Abstract
This study was conducted to investigate the occurrence of multiple-antibiotic resistance among 261 clinical isolates of Salmonella enterica serotype Paratyphi B strains collected between 2000 and 2003 through the network of the French National Reference Center for Salmonella. The 47 multidrug-resistant (MDR) isolates identified (18%), were characterized on the basis of the presence of several resistance genes (blaTEM, blaPSE-1, blaCTX-M, floR, aadA2, qacEΔ1, and sul1), the presence of Salmonella genomic island 1 (SGI1) by PCR mapping and hybridization, and the clonality of these isolates by several molecular (ribotyping, IS200 profiling, and pulsed-field gel electrophoresis [PFGE]) and phage typing methods. The results of PCR and Southern blot experiments indicated that 39 (83%) of the 47 S. enterica serotype Paratyphi B biotype Java MDR isolates possessed the SGI1 cluster (MDR/SGI1). Among these 39 MDR/SGI1 isolates, only 3 contained variations in SGI1, SGI1-B (n = 1) and SGI1-C (n = 2). The 39 MDR/SGI1 isolates showed the same specific PstI-IS200 profile 1, which contained seven copies from 2.6 to 18 kb. Two PstI ribotypes were found in MDR/SGI1 isolates, RP1 (n = 38) and RP6 (n = 1). Ribotype RP1 was also found in two susceptible strains. Analysis by PFGE using XbaI revealed that all the MDR/SGI1 isolates were grouped in two related clusters, with a similarity percentage of 82%. Isolation of MDR/SGI1 isolates in France was observed mainly between the second quarter of 2001 and the end of 2002. The source of the contamination has not been identified to date. A single isolate possessing the extended-spectrum β-lactamase blaCTX-M-15 gene was also identified during the study.
Two clinical syndromes have been associated with Salmonella enterica subspecies enterica serotype Paratyphi B (1,4 [5], 12:b:1,2): enteric fever and self-limited gastroenteritis. Kauffmann (16) suggested the use of dextrorotatory tartrate (d-tartrate; the same as l-tartrate) to differentiate between strains of septicemic and enteric origins. Recently, the fermentation of d-tartrate has been found in good correlation with molecular markers (sopE1 and avrA virulence genes detected by PCR) associated with systemic or enteric pathovars (27). As the biochemical tests for d-tartrate fermentation sometimes give incorrect or unreliable results after up to 7 days of culture, a rapid and reliable multiplex PCR assay has been developed by Malorny et al. (20). The d-tartrate-fermenting variant (dT+), which is called biotype Java, is isolated from both humans and animals. In the human host, this variant causes gastroenteritis, while the non-d-tartrate-fermenting variant (dT−) usually provokes typhoid fever-like disease.
Recently, the dT+ variant has become increasingly important. It has been associated with human outbreaks in France in 1996 (273 cases caused by an unpasteurized goats' milk cheese) (5) and in Canada in 1999 (43 cases due to contaminated alfalfa sprouts) and in 2000 (7 cases linked to aquariums) (12, 33). A particular clone of biotype Java with multiple-antibiotic resistance is increasingly recovered in poultry and poultry products in Germany and in The Netherlands, where S. enterica serotype Paratyphi B is the predominating serotype (22, 36). This multidrug-resistant (MDR) clone has possibly led to human cases in The Netherlands and Scotland (4, 36). All isolates belonging to this clone revealed resistance to streptomycin, spectinomycin, and trimethoprim that was mediated by a chromosomally located class 2 integron carrying a dfrA1-sat1-aadA1 array of gene cassettes (23). Additional resistances were also frequently found to sulfonamides (71% of German strains isolated between 1995 and 2001), nalidixic acid (62%), and ampicillin (51%) (23).
In 1997, the chromosomal acquisition of Salmonella genomic island 1 (SGI1) was identified in an S. enterica serotype Paratyphi B biotype Java strain isolated in a tropical fish from Singapore (21). SGI1, first described to occur in serotype Typhimurium DT104, is a 43-kb structure located between chromosomal genes thdF and int2 (2). The int2 gene, located upstream of the yidY gene, is part of a cryptic retronphage sequence reported to date to occur only in serotype Typhimurium (2). In other S. enterica serotypes, SGI1 is located between Salmonella genes thdF and yidY (3, 6, 7, 8, 21). The multidrug resistance region is located on a 14-kb region at the 3′ end of the structure. The resistance genes floR and tet(G) are bracketed by two class I integrons, one carrying an aadA2 cassette (1.0 kb) and the other a blaPSE-1 cassette (1.2 kb). Strains containing SGI1 are usually resistant to ampicillin (and amoxicillin), streptomycin, spectinomycin, chloramphenicol (and florfenicol), sulfonamides, and tetracycline (R-type [resistance type] ASSpCTeSul). However, strains containing SGI1 variants (classified as SGI1-A to SGI1-H) conferring different antibiotic resistance profiles have recently been described to occur in various serotypes of S. enterica (3, 6, 7, 8). Since 2001, the emergence of SGI1 within serotype Paratyphi B biotype Java has been observed in Canada (24) and in Great Britain (35). This study was conducted to investigate the occurrence of multidrug resistance among 261 clinical isolates of S. enterica serotype Paratyphi B collected through the network of the French National Reference Center for Salmonella (NRC-Salm) located at the Institut Pasteur, Paris, France, between 2000 and 2003. The MDR isolates were characterized for the presence of several resistance genes and of SGI1 and for their clonality by several molecular and phage typing methods.
MATERIALS AND METHODS
Bacterial strains.
The 47 MDR S. enterica serotype Paratyphi B clinical isolates and the 9 strains used for comparison are listed in Table 1. All the isolates were submitted to the NRC-Salm from January 2000 to December 2003 through its network comprising approximately 1,500 clinical microbiology laboratories (about 30% of all the French Medical laboratories). Multiple-antibiotic resistance was defined by resistance to at least two antibiotic classes.
TABLE 1.
Phenotypic and molecular characteristics of MDR S. enterica serotype Paratyphi B isolates and comparison strainsa
| Strains | Antimicrobial resistance patternb | Isolate(s) | Source (no. of samples) | Agec of the patients (n) | Sex of the patients (n) | Date of isolation | Antibiotic and antiseptic resistance gene(s) tested by PCR | Size(s) of class I integron(s) (kb) | SGI1 junction PCR resultd
|
Ribotype (no. of isolates) | IS200 type (no. of isolates) | PFGE typec (no. of isolates) | Phage typef (no. of isolates) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left
|
Right
|
||||||||||||||
| thdF/int | S044/yidY | S044/int2 | |||||||||||||
| S. enterica serotype Paratyphi B strains | ASSpCTeSul | 00-1062, 00-6045, 00-7295, 00-8941, 01-2407, 01-4034, 01-4259, 01-5072, 01-5498, 01-6505, 01-9984, 01-9997, 02-0616, 02-0872, 02-0972, 02-1286, 02-1386, 02-2529, 02-3871, 02-3872, 03-0178, 02-4026, 02-4755, 02-5458, 02-6559, 02-9157, 02-9491, 02-9939, 03-8082, 03-9114, 03-9114 | Stools (30) | I (7) | M (10) | Feb 2000 to Dec 2003 | blaPSE-1, floR, sul1, aadA2, qacEΔ1 | 1.0, 1.2 | + | + | − | RP1 (30) | IP1 (31) | X1.1 (16) | RDNC [3b] (19) |
| NK (1) | II (12) | F (20) | |||||||||||||
| III (6) | NK (1) | RP6 (1) | X2 (1) | RDNC [1] (4) | |||||||||||
| IV (4) | X4 (4) | RDNC [1010] (3) | |||||||||||||
| V (2) | X5 (4) | 3b var.2 (2) | |||||||||||||
| X6 (2) | 1010 (2) | ||||||||||||||
| X7 (3) | 1 var.3 (1) | ||||||||||||||
| UT (1) | |||||||||||||||
| ASSpCTeSulTmp | 02-3609, 02-3869, 02-3870, 02-3873, 02-9684 | Stools (5) | I (1) | M (2) | June 2002 to Dec 2002 | blaTEMh, blaPSE-1, floR, sul1, aadA2, qacEΔ1 | 1.0, 1.2 | + | + | − | RP1 (5) | IP1 (5) | X1.2 (4) | RDNC [3b] (4) | |
| II (2) | F (3) | ||||||||||||||
| III (2) | X5 (1) | Not typeable (1) | |||||||||||||
| ASul | 02-1062 | Stools | IV | M | Feb 2002 | blaTEM | ND | − | − | − | RP2 | IP4 | X3 | Dundee | |
| 02-5269 | Stools | II | F | Aug 2002 | blaPSE-1, sul1, qacEΔ1 | 1.2 | + | + | − | RP1 | IP1 | X4 | 1010 | ||
| SSpSulTmpNal | 02-3201, 02-3487 | Stools (2) | IV (1) | F (2) | May 2002 to June 2002 | Negative for genes tested | ND | − | − | − | RP3 (2) | NT (2) | X8 (2) | Not typeable (2) | |
| NK (1) | |||||||||||||||
| ASulTmp | 02-5932, 03-7087 | Blood (1) | IV (2) | F (1) | Aug 2002 to Sept 2003 | blaTEM | ND | − | − | − | RP2 (2) | IP4 (2) | X3 (2) | 1 var.3 (1) | |
| Stools (1) | NK (1) | 1 var.4 (1) | |||||||||||||
| SSpTeSul | 02-3610 | Stools | II | F | June 2002 | sul1, aadA2, qacEΔ1 | 1.0 | + | + | − | RP1 | IP1 | X1.1 | RDNC [3b] | |
| SSpSul | 01-9585 | Stools | II | M | Nov 2001 | sul1, aadA2, qacEΔ1 | 1.0 | + | + | − | RP1 | IP1 | UT | 3bvar.2 | |
| SSpToGSulTmp | 02-9348 | Stools | II | M | Nov 2002 | Negative for genes tested | − | − | − | − | RP3 | IP5 | NT | Dundee | |
| ASSpSulTmpNal | 01-0582 | Stools | IV | M | Feb 2001 | blaTEM | ND | − | − | − | RP3 | IP5 | X8 | Dundee var.1 | |
| ACroTmpg | 01-7995 | Stools | II | M | Oct 2001 | blaCTX-M-15-like | ND | NT | NT | − | RP2 | IP7 | NT | Taunton | |
| Comparison strains | Susceptible | 00-2521, 02-0495, 02-3857, 02-7168, 03-1116 | Stools (5) | II (1) | F (3) | Apr 2000 to Feb 2003 | RP1 (3) | IP2 (1) | X12 (1) | Dundee (2) | |||||
| III (2) | M (2) | RP2 (2) | IP3 (2) | X13 (1) | 1 var.3 (2) | ||||||||||
| IV (2) | IP4 (1) | X14 (1) | Taunton (1) | ||||||||||||
| Paratyphi B dT+ reference | IP7 (1) | NT (2) | |||||||||||||
| strain 5K | RP4 | IP6 | X9 | NT | |||||||||||
| Paratyphi B reference strain 7K | RP5 | IP7 | X10 | NT | |||||||||||
| ASSpSulTmpNal | Paratyphi B dT+ strain 1543/01 (poultry, Germany) | RP3 | IP5 | X8 | Dundee var.1 | ||||||||||
| ASSpCTeSul | Serotype Typhimurium isolate 02-5494 | blaPSE-1, floR, sul1, aadA2, qacEΔ1 | 1.0, 1.2 | + | − | + | RP7 | IP8 | X15 | DT104 | |||||
Abbreviations: ND, none detected; NT, not tested; NK, not known.
Abbreviations for resistance patterns: A, amoxicillin; Cro, ceftriaxone; S, streptomycin; Sp, spectinomycin; To, tobramycin; G, gentamicin; C, chloramphenicol; Te, tetracycline; Sul, sulfonamides; Tmp, trimethroprim; Nal, nalidixic acid.
Age group I, <1 yr; II, 1 to 5 yr; III, 6 to 14 yr; IV, 15 to 64 yr; V, >65 yr.
+, product obtained with primers specific for the indicated genes; −, no product obtained.
UT, untypeable (lysis).
RDNC, react but does not conform (phage type group shown in brackets).
Extended-spectrum β-lactamase-producing isolate.
No product was obtained with blaTEM primers for isolate 02-9684.
S. enterica serotype Paratyphi B biotype Java strain 1543/01, isolated from poultry in Germany, was kindly provided by A. Miko (Laboratory for Molecular Biology and National Salmonella Reference Laboratory, Bundesinstitut für Risikobewertung, Berlin, Germany). The reference strains 7K (serotype Paratyphi B) and 5K (serotype Paratyphi B biotype Java) were from the World Health Organization Collaborative Center for Reference and Research on Salmonella (Institut Pasteur).
Serotyping.
Isolates were serotyped on the basis of somatic O and phase 1 and phase 2 flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and World Health Organization Collaborative Center for Reference and Research on Salmonella) according to the White-Kauffmann-Le Minor scheme (26).
Biotyping.
Utilization of d-tartrate was assessed by determining the ability of the strains to use d-tartrate as the sole carbon source in minimal agar within 7 days. To the basal medium described by Tanaka et al. (34) containing inorganic components and casein hydrolysate was added a filtered d-tartrate [l-(+)-tartaric acid; Merck, Darmstadt, Germany] solution to a final concentration of 0.2% (wt/vol). The inoculum was prepared by suspending several colonies in water to a density of about 108 bacteria per ml. A 100-μl aliquot of this suspension was inoculated into the cotton wool-stoppered test tubes containing 2.5 ml of the medium. The cultures were incubated at 37°C for 7 days aerobically without shaking. Two control media were inoculated simultaneously: a medium containing 0.2% glucose instead of d-tartrate solution and a medium without a carbon source.
Antimicrobial susceptibility testing.
All isolates were screened for resistance to 32 antimicrobials by the disk diffusion method on Mueller-Hinton agar (Bio-Rad) according to the guidelines of the Antibiogramm Committee of the French Society for Microbiology (32). Disks with the following antibiotics (Bio-Rad) were tested: amoxicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cephalothin, cefamandole, cefoperazone, cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, moxalactam, imipenem, streptomycin, spectinomycin, kanamycin, tobramycin, netilmicin, gentamicin, amikacin, isepamicin, nalidixic acid, pefloxacin, ciprofloxacin, sulfonamides, trimethoprim, chloramphenicol, and tetracycline. The extended-spectrum β-lactamase (ESBL) phenotype was detected by using the double-disk synergy diffusion test (15). Escherichia coli ATCC 25922 was used as a control.
PCR amplification of antimicrobial resistance genes and class I integrons, and sequence analysis.
Total DNA was extracted using the InstaGene matrix kit (Bio-Rad) in accordance with the manufacturer's recommendations. PCR amplifications of blaTEM, blaPSE-1, blaCTX-M, floR, aadA2, qacEΔ1, and sul1 were performed using primers listed in Table 2. All amplifications were performed on 50-μl samples as described previously (37, 38). The cycling conditions included 10 min of denaturation at 94°C (1 cycle); 30 s of denaturation at 94°C, 30 s of annealing at 50°C (53°C for qacEΔ1, 55°C for aadA2, 62°C for floR, and 63°C for sul1), and 1 min of polymerization at 72°C (35 cycles), followed by 10 min of extension at 72°C.
TABLE 2.
Oligonucleotide primers used in this studya
| Target | Primer name | Oligonucleotide sequence (5′→3′) | Reference | PCR product size (bp) |
|---|---|---|---|---|
| Antibiotic resistance genes | ||||
| aadA2 | aadA2-F | TGTTGGTTACTGTGGCCGTA | 1 | 380 |
| aadA2-R | GCTGCGAGTTCCATAGCTTC | |||
| blaTEM | TEM-F | ATAAAATTCTTGAAGACGAAA | 19 | 1,080 |
| TEM-R | GACAGTTACCAATGCTTAATC | |||
| blaPSE-1 | PSE-1-F | CGCTTCCCGTTAACAAGTAC | 30 | 795 |
| PSE-1-R | CTGGTTCATTTCAGATAGCG | |||
| CTX-M consensus | CTX-M-F | CRATGTGCAGYACCAGTAA | 38 | 541 |
| CTX-M-R | CGCRATATCRTTGGTGGTG | |||
| floR | flo-F | ACCCGCCCTCTGGATCAAGTCAAG | 18 | 856 |
| flo-R | CAAATCACGGGCCACGCTGTATC | |||
| sul1 | Sul-1-F | CTTCGATGAGAGCCGGCGGC | 30 | 417 |
| Sul-1-R | GCAAGGCGGAAACCCGCGCC | |||
| qacEΔ1 | QaceD1-F | ATCGCAATAGTTGGCGAAGT | 30 | 206 |
| QaceD1-R | CAAGCTTTTGCCCATGAAGC | |||
| Class I integron | 5′-CS | GGCATCCAAGCAGCAAGC | 17 | Various |
| 3′-CS | AAGCAGACTTGACCTGAT | |||
| Molecular typing method target IS200 | IS200-F | CCTAACAGGCGCATACGATC | 22 | 557 |
| IS200-R | ACATCTTGCGGTCTGGCAAC | |||
| Salmonella genomic island I | ||||
| Left junction | ||||
| thdF | U7-L12 | ACACCTTGAGCAGGGCAAG | 2 | 500 |
| int | LJ-R1 | AGTTCTAAAGGTTCGTAGTCG | ||
| Right junction | ||||
| S044 | 104-RJ | TGACGAGCTGAAGCGAATTG | 2 | |
| int2 | C9-L2 | AGCAAGTGTGCGTAATTTGG | 515 | |
| yidY | 104-D | ACCAGGGCAAAACTACACAG | 500 | |
| Set A | ||||
| int | IntSG1 | AGGTATCAGTAAACAAGCGT | This study | 1,594 |
| xis | XisSGI | CTGTAGACGTGAATGAAAC | ||
| Set B | ||||
| IS6100 | DBT1 | TGCCACGCTCAATACCGAC | 2 | 631 |
| S044 | MDRB | GAATCCGACAGCCAACGTTCC | ||
| Set C | ||||
| tet(G) | tetG-L | CAGCTTTCGGATTCTTACGG | 25 | 844 |
| tet(G) | tetG-R | GATTGGTGAGGCTCGTTAGC | ||
| Set D | ||||
| IS6100 | IS6100-L | ATGCTTGGCGGAGATTGGAC | This study | 791 |
| IS6100 | IS6100-R | TCAGGCGGCTGCTGCGAAAT | ||
| Set E (probe p1-9-like) | ||||
| S023 | HEL2 | CGAATAATCCGTATCCAGAGC | This study | 1,180 |
| S024 | EXOR2 | TTACTGAAACCCGGCAATCAAG | ||
| Set F (probe QS) | ||||
| qacEΔ1 | QS-1 | ATGAAAGGCTGGCTTTTTCTTG | 2 | 721 |
| sul1 | QS-2 | TGAGTGCATAACCACCAGCC |
Abbreviations: Y, C or T; R, A or G.
For amplification of class I integrons, primers 5′ CS and 3′ CS were used (Table 2), as described previously (37). The purified PCR fragments were sequenced on both strands by Genome Express (Meylan, France) using an ABI 100 DNA sequencer (Applied Biosystems, Foster City, CA). The nucleotide sequence was analyzed with the Lasergene software (Dnastar, Madison, WI). The BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov) was used for databases searches.
Detection of Salmonella genomic island 1 by PCR mapping and hybridization.
Total DNA used for PCR amplifications was extracted as described above. PCR amplifications were performed with several sets of primers listed in Table 2 to assess the organization of the SGI1. PCR mapping explored the presence of left and right junctions of SGI1 with the chromosome, of the int-xis region (located at the 5′ end of SGI1), of the S023-S024 region (corresponding to a central region of SGI1, upstream of the antibiotic genes cluster), of the qacEΔ1-sul1 region (located 4 kb upstream of the 3′ end of the SGI1 antibiotic genes cluster), of the IS6100-S044 region (located at the 3′ end of the SGI1 antibiotic genes cluster), and of tet(G) and IS6100 (both located in the SGI1 antibiotic genes cluster). PCR amplifications were done as described above with 30 s of annealing at 50°C for set A (int-xis region) and set F (qacEΔ1-sul1 region), 55°C for the left and right junctions of SGI1, set B (IS6100-S044 region), set C [tet(G) gene], set D (IS6100), and set E (S023-S024 region).
The presence of SGI1 was confirmed by Southern blotting. Genomic DNA was extracted from brain heart infusion broth cultures as previously described (14) and was cleaved by the restriction enzyme XbaI (Roche Diagnostics). Southern blotting of XbaI-cleaved DNA was performed as previously described (28). Probe p1-9-like (1,180 bp) and probe QS (721 bp) were generated by PCR amplification using set F and set G, respectively. The PCR products were purified with a QIAquick PCR purification kit (QIAGEN, Courtaboeuf, France). Preparation of dUTP-11 fluorescein-labeled PCR-generated probes, prehybridization, and hybridization was done using the ECL random prime labeling and detection systems (version II; Amersham Biosciences) in accordance with the manufacturers. Membranes were exposed to X-ray film (ECL Hyperfilm; Amersham Biosciences).
IS200 profiling.
Southern blotting of PstI-cleaved genomic DNA was performed as described previously (28). The probe was generated by PCR amplification of a 557-bp internal fragment of the IS200 element, using the IS200-F and IS200-R primers (Table 2) as described previously (22). Preparation of the dUTP-11 fluorescein-labeled PCR-generated IS200 probe, prehybridization, and hybridization were done as described above.
Ribotyping.
The membranes used for IS200 profiling were reprobed with the digoxigenin (DIG)-labeled rRNA OligoMix5 probe (28). The DIG-labeled oligonucleotide probe was from MWG-Biotech (Ebersberg, Germany). Hybridization was performed as described previously (28), with minor modifications. Hybridization was performed at 54°C overnight. Immunoenzymatic detection was performed by using the DIG nucleic acid detection kit (Roche Diagnostics) according to the supplier's instructions. Ribopatterns were automatically compared using Taxotron software (Institut Pasteur). A maximum fragment size variation of 5.0% was accepted.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was carried out with a CHEF-DR III system (Bio-Rad) as described previously (37). The running conditions were 6 V/cm at 12°C for 20 h, with pulse times ramped from 2.2 to 63.8 s. BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to compare the PFGE profiles. The bands generated were analyzed by using the Dice coefficient and the unweighted pair group method with arithmetic averages.
Phage typing.
Phage typing of S. enterica serotype Paratyphi B isolates was done according to a standardized methodology (11). Phage suspensions were kindly provided by the Health Protection Agency, Colindale, United Kingdom.
RESULTS
Antimicrobial susceptibilities of S. enterica serotype Paratyphi B isolates.
Among the 261 isolates of S. enterica serotype Paratyphi B addressed to NRC-Salm from 2000 to 2003, 214 (82%) were susceptible to all antimicrobial agents tested. Thirty-one (11.9%) isolates displayed the pentaresistance phenotype ASSpCTeSul, and five (1.9%) displayed the R-type ASSpCTeSulTmp (Table 1). A single isolate of R-type ACroTmp produced an ESBL. Other isolates (n = 10) exhibited various antimicrobial resistance phenotypes: ASSpSulTmpNal (n = 1), SSpSul (n = 1), ASul (n = 2), SSpSulTmpNal (n = 2), SSpTeSul (n = 1), ASulTmp (n = 2), and SSpToGSulTmp (n = 1). All 47 MDR isolates belonged to biotype Java, except the ESBL-producing isolate 01-7995 (data not shown). When studied by PCR using primers specific for antibiotic and antiseptic resistance genes, the 31 isolates of R-type ASSpCTeSul and the five isolates of R-type ASSpCTeSulTmp were positive for blaPSE-1, floR, sul1, aadA2, and qacEΔ1 (Table 1), as described for S. enterica serotype Typhimurium DT104 harboring SGI1 (3). Two isolates (01-9585 and 02-3610) of R-types SSpSul and SSpTeSul were positive for sul1, aadA2, and qacEΔ1, as known for S. enterica serotype Typhimurium DT104 harboring SGI1-C (3). A single isolate (02-5269) of R-type ASul possessed only blaPSE-1, sul1, and qacEΔ1, as known for S. enterica serotype Typhimurium DT104 harboring SGI1-B (3). Four isolates of R-type ASSpCTeSulTmp also possessed blaTEM. This blaTEM gene was found in other isolates (01-0582, 02-1062, 02-5932, and 03-7087) of R-types ASSpSulTmpNal, ASul, and ASulTmp. PCR analysis with the blaCTX-M consensus primers showed the presence of a 540-bp fragment in the ESBL-producing isolate 01-7995. The sequence from both strands of PCR product was 99% identical to the respective segment of blaCTX-M-15 (GenBank accession number AY0044436).
The presence of class I integrons was tested by PCR using specific primers targeting the conserved 5′ and 3′ segments of the integron. The 31 isolates of R-type ASSpCTeSul and the five isolates of R-type ASSpCTeSulTmp carried integrons with amplicons of 1.0 and 1.2 kb (Table 1), as known for S. enterica serotype Typhimurium DT104 (3). In isolate 01-9585 of R-type SSpSul and isolate 02-3610 of R-type SSpTeSul, a class I integron generating a PCR product of 1.0 kb was found. Isolate 02-5269 of R-type ASul contained a 1.2-kb integron. No class I integron was detectable in other tested isolates.
Study of SGI1-like cluster.
To assess the presence of SGI1 and its location in the chromosome, PCR was performed on all MDR isolates by using primers corresponding to the left and right junctions of SGI1 in the chromosome. Results are listed in Table 1. SGI1 was detected in 39 S. enterica serotype Paratyphi B isolates between the thdF and the yidY genes of the chromosome. All the MDR isolates lacked the retron sequence found only downstream of SGI1 in serotype Typhimurium DT104 (3).
To ensure that SGI1 was intact, PCR mapping with six sets of primers (Tables 2 and 3) was carried out on eight selected isolates (02-0616, 02-5269, 02-3609, 02-3610, 02-3869, 02-3870, 02-3873, and 02-9684) of R-types ASSpCTeSul, ASSpCTeSulTmp, ASul, and SSpTeSul. DNA from six isolates gave positive results for all primer sets, which suggests that SGI1 was intact in these isolates. DNA from the two isolates of R-types ASul and SSpTeSul did not give a PCR product using set C primers targeting the tet(G) gene.
TABLE 3.
Results of the PCR mapping of SGI1 from DNA of eight selected MDR/SGI1 S. enterica serotype Paratyphi B isolates and one serotype Typhimurium DT104 strain
| Isolate | Antibiotic resistance pattern | PCR mapping resultb
|
|||||
|---|---|---|---|---|---|---|---|
| Set A (IntSG1/ XisSGI) | Set B (DBT1/ MDRB) | Set C (tetG-L/ tetG-R) | Set D (IS6100-L/ IS6100-R) | Set E (HEL2/ EXOR2) | Set F (QS-1/ QS-2) | ||
| 02-5269 | ASul | + | + | − | + | + | + |
| 02-0616 | ASSpCTeSul | + | + | + | + | + | + |
| 02-3610 | SSpTeSul | + | + | − | + | + | + |
| 02-3609 | ASSpCTeSulTmp | + | + | + | + | + | + |
| 02-3869 | ASSpCTeSulTmp | + | + | + | + | + | + |
| 02-3870 | ASSpCTeSulTmp | + | + | + | + | + | + |
| 02-3873 | ASSpCTeSulTmp | + | + | + | + | + | + |
| 02-9684 | ASSpCTeSulTmp | + | + | + | + | + | + |
| Comparison straina | |||||||
| 02-5494 | ASSpCTeSul | + | + | + | + | + | + |
The comparison strain is an S. enterica serotype Typhimurium DT104 strain.
+, product obtained with set of primers indicated; −, no product obtained.
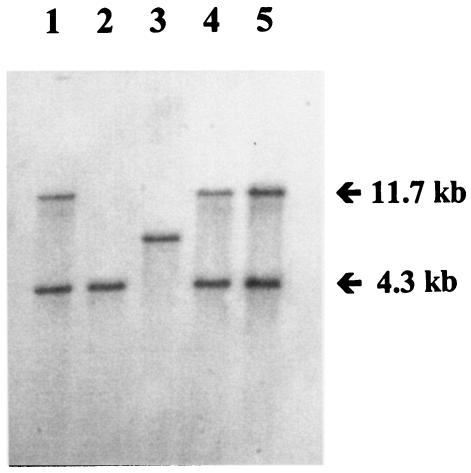
The presence of all of SGI1 was also confirmed by Southern blotting of XbaI-digested genomic DNA with the PCR-generated p1-9-like probe and the QS probe in four MDR/SGI1 serotype Paratyphi B isolates carrying class I integrons of 1.0 kb, or 1.2 kb or 1.0 and 1.2 kb, as well as in a DT104 S. enterica serotype Typhimurium strain carrying SGI1 used for comparison (Fig. 1). The p1-9-like probe revealed two XbaI fragments of the expected 9- and 4-kb sizes in the four MDR/SGI1 serotype Paratyphi B strains, whatever the resistance phenotype, as obtained in the serotype Typhimurium DT104 strain (data not shown). The QS probe revealed two XbaI fragments of the expected 4.3- and 11.7-kb sizes in isolates 02-616 and 02-3609 of R-types ASSpCTeSul and ASSpCTeSulTmp, respectively, as in the serotype Typhimurium DT104 strain (Fig. 1). These results indicated that the trimethoprim resistance in isolate 02-3609 was not due to the presence of the orf513/dfrA10 region, an additional fragment found in SGI1-D of S. enterica serotype Agona (3). Probing of isolate 02-3610 of R-type SSpTeSul with the QS probe showed hybridization products of approximately 6.5 kb, suggesting that only one copy of qacEΔ1-sul1 was present, and it was localized to the same XbaI fragment that contains aadA2, as found in SGI1-C of S. enterica serotypes Typhimurium and Agona (3). The 4.3-kb hybridization fragment obtained for the R-type ASul isolate 02-5269 with the QS probe suggested the presence of only one copy of qacEΔ1-sul1, localized to the same XbaI fragment that contains blaPSE-1, as found in SGI1-B of S. enterica serotype Typhimurium (3).
FIG. 1.
Southern blot hybridization with the QS probe of XbaI-digested genomic DNA of S. enterica serotype Typhimurium DT104 strain 02-5494 (lane 1) and MDR/SGI1 serotype Paratyphi B isolates 02-5269 (lane 2), 02-3610 (lane 3), 02-616 (lane 4), and 02-3609 (lane 5).
Results of PCR and Southern blot experiments indicated that among 47 S. enterica serotype Paratyphi B MDR isolates collected in France from 2000 to 2003, 39 (83%) possessed the SGI1 cluster. Among these 39 isolates, only 3 contained variants of SGI1, SGI1-B (n = 1) and SGI1-C (n = 2).
Molecular and phage typing.
The results of molecular and phage typing are summarized in Table 1. Ribotyping performed with the restriction endonuclease PstI revealed six different patterns among the S. enterica Paratyphi B isolates, designated RP1 to RP6 (Table 1 and data not shown). Profile RP1, the most common, was found in 38 of 39 MDR/SGI1 isolates. RP1 was also observed in three susceptible strains. Profile RP6 was seen in a single isolate (02-4026) harboring SGI1. Profile RP2 was found in three blaTEM-containing isolates, the ESBL-producing isolate and two susceptible isolates. Profile RP3 was found in the poultry strain isolated in Germany used for comparison and in four French isolates exhibiting diverse R-types with a core spectrum of antibiotic resistance determinants for streptomycin, spectinomycin, and trimethoprim.
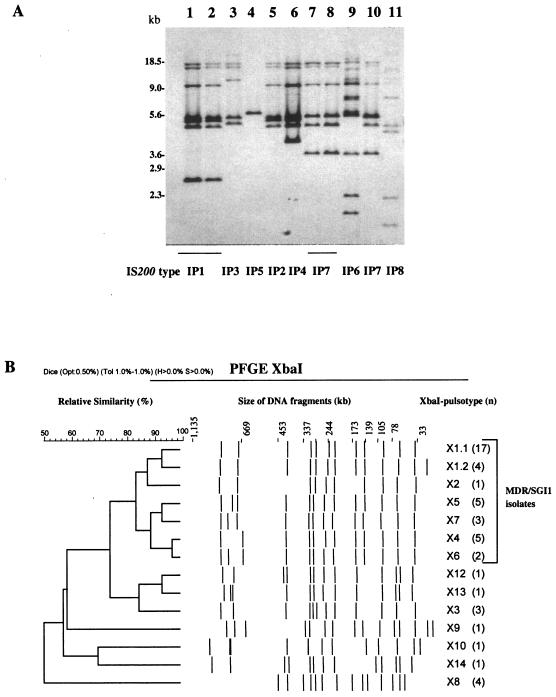
IS200 profiling has been carried out using PstI. Since PstI does not cut within the IS200 element, the number of probe-positive fragments allowed an estimation of IS200 copy number. We observed in S. enterica serotype Paratyphi B isolates seven different IS200 profiles, which were termed IP1 to IP7 (Fig. 2A). The 39 MDR/SGI isolates showed the same profile IP1, which contained seven copies from 2.6 to 18 kb. Profile IP1 was not found in other isolates. Profile IP5, with only one band of approximately 5.6 kb, was linked with the RP3 ribotype. IS200 profiles IP7, IP4, and IP6 have been detected in strains of various antimicrobial resistance phenotypes and which displayed an RP2, RP4, or RP5 ribotype. Profile IP7 was found only in serotype Paratyphi B dT− strains.
FIG. 2.
A. IS200 profiles of representative multidrug-resistant S. enterica serotype Paratyphi B isolates and comparison strains. B. Dendrogram generated by BioNumerics showing the results of cluster analysis of the 14 XbaI-PFGE patterns observed among 49 S. enterica serotype Paratyphi B isolates typed. Similarity analysis was performed using the Dice coefficient, and clustering was by the unweighted pair group method with arithmetic averages. The different PFGE types and corresponding numbers of isolates are indicated.
PFGE using XbaI subtyped 49 Paratyphi B strains into 14 pulsotypes designated X1.1, X1.2, X3 to X10, and X12 to X14 (Fig. 2B). The XbaI patterns were characterized by 10 to 15 fragments ranging in size from <30 kb to >700 kb. Analysis of the dendrogram revealed that all the MDR/SGI1 isolates were grouped in two related clusters, with a similarity percentage of 82% (Fig. 2B). Pulsotype X8, which showed the lowest similarity value (50%), was linked to RP3 and IP5 profiles.
Phage typing performed by using Felix and Callow's international system revealed that some MDR/SGI1 isolates belonged to phage types (PTs) 3b var.2 (n = 2), 1010 (n = 2), and 1 var.3 (n = 1), while most were unclassified (they reacted but did not conform) (Table 1). However, these unclassified isolates displayed lysis reactions associated with the PT 3b group (n = 24), PT 1 group (n = 4), or PT 1010 group (n = 2).
DISCUSSION
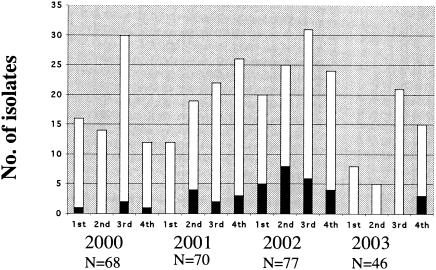
The present study documented the occurrence and the molecular mechanisms of multidrug resistance among the 261 human S. enterica serotype Paratyphi B isolates collected in France between 2000 and 2003, through the representative NRC-Salm network. During this period, this serotype ranked between the 12th and the 15th positions of the most prevalent serotypes in France. Among the 47 MDR isolates, 36 harbored all of SGI1, and three harbored variants of SGI1. These two variants, SG1-C and SG1-B, were characterized for the first time in this study in S. enterica serotype Paratyphi B biotype Java. The presence of SGI1 or variants of SGI1 in several S. enterica serotypes (Typhimurium, Agona, Albany, Newport, Meleagridis, Newport, and Paratyphi B biotype Java) at the same chromosomal location (between thdF and yidY or int2) suggests horizontal transfer of this region through site-specific recombination (3, 6, 7, 8, 9, 21). Transduction by phages or self-transmission of the SGI1 structure have been proposed to explain insertion of SGI1 (7). Generation of variants of SGI1 has been postulated to have occurred by recombination between homologous regions of the MDR region (3). The emergence of SGI1 harboring S. enterica serotype Paratyphi B isolates was documented recently in Canada and in Great Britain with isolates collected from 1998 to 2002 (24, 35). The source of the contamination has not been identified in both studies. In France, the distribution of MDR/SGI1 isolates over time is shown in Fig. 3. MDR/SGI1 isolates were observed mainly between the second quarter of 2001 and the end of 2002. In 2002, an epidemiological investigation was conducted after the identification of a cluster of nine cases in the Hauts-de-Seine area in June and July 2002. Using standardized questionnaires for investigating food-borne diseases, no link between the six cases investigated has been found (E. Espié, personal communication). A search for similar MDR/SGI1 strains isolated from food or animals in France was unsuccessful. Antibiotic-resistance screening results of 42 S. enterica Paratyphi B food isolates, collected through the French Food Agency Salmonella Network between 2001 and 2003, did not reveal any SGI1-associated antimicrobial resistance patterns. The first isolate of S. enterica serotype Paratyphi B harboring SGI1 has been identified in a tropical fish imported from Singapore in 1997 (21). Since then, no other food or environmental SGI1-carrying isolates explaining the emergence of such strains in North America or Europe have been identified. A typing study using four methods (ribotyping, IS200 profiling, PFGE, and phage typing) revealed that IS200 profiling was the method of choice for the screening of MDR/SGI1 isolates. PFGE allowed a better discrimination between MDR/SGI1 isolates, but discrimination between MDR/SGI1 isolates and other MDR isolates was less evident than using IS200 profiling. Analysis of IS200 profiles showed that the unique MDR/SGI1 IS200 pattern in our study, IP1, is indistinguishable from Spj-IP2.1 given by an environmental strain isolated in 1990 in Indonesia (10). Have the MDR SG1 serotype Paratyphi B strains originated from Southeast Asia? A study of antimicrobial resistance and fingerprinting by PFGE was carried out on 86 isolates of serotype Paratyphi B collected in Malaysia in 1982 to 1983, 1992, and 1996 to 2002 (13). The absence of an investigation of resistance genes or SGI1 and the difficulty of comparing PFGE fingerprints did not allow us to detect MDR/SGI1 isolates in the publication. Therefore, further molecular typing and molecular characterization of antibiotic resistance gene studies of strains collected in Asia are necessary to appreciate the prevalence of MDR/SGI1 isolates in that part of the world. Human salmonellosis caused by S. enterica serotype Paratyphi B biotype Java after exposure to exotic turtles or tropical fish has been described in North America and in Europe (12, 29, 31, 39). Inadequate use of antibiotics by breeders and wholesalers in the prophylactic treatment of exotic pets to prevent bacterial diseases may have contributed to the development or the selection of such MDR Salmonella strains. As the search for contact with exotic pets is not included in standardized French questionnaire, it would be very interesting to add this question for further investigation of cases associated with MDR/SGI1 S. enterica serotype Paratyphi B isolates.
FIG. 3.
Total number of S. enterica serotype Paratyphi B isolates (white bars) and the number of multidrug-resistant S. enterica serotype Paratyphi B isolates (black bars) identified at the NRC-Salm between 2000 and 2003, by quarter.
An emerging MDR S. enterica serotype Paratyphi B biotype Java clone, which does not harbor SGI1, has been identified in poultry and poultry products in Germany and The Netherlands, since the second half of the 1990s (22, 36). In The Netherlands the proportion of S. enterica serotype Paratyphi B biotype Java strains isolated from poultry drastically rose from 3% in 1995 to 33% in 2000 and 61% in 2002, while quinolone (flumequine) resistance rose from 0.3% in 1996 to 39% in 2002 (36). Human infections with this clone have been documented in Scotland, and it has been strongly suggested that imported poultry meat from Holland was the source of infection (4). The potential for poultry meat to act as a vehicle for this MDR clone is a cause of concern for public health. In the present study, X8/IP5 fingerprints were indistinguishable from X8/ISP9 fingerprints observed for this predominating clone in poultry in Germany (22). The prevalence of this non-SGI1 MDR S. enterica serotype Paratyphi B biotype Java clone displaying fingerprints RP3/IP5/X8 among human clinical isolates in France is weak. Isolates belonging to this clone had a prevalence of 1.5% of all the isolates collected between 2000 and 2003 in France and represented 4 of the 46 MDR (8.7%) isolates during the same period.
In conclusion, it is important to continue monitoring multidrug resistance in serotype Paratyphi B, which like serotype Typhimurium DT104 and several other serotypes revealed the property of acquisition and dissemination of the SGI1 cluster. The use of molecular methods for the characterization of antimicrobial mechanisms and for the subtyping of the isolates is important for a better understanding of the epidemiology of such MDR serotype Paratyphi B biotype Java strains.
Acknowledgments
We thank all the corresponding laboratories of the French National Reference Center Salmonella network, E. Espié (Institut de Veille Sanitaire), and R. Lailler (Agence Française de Sécurité Sanitaire des Aliments) for their collaboration.
REFERENCES
- 1.Bito, A., and M. Susani. 1994. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother. 38:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. J., H. Mather, L. M. Browning, and J. E. Coia. 2003. Investigation of human infections with Salmonella enterica serovar Java in Scotland and possible association with imported poultry. Euro. Surveill. 8:35-40. [DOI] [PubMed] [Google Scholar]
- 5.Desenclos, J. C., P. Bouvet, E. Benz-Lemoine, F. Grimont, H. Desqueyroux, I. Rebiere, and P. A. D. Grimont. 1996. Large outbreak of Salmonella enterica serotype paratyphi B infection caused by a goats' milk cheese, France, 1993: a case finding and epidemiological study. BMJ 312:91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doublet, B., F. X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside transferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebner, P., K. Garner, and A. Mathew. 2004. Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Meleagridis. J. Antimicrob. Chemother. 53:1004-1009. [DOI] [PubMed] [Google Scholar]
- 10.Ezquerra, E., A. Burnens, C. Jones, and J. Stanley. 1993. Genotypic typing and phylogenetic analysis of Salmonella paratyphi B and S. java with IS200. J. Gen. Microbiol. 139:2409-2414. [DOI] [PubMed] [Google Scholar]
- 11.Felix, A., and B. R. Callow. 1951. Paratyphoid B-Vi phage-typing. Lancet ii:10-14. [DOI] [PubMed] [Google Scholar]
- 12.Gaulin, C., C. Vincent, L. Alain, and J. Ismail. 2002. Outbreak of Salmonella paratyphi B linked to aquariums in the province of Quebec, 2000. Can. Commun. Dis. Rep. 28:89-93. [PubMed] [Google Scholar]
- 13.Goh, Y. L., R. Yasin, S. D. Puthucheary, Y. T. Koh, V. K. Lim, Z. Taib, and K. L. Thong. 2003. DNA fingerprinting of human isolates of Salmonella enterica serotype Paratyphi B in Malaysia. J. Appl. Microbiol. 95:1134-1142. [DOI] [PubMed] [Google Scholar]
- 14.Grimont, F., and P. A. D. Grimont. 1995. Determination of rDNA gene restriction patterns. Methods Mol. Biol. 46:181-200. [DOI] [PubMed] [Google Scholar]
- 15.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 16.Kauffmann, F. 1955. Zur Differentialdiagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. tZ. Hyg. 141:546-550. [PubMed] [Google Scholar]
- 17.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstedt, B. A., E. Heir, I. Nygard, and G. Kapperud. 2003. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J. Med. Microbiol. 52:141-149. [DOI] [PubMed] [Google Scholar]
- 19.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-559. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 20.Malorny, B., C. Bunge, and R. Helmuth. 2003. Discrimination of d-tartrate-fermenting and -nonfermenting Salmonella enterica subsp. enterica isolates by genotypic and phenotypic methods. J. Clin. Microbiol. 41:4292-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miko, A., B. Guerra, A. Schroeter, C. Dorn, and R. Helmuth. 2002. Molecular characterization of multiresistant d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miko, A., K. Pries, A. Schroeter, and R. Helmuth. 2003. Multiple-drug resistance in d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates from poultry is mediated by class 2 integrons inserted into the bacterial chromosome. Antimicrob. Agents Chemother. 47:3640-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey, M. R., D. Boyd, A. Cloeckaert, R. Ahmed, and L. K. Ng. 2004. Emergence of multidrug-resistant Salmonella Paratyphi B dT+, Canada. Emerg. Infect. Dis. 10:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, L. K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 4:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. World Health Organization Collaborating Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 27.Prager, R., W. Rabsch, W. Streckel, W. Voigt, E. Tietze, and H. Tschape. 2003. Molecular properties of Salmonella enterica serotype Paratyphi B distinguish between its systemic and its enteric pathovars. J. Clin. Microbiol. 41:4270-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regnault, B., F. Grimont, and P. A. D. Grimont. 1997. Universal ribotyping method using a chemically labelled oligonucleotide probe mixture. Res. Microbiol. 148:649-659. (Erratum, 149:73, 1998.) [DOI] [PubMed] [Google Scholar]
- 29.Riley, A., M. Hanson, and C. Ramsay. 1992. Tropical fish as a source of Salmonella java infection. Commun. Dis. Environ. Health Scotland 26:4-5. [Google Scholar]
- 30.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 31.Senanayake, S. N., M. J. Ferson, S. J. Botham, and R. T. Belinfante. 2004. A child with Salmonella enterica serotype Paratyphi B infection acquired from a fish tank. Med. J. Aust. 180:250. [DOI] [PubMed] [Google Scholar]
- 32.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué 2000-2001. Pathol. Biol. 48:832-871. [PubMed] [Google Scholar]
- 33.Stratton, J., L. Stefaniw, L., K. Grimsrud, D. H. Werker, A. Ellis, E. Ashton, L. Chui, E. Blewett, R. Ahmed, C. Clark, F. Rodgers, L. Trottier, and B. Jensen. 2001. Outbreak of Salmonella paratyphi B var java due to contaminated alfalfa sprouts in Alberta, British Columbia and Saskatchewan. Can. Commun. Dis. Rep. 27:133-137. [PubMed] [Google Scholar]
- 34.Tanaka, S., S. A. Lerner, and E. C. C. Lin. 1967. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked deshydrogenase for the utilization of mannitol. J. Bacteriol. 93:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Threlfall, J., B. Levent, K. L. Hopkins, E. de Pinna, L. R. Ward, and D. J. Brown. 2005. Multidrug-resistant Salmonella Java. Emerg. Infect. Dis. 11:170-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Pelt, W., H. van der Zee, W. J. Wannet, A.W. van de Giessen, D. J. Mevius, N. M. Bolder, R. E. Komijn, and Y. T. van Duynhoven. 2003. Explosive increase of Salmonella Java in poultry in the Netherlands: consequences for public health. Euro. Surveill. 8:31-35. [DOI] [PubMed] [Google Scholar]
- 37.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weill, F. X., R. Lailler, K. Praud, A. Kérouanton, L. Fabre, A. Brisabois, P. A. D Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodward, D. L., R. Khakhria, and W. M. Johnson. 1997. Human salmonellosis associated with exotic pets. J. Clin. Microbiol. 35:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]