Abstract
The metallo-β-lactamases fall into two groups: Ambler class B subgroups B1 and B2 and Ambler class B subgroup B3. The two groups are so distantly related that there is no detectable sequence homology between members of the two different groups, but homology is clearly detectable at the protein structure level. The multiple structure alignment program MAPS has been used to align the structures of eight metallo-β-lactamases and five structurally homologous proteins from the metallo-β-lactamase superfamily, and that alignment has been used to construct a phylogenetic tree of the metallo-β-lactamases. The presence of genes from Eubacteria, Archaebacteria, and Eukaryota on that tree is consistent with a very ancient origin of the metallo-β-lactamase family.
β-Lactam antibiotics are the most widely used antibiotics, and the major cause of resistance to β-lactam antibiotics is the presence of β-lactamases, enzymes that inactivate β-lactam antibiotics by hydrolyzing the β-lactam bond in those drugs. β-Lactamases fall into two groups that share no structural or sequence homology: the serine β-lactamases, which employ an active-site serine to catalyze hydrolysis, and the metallo-β-lactamases, which require a bivalent metal ion (Zn2+) in the active site (6). The most commonly used classification system for β-lactamases, the Ambler system (1), assigns the metallo-β-lactamases to Ambler group B and divides them into subgroups B1, B2, and B3 (27). Subgroups B1 and B2 really form a single group within which sequences share sequence homology (18, 19), with subgroups B1 and B2 forming two distinct clades with the group. Although enzymes of subclasses B1 and B2 are descended from a common ancestor, enzymes of subclasses B1 and B3 possess a broad spectrum of activity toward β-lactam molecules while enzymes of subclass B2 are characterized by a narrow activity spectrum (3). Subgroup B3 shares structural homology (12), but not sequence homology, with subgroup B1+B2 (18, 19). The structures of eight metallo-β-lactamases, five subgroup B1, one subgroup B2, and two subgroup B3, are available and have recently been used to define a standard numbering system that can be applied to all metallo-β-lactamases (14). The general structure architecture is similar for all three subclasses, in particular among enzymes of subclasses B1 and B2. However, there are relevant localized structural features which explain the antibiotic spectrum profile differences of the three subclasses (13).
Recent studies have elucidated the phylogenetic relationships of metallo-β-lactamases and their homologs within subgroup B1+B2 and within subgroup B3 (18, 19). Because it is not possible to construct valid sequence-based phylogenies that include sequences that do not exhibit sufficient sequence homology, it is not possible to construct a single sequence-based phylogeny that illustrates the historical relationships among all of the metallo-β-lactamases. The purpose of this study is to elucidate those relationships on the basis of a structure-based phylogeny.
MATERIALS AND METHODS
Structures were aligned using the program MAPS of Guoguang Lu. MAPS (which stands for multiple alignment of protein structures) is an automated program for comparisons of multiple protein structures and is an extension of the program TOP (24). From several homologous proteins with common structural similarities, the program can automatically superimpose the three-dimensional models, detect which residues are structurally equivalent among all the structures, and provide the residue-to-residue alignment. The structurally equivalent residues are defined according to the approximate position of both main chain and side chain atoms of all the proteins. According to structure similarity, the program calculates a score of structure diversity, which can be used to build a phylogenetic tree. The VAST program (15) hosted by the National Center for Biotechnology Information web server (http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html) and the DALI program hosted by the EMBL-EBI web server (http://www.ebi.ac.uk/dali/index.html) were used to identify structural neighbors of metallo-β-lactamases. Phylogenetic trees were estimated by the neighbor-joining method using PAUP* 4.0b10 (28).
The Jackknife method was used to assess the confidence in the topology of the trees. Jackknife first randomly deletes a user-specified fraction of the sites in an alignment to produce a pseudoalignment. That pseudoalignment is then used to construct a tree. The process of deletion, construction of a pseudoalignment, and construction of a tree is repeated a specified number of times. Finally, Jackknife calculates the fraction of trees in which the descendants of a particular node are together in the same clade. That fraction, expressed as a percentile value, is a measure of the confidence in the topology of that tree.
RESULTS AND DISCUSSION
Phylogenies are always based on comparisons of homologous characters, whether those characters are morphological characters, structural characters, restriction fragments, nucleotides, or amino acids. Those that are based on nucleic acids or protein sequences depend upon a multiple alignment in which the rows are the sequences and the columns are homologous nucleotides or amino acids. Programs such as CLUSTAL (20, 30, 31) introduce gaps into the individual sequences in order to place homologous amino acids into columns. If some of the sequences have diversified so much that no significant sequence similarity can be detected by programs such as BLAST (29), not only is alignment of those sequences meaningless but also the presence of those sequences can disrupt the proper alignment of sequences that are homologous to each other. Thus, it is not possible to put sequences from subgroup B1+B2 and sequences from subgroup B3 into the same multiple sequence alignment.
Sequence, however, is not the only basis for identifying homologous amino acids in distantly related proteins. The structures of proteins from subgroup B1+B2 are sufficiently similar to the structures of proteins from subgroup B3 that it is possible to identify homologous amino acids on the basis of their occupying virtually identical positions within the protein structures (14). The structures of eight metallo-β-lactamases, five subgroup B1, one subgroup B2, and two subgroup B3, are available and have recently been used to define a standard numbering system that can be applied to all metallo-β-lactamases (14).
The metallo-β-lactamases are members of a large, diverse, superfamily of proteins that share a similar four-layered αβ/βα structure (8). The sequences of the superfamily members are highly divergent, and members are related to hydrolysis processes, redox processes, DNA repair and uptake, and RNA processing (2, 9). The superfamily includes the human glyoxylase II (Gox) and the Desulfovibrio gigas rubredoxin oxygen-oxidoreductase (Roo) proteins (14), whose structures have been reported (7, 11, 14). VAST and DALI programs identified two additional structural neighbors of metallo-β-lactamases that have been included in this structure-based phylogeny of the metallo-β-lactamases: a methyl parathion hydrolase from Pseudomonas sp. strain WBC-3 (Pah) and a Zn-dependent hydrolase, protein Tm0207, of Thermotoga maritima (Tm). Recently, the structure of the teichoic acid phosphorylcholine esterase from Streptococcus pneumoniae (CbpE) was solved (G. Garau, A. M. Di Guilmi, and O. Dideberg, unpublished data). This structure proved to have a four-layered αβ/βα structure, and so we have included also this protein in our structure-based phylogeny study. Table 1 lists the structures included in this phylogeny together with the structure and the protein database accession numbers.
TABLE 1.
Structures and accession numbers
| Protein | Description | Source | PDB codea | Åb | Sequence code |
|---|---|---|---|---|---|
| BcII | Metallo-β-lactamase subclass B1 | Bacillus cereus | 1BVT | 1.85 | AAA22276 |
| Imp-1 | Metallo-β-lactamase subclass B1 | Pseudomonas aeruginosa | 1JJE | 1.80 | P52699 |
| CcrA | Metallo-β-lactamase subclass B1 | Bacteroides fragilis | 2BMI | 2.00 | AAA22904 |
| Vim-2 | Metallo-β-lactamase subclass B1 | Pseudomonas aeruginosa | 1KO3 | 1.90 | AAF61483 |
| BlaB | Metallo-β-lactamase subclass B1 | Chryseobacterium meningosepticum | 1M2X | 1.50 | BAA07084 |
| CphA | Metallo-β-lactamase subclass B2 | Aeromonas hydrophila | 1X8G | 1.70 | CAA40386 |
| L1 | Metallo-β-lactamase subclass B3 | Stenotrophomonas maltophilia | 1SML | 1.70 | CAA52968 |
| Fez-1 | Metallo-β-lactamase subclass B3 | Legionella gormanii | 1KO7 | 1.65 | CAB96921 |
| Roo | Rubredoxin oxygen-oxidoreductase | Desulfovribrio gigas | 1E5D | 2.50 | AAG34792 |
| Gox | Hydroxyacylglutathione hydrolase (glyoxalase II) | Human | 1QH5 | 1.45 | Q16775 |
| Pah | Methyl parathion hydrolase | Pseudomonas sp. | 1P9E | 2.40 | AAP06948 |
| CbpE | Teichoic acid phosphorylcholine esterase | Streptococcus pneumoniae | 1WRA | 2.00 | CAC29434 |
| Tm | αβ/αβ fold; unknown function | Thermotoga maritima | 1VJN | 2.00 | NP_228022 |
Protein Data Base chain code. PDB chain codes are unique identifiers of entries in the PDB structure database.
Resolution of solved structures.
Pairwise BLAST alignments of the protein sequences of these structures showed that sequences within subgroup B1+B2 aligned with each other and sequences within subgroup B3 aligned with each other, but no significant similarity was found among any other pairs of sequences. Thus, the proteins Roo, Gox, Pah, CbpE, and Tm are all outside of the two metallo-β-lactamase groups.
The structures were aligned by the program MAPS, an extension of the program TOP (24), for multiple alignment of protein structures. According to structure similarity, the program calculates the sequence identities of aligned residues, the root mean square of Cα atoms among structures (see Table 2, below), and a score of structure diversity (see Table 3, below). The structure diversity score is a measure of the root mean square (RMS) deviations of the distances between the matched Cα atoms normalized to the numbers of matching amino acids in a pair of aligned structures. Table 3 shows the structure diversity scores.
TABLE 2.
Sequence identities of aligned residues and RMS differences among proteins
| Protein | Sequences identity or RMS difference among structuresa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BcII | Imp-1 | CcrA | Vim-2 | BlaB | CphA | L1 | Fez-1 | Roo | Gox | Pah | CbpE | Tm | |
| BcII | 0 | 1.13 | 0.96 | 1.05 | 1.15 | 1.10 | 1.63 | 1.65 | 1.65 | 1.58 | 1.45 | 1.69 | 1.75 |
| Imp-1 | 0.36 | 0 | 1.19 | 1.26 | 1.41 | 1.28 | 1.78 | 1.67 | 1.74 | 1.58 | 1.47 | 1.44 | 2.07 |
| CcrA | 0.37 | 0.36 | 0 | 1.09 | 1.21 | 1.21 | 1.59 | 1.66 | 1.75 | 1.45 | 1.49 | 1.67 | 1.65 |
| Vim-2 | 0.39 | 0.35 | 0.33 | 0 | 1.19 | 1.07 | 1.62 | 1.72 | 1.78 | 1.59 | 1.55 | 1.79 | 1.58 |
| BlaB | 0.37 | 0.33 | 0.29 | 0.28 | 0 | 1.24 | 1.68 | 1.70 | 1.78 | 1.78 | 1.45 | 1.62 | 1.36 |
| CphA | 0.32 | 0.24 | 0.29 | 0.28 | 0.29 | 0 | 1.70 | 1.78 | 1.79 | 1.62 | 1.65 | 1.56 | 1.55 |
| L1 | 0.22 | 0.17 | 0.16 | 0.17 | 0.18 | 0.20 | 0 | 1.31 | 1.36 | 1.41 | 1.53 | 1.48 | 1.44 |
| Fez-1 | 0.14 | 0.16 | 0.12 | 0.13 | 0.18 | 0.15 | 0.33 | 0 | 1.47 | 1.40 | 1.38 | 1.71 | 1.55 |
| Roo | 0.17 | 0.20 | 0.17 | 0.19 | 0.18 | 0.20 | 0.18 | 0.16 | 0 | 1.56 | 1.42 | 1.52 | 1.79 |
| Gox | 0.24 | 0.22 | 0.26 | 0.21 | 0.22 | 0.23 | 0.21 | 0.21 | 0.21 | 0 | 1.35 | 1.26 | 1.52 |
| Pah | 0.23 | 0.22 | 0.19 | 0.19 | 0.15 | 0.18 | 0.31 | 0.24 | 0.18 | 0.26 | 0 | 1.45 | 1.68 |
| CbpE | 0.18 | 0.19 | 0.21 | 0.23 | 0.11 | 0.10 | 0.12 | 0.11 | 0.15 | 0.27 | 0.27 | 0 | 1.54 |
| Tm | 0.17 | 0.17 | 0.13 | 0.14 | 0.18 | 0.16 | 0.10 | 0.15 | 0.19 | 0.18 | 0.25 | 0.20 | 0 |
Sequence identities are in the lower left triangle; RMS differences are in the upper right triangle.
TABLE 3.
Structural diversity scores
| Protein | Structural diversity score between structures
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BCII | IMP-1 | CcrA | VIM-2 | BlaB | CphA | L1 | FEZ-1 | Roo | Gox | Pah | CbpE | Tm | |
| BCII | 0 | ||||||||||||
| IMP-1 | 0.49 | 0 | |||||||||||
| CcrA | 0.39 | 0.48 | 0 | ||||||||||
| VIM-2 | 0.42 | 0.54 | 0.45 | 0 | |||||||||
| BlaB | 0.46 | 0.63 | 0.48 | 0.49 | 0 | ||||||||
| CphA | 0.55 | 0.57 | 0.54 | 0.50 | 0.57 | 0 | |||||||
| L1 | 1.05 | 1.36 | 0.97 | 1.19 | 1.11 | 1.42 | 0 | ||||||
| FEZ-1 | 1.20 | 1.31 | 1.14 | 1.22 | 1.14 | 1.44 | 0.44 | 0 | |||||
| Roo | 1.31 | 1.51 | 1.42 | 1.41 | 1.69 | 1.41 | 1.06 | 0.98 | 0 | ||||
| Gox | 1.51 | 1.59 | 1.36 | 1.55 | 1.57 | 1.98 | 1.23 | 1.55 | 1.47 | 0 | |||
| Pah | 1.54 | 1.35 | 1.43 | 1.88 | 1.50 | 1.54 | 1.66 | 1.53 | 1.20 | 1.50 | 0 | ||
| CbpE | 2.10 | 2.18 | 2.12 | 3.59 | 5.00 | 2.59 | 6.66 | 2.60 | 1.91 | 2.34 | 2.16 | 0 | |
| Tm | 5.46 | 8.11 | 5.28 | 8.44 | 6.23 | 7.41 | 5.09 | 5.73 | 4.86 | 3.27 | 5.37 | 2.49 | 0 |
Other normalizations of RMS distances in structural alignments have been used to construct structure-based phylogenies (4, 5, 16, 22, 26). In those studies the normalized RMS distances were used as the values in a distance matrix and distance-based phylogenies were constructed using unweighted pair group method with averages (UPGMA) or neighbor-joining methods. It is both tempting and intuitive to assume that the more closely related two proteins are, the closer the positions of the Cα atoms of homologous amino acids will be. For phylogenetic purposes, that assumption would be valid only if the physical distances between corresponding Cα atoms were linearly proportional to the genetic distances, i.e., to the number of changes among the alignable amino acids. Most published studies (4, 5, 16, 26) that include structure-based phylogenies simply assert that normalized RMS distances can be used for phylogenetic reconstruction without presenting a basis for that assertion. We are aware of only two studies (21, 22) that have explicitly addressed the issue of RMS physical distances versus phylogenetic distances and concluded that structure-based phylogenetic trees are in good agreement with sequence-based trees. One of those studies (22) explicitly found linear relationships between normalized RMS distances and sequence-based phylogenetic distances, even though there is a great deal of scatter about those lines and the regressions do not pass through the origin.
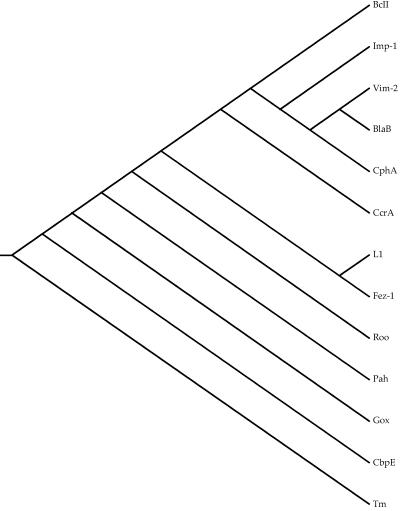
Figure 1 shows a neighbor-joining tree for metallo-β-lactamases that is derived from the structure diversity scores in Table 3. That tree places the subgroup B2 structure, CphA, deep within subgroup B1, a topology that disagrees with all sequence-based trees of those enzymes (18, 19, 27). So, for metallo-β-lactamases we cannot use either a sequence-based phylogeny or an RMS distances-based phylogeny. An alternative approach is to use the structures only to generate a reliable alignment of homologous amino acids and to use the resulting amino acid alignment to construct a phylogenetic tree (17).
FIG. 1.
Neighbor-joining tree based on structural diversity scores.
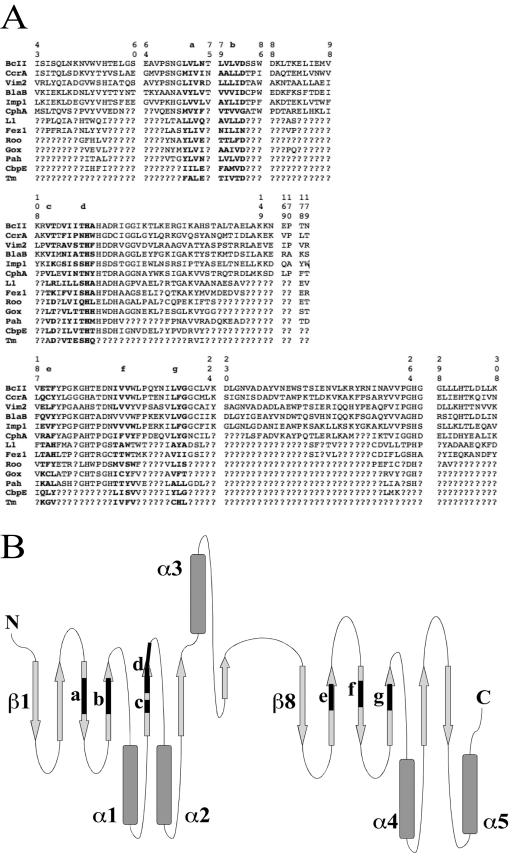
The MAPS output includes a list of structure fragments in which each amino acid in a fragment is aligned across all structures. Those fragments can be combined to form a multiple alignment in which all of the amino acids in a column are homologous based on occupying the same positions in their respective structures. MAPS alignment of all 13 structures listed in Table 1 produces a set of fragments that, when combined, result in an alignment that is only 30 residues long. There is considerable uncertainty associated with a phylogeny based on only 30 amino acids. Accuracies of phylogenetic methods tend to increase as the amount of data increases, and so a longer alignment was developed as follows.
Fragments from the MAPS alignment of the subgroup B1 structures were combined to construct an alignment 173 residues long. A MAPS alignment of subgroup B1+B2 (CphA) yielded a structural alignment of 157 amino acids. The aligned amino acids of CphA were written below the initial alignment of 173 amino acids, and the missing data symbol, “?,” was written in those columns where CphA did not align with the subgroup B1 amino acids. Similarly, MAPS alignments of B1, B2, and B3, of B1, B2, B3, and Roo, of B1, B2, B3, and Gox, etc., were used to build up the alignment shown in Fig. 2A. That alignment includes 173 characters for subgroup B1, 157 characters for B2, 108 characters for B3, 78 characters for Roo, 75 characters for Gox, 76 characters for Pah, 55 characters for CbpE, and 30 characters for Tm. There are six sequence fragments which cover the entirety of all sequences (Fig. 2A, fragments a to g), and the positions of these sequence fragments in the general metallo-β-lactamase topology (11) are illustrated in Fig. 2B.
FIG. 2.
(A) Structure-based multiple alignment of homologous amino acids. The alignment is separated into the fragments returned by MAPS for ease of viewing. The six sequence fragments of structurally conserved positions (from a to g), which cover the entirety of all sequences, are shown in boldface. Fragment ends are numbered according to the standard numbering scheme for class B β-lactamases (14). Question marks indicate positions that are not structurally conserved. (B) Position of the sequence fragments (from a to g) in the general metallo-β-lactamase topology (13).
The validity of conclusions drawn from any structural comparison is critically dependent on a proper assessment of the reliability of the individual structural models which are being compared and of the degree to which any observed differences are supported by experimental data (23). In our study on metallo-β-lactamases, all the structures we have analyzed have good and comparable resolution (Table 2). Only Roo and Pah structures have a resolution greater than 2.0 Å. The RMS differences among these structures are on the order of 1.0 to 2.0 Å (Table 2), while the accuracy of atomic positions in X-ray structures obtained at about 2.0 Å can be estimated in the range of 0.1 to 0.3 Å (10, 32). While we do not have confidence in the structural diversity values as distances for constructing a phylogeny, for metallo-β-lactamases the RMS differences among structures cannot be considered artifacts of the X-ray structure determination protocol, and the structural diversity values are really valuable for clustering purposes. Clustering identifies groups of related sequences but does not elucidate orders of descent from a common ancestor. Table 3 shows that subgroup B1+B2 forms a cluster within which structural diversity scores range from 0.39 to 0.57, and the two subgroup B3 structures, L1 and Fez-1, have a structural diversity score of 0.44. Metallo-β-lactamase structures have scores ranging from 0.98 to 1.98 when compared with Roo, Gox, or Pah and from 2.1 to 6.66 when compared with CbpE. When compared with Tm the scores range from 5.09 to 7.41. Those scores are consistent with Tm being the most distantly related of the structures, i.e., Tm is a legitimate outgroup to the remaining sequences.
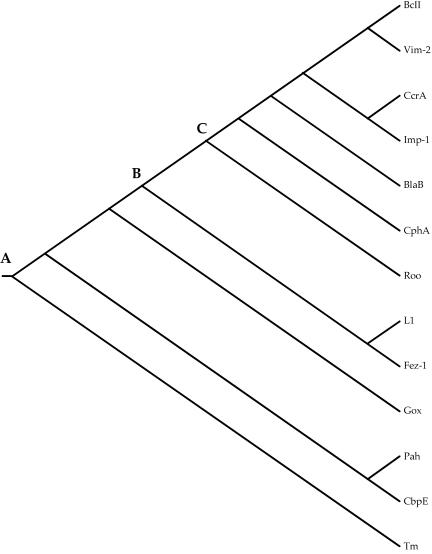
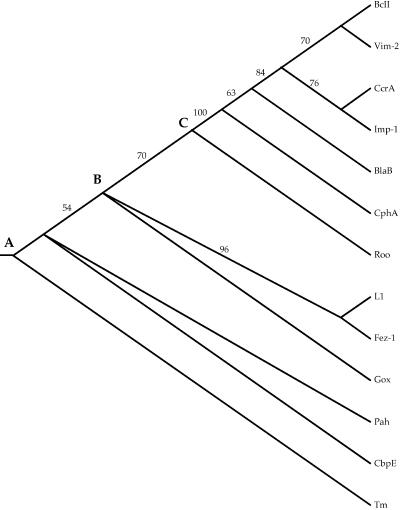
PAUP* 4.0 was used to construct a neighbor-joining phylogeny from the alignment shown in Fig. 2. That phylogeny, rooted with Tm, is shown in Fig. 3. Because only a fraction of the sites in the proteins were used to construct the phylogeny, any branch lengths estimated would be meaningless and are therefore not indicated on that tree. Finally, the Jackknife method, with 25% deletion and 5,000 replicates, was used to assess the reliability of that tree. Figure 4 shows the Jackknife tree. A polytomy, three or more branches descended from a single node, indicates statistical uncertainty about the order of descent from that node.
FIG. 3.
Neighbor-joining tree based on structural multiple alignment shown in Fig. 2.
FIG. 4.
Jackknife neighbor-joining tree based on 5,000 replicate samples in which a random 25% of the sites in the alignment shown in Fig. 2 were deleted. Numbers are the percentages of replicates in which the descendants of a given node were present.
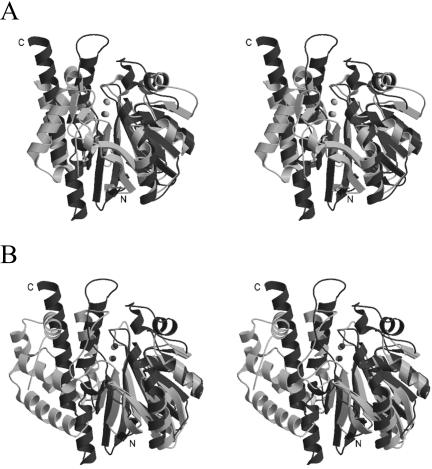
Comparison of the fold of Fez-1 with that of Roo and Gox is represented in the ribbon in Fig. 5. The overall general architecture of these enzymes is similar. Relevant differences involve mainly last secondary structural elements at the C terminus domain. All residues involved in the metal coordination spheres of metallo-β-lactamases are conserved in Gox (Table 4). With respect to Gox, metallo-β-lactamases have no bridging aspartate between the two zinc ions. Fez-1 and Roo differ in few residues of the active sites groove and in the nature of the metal center, zinc for the former and iron for the latter (Table 4). As a consequence, the β-lactamase-like domain of rubredoxin oxidoreductase loses the hydrolytic activity of metallo-β-lactamases, acquiring the dioxygen reduction activity. The structure-based phylogeny shows that the metallo-β-lactamases and the β-lactamase-like domain of rubridoxin oxidoreductase descended from a common ancestor at node A. Figure 4 also suggests that enzymes of subclass B3 (Fez-1 and L1) are closer to Gox, in good agreement with minor differences in the active site. The subgroup B3 enzymes diverged from subgroup B1+B2 and Roo at node B, and later, at node C, Roo diverged from metallo-β-lactamases subgroup B1+B2. The structure-based phylogeny is consistent with sequence-based phylogenies in which subgroup B1 enzymes constitute a monophyletic clade that is distinct from the subgroup B2 clade, here represented by CphA. B1 enzymes show the existence of a binuclear zinc active site, while CphA is a mononuclear enzyme and the zinc ion is located in site 2 (Table 4). CphA has an active site specifically modeled for carbapenems (13). Division of B1 and B2 clades might have been a consequence of the appearance of the C-5 and C-6 trans-configuration in the β-lactam skeleton, which is typical of stable carbapenems (25). It is also consistent with the conclusion of an earlier study (18) that the ability to hydrolyze the β-lactam bond arose independently in subgroup B1+B2 and in subgroup B3.
FIG. 5.
Stereoview diagram showing the superimposition of Fez-1 (dark gray) and Roo (light gray) structures (A) and Fez-1 (dark gray) and Gox (light gray) (B).
TABLE 4.
Residues involved in metal coordination
| Site | Residues involved in coordination
|
||||
|---|---|---|---|---|---|
| BcIIa | CphAab | Fez-1 | Rooc | Gox | |
| 1 | H116, H118, H196 | N116, H118, H196 | H116, H118, H196 | H79, E81, H146, (D165)d | H54, H56, H110, (D134)d |
| 2 | D120, C221, H263 | D120, C221, H263 | D120, H121, H263 | D83, —, H226, (D165)d | D58, H59, H173, (D134)d |
BBL numbering (11).
Monozinc enzyme with zinc ion located in site 2.
Di-iron enzyme.
Bridging aspartate not present in zinc-β-lactamases.
Subgroup B1+B2 enzymes include both true metallo-β-lactamases, i.e., proteins that can hydrolyze β-lactam antibiotics, and closely related proteins that have no β-lactamase activity (18). It was estimated that the subgroup B1+B2 proteins arose shortly after the proteobacteria diverged from the gram-positive bacteria, about 2.2 billion years ago, and that the true metallo-β-lactamases of subgroup B3 arose earlier than 2.2 billion years ago (18). Indeed, subgroup B3 homologs and Roo-like proteins are present in both Eubacteria and Archaebacteria. Similarly to metallo-β-lactamases, glyoxalase II has a dinuclear active site. Its action is to convert 2-oxoaldehydes into 2-hydroxy acids in the presence of glutathione. The position of the human protein Gox in this structure-based phylogeny is consistent with a very ancient origin of the metallo-β-lactamase family.
Acknowledgments
We are grateful to Joe Felsenstein, University of Washington, to Tony Dean, University of Minnesota, and to Otto Dideberg, Institut de Biologie Structural of Grenoble (France), for very helpful discussions.
This work was supported by a grant from the European Union (HPRN-CT-2002-00264).
REFERENCES
- 1.Ambler, R. P. 1980. The structure of beta-lactamases. Philos. Trans. R. Soc. London B 289:321-331. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L. 1998. An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol. 1:0008. [PubMed] [Google Scholar]
- 3.Boschi, L., P. S. Mercuri, M. L. Riccio, G. Amicosante, M. Galleni, J. M. Frere, and G. M. Rossolini. 2000. The Legionella (Fluoribacter) gormanii metallo-beta-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 beta-lactamases. Antimicrob. Agents Chemother. 44:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitling, R., D. Laubner, and J. Adamski. 2001. Structure-based phylogenetic analysis of short-chain alcohol dehydrogenases and reclassification of the 17β-hydroxysteroid dehydrogenase family. Mol. Biol. Evol. 18:2154-2161. [DOI] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M. 2000. Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol. 50:39-44. [DOI] [PubMed] [Google Scholar]
- 6.Bush, K. 1998. Metallo-beta-lactamases: a class apart. Clin. Infect. Dis. 27(Suppl. 1):S48-S53. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, A. D., M. Ridderstrom, B. Olin, and B. Mannervik. 1999. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure Fold Des. 7:1067-1078. [DOI] [PubMed] [Google Scholar]
- 8.Carfi, A., S. Pares, E. Duee, M. Galleni, C. Duez, J. M. Frere, and O. Dideberg. 1995. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daiyasu, H., K. Osaka, Y. Ishino, and H. Toh. 2001. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett. 503:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Daopin, S., D. Davies, M. Schlunegger, and M. Grütter. 1994. Comparison of two crystal structures of TGF-2: the accuracy of refined protein structures. Acta Crystallogr. D 50:85-92. [DOI] [PubMed] [Google Scholar]
- 11.Frazao, C., G. Silva, C. M. Gomes, P. Matias, R. Coelho, L. Sieker, S. Macedo, M. Y. Liu, S. Oliveira, M. Teixeira, A. V. Xavier, C. Rodrigues-Pousada, M. A. Carrondo, and J. Le Gall. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7:1041-1045. [DOI] [PubMed] [Google Scholar]
- 12.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J. M. Frere. 2001. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garau, G., C. Bebrone, C. Anne, M. Galleni, J. M. Frere, and O. Dideberg. 2005. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345:785-795. [DOI] [PubMed] [Google Scholar]
- 14.Garau, G., I. Garcia-Saez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J. M. Frere, and O. Dideberg. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibrat, J. F., T. Madej, and S. H. Bryant. 1996. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6:377-385. [DOI] [PubMed] [Google Scholar]
- 16.Hall, B. G., and M. Barlow. 2004. Evolution of the serine β-lactamases: past, present and future. Drug Resist. Update 7:111-123. [DOI] [PubMed] [Google Scholar]
- 17.Hall, B. G., and M. Barlow. 2003. Structure-based phylogenies of the serine β-lactamases. J. Mol. Evol. 57:255-260. [DOI] [PubMed] [Google Scholar]
- 18.Hall, B. G., S. J. Salipante, and M. Barlow. 2004. Independent origins of the subgroup (B1+B2) and the subgroup B3 metallo-β-lactamases. J. Mol. Evol. 59:132-140. [DOI] [PubMed] [Google Scholar]
- 19.Hall, B. G., S. J. Salipante, and M. Barlow. 2003. The metallo-β-lactamases fall into two distinct phylogenetic groups. J. Mol. Evol. 57:249-254. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, M. S., A. Sali, and T. L. Blundell. 1990. Phylogenetic relationships from three-dimensional protein structures. Methods Enzymol. 183:670-690. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, M. S., M. J. Sutcliffe, and T. L. Blundell. 1990. Molecular anatomy: phyletic relationships derived from three-dimensional structures of proteins. J. Mol. Evol. 30:43-59. [DOI] [PubMed] [Google Scholar]
- 23.Kleywegt, G. 1999. Experimental assessment of differences between related protein crystal structures. Acta Crystallogr. D 55:1878-1884. [DOI] [PubMed] [Google Scholar]
- 24.Lu, G. 2000. TOP: a new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr. 33:176-183. [Google Scholar]
- 25.Neu, H. C. 1992. Structure-activity relationships: biological, p. 101-128. In M. I. Page (ed.), The chemistry of β-lactams. Blackie, Glasgow, United Kingdom.
- 26.O'Donoghue, P., and Z. Luthey-Schulten. 2003. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 67:550-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing beta-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b4a. Sinauer Associates, Sunderland, Mass.
- 29.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tickle, I., R. Laskowski, and D. Moss. 1998. Error estimates of protein structure coordinates and deviations from standard geometry by full-matrix refinement of γB- and βB2-crystallin. Acta Crystallogr. D 54:243-252. [DOI] [PubMed] [Google Scholar]