] [Google Scholar]
- 16.Master, R. N. 1992. Mycobacteriology, p. 3.0.1-3.1.6.4. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.
- 17.Metchock, B. G., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 18.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved standards. Vol. 23, no. 18. M24-A. NCCLS, Wayne, Pa. [PubMed]
- 19.Perlman, D. C., W. M. El Sadr, L. B. Heifets, E. T. Nelson, J. P. Matts, K. Chirgwin, N. Salomon, E. E. Telzak, O. Klein, B. N. Kreiswirth, J. M. Musser, R. Hafner, et al. 1997. Susceptibility to levofloxacin of Myocobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. AIDS 11:1473-1478. [DOI] [PubMed] [Google Scholar]
- 20.Rossau, R., H. Traore, H. De Beenhouwer, W. Mijs, G. Jannes, P. De Rijk, and F. Portaels. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saiki, R. K., P. S. Walsh, C. H. Levenson, and H. A. Erlich. 1989. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 86:6230-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqi, N., M. Shamim, S. Hussain, R. K. Choudhary, N. Ahmed, Prachee, S. Banerjee, G. R. Savithri, M. Alam, N. Pathak, A. Amin, M. Hanief, V. M. Katoch, S. K. Sharma, and S. E. Hasnain. 2002. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob. Agents Chemother. 46:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sindelar, G., X. Zhao, A. Liew, Y. Dong, T. Lu, J. Zhou, J. Domagala, and K. Drlica. 2000. Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrob. Agents Chemother. 44:3337-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sougakoff, W., N. Lemaitre, E. Cambau, M. Szpytma, V. Revel, and V. Jarlier. 1997. Nonradioactive single-strand conformation polymorphism analysis for detection of fluoroquinolone resistance in mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 16:395-398. [DOI] [PubMed] [Google Scholar]
- 25.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan, E. A., B. N. Kreiswirth, L. Palumbo, V. Kapur, J. M. Musser, A. Ebrahimzadeh, and T. R. Frieden. 1995. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet 345:1148-1150. [DOI] [PubMed] [Google Scholar]
- 27.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tortoli, E., P. Cichero, C. Piersimoni, M. T. Simonetti, G. Gesu, and D. Nista. 1999. Use of BACTEC MGIT 960 for recovery of mycobacteria from clinical specimens: multicenter study. J. Clin. Microbiol. 37:3578-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veziris, N., C. Truffot-Pernot, A. Aubry, V. Jarlier, and N. Lounis. 2003. Fluoroquinolone-containing third-line regimen against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Chemother. 47:3117-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watterson, S. A., S. M. Wilson, M. D. Yates, and F. Drobniewski. 1998. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 36:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, K. J., R. Chan, and L. J. Piddock. 1996. gyrA of ofloxacin-resistant clinical isolates of Mycobacterium tuberculosis from Hong Kong. J. Antimicrob. Chemother. 37:1032-1034. [DOI] [PubMed] [Google Scholar]
- 32.Xu, C., B. N. Kreiswirth, S. Sreevatsan, J. M. Musser, and K. Drlica. 1996. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J. Infect. Dis. 174:1127-1130. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-254. In G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. American Society for Microbiology, Washington, D.C.
- 34.Zhou, J., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517-525. [DOI] [PubMed] [Google Scholar]