Abstract
Background
In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) developed standardized variant curation guidelines for Mendelian disorders. Although these guidelines have been widely adopted, they are not gene- or disease-specific. To mitigate classification discrepancies, the Clinical Genome Resource FBN1 variant curation expert panel (VCEP) was established in 2018 to develop adaptations to the ACMG/AMP criteria for FBN1 in association with Marfan syndrome.
Methods
The specific recommendations were developed through literature review, surveys, online expert panel discussions, and pilot testing of a set of 60 different variants. Consensus among experts was considered reached if at least 75% of the members agreed with a given rule specification. The final set of rules received approval from the ClinGen Sequence Variant Interpretation Working Group.
Results
The developed specifications introduce modifications to 14 of the 28 ACMG/AMP evidence criteria and deem 6 criteria non-applicable. Some of these specifications include refining the minor allele frequency thresholds, creating a FBN1-specific flowchart for PVS1, defining functional domains of the protein, developing a point-based system of counting probands and instances of de novo occurrences, recommending a points-based method of accounting for family segregation data, and clarifying the applicable functional assays that should be considered. To date, this VCEP has curated 120 variants which have been deposited to ClinVar with the 3-star review status.
Conclusions
Establishing specific adaptations for FBN1 has provided a framework to foster greater classification concordance among clinical laboratories, ultimately improving clinical care for patients with Marfan syndrome.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01423-3.
Keywords: Marfan syndrome, FBN1, ACMG-AMP guidelines, Variant interpretation, Variant curation
Background
Pathogenic variation in the fibrillin-1 gene (FBN1, MIM *134,797) is associated with Marfan syndrome (MIM #154,700), an autosomal dominant multisystemic connective tissue disorder characterized by a broad and variable phenotypic spectrum involving the cardiovascular, ocular, and skeletal systems. The cardinal characteristics are thoracic aortic aneurysm and dissection (TAAD) and ectopia lentis. Other cardiovascular features, present in a variable number of patients, include mitral valve prolapse, cardiomyopathy, and arrhythmias [1], while additional ocular features such as myopia, astigmatism, and flat cornea may be present [2]. Musculoskeletal manifestations include arachnodactyly, protrusio acetabuli, pectus anomalies, scoliosis, and others, and are usually essential to identify patients with Marfan syndrome.
Fibrillin-1 is a critical structural protein of the extracellular matrix, present in both elastic and non-elastic tissues. Besides its structural role, fibrillin-1 plays a crucial function in mechanosensing and mechanotransduction of environmental changes [3], interacting with various microfibril-associated proteins, growth factors, and cell membrane receptors. Together with its structural significance, fibrillin-1 regulates the bioavailability of TGF-β and therefore direct involvement in processes including inflammation, fibrosis, and matrix metalloproteinase activation results in the characteristic phenotype including the aortic wall weakening [4].
Marfan syndrome is among the more common of the “rare diseases,” with an estimated prevalence between 1:5000 and 1:10,000 [5]. Over 3000 different (likely) pathogenic variants spanning all 65 exons of FBN1 have been reported as causative for Marfan syndrome, a considerable proportion of which have only been reported in a single individual or family [6, 7]. These comprise variants resulting in haploinsufficiency (i.e. nonsense, frameshift, splicing, and gross deletions) and those thought to confer a dominant negative effect, namely by altering cysteine residues or other conserved amino acids in the encoded epidermal growth factor (EGF)-like, calcium-binding EGF-like, TGF-β-binding protein-like, and hybrid domains [8].
The diagnosis of Marfan syndrome can be made clinically without molecular testing via the revised Ghent criteria [9]. These criteria take into consideration the presence of aortic root dilation or history of a dissection, the presence of ectopia lentis, and the systemic score, the latter of which is a combination of mostly skeletal and some non-skeletal features. Genetic testing can dramatically aid in establishing a diagnosis prior to development of the full clinical phenotype. At least two studies have highlighted the underdiagnosis of Marfan syndrome when a genotype-first approach was used in population databases [10, 11]. This is especially important due to the frequent morbidity or mortality from undiagnosed TAAD [12, 13]. Molecular diagnoses also assist clinicians in their medical management decisions by informing the need for earlier clinical monitoring and medical interventions such as prophylactic aortic surgery [14, 15]. Obtaining a molecular diagnosis, especially for patients who do not meet Marfan syndrome clinical diagnostic criteria, allows differentiation from other connective tissue disorders with overlapping phenotypes such as Loeys-Dietz syndrome, Shprintzen-Goldberg syndrome, Meester-Loeys syndrome, and congenital contractural arachnodactyly, as well as the many other genetic etiologies of heritable thoracic aortic disease. Further, and critically, identification of a causative pathogenic variant permits vital cascade screening of potentially affected biological family members and enables more comprehensive counselling about reproductive decision making and family planning options, including prenatal and preimplantation diagnoses [16].
For several years, laboratories have been largely utilizing the same generic framework for interpretation and classification of sequence variants published jointly by the American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathology (AMP) [17, 18]. However, significant discrepancies in variant classifications persist due to differences in how laboratories assess evidence and decide to modify and apply the criteria described in the ACMG-AMP guidelines [19, 20]. Mitigating the possibility for confusion, misdiagnosis, or mismanagement caused by inter-laboratory variant classification discrepancies and reducing the frequency with which novel and recurrent variants are classified as of uncertain significance (VUS) are eminently desirable. With these goals, numerous Clinical Genome Resource (ClinGen) variant curation expert panels (VCEPs) have been created to develop gene- or disease-specific modifications to the ACMG-AMP criteria to foster more tailored, accurate, and standardized variant interpretations [21–24].
The high prevalence of Marfan syndrome and the clinical significance of establishing its diagnosis underline the importance of addressing these limitations in interpretation. Further, it emphasizes the potential impact that improvements to the utility that FBN1 analyses can bring and the need to develop guidance on FBN1 variant interpretation. To bridge the existing gap in interpretation and classification practices, Muiño-Mosquera et al. developed their own FBN1-specific adjustments to the ACMG-AMP criteria within a single institution [8]. Recognizing the importance for broader expertise and consensus, an international group of experts specializing in FBN1 and Marfan syndrome was then engaged to further refine these specifications. As a result, the ClinGen’s FBN1 VCEP was established aiming to enhance the management of this extensive patient population with two primary objectives. Firstly, the panel aimed to develop consensus recommendations for best practices of FBN1 variant interpretation, ensuring wider dissemination and implementation among relevant stakeholders. Secondly, leveraging the collective expertise in FBN1 and Marfan syndrome, the panel sought to provide expert curations of previously identified FBN1 variants.
Methods
ClinGen FBN1 variant curation expert panel
The FBN1 VCEP membership represents a multidisciplinary group of medical geneticists and (paediatric) cardiologists, research scientists, molecular genetic diagnostic scientists, and genetic counsellors with a wealth of experience and expertise surrounding FBN1, Marfan syndrome, and related connective tissue disorders. Membership currently comprises nine institutions, three of which are designated as the “core” team (Ghent University Hospital, Hôpital Bichat, Mayo Clinic), and spans five countries (Belgium, Canada, France, Japan, USA).
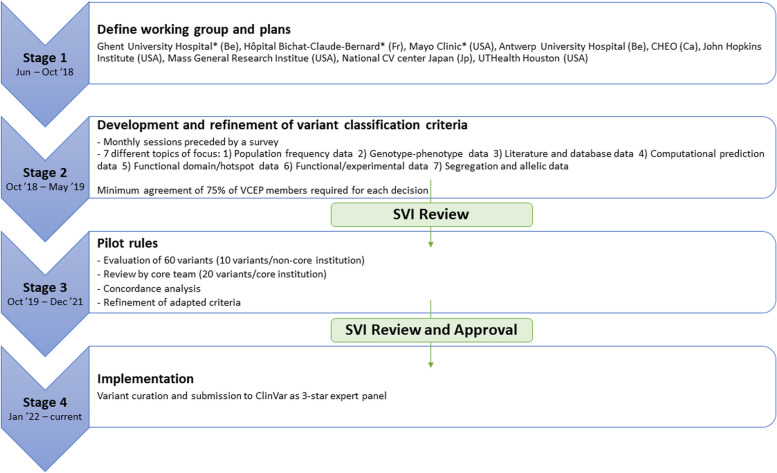
Beginning with the first meeting in November 2018, the VCEP met once every month until the finalization of the rules. Before every meeting, a survey was conducted to gather feedback about the initial proposal for rule specifications from Muiño-Mosquera et al. [8] The survey consisted of multiple-choice questions, with each question providing an option for commentary to include any rationale not covered by the given choices. The survey results informed subsequent discussion and allowed for additional clarification during group discussions. During each meeting, an online voting system was used to reach a consensus. Points of disagreement were discussed until at least 75% agreement was achieved. In most cases, this required reappraisal of the literature or internal data. Once a set of rule specifications were preliminarily agreed upon by the panel, they were piloted by the VCEP to identify possible pitfalls and inconsistencies and thus prompt further refinements. The iterative development of the rule specifications involved guidance from the ClinGen Sequence Variant Interpretation Working Group (SVI) as well as exploitation of other previous VCEP modifications, including reference of the RASopathy VCEP for the PS4 criterion, the Hearing Loss VCEP for the PP1 criterion, and the MYH7 VCEP for the population frequency cut-off values [21–23]. The ultimate version of the rule specifications was voted and agreed on by a majority of the VCEP members prior to submitting and receiving final approval from the ClinGen SVI. A minimum of 75% agreement among members was deemed necessary to approve a specification, also at this stage. The process of creation and rule development of the FBN1 VCEP is represented in Fig. 1 and followed the established ClinGen’s framework [25].
Fig. 1.
Overview of the process used for adapting the ACMG/AMP criteria to FBN1: the procedure was staged in four phases as defined in the figure. Stages 2 and 3 were followed by evaluation and feedback from the ClinGen Sequence Variant Interpretation Working Group (SVI). Stage 4 consists of an ongoing process of variant curation and submission to ClinVar for public accessibility. *Core members. Abbreviations: Be: Belgium, Fr: France, USA: United States of America, Ca: Canada, Jp: Japan
The ontology used for curation is Marfan syndrome (MONDO:0007947) with autosomal dominant inheritance (HP:0000006). Other diseases associated with FBN1, like stiff skin syndrome (MONDO:0008492) or geleophysic dysplasia (MONDO:0013612), were not considered for the specifications. The ClinGen’s framework specifically abrogates for creating specifications and performing variant curations for a single gene-disease relationship. There is only one biologically relevant FBN1 transcript; consequently, all variants are curated according to their annotation on the MANE Select transcript NM_000138.5. All curations are published in the ClinGen Variant Curation Interface under the FBN1 affiliation and in the ClinVar repository as reviewed by an expert panel (3 stars).
Piloting the rule specifications
Each of the nine constitutive VCEP institutions were asked to contribute 10 FBN1 variants for possible inclusion in the pilot study that had been identified in patients evaluated at that clinic or tested at that laboratory with a clinical diagnosis or suspicion based on clinical features and/or family history. Notably, as these variants may have been identified at any point in time, testing methodologies at these reputable clinical and research laboratories include various molecular techniques that were standard of practice at that point in time (e.g. denaturing high-performance liquid chromatography [dHPLC], Sanger sequencing and/or next-generation sequencing, with orthogonal variant confirmation when appropriate). Each institution was asked to submit five missense, two frameshift, one nonsense, one splicing, and one in-frame insertion/deletion (indel) variant, including at least two (likely) benign, two VUSs, and two (likely) pathogenic. From that subset of 92 variants, a total of 60 variants were deliberately chosen by the “core” team on the basis of creating a set with challenging interpretations that represent as wide a range of characteristics as possible. This included the variant type (missense, frameshift, nonsense, splicing, in-frame deletion or insertion, synonymous), the amount of available evidence and variety of evidence types (e.g. probands with various degrees of phenotype information available, familial vs. de novo variants, experimental data, variants affecting different putative functional domains/conserved positions), having both recurrent and unique variants, and having each of the five classifications of clinical significance represented in order to comprehensively test the rule modifications.
The six non-core institutions were each assigned 10 variants to interpret and classify using the FBN1-specific rules, with institutions’ internal case-level data supplementing the publicly available data for interpretation. The variants’ original classifications and their reclassifications derived using the VCEP rule specifications were compared. All 60 variants were then reassessed by the three core institutions, with each responsible for evaluating 20 variants. Classification discordance between core and non-core institutions was then calculated, and the reasons for discordance were noted for additional discussion and refinement with the full VCEP and SVI. Variants with discordant classifications were re-interpreted following these discussions and any amendments to the criteria specifications that were made. If internal data at one VCEP institution was key to a variant’s discordant classifications, that data was shared so that the interpretation could be repeated with equivalent information available.
Results
Disease-specific adaptations of ACMG-AMP Classification Criteria
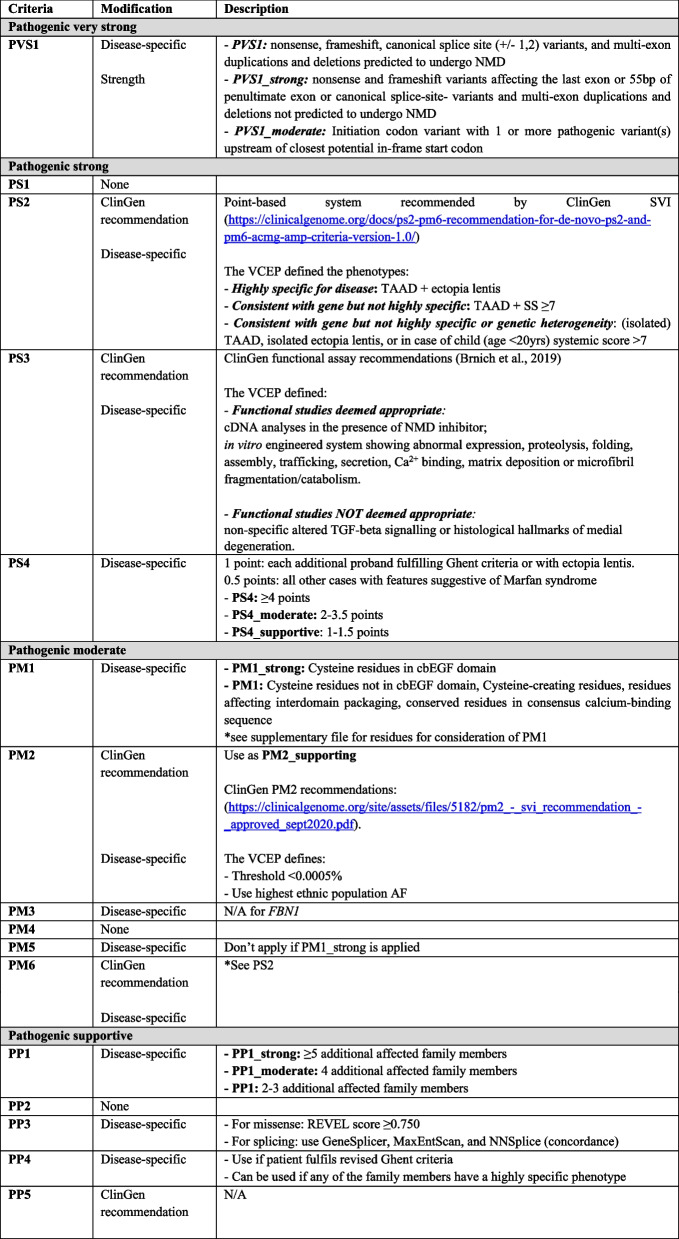
Of the 28 individual ACMG-AMP criteria, FBN1-specific modifications to utilization and/or strength level were introduced for 14 criteria. Six criteria (PM3, PP5, BS2, BP1, BP3, BP6) were deemed to not be applicable for variants in FBN1. Additionally, one change to the ACMG-AMP combining rules was instituted to allow sufficiently rare loss of function variants that satisfy both PVS1 and PM2_Supporting criteria to reach a likely pathogenic classification; this practice accords with recommendations from the ClinGen SVI [26] and has been previously implemented by other VCEPs(23,24). A summary of the specifications can be found in Table 1.
Table 1.
Overview of the adapted criteria from the ACMG/AMP guidelines to FBN1
Null variant in a gene where loss-of-function is a known mechanism of disease (PVS1)
Haploinsufficiency is well known as one of the mechanisms of pathogenesis for FBN1 and Marfan syndrome [27, 28] and the gnomAD probability of being loss-of-function intolerant (pLI) and loss-of-function observed/expected upper bound fraction (LOEUF) scores are 1 and 0.105, respectively; therefore, PVS1 is applicable for all putative loss-of-function variants (i.e. nonsense, frameshift, consensus splice site, large deletions). The VCEP made minor FBN1-specific modifications to Abou Tayoun et al.’s PVS1 decision tree developed to guide application of PVS1 at varying strengths(29) as shown in Fig. 2.
Fig. 2.
Flowchart for the adapted PVS1 criterion for null variants: the FBN1 VCEP made minor modifications to the original PVS1 decision tree developed by ClinGen [29]. The only biological relevant transcript for FBN1 is NM_000138.5. Additionally, two aspects need to be taken into consideration: (1) NMD (nonsense-mediated mRNA decay) is predicted to occur when a stop codon is integrated in the FBN1 sequence, except for stop codons in the last exon or the last 50–55 nucleotides of the penultimate exon. (2) A critical region is defined using the same criteria as for the PM1 and PM1_Strong criteria
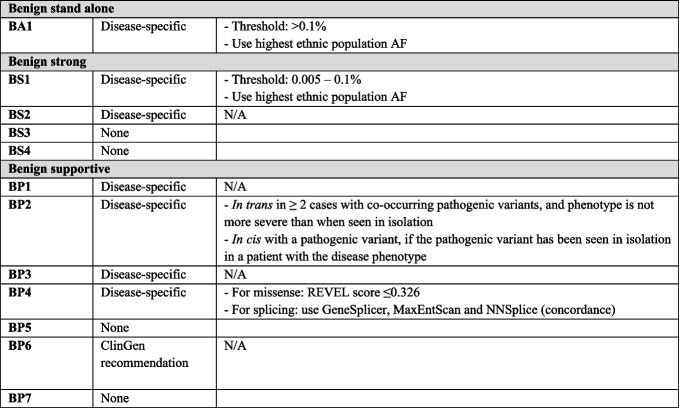
Assessment of variant minor allele frequency (BA1, BS1, PM2)
The optimal threshold for variant minor allele frequency was calculated according to the recommendations published by Whiffin et al. [30], including the maximum estimated prevalence of Marfan syndrome of 1:5000 individuals (1:10,000 chromosomes) [31], a presumed possible penetrance of TAAD of 80% [32], the contribution of FBN1 to Marfan syndrome of 90% [9], and an extremely conservative estimate that no pathogenic variant is responsible for more than 40% of cases of Marfan syndrome. PM2 (variant is absent or rare in the general population) was reduced in strength to PM2_Supporting, consistent with a long-standing recommendation from the ClinGen SVI [33]. The threshold for use of PM2_Supporting was established as a minor allele frequency less than 0.0005% (0.000005). For BA1 and BS1, the optimal frequency thresholds were determined to be 0.1% (0.01) and 0.005% (0.00005), respectively. No known FBN1 pathogenic variants have frequencies exceeding 0.005% in gnomAD; thus, no pathogenic variants would be inappropriately assigned BS1 in support of benignity. The VCEP recommends that for assessment of minor allele frequency data in gnomAD the highest ancestral population frequency should be utilized, with the stipulations that the bottlenecked (i.e. European [Finnish], Ashkenazi Jewish) and “Other” populations should not be utilized, and that any ancestral population being considered should have at least 2000 alleles studied at that position.
Increased variant prevalence in cases versus controls (PS4)
Due to the rarity of FBN1 pathogenic variants and the inherent inability to perform case–control studies for variants, a points-based system of counting probands with the variant of interest was developed, consistent with the practice employed by many other ClinGen VCEPs associated with conditions with autosomal dominant inheritance [34–37]. Probands reported in internal or public databases or published in the primary literature documented to have ectopia lentis and/or a Marfan syndrome diagnosis based on the revised Ghent criteria [9] are each awarded 1 point. Probands who do not meet the revised Ghent criteria and do not have ectopia lentis (e.g. have isolated TAAD or a systemic score greater than or equal to 7 without TAAD) or whose clinical phenotypes are incompletely described are awarded 0.5 points. The sum of the proband points corresponds to a given PS4 strength: 1–1.5 points is sufficient for PS4_Supporting, 2–3.5 points is sufficient for PS4_Moderate, and 4 or more points is sufficient for PS4. To avoid inappropriate use of the PS4 criterion for variants commonly seen in the general population, PS4 should not be applied at any strength for variants that are frequent enough in gnomAD to apply the BA1 or BS1 or criteria.
Mutational hot spot or well-studied functional domain without benign variation (PM1)
Cysteine residues form disulfide bonds throughout the fibrillin-1 protein and are therefore well-established as critical to the stability and function of the protein [38, 39]. Disulfide bonds in the calcium-binding epidermal growth factor (cbEGF)-like domains are especially crucial, and variants that alter one of these cysteine residues are among the most prevalent of disease-causing variation in FBN1 [39]. Therefore, for cysteine-removing variants in any of the 43 cbEGF-like domains, PM1 was increased in strength to PM1_Strong; this encompasses 258 cysteine residues encoded throughout the gene. For cysteine substitutions in the other domains (i.e. EGF-like, TGF-β-binding protein-like, and hybrid) of the gene and for cysteine-creating variants in cbEGF domains, PM1 is applicable at its original moderate strength.
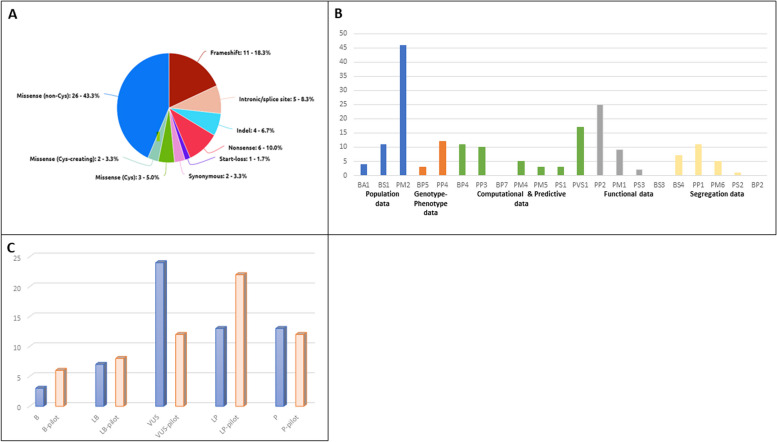
There are numerous other functionally and structurally important non-cysteine residues appropriate for application of PM1 based on their role in either interdomain packaging (e.g. glycine residues in cbEGF-like domains that are positioned between the second and third cysteines or between the third and fourth cysteines, the latter of which requires the presence of an upstream cbEGF-like domain to be applicable), calcium binding (e.g. variants altering the conserved residues in the consensus calcium-binding sequence [D]-X-[D/N]-[E/H]-Xm-[D/N]-Xn-[Y/F]), or sites of possible β-hydroxylation (the second [D/N] of the consensus calcium-binding sequence). The VCEP notes that an asparagine-to-serine (N > S) substitution at the second of the aspartic acid or asparagine (D/N) positions might be tolerated based on the frequency of this type of missense variant in gnomAD [40], and PM1 should therefore not be applied in these instances. Including the aforementioned variant types and the cysteine-involved variants outside of the cbEGF-like domains, PM1 can be applied for variants at 375 different amino acid positions. In total, the PM1 criterion can be used at either moderate or strong level for 22.0% (633/2871) of amino acid positions across FBN1 (Supplementary Table 1). Indeed, the use of PM1 for the above types of non-cysteine variants was a critical factor to the reduction of VUS in favour of likely pathogenic/pathogenic classifications in the pilot study (Fig. 4).
Fig. 4.
Results of the FBN1 VCEP pilot study. A Distribution of variant types included in the study. B Criteria evaluated during the pilot study. Each colour in the X-axis represents a different category of data as defined by the text underneath. The Y-axis shows the number of times each criterion was evaluated. C Comparison of the classification of the pilot variants according to the referring laboratory (blue) and the classification according to the FBN1 VCEP (orange)
De novo events (PS2, PM6)
The ClinGen SVI has universal recommendations for a points-based application of the PS2 and PM6 criteria that involves consideration of the phenotype of either the patient in question or previously reported probands with de novo inheritance [41]. Application requires an evaluation of the extent to which an individual’s phenotype is specific for a certain gene and how much genetic heterogeneity exists for that phenotype. The FBN1 VCEP recommends utilizing the same points-based framework for utilization of PS2 and PM6 and developed tiers of phenotype specificity and genetic heterogeneity to utilize this SVI-derived system for instances of de novo FBN1 variation. ClinGen’s “Phenotype highly specific for gene” category was defined for FBN1 as the presence of TAAD and ectopia lentis. The “Phenotype consistent with gene but not highly specific” category was defined as the presence of TAAD and a systemic score greater than or equal to seven. Finally, probands with isolated TAAD, isolated ectopia lentis, or in an individual younger than 20 years of age for whom TAAD may still develop later in life, a systemic score greater than or equal to seven were selected for the SVI’s “Phenotype consistent with gene but not highly specific and high genetic heterogeneity” category.
Multiple segregations of a variant with phenotype in affected family members (PP1)
Jarvik & Browning previously published Bayesian-derived guidelines for consideration of a variant’s co-segregation with disease in a family [42]. The VCEP initially incorporated this guidance but ultimately determined that the less stringent framework utilized by the Hearing Loss VCEP for autosomal dominant hearing loss [23] was more appropriate for Marfan syndrome. As such, PP1_Supporting is met by the presence of two to three segregations of a given variant with clinical features of Marfan syndrome, PP1_moderate is met with four segregations, and five or more segregations fulfills the PP1_Strong criterion.
Functional evidence supportive of a damaging effect or no effect (PS3, BS3)
Evaluation of functional data should follow the ClinGen SVI’s framework for application of PS3 and BS3 [43]. Consistent with this recommendation, the VCEP specifies the types of experimental assays that are valid for assessment of FBN1 variants as shown in Fig. 3. The assays deemed appropriate include complementary DNA (cDNA) analyses performed in the presence of a nonsense-mediated decay (NMD) inhibitor showing an altered FBN1 RNA sequence, and in vitro engineered systems showing altered FBN1 protein or RNA expression, proteolysis, folding, assembly, trafficking, secretion, calcium (Ca2+)-binding, matrix deposition, and microfibril fragmentation or catabolism. Functional studies deemed inappropriate for application of PS3 or BS3 include assays that identify non-specifically altered TGF-β signalling or histological hallmarks of medial degeneration, as these are also seen with variation in several other genes associated with hereditary TAAD and are not specific to FBN1 and Marfan syndrome.
Fig. 3.
Flowchart for the adapted PS3/BS3 criteria for functional evidence of damaging effect: the FBN1 VCEP made minor modifications to the original decision tree developed by ClinGen [43]. The most relevant specification is step 2 for which the VCEP defines the assays deemed appropriate for consideration
Computational evidence supporting a deleterious effect or no effect on the gene or gene product (PP3, BP4)
The repeated demonstration of the meta-predictor REVEL’s [44] high performance, its positive and negative predictive value for FBN1 missense variants compared to others, and its availability and ease of use resulted in its recommendation as the computational pathogenicity prediction algorithm for evaluation of FBN1 variants. Based on analyses of known pathogenic missense variants and their respective REVEL scores, PP3 was determined to be applicable for variants with REVEL scores greater than or equal to 0.75. Following the comprehensive analysis by Tian et al. [45], BP4 was determined to be applicable for missense variants with REVEL scores less than or equal to 0.326. For variants with potential impacts to splicing, either PP3 or BP4 should be applied when the computational splicing prediction algorithms GeneSplicer [46], MaxEntScan [47], and NNSplice [48] are concordant on their predictions of either an impact or lack of predicted impact on splicing, respectively.
Patient’s phenotype or family history is highly specific for a disease with a single genetic aetiology (PP4)
Application of the PP4 criterion accounts for an individual under investigation who manifests a phenotype and/or has a family history highly specific to a single gene. The FBN1 VCEP opted to utilize the revised Ghent criteria for diagnosis of Marfan syndrome [9], stating that PP4 should be applied for variants identified in individuals who meet these well-established and highly specific clinical diagnostic criteria. The VCEP also advises that a laboratory should be cautious but may use their discretion regarding the requirement of a clinical diagnosis in instances in which a variant is identified in a young patient with a highly suspicious phenotype in whom some of the characteristic features of Marfan syndrome may not have yet manifested (e.g. an infant with ectopia lentis and systemic features but a systemic score less than seven and no TAAD).
Variant co-occurs with a pathogenic variant for a fully penetrant disorder (BP2)
The VCEP states that in order to apply the BP2 criterion, one of two scenarios must be fulfilled. The first is that the variant under investigation must have been found in trans with a pathogenic FBN1 variant in at least two distinct cases without the patients manifesting more severe phenotypes than when the variant is present in isolation. Second, BP2 can be applied if a variant under investigation has been shown in cis with a pathogenic variant, with the requirement that the pathogenic variant has been previously identified in isolation in an individual with a phenotype consistent with Marfan syndrome.
Pilot testing of rule specification
The pilot study cohort (n = 60) comprised a wide variety of variant types and characteristics to ensure that a wide breadth of possible evidence types and associated evidence criteria would be addressed during the pilot (Fig. 4A, B). The cohort included multiple variants with each of the five classifications (benign, likely benign, VUS, likely pathogenic, and pathogenic) according to the submitting VCEP institutions (Fig. 4C).
Results of the pilot study
In comparing the original institution-made classifications with those achieved using the FBN1 specifications (Fig. 4C), the number of VUSs reduced from 24 to 12, and the number of benign (3 to 6), likely benign (7 to 8), and likely pathogenic (13 to 22) classifications all increased, with a slight decrease in pathogenic classifications (13 to 12). There were three criteria deemed applicable for FBN1 that were not used for any variants in the pilot (Fig. 4B): BP7 (synonymous variant with no predicted impact on splicing and occurs at a poorly conserved nucleotide), as there were only two synonymous variants included, and they had predicted aberrant splicing impacts; BS3 (functional study demonstrates no impact of the variant), which will rarely be considered in practice because very few publications exist that experimentally demonstrate an FBN1 variant’s lack of impact; and BP2 (variant co-occurs with a pathogenic variant for a fully penetrant disorder), due to the decision to use the similar BP5 criterion (variant found in a case with an alternate molecular basis for disease) for multiple cases instead.
Concordance between the non-core VCEP institutions and the core team for variant classifications obtained using the FBN1 specifications was 85.0% (51/60). Of the nine variants with discordant classifications, five represented potentially clinically significant discordance (e.g. VUS vs. likely benign, VUS vs. likely pathogenic) and four represented differences in the degree of confidence (e.g. benign vs. likely benign, pathogenic vs. likely pathogenic). The most prominent causes of classification discordance included differences in usage of population cut-offs due to specifications about the appropriate population in gnomAD [49] to be used for minor allele frequency evaluation, different standards for the application of PP4 (phenotype is highly specific for a single gene) for affected probands, differences in application of PM1 (mutational hotspot or functional domain without benign variation), discordant criteria strength modifications, and discrepancies in classification practices when faced with conflicting variant evidence. These differences were targeted for resolution in the subsequent iteration of the specifications. While each VCEP institution may have encountered slightly different issues with the specifications due to receiving different pilot variants to interpret and classify, in general, most sources of discordance or potential confusion were experienced by each institution. More detail on the sources of discordance experienced during the pilot study are presented in Supplementary Table 2. Of note, the VCEP classifications reported in Supplementary Table 2 represent the classification determined during the pilot process; these may or may not reflect the ultimate classification obtained when the same variants are formally curated and submitted to ClinVar following ClinGen SVI approval of these specifications, such as in the case newly available data. The final classification as formally curated and published in ClinVar is also shown for the relevant variants in Supplementary Table 2.
Discussion
The process of defining the ACMG-AMP criteria with respect to a single gene or disease is an undertaking of considerable complexity, and any refinements are contingent upon a deep understanding of the gene and its role in pathogenesis. Extensive clinical and bench research has contributed valuable insights into the relationship between FBN1 variation and Marfan syndrome facilitated the work of the FBN1 VCEP. The comprehensive understanding of the mutational spectrum, penetrance, and disease prevalence enabled the establishment of appropriate discriminatory minor allele frequency cut-offs. These cut-offs help to effectively identify benign variants and exclude them from further analysis. Additionally, the identification of 22.0% of amino acid positions in the encoded protein as likely functionally important, supported the PM1 evidence code, thereby greatly aiding in the classification of missense variants. These variants may not have an obvious detrimental impact on the gene product compared to variants resulting in haploinsufficiency. Baudhuin et al. emphasized the significance of incorporating gene-specific knowledge into the interpretation of FBN1 variants in ClinVar [40]. This includes considering minor allele frequency cut-offs and recognizing the importance of conserved and functionally important non-cysteine residues throughout the gene, the latter of which was critical in the interpretation and classification of non-cysteine missense variants. The specifications outlined in this publication address these concerns and aim to minimize the potential for misclassification.
The discordance in classification observed during the pilot study can be attributed to variations in the interpretations and application of certain evidence criteria. These discrepancies include differences in minor allele frequency thresholds, determining functional domains or mutational hotspots, and evaluating relevant phenotypes and their specificity to FBN1. Amendola et al. have reported similar findings when assessing the usage of the ACMG-AMP criteria across multiple laboratories [19, 20]. Their research revealed that modifications to criteria strength levels and selective application of certain criteria, specifically those involving subjective judgement or discretion, resulted in discordant classifications. In the case of FBN1, the VCEP has successfully worked towards minimizing the potential differences in laboratory-specific utilization of the ACMG-AMP criteria, thereby reducing classification discordance.
The pilot study demonstrated these rule specifications’ utility for reducing the quantity of variants given VUS classifications in favour of more benign, likely benign, and likely pathogenic classifications. The resultant increase in the rate of diagnostic genetic testing will have significant medical management and family planning implications for probands and their family members [12, 14–16]. The reduction of VUS in favour of (likely) benign genetic testing results is also desirable, as negative results provide clinicians with justification for pursuing additional genetic testing and eliminate the need to spend both time and financial resources on pursuing segregation analyses for VUSs that may be of relatively low clinical suspicion [50]. Therefore, it is of utmost importance to recontact the clinicians who requested the genetic investigations to pass on the information regarding variant reclassification to their patients. However, the protocols established for this purpose will depend on each institution and fall outside the scope of this work. Further, the return of VUSs from genetic testing has been demonstrated to have variable psychological impacts on some patients [51]; the reduction of VUSs could feasibly have the additional benefit of reducing occurrences of psychological distress for some individuals and families.
The collaborative nature of the VCEP’s curation effort highlights the importance of improved sharing of data and processes between laboratories or other institutions involved in variant classification. As each VCEP institution contributes their own internal data for individual variant classification, the nine institutions are effectively sharing data in an effort to reach a consensus classification. This is highly analogous to the work of Harrison et al. who demonstrated that data sharing between clinical laboratories, particularly of clinical data related to probands and their families, was extremely effective in reducing classification discrepancies and resulted in vastly improved classification concordance [52, 53]. VCEPs are uniquely suited for this type of high-impact data sharing; most VCEPs’ constituent institutions and experts have prolific histories of managing individuals with the disease(s) of interest and thus possess an abundance of useful clinical data, and there are already-established lines of communication and methods for data sharing that ease the potential burden associated with this process. Successful collaborations to reach consensus classifications also emphasize the importance at the institutional level of contributing to data sharing initiatives like ClinVar [7], DECIPHER [54], Leiden Open Variation Database [55], and Universal Mutation Database [56] so that laboratorians and clinicians can access as much relevant clinical data as possible and employ these interpretation specifications to their fullest extent.
Limitations
We recognize that several of the criteria, which can be crucially important to a variant’s curation, are dependent on the presence of robust clinical information, including for assessment of the applicability and strength of PS4, PS2/PM6, and PP1, and the appropriateness of PP4. Diagnostic laboratories do not always have detailed clinical data for a patient or family when assessing a variant, and these specifications cannot provide clarity beyond the available data. Further, due to the variable expressivity and often age-dependent penetrance of Marfan syndrome features, there will always be an inherent limitation in the interpretation of some FBN1 variants based on the patients’ clinical presentations, as well as a potential bias in evaluating variant re-interpretation surrounding the ages between initial and re-interpretation. Additionally, these criteria have been developed specifically to curate variants in FBN1 causing Marfan syndrome and may not be applicable to other diseases caused by variation in FBN1.
Conclusions
The FBN1 VCEP introduced 14 modifications to the original 28 ACMG-AMP variant classification criteria. Establishing these specific adaptations for FBN1 provides a framework to improve classification concordance among clinical laboratories which will ultimately result in an improvement of clinical care for patients with Marfan syndrome.
The VCEP will maintain a monthly meeting schedule to review variants that have been pre-curated by one of two biocurators in collaboration with a rotating VCEP institution. The final classification of these variants will be determined through group consensus among all members. The primary focus of curation efforts will be on variants with conflicting interpretations in ClinVar. Currently, the VCEP has completed curation and ClinVar submission for 120 FBN1 variants, with an estimated annual curation rate of 120 variants. We acknowledge that upcoming updates to the technical standards for sequence variant interpretation will require adjustments to these rule specifications to align with standard practices. This challenge is faced by all VCEPs, and guidance from the ClinGen SVI will likely be relied upon. We are confident that the current rule specifications can be easily transferred to the new framework, as the fundamental principles for FBN1 variant assessment will remain unchanged. Furthermore, this forthcoming publication will provide an opportunity for reassessment of the FBN1 specifications as outlined here. The FBN1-specific guidelines for variant classification and curation have already demonstrated success in the pilot phase and in practice. They have introduced a standardized interpretation framework and improved classification agreement among clinical laboratories, ultimately leading to enhanced clinical care for this patient population. However, variant curation is an evolving effort in continuous need for re-assessment and this process will persist over time.
Supplementary Information
Acknowledgements
The FBN1 VCEP would like to thank Lisa Kurtz and Courtney Thaxton for their contribution in coordinating stage 1–3 of this process.
Abbreviations
- ACMG/AMP
American College of Medical Genetics and Genomics and the Association for Molecular Pathology
- ClinGen
Clinical Genome Resource
- EGF
Epidermal growth factor
- SVI
ClinGen Sequence Variant Interpretation Working Group
- TAAD
Thoracic aortic aneurysm and dissection
- VCEP
Variant curation expert panel
- VUS
Variant of unknown significance
Authors’ contributions
MR, JDB, and LMM organized the panel meetings in stages 1–3. They built and analysed the online surveys. JDB and LMM organized and coordinated the pilot study. All authors participated in the monthly discussions and contributed variants to the pilot study. LB, KK, MLK, PA, and NH reviewed all surveys and the results of the pilot study as part of the core team. AD and LMM drafted the manuscript and the rebuttal to the reviewers. All authors read and approved the final manuscript.
Funding
JM is supported by the Research Foundation Flanders and the Marfan Foundation. During the specification development MR was supported by a postdoctoral grant of the Research Foundation Flanders. JDB is supported by a grant for Medical Research from the Baillet Latour Fund, a Concerted Research Action grant from the Ghent University Special Research Fund and a Senior Clinical Fellowship grant of the Research Foundation Flanders. LMM is supported by the Fund for Innovation and Clinical Research of the Ghent University Hospital.
Data availability
The datasets supporting the conclusions of this article are included within the article and supplementary files. All curated variants have been deposited in the ClinVar database7 with 3-star “Expert panel” review status (https://www.ncbi.nlm.nih.gov/clinvar/). The results of the conducted surveys and the specific report on the pilot study process and progress can be made available upon request. Some data included in the original files could compromise individual privacy.
Declarations
Ethics approval and consent to participate
This research was performed according to the principles of the Helsinki Declaration. It only includes de-identified data, and therefore ethical approval is not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
J. De Backer and L. Muiño-Mosquera are joint last authors.
References
- 1.Demolder A, von Kodolitsch Y, Muiño-Mosquera L, De Backer J. Myocardial function, heart failure and arrhythmia in marfan syndrome: a systematic literature review. Diagn Basel Switz. 2020;10(10):751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esfandiari H, Ansari S, Mohammad-Rabei H, Mets MB. Management strategies of ocular abnormalities in patients with Marfan syndrome: current perspective. J Ophthalmic Vis Res. 2019;14(1):71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sengle G, Sakai LY. The fibrillin microfibril scaffold: A niche for growth factors and mechanosensation? Matrix Biol J Int Soc Matrix Biol. 2015;47:3–12. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Huang J, Liu Y. The extracellular matrix glycoprotein fibrillin-1 in health and disease. Front Cell Dev Biol. 2023;11:1302285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milewicz DM, Braverman AC, De Backer J, Morris SA, Boileau C, Maumenee IH, et al. Marfan syndrome. Nat Rev Dis Primer. 2021;7(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collod-Béroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22(3):199–208. [DOI] [PubMed] [Google Scholar]
- 7.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muiño-Mosquera L, Steijns F, Audenaert T, Meerschaut I, De Paepe A, Steyaert W, et al. Tailoring the American college of medical genetics and genomics and the association for molecular pathology guidelines for the interpretation of sequenced variants in the FBN1 gene for Marfan syndrome: proposal for a disease- and gene-specific guideline. Circ Genomic Precis Med. 2018;11(6):e002039. [DOI] [PubMed] [Google Scholar]
- 9.Loeys BL, Dietz HC, Braverman AC, Callewaert BL, De Backer J, Devereux RB, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47(7):476–85. [DOI] [PubMed] [Google Scholar]
- 10.Wenger BM, Patel N, Lui M, Moscati A, Do R, Stewart DR, et al. A genotype-first approach to exploring Mendelian cardiovascular traits with clear external manifestations. Genet Med Off J Am Coll Med Genet. 2021;23(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klemenzdottir EO, Arnadottir GA, Jensson BO, Jonasdottir A, Katrinardottir H, Fridriksdottir R, et al. A population-based survey of FBN1 variants in Iceland reveals underdiagnosis of Marfan syndrome. Eur J Hum Genet EJHG. 2024;32(1):44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groth KA, Stochholm K, Hove H, Andersen NH, Gravholt CH. Causes of mortality in the Marfan syndrome(from a Nationwide Register Study). Am J Cardiol. 2018;122(7):1231–5. [DOI] [PubMed] [Google Scholar]
- 13.Bakalli A, Bekteshi T, Basha M, Gashi A, Bakalli A, Ademaj P. Late diagnosis of Marfan syndrome with fatal outcome in a young male patient: a case report. Cases J. 2009;9(2):8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Kodolitsch Y, Rybczynski M, Vogler M, Mir TS, Schüler H, Kutsche K, et al. The role of the multidisciplinary health care team in the management of patients with Marfan syndrome. J Multidiscip Healthc. 2016;9:587–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isselbacher EM, Preventza O, Hamilton Black J 3rd, Augoustides JG, Beck AW, Bolen MA, et al. 2022 ACC/AHA Guideline for the Diagnosis and Management of Aortic Disease: A Report of the American Heart Association/American College of Cardiology Joint Committee on Clinical Practice Guidelines. Circulation. 2022;146(24):e334–482. [DOI] [PMC free article] [PubMed]
- 16.Arslan-Kirchner M, von Kodolitsch Y, Schmidtke J. The importance of genetic testing in the clinical management of patients with Marfan syndrome and related disorders. Dtsch Arzteblatt Int. 2008;105(27):483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med Off J Am Coll Med Genet. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niehaus A, Azzariti DR, Harrison SM, DiStefano MT, Hemphill SE, Senol-Cosar O, et al. A survey assessing adoption of the ACMG-AMP guidelines for interpreting sequence variants and identification of areas for continued improvement. Genet Med Off J Am Coll Med Genet. 2019;21(8):1699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016;98(6):1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amendola LM, Muenzen K, Biesecker LG, Bowling KM, Cooper GM, Dorschner MO, et al. Variant Classification Concordance using the ACMG-AMP Variant Interpretation Guidelines across Nine Genomic Implementation Research Studies. Am J Hum Genet. 2020;107(5):932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelb BD, Cavé H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H, et al. ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet Med Off J Am Coll Med Genet. 2018;20(11):1334–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med Off J Am Coll Med Genet. 2018;20(3):351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 2018;39(11):1593–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKnight D, Bean L, Karbassi I, Beattie K, Bienvenu T, Bonin H, et al. Recommendations by the ClinGen Rett/Angelman-like expert panel for gene-specific variant interpretation methods. Hum Mutat. 2022;43(8):1097–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen–the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison SM, Biesecker LG, Rehm HL. Overview of Specifications to the ACMG/AMP Variant Interpretation Guidelines. Curr Protoc Hum Genet. 2019;103(1):e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrijver I, Liu W, Odom R, Brenn T, Oefner P, Furthmayr H, et al. Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet. 2002;71(2):223–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Judge DP, Biery NJ, Keene DR, Geubtner J, Myers L, Huso DL, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest. 2004;114(2):172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiffin N, Minikel E, Walsh R, O’Donnell-Luria AH, Karczewski K, Ing AY, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med Off J Am Coll Med Genet. 2017;19(10):1151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Judge DP, Dietz HC. Marfan’s syndrome. Lancet Lond Engl. 2005;366(9501):1965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman MJ, Devereux RB, Preiss LR, Asch FM, Eagle KA, Holmes KW, et al. Associations of Age and Sex With Marfan Phenotype: The National Heart, Lung, and Blood Institute GenTAC (Genetically Triggered Thoracic Aortic Aneurysms and Cardiovascular Conditions) Registry. Circ Cardiovasc Genet. 2017;10(3):e001647. [DOI] [PMC free article] [PubMed]
- 33.ClinGen Sequence Variant Interpretation Working Group. PM2: Recommendation for Absence/Rarity Criterion PM2 (Version 1.0). Available from: https://clinicalgenome.org/working-groups/sequence-variant-interpretation/. Cited 2023 Dec 28.
- 34.Fortuno C, Lee K, Olivier M, Pesaran T, Mai PL, de Andrade KC, et al. Specifications of the ACMG/AMP variant interpretation guidelines for germline TP53 variants. Hum Mutat. 2021;42(3):223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston JJ, Dirksen RT, Girard T, Hopkins PM, Kraeva N, Ognoon M, et al. Updated variant curation expert panel criteria and pathogenicity classifications for 251 variants for RYR1-related malignant hyperthermia susceptibility. Hum Mol Genet. 2022;31(23):4087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatton JN, Frone MN, Cox HC, Crowley SB, Hiraki S, Yokoyama NN, et al. Specifications of the ACMG/AMP Variant Classification Guidelines for Germline DICER1 Variant Curation. Chen JM, editor. Hum Mutat. 2023;2023:9537832. [DOI] [PMC free article] [PubMed]
- 37.Luo X, Maciaszek JL, Thompson BA, Leong HS, Dixon K, Sousa S, et al. Optimising clinical care through CDH1-specific germline variant curation: improvement of clinical assertions and updated curation guidelines. J Med Genet. 2023;60(6):568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson PN, Arteaga-Solis E, Baldock C, Collod-Béroud G, Booms P, De Paepe A, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43(10):769–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faivre L, Collod-Beroud G, Loeys BL, Child A, Binquet C, Gautier E, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81(3):454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudhuin LM, Kluge ML, Kotzer KE, Lagerstedt SA. Variability in gene-based knowledge impacts variant classification: an analysis of FBN1 missense variants in ClinVar. Eur J Hum Genet EJHG. 2019;27(10):1550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ClinGen Sequence Variant Interpretation Working Group. PS2/PM6: Recommendation for de novo PS2 and PM6 ACMG/AMP criteria (Version 1.1) [Internet]. [cited 2023 Dec 28]. Available from: https://clinicalgenome.org/working-groups/sequence-variant-interpretation/.
- 42.Jarvik GP, Browning BL. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet. 2016;98(6):1077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian Y, Pesaran T, Chamberlin A, Fenwick RB, Li S, Gau CL, et al. REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci Rep. 2019;9(1):12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29(5):1185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol J Comput Mol Cell Biol. 2004;11(2–3):377–94. [DOI] [PubMed] [Google Scholar]
- 48.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol J Comput Mol Cell Biol. 1997;4(3):311–23. [DOI] [PubMed] [Google Scholar]
- 49.Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625(7993):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrett LT, Hickman N, Jacobson A, Bennett RL, Amendola LM, Rosenthal EA, et al. Family studies for classification of variants of uncertain classification: current laboratory clinical practice and a new web-based educational tool. J Genet Couns. 2016;25(6):1146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mighton C, Shickh S, Uleryk E, Pechlivanoglou P, Bombard Y. Clinical and psychological outcomes of receiving a variant of uncertain significance from multigene panel testing or genomic sequencing: a systematic review and meta-analysis. Genet Med Off J Am Coll Med Genet. 2021;23(1):22–33. [DOI] [PubMed] [Google Scholar]
- 52.Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med Off J Am Coll Med Genet. 2017;19(10):1096–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison SM, Dolinksy JS, Chen W, Collins CD, Das S, Deignan JL, et al. Scaling resolution of variant classification differences in ClinVar between 41 clinical laboratories through an outlier approach. Hum Mutat. 2018;39(11):1641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans using ensembl resources. Am J Hum Genet. 2009;84(4):524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT. LOVD v20: the next generation in gene variant databases. Hum Mutat. 2011;32(5):557–63. [DOI] [PubMed] [Google Scholar]
- 56.Béroud C, Collod-Béroud G, Boileau C, Soussi T, Junien C. UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum Mutat. 2000;15(1):86–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and supplementary files. All curated variants have been deposited in the ClinVar database7 with 3-star “Expert panel” review status (https://www.ncbi.nlm.nih.gov/clinvar/). The results of the conducted surveys and the specific report on the pilot study process and progress can be made available upon request. Some data included in the original files could compromise individual privacy.