Abstract
A large 10-mer phage peptide library was panned against whole Escherichia coli cells, and an antimicrobial peptide (QEKIRVRLSA) was selected. The peptide was synthesized in monomeric and dendrimeric tetrabranched form (multiple antigen peptide [MAP]), which generally allows a dramatic increase of peptide stability to peptidases and proteases. The antibacterial activity of the dendrimeric peptide against E. coli was much higher than that of the monomeric form. Modification of the original sequence, by residue substitution or sequence shortening, produced three different MAPs, M4 (QAKIRVRLSA), M5 (KIRVRLSA), and M6 (QKKIRVRLSA) with enhanced stability to natural degradation and antimicrobial activity against a large panel of gram-negative bacteria. The MICs of the most potent peptide, M6, were as low as 4 to 8 μg/ml against recent clinical isolates of multidrug-resistant Pseudomonas aeruginosa and members of the Enterobacteriaceae. The same dendrimeric peptides showed high stability to blood proteases, low hemolytic activity, and low cytotoxic effects on eukaryotic cells, making them promising candidates for the development of new antibacterial drugs.
Antibiotic resistance has become a global public health problem impairing the efficacy of antimicrobial chemotherapy, and the emergence of multidrug-resistant bacteria has led to situations in which few or no treatment options remain for infections caused by certain microorganisms (29). It has therefore become increasingly important to develop new antibiotics. Antimicrobial peptides are a new family of antibiotics that have stimulated research and clinical interest (30). Most antibacterial peptides are components of the innate immunity of animals and plants against microbial infections (4, 32). They generally consist of 6 to 50 amino acid residues and have a positive net charge (7, 13). These cationic peptides seem to interact selectively with anionic bacterial membranes (14, 16, 17), although different mechanisms of killing microbes may be used by different peptides under different conditions (9). A number of antibacterial peptides have been shown to bind to negatively charged lipopolysaccharide and to render the outer membrane of gram-negative bacteria permeable. Lipopolysaccharide-binding cationic peptides can competitively displace the native divalent cations Ca2+ and Mg2+, disrupting their stabilizing effect and altering the normal barrier property of the outer membrane (11).
The use of peptides in vivo has largely been limited by their short half-lives, since peptides are mainly broken down by endogenous proteases and peptidases. Peptide half-life is, therefore, the bottleneck in the development of new peptide drugs. Many laboratories throughout the world use different strategies to increase peptide stability. These include introduction of d-amino acids or pseudo-amino acids (1) and peptide cyclization (10).
In a previous study (6), we reported that the synthesis of bioactive peptides in dendrimeric form (multiple antigen peptide [MAP]) can result in increased half-life due to acquired resistance to protease and peptidase activity. These dendrimeric molecules, first developed in the 1980s (21, 26), have a peptidyl core of radially branched lysine residues onto which peptide sequences can be added using standard solid-phase chemistry.
Phage display is a powerful tool for selecting peptides or proteins with specific binding properties from a large number of variants (22, 25). Here, we describe the characterization of novel peptides with antimicrobial activity, by phage library selection and rational modification, synthesized in dendrimeric MAP4 form on a core of three lysines.
MATERIALS AND METHODS
Construction of the peptide phage library.
The decapeptide library DIP3 expressed on the minor phage coat protein pIII of M13 bacteriophage was constructed modifying the vector pDN332 (20). Phage peptides are encoded by a DNA insert amplified from synthetic degenerate oligonucleotide [5′-CTCGCGGCCCAGCCGGCCATGGCC(NNK)10GGTGCAGGCGCCACTGTTGAAAGTTG-3′], which contains an NcoI restriction site at its 5′ terminals. The codon scheme used was NNK, where N is an equimolar representation of all four bases and K is an equimolar representation of G and T. This scheme uses 32 codons to encode 20 amino acids, reducing the frequency of termination codons to 1 (TAG).
The degenerate oligonucleotide and pIII-encoding gene were PCR amplified separately. The PCR fragments were agarose gel purified and assembled by PCR with an equimolar mixture of Biot1 primer (5′-CTCGCGGCCCAGCCGGCC-3′) and EcopIIIfor primer (5′-CGGAATTCTTAAGACTCCTTATTACGCAG-3′); the latter has an EcoRI site for cloning. The single recombinant gene was double digested with restriction enzymes NcoI and EcoRI (Takara) and ligated (5 μg) into the phagemid vector pDN332 (4 to 7 μg). The ligated DNA was then purified in Qiaquick PCR purification columns (QIAGEN) and electroporated into TG1 Escherichia coli electrocompetent cells. Each electroporation entailed the introduction of 100 ng of ligation mixture into 50 μl of high-efficiency TG1 E. coli competent cells (transformation efficiency, >109 CFU). The cell library showed a complexity of ∼1.2 × 109 different clones. The presence of the recombinant fusion gene in the phagemid was assessed by PCR screening. A total of 50% of the clones analyzed showed the expected molecular weight insert. Phage particles were rescued from the library after infection with VCS-M13 helper phage (>1011 transforming units [TU]/ml; Stratagene) and 20% polyethylene glycol 6000-2.5 M NaCl (PEG/NaCl) precipitation.
Phage peptide selection.
Library screening was performed using three rounds of affinity selection. Selection against E. coli whole cells was performed by incubating 1014 phages with approximately 8 × 107 TG1 E. coli cells (optical density at 600 nm [OD600] = 0.1) in phosphate-buffered saline (PBS; 1 ml) for 60 min at room temperature with gentle agitation. Bacteria and phages were pelleted by spinning for 3 min at 17,000 × g, and the pellet was washed 10 times with PBS-Tween 0.1% in the first round of selection and with PBS-Tween 0.5% in subsequent rounds. Bound phages were pelleted with cells and eluted nonspecifically with 1 ml of 0.2 M glycine-HCl (pH 2.2) with gentle shaking at room temperature for 5 min, followed by neutralization with 150 μl of 1 M Tris-HCl (pH 9.0). A total of 100 μl of eluted phages was used to infect 10 ml of exponentially growing TG1 E. coli cells for 30 min at 37°C. After infection, the bacteria were pelleted by being spun for 10 min at 6,000 × g, resuspended in 1 ml of 2×TY medium (16 g/liter tryptone, 10 g/liter yeast extract, and 5 g/liter NaCl; pH adjusted to 7.4), and spread on a large agar plate containing ampicillin (100 μg/ml)-glucose (1%). After overnight incubation at 30°C, the colonies were recovered from the agar plate with a glass spreader, and 5 to 10 ml of 2×TY buffer was added until a homogenous suspension was obtained. A total of 100 ml of 2×TY-ampicillin (100 μg/ml)-glucose (1%) was inoculated with 100 μl of bacterial suspension to yield an OD600 of 0.05 to 0.1. After growth to OD600 of 0.4 to 0.5 at 37°C, 10 ml of culture was infected with 100 μl of VCS-M13 helper phage (>1011 TU/ml, Stratagene). The infected bacteria were spun down at 6,000 × g for 10 min, the pellet was gently resuspended in 100 ml of 2× TY-ampicillin (100 μg/ml)-kanamycin (25 μg/ml) and shaken overnight at 30°C. The phages were concentrated by precipitation with PEG 6000/NaCl and resuspended in 2 ml PBS. Phage preparation and further panning were repeated as previously described.
Screening positive phages by ELISA.
Phage enzyme-linked immunosorbent assay (ELISA) was performed with bacteria by coating the wells with 100 μl of a suspension of 2.5 × 108 CFU/ml E. coli cells in PBS and incubating the cells for 1 h at 37°C, followed by overnight incubation at 4°C as previously described (10). A total of 94 colonies from the third-round were picked and inoculated into 150 μl 2×TY-ampicillin (100 μg/ml)-glucose (1%) in a 96-well plate. Individual clones were propagated with gentle agitation at 37°C for 3 h. Then, 25 μl 2× TY-ampicillin (100 μg/ml)-glucose (1%) containing VCS-M13 helper phage (109 TU) was added to each well. After incubation for 30 min at 37°C, the plate was spun at 600 × g for 10 min, and the supernatant was aspirated. The bacterial pellet was resuspended in 200 μl 2×TY-ampicillin (100 μg/ml)--kanamycin (25 μg/ml) and grown overnight at 30°C. The amplified phage particles were separated from cells by centrifugation at 800 × g for 20 min and incubated in TG1 E. coli-coated ELISA wells for 1 h at 37°C. The wells were blocked with 3% bovine serum albumin in PBS buffer for 1 h and washed with PBS.
Anti-M13 peroxidase conjugate antibody (Amersham) was added at a dilution of 1/2,500 in 3% bovine serum albumin-PBS. After further washing, antibody binding was detected by adding TMB substrate. Optical absorbance was recorded at 450 nm. Positive phage clones were selected randomly for DNA sequencing.
Peptide synthesis.
Monomeric peptide was synthesized as peptide amide by an automated synthesizer (MultiSynTech, Witten, Germany) on a Rink Amide MBHA resin (Nova Biochem) using 9-fluorenylmethoxycarbonyl chemistry and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate/1,3-diisopropylethylamine activation. MAPs were synthesized on (9-fluorenylmethoxy carbonyl)4-Lys2-Lys-βAla Wang resin. Side-chain-protecting groups were tert-butyl ester for Glu, trityl for Gln, tert-butoxycarbonyl for Lys and Trp, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl for Arg, and tert-butyl ether for Ser. Peptides were then cleaved from the resin and deprotected by treatment with trifluoroacetic acid-containing water and triisopropylsilane (at a 95/2.5/2.5 ratio). Crude peptides were purified by reversed-phase chromatography with a Vydac C18 column. Identity and purity of final products were confirmed by Ettan matrix-assisted laser desorptionionization-time of flight mass spectrometry (MS) (Amersham Biosciences).
Evaluation of the antibacterial activity of peptides.
TG1 E. coli was used in preliminary studies of antimicrobial activity of some peptides as follows. Peptide dilutions in 10 mM sodium phosphate buffer (pH 7.4) were added (25 μl) to 25 μl of early-growth-phase TG1 E. coli cells (final concentration, 8 × 107 CFU/ml) in 2×TY medium, and the mixtures were incubated at 37°C for 75 min. CFU counts were then determined by plating dilutions of each mixture in 2×TY medium. A test without added peptide was always carried out in parallel as a control.
Reference strains (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococus aureus ATCC 25923, Enterococcus faecalis ATCC 29213, and Chryseobacterium meningosepticum CCUG 4310) and several recent clinical isolates (including multidrug-resistant ones) of various species (Table 1) were used for conventional susceptibility testing experiments. MIC was determined by a standard microdilution assay as recommended by CLSI (formerly NCCLS) using cation-supplemented Mueller-Hinton (MH) broth (Oxoid, Ltd., Basingstoke, United Kingdom) and a bacterial inoculum of 5 × 104 CFU per well in a final volume of 100 μl. Results were recorded by visual inspection after 24 h of incubation at 37°C. Minimum bactericidal concentration (MBC), defined as the concentration at which ≥99.9% of the bacterial inoculum is killed, was determined as recommended by CLSI (18) after MIC testing.
TABLE 1.
MICs of antimicrobial peptides for various gram-negative and gram-positive bacteria
| Species and strain | Relevant feature(s)a | MIC (μg/ml) of:
|
||
|---|---|---|---|---|
| M4 | M5 | M6 | ||
| Escherichia coli ATCC 25922 | Reference strain | 128 | 16 | 8 |
| Escherichia coli W99FI0077 | FQr ESCr (ESBL/SHV type) | 16 | 128 | 8 |
| Escherichia coli W03BG0025 | FQr AGr ESCr (ESBL/CTX-M-15) | NDb | ND | 8 |
| Escherichia coli W03NO0013 | FQr ESCr (ESBL/CTX-M-1) | ND | ND | 8 |
| Pseudomonas aeruginosa ATCC 27853 | Reference strain | 32 | 16 | 4 |
| Pseudomonas aeruginosa 885149 | FQr AGr ESCr CPr (MBL/IMP-13) | 64 | 32 | 8 |
| Pseudomonas aeruginosa 891 | FQr AGr ESCr CPr (MBL/VIM-2) | 64 | 16 | 8 |
| Pseudomonas aeruginosa VA463/98 | FQr AGr ESCr (ESBL/PER-1) | ND | ND | 4 |
| Klebsiella pneumoniae W99FI0057 | ESCr (ESBL/SHV type) | 64 | >128 | 4 |
| Klebsiella pneumoniae W03NO0078 | ESCr (ESBL/CTX-M-1) | ND | ND | 16 |
| Klebsiella pneumoniae W03BG0019 | AGr ESCr (ESBL/CTX-M-15) | ND | ND | 8 |
| Klebsiella oxytoca W99FI00049 | ESCr (ESBL/SHV-12) | ND | ND | 64 |
| Proteus mirabilis W99FI0089 | FQr | ND | ND | >128 |
| Proteus mirabilis W03VA1144 | FQr AGr ESCr (ESBL/PER-1) | ND | ND | 64 |
| Enterobacter aerogenes W03BG0067 | AGr ESCr (ESBL/SHV-5) | ND | ND | 8 |
| Enterobacter cloacae W03AN0041 | ESCr (ESBL/SHV-12) | ND | ND | 4 |
| Morganella morganii W03VA1342 | FQr ESCr (ESBL/CTX-M-1) | ND | ND | >128 |
| Acinetobacter baumannii AB1MG | FQr AGr ESCr (ESBL/TEM-92) | ND | ND | 16 |
| Acinetobacter baumannii AB7MG | FQr AGr ESCr | ND | ND | 32 |
| Citrobacter freundii W99FI00007 | ESCr (ESBL/SHV-12) | ND | ND | 16 |
| Chryseobacterium meningosepticum CCUG4310 | Reference strain | ND | ND | >128 |
| Burkholderia cepacia SMC71 | FQr AGr ESCr | ND | ND | 64 |
| Serratia marcescens W99FI0111 | FQr AGr ESCr (ESBL/SHV-5) | ND | ND | >128 |
| Stenotrophomonas maltophilia PT4/99 | Wild-type profile | ND | ND | >128 |
| Providencia stuartii W03FI0001 | AGr ESCr (ESBL/PER-1) | ND | ND | >128 |
| Staphylococcus aureus ATCC 25923 | Reference strain | 64 | 128 | >128 |
| Staphylococcus aureus MIU-68A | MS | >128 | 128 | 128 |
Except for reference strains, the other strains were clinical isolates. Relevant resistance phenotypes and known resistance mechanisms are indicated. FQr, resistance to fluoroquinolones (ciprofloxacin); AGr, resistance to aminoglycosides (gentamicin, amikacin, and/or tobramycin); ESCr, resistance to extended-spectrum cephalosporins (cefotaxime, ceftazidime, and/or cefepime); CPr, resistance to carbapenems (imipenem and/or meropenem); ESBL, extended-spectrum β-lactamase; MBL, metallo-β-lactamase; MS, methicillin susceptible.
ND, not determined.
Assay of bactericidal activity in time-kill experiments was carried out as follows. The peptide was added, at the desired concentration, to exponentially growing cultures of the test strain in MH broth containing a total inoculum of 5 × 107 CFU (1 × 107 CFU/ml) at 37°C. Samples were drawn at different times, and suitable dilutions were plated on MH agar to score the residual number of CFU. A culture without peptide was always grown in parallel as a control.
Hemolysis assay.
Hemolysis of fresh human erythrocytes was determined using the method of Parpart et al. (19). Briefly, a calibration curve was constructed by suspending fresh human erythrocytes in phosphate buffer (pH 7.4; 110 mM sodium phosphate) with various concentrations of NaCl and incubated for 30 min at room temperature. Samples were centrifuged at 500 × g for 5 min, and hemoglobin release was monitored by measuring the absorbance of supernatants at 540 nm. The absorbance obtained with 0.1% NaCl corresponded to 100% lysis and that with 1% NaCl to 0% lysis. Peptides dissolved in PBS were added to a human erythrocyte solution at several concentrations. The resulting suspension was incubated separately at 37°C for 2 h and 24 h. Release of hemoglobin was monitored by measuring the absorbance of the suspernatant at 540 nm after centrifuging; the percentage of hemolysis was calculated by using the calibration curve.
Cytoplasmic membrane permeabilization assay.
ML-35 E. coli, a lactose permease-deficient strain with constitutive production of β-galactosidase, was used in the cytoplasmic membrane permeabilization assay. Inner membrane permeability was determined by measuring β-galactosidase activity in ML-35 E. coli cells with p-nitrophenyl-β-d-galactopyranoside as a substrate. Logarithmic-phase bacteria were washed in 10 mM sodium phosphate (pH 7.4) containing 100 mM NaCl and resuspended in 0.7 ml of the same buffer containing 1.5 mM p-nitrophenyl-β-d-galactopyranoside. At time zero, different amounts of peptide were added, and the production of p-nitrophenol over time was monitored spectrophotometrically at 420 nm.
Peptide stability in serum and plasma.
A total of 10 μl of a 10 mM solution of peptides was incubated at 37°C with 10 μl human plasma and serum. Samples were collected after 2 or 24 h of incubation, precipitated with 150 μl methanol, and centrifuged for 2 min at 10,000 × g. The crude solution was then analyzed by high-performance liquid chromatography (HPLC) and MS. HPLC was performed on all samples with a Vydac C18 column, and the crude solutions were diluted five times with 0.1% trifluoroacetic acid before injection and monitored at 280 nm.
Cell toxicity assay.
The toxicity of peptides for the mouse macrophage cell line J774.A1 and the human HaCaT keratinocytes was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT; Sigma-Aldrich) cytotoxicity assay. J774.A1 and HaCaT cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 5% fetal bovine serum (BioWhittaker), glutamine (2 mM), and antibiotics and plated in triplicate wells at 6 × 104 and 3 × 104 cells/well, respectively. Peptides, previously filtered with a 0.2-μm filter disk (Whatman), were added at various concentrations and incubated overnight with the cells at 37°C in a 5% CO2 atmosphere. MTT (final concentration, 0.5 mg/ml) was then added for 90 min at 37°C. Solubilization of formazan, formed by the metabolic reduction of MTT, was obtained using lysis buffer (10% sodium dodecyl sulfate-45% dimethyl formamide, adjusted to pH 4.5 with glacial acetic acid). The plates were read with an automatic ELISA plate reader. Cultures exposed to peptides or exposed to the medium alone were measured spectrophotometrically at dual wavelengths (595 to 650 nm).
RESULTS
Selection of peptides with antimicrobial activity and synthesis in monomeric and dendrimeric form.
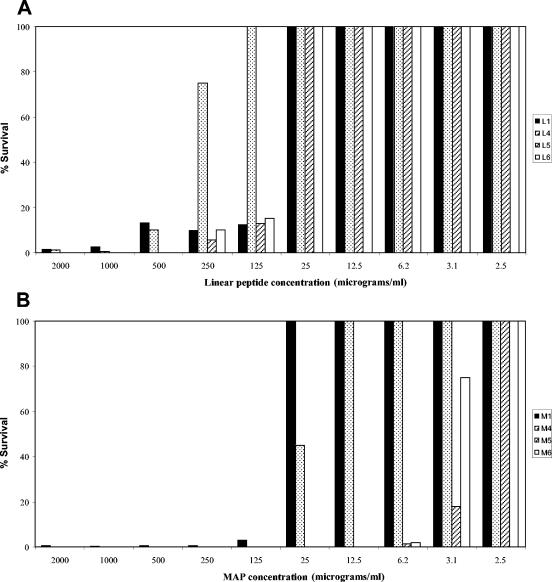
We isolated a novel 10-mer peptide (L1) from a high-complexity peptide phage library constructed in our laboratory. The biopanning process consisted of three rounds of library affinity selection on whole TG1 E. coli cells. Bound phages were eluted nonspecifically by conventional methods (see Materials and methods), and their DNA was sequenced. Several clones showed a unique sequence with a characteristic pattern of alternate hydrophobic and hydrophilic residues. The sequence QEKIRVRLSA showed features common to a variety of antimicrobial peptides: a net positive charge (+2) and a total residue hydrophobic ratio of 40%. However, no obvious similarity to other sequences was found in the Antimicrobial Sequences Database (http://www.bbcm.univ.trieste.it/∼tossi/antimic.html), and no significant similarities were found in other conventional databases for proteins. Although phage particles displaying the sequence QEKIRVRLSA did not show any antibacterial activity against TG1 E. coli cells (not shown), the linear monomeric peptide L1 (synthesized as described in Materials and Methods) almost completely prevented (1.5% survival) colony growth of TG1 E. coli at a concentration of 2 mg/ml and remained effective up to a concentration of 125 μg/ml (12.2% survival) (Fig. 1A). The dendrimeric MAP4 form of the same sequence (peptide M1), allowed 3.2% survival at 125 μg/ml (Fig. 1B). A noncorrelated dendrimeric tetra-branched peptide, used under the same conditions, allowed the same survival of untreated cells (100% survival up to 2 mg/ml; not shown).
FIG. 1.
(A) Antibacterial activity of monomeric linear peptides L1 (columns in black), L4 (columns with dots), L5 (columns with cross-hatching), and L6 (columns in white). (B) Antibacterial activity of tetrabranched MAP4 form M1 (columns in black), M4 (columns with dots), M5 (columns with cross-hatching), and M6 (columns in white). Experiments were performed by incubating E. coli TG1 cells (8 × 107 CFU/ml) with the indicated amounts of peptide. The survival percentage is the number of living colonies with respect to the number of colonies in controls without peptides.
Modification of peptide sequence and antimicrobial activity of modified peptides.
Since lead L1 and M1 peptides showed a drop in activity over time once resuspended in solution, we examined possible amino acid modification by MS (not shown). We saw that the peptide lost a water molecule in solution, possibly due to amide bond formation between adjacent side chains of Glu2 and Lys3, rendering the peptide unstable in solution.
The original sequence QEKIRVRLSA was consequently modified to obviate this problem. The new peptide amino acid sequences were QAKIRVRLSA (peptide 4), KIRVRLSA (peptide 5), and QKKIRVRLSA (peptide 6), all having a greater cationic charge than the lead sequence. In dendrimeric MAP4 form, those peptides (M4, M5, and M6) showed greater stability and activity than the linear homologs (L4, L5, and L6) when tested against TG1 E. coli cells (Fig. 1A and B). Notably, M5 and M6 prevented TG1 E. coli colony growth at concentrations down to 6.25 μg/ml, whereas M1 and M4 appeared less effective at the same concentrations (Fig. 1B).
MICs of M4, M5, and M6 were determined for the reference strains S. aureus ATCC 25923, E. coli ATCC 25922, and P. aeruginosa ATCC 27853 and for a number of recent clinical isolates of the same or different species (Table 1). All three peptides showed relatively poor activity against S. aureus, appearing to be more active against the gram-negative strains (MICs, 4 to 128 μg/ml), with M6 the most active against all species. MICs of M6 were then determined against a larger panel of bacteria (Table 1). These experiments confirmed the good activity of M6 (MICs, 4 to 16 μg/ml) against E. coli, Klebsiella pneumoniae, Enterobacter spp., and P. aeruginosa, including clinical isolates showing a multidrug-resistant phenotype. A somewhat lower activity (MICs, 16 to 32 μg/ml) was observed against Citrobacter freundii and Acinetobacter baumannii and even lower activity against Proteus mirabilis, Morganella morganii, Providencia stuartii, Stenotrophomonas maltophilia, Burkholderia cepacia, and Chryseobacterium meningosepticum (Table 1).
MBCs of M6 tested against E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were identical to MICs, indicating bactericidal activity of the MAP.
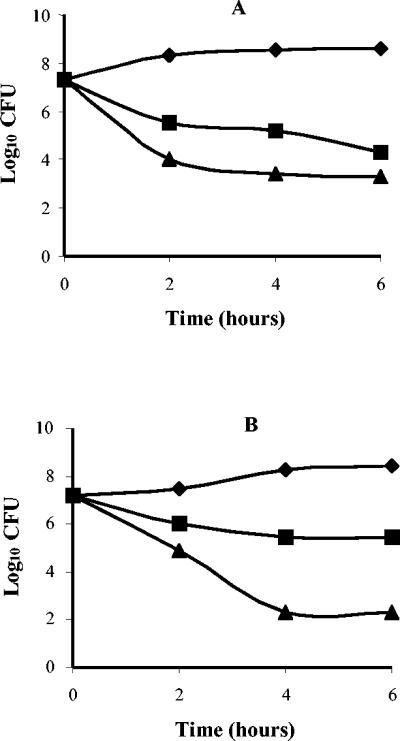
In time-kill experiments, M6 exhibited rapid bactericidal activity against E. coli ATCC 25922 and P. aeruginosa ATCC 27853, reducing an inoculum of >107 CFU by >99.9% in 4 h at a concentration of 32 or 16 μg/ml, respectively (Fig. 2). Bactericidal activity appeared to be concentration dependent, especially with P. aeruginosa.
FIG. 2.
Time-kill kinetics of M6 against E. coli ATCC 25922 (A) and P. aeruginosa ATCC 27853 (B). Symbols: ♦, growth control; ▪, 2× MIC (16 μg/ml for E. coli ATCC 25922 and 8 μg/ml for P. aeruginosa ATCC 27853); ▴, 4× MIC (32 μg/ml for E. coli ATCC 25922 and 16 μg/ml for P. aeruginosa ATCC 27853).
Peptide stability in plasma and serum.
We previously demonstrated that peptides synthesized in a dendrimeric form can retain biological activity and become resistant to proteases, thus dramatically increasing their in vivo half-life (6). We verified the stability in human plasma and serum of our antimimicrobial peptides in monomeric (only L1) and dendrimeric forms. A total of 10 μl of monomeric and dendrimeric peptides was incubated with the same volume of human plasma or serum at 37°C for 2 or 24 h. The reaction was stopped by the addition of 0.15 ml methanol and 0.75 ml 0.1% trifluoroacetic acid. The mixture was analyzed by HPLC to follow the presence of uncleaved monomeric and dendrimeric peptides. The presence of uncleaved peptides was confirmed by MS (not shown).
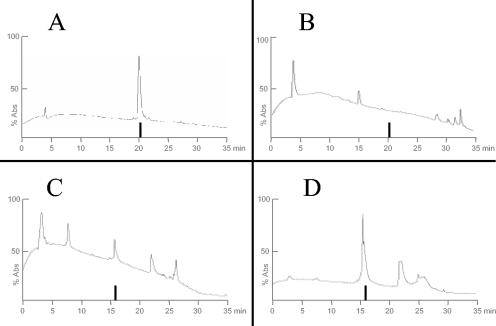
Monomeric peptide L1 was completely degraded within 2 h in serum, whereas the dendrimeric form of the same peptide (M1) was still detected after 24 h in plasma and serum (Fig. 3). Comparable results were obtained with dendrimeric peptides M4, M5, and M6 (not shown).
FIG. 3.
HPLC profiles of linear (L1) and dendrimeric (M1) peptides in serum. (A) L1 in serum at 0 h. (B) L1 after incubation in serum for 2 h; peptide is no longer detectable. (C) M1 in serum at 0 h. (D) M1 after incubation in serum for 24 h; the peptide is still present. The vertical bar indicates peptide retention time. x axis, retention time (min); y axis, absorbance at 220 nm.
Cytototoxicity.
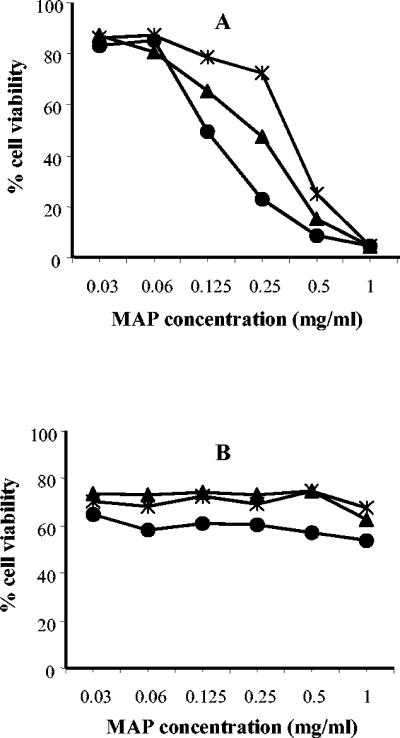
The toxicity of M4, M5, and M6 towards mouse macrophage cells J774.A1 was tested by MTT and is shown in Fig. 4A. Treatment of cells overnight with 30 μg/ml of M4, M5,or M6 did not substantially affect cell viability, whereas a drop in cell viability was evident after treatment with peptide M4 at concentrations of ≥250 μg/ml and with peptides M5 and M6 at ≥125 μg/ml. The same dendrimeric peptides showed low toxicity for human keratinocyte HaCaT cells (Fig. 4B), even when used at high concentrations (1 mg/ml).
FIG. 4.
Toxicity of M4 (*), M5 (▴), and M6 (•) dendrimeric peptides for mouse macrophage cell line J774.A1 (A) and human HaCaT keratinocytes (B). Cell viability was measured by a MTT colorimetric assay. Data points represent means of three replicates.
The hemolytic activity of M4, M5, and M6 was also evaluated. Hemolysis of fresh human erythrocytes was determined at peptide concentrations ranging from 1 to 120 μg/ml. At concentrations of 120 μg/ml, all dendrimeric peptides showed very poor hemolytic activity (<1%) after an incubation of 2 h and slightly higher levels of activity (around 7 to 8%) after an incubation of 24 h (not shown).
Mechanism of action.
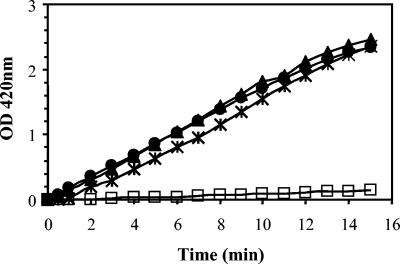
The permeabilization assays showed that peptides M4, M5, and M6 permeabilize the bacterial inner membrane, unmasking cytoplasmic β-galactosidase in ML-35 E. coli permease-negative mutant (Fig. 5). The activity of dendrimeric peptides against the inner membrane was evaluated at concentrations of 16, 32, and 64 μg/ml. All dendrimeric peptides permeabilized bacterial inner membrane at 16 μg/ml. Permeabilization occurred after a lag of <1 min, and the rate of permeabilization depended on peptide concentrations (not shown).
FIG. 5.
Kinetics of the membrane permeabilization of ML-35 E. coli by M4 (*), M5 (▴), M6 (•), and untreated cells (□). Bacteria were treated with 16 μg of dendrimeric peptides/ml.
Improvement in M6 peptide activity.
To gain further information on sequence-function correlations of antimicrobial peptides, dendrimeric mutants of the M6 sequence (QKKIRVRLSA) were synthesized and tested for their ability to kill E. coli ATCC 25922, P. aeruginosa ATCC 27853, and S. aureus ATCC 25922, in comparison with M6 (Table 2).
TABLE 2.
MICs of alanine scanning and Arg-to-Lys substitution in M6
| Peptide | Amino acid sequencea | MIC (μg/ml) of:
|
||
|---|---|---|---|---|
| E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | S. aureus ATCC 29213 | ||
| M6 | QKKIRVRLSA | 4 | 4 | >64 |
| M31 | AKKIRVRLSA | 16 | 16 | >64 |
| M32 (=M4) | QAKIRVRLSA | 128 | 32 | >64 |
| M33 | QKAIRVRLSA | 8 | 8 | >64 |
| M34 | QKKARVRLSA | >64 | 64 | >64 |
| M35 | QKKIAVRLSA | 32 | 8 | >64 |
| M36 | QKKIRARLSA | >64 | >64 | >64 |
| M37 | QKKIRVALSA | 16 | 16 | >64 |
| M38 | QKKIRVRASA | >64 | >64 | >64 |
| M39 | QKKIRVRLAA | 16 | 16 | >64 |
| M28 | QRKIRVRLSA | 2 | 4 | >64 |
| M29 | QKRIRVRLSA | 32 | 32 | >64 |
| M30 | QRRIRVRLSA | 16 | 64 | >64 |
The replaced Ala and Arg residues are in boldface type.
The first set of mutant peptides (M31 to M39), synthesized by alanine mutagenesis, showed lower levels of activity against E. coli and P. aeruginosa than M6, and activity against S. aureus did not improve (Table 2). Interestingly, the substitutions that most affected the antimicrobial activity (M34, M36, and M38) (Table 2) were replacements of the hydrophobic residues Val and Leu or of the polar residue Ser with Ala.
Furthermore, since recent studies reported the effect of different basic residues on antibacterial peptide activity and cell selectivity (28, 31), we also substituted lysines in M6 sequence with arginines and synthesized the following peptides: QKRIRVRLSA (M28), QRKIRVRLSA (M29), and QRRIRVRLSA (M30). The M28 peptide showed a slight reduction in MIC for E. coli with respect to M6, whereas its activity against the other bacteria was unmodified. Substitution of Lys at position 3 (M29) and substitution of both Lys (M30) worsened antimicrobial activity (Table 2).
DISCUSSION
The development of synthetic and biological combinatorial libraries is a major advance in research into new lead compounds for drug development. Although their use for the identification of novel antimicrobial agents requires further investigation and implementation, the present study, together with previous reports (2, 3), indicates that combinatorial techniques can play an important role in this area.
Our work is based on traditional selections of a large combinatorial phage peptide library panned against whole E. coli cells and the isolation and modification of a specific sequence (QEKIRVRLSA), which showed antimicrobial activity once synthesized as a soluble peptide. As long as the sequence was displayed on the phage particles, it bound E. coli cells but did not affect their viability. The possibility that antimicrobial peptides can be selected by phage libraries and amplified in E. coli cells without affecting bacterial viability during the selection phases is extremely promising for the discovery of new peptide sequences with antimicrobial activity. Peptides seem to be unable to accomplish antimicrobial activity as long as they are linked to the phage particle by fusion with the phage pIII protein. This may be due to interference by the phage particle with the carpeting mechanism on the bacterial membrane or with peptide entrance into the cell cytoplasm. Moreover, it might be related to the low concentration of peptides on phages, which might not be high enough to kill bacteria.
The tetrabranched form (MAP) of QEKIRVRLSA (peptide M1) was found to be more effective than the linear homolog when tested on TG1 E. coli cells. We previously demonstrated that branched peptides are much more active in vivo than corresponding monomeric sequences (15), due to their high resistance to proteases and peptidases of biological fluids (6). The higher antimicrobial activity of MAPs with respect to monomeric peptides had already been described for short peptides (27) and may be related to the multimeric nature of the molecule. Multiple peptide copies in very close proximity not only may allow multivalent binding but also may generate a higher local concentration of the antimicrobial peptide, thus increasing the destabilizing effect on the bacterial membrane.
In this study, the M1 peptide was unstable in solution. The original peptide sequence was consequently modified to produce the following peptides synthesized either as monomers or as tetrabranched MAPs: QAKIRVRLSA, KIRVRLSA, and QKKIRVRLSA. These peptides showed high stability to prolonged storage and, particularly when synthesized in MAP4 form, also a better antimicrobial activity when tested with TG1 E. coli cells (Fig. 1B).
The best antimicrobrial peptides known in the literature kill susceptible bacteria in vitro, including certain antibiotic-resistant strains, with MICs ranging from 1 to 8 μg/ml (12). Our dendrimeric peptides M4, M5, and M6 showed antimicrobial activity against a panel of gram-negative bacteria. In particular, M6 was the most effective against the gram-negative bacteria tested, with a MIC of 8 to 4 μg/ml for E. coli ATCC 25922, P. aeruginosa ATCC 27853, and a clinical isolate of K. pneumoniae. Moreover, the MBC/MIC ratio of 1 indicated bactericidal activity against E. coli ATCC 25922 and P. aeruginosa ATCC 27853 (18).
The lack of effect of M4, M5, and M6 on gram-positive S. aureus was probably due to the different structure and composition of the membrane with respect to gram negative bacteria.
The very low cytotoxicity of the peptides for keratinocytes, even after prolonged exposure, makes these molecules good candidates for topical use. We tested the cytotoxicity of M4, M5, and M6 on HaCaT immortalized keratinocytes (5), because they closely resemble normal keratinocytes in growth and differentiation (8). On the contrary, a significantly higher cytotoxicity was observed for J774.A1 murine macrophages tested in the same way. Cytotoxic effects were evident at concentrations of around 0.5 to 1 mg/ml, whereas at lower concentrations the cytotoxic response was quite similar to that measured in HaCaT cells. The difference in toxicity between keratinocytes and macrophages also relies on the phagocytic ability of the latter, which could improve internalization of the peptide by these cells. Nonetheless, since peptide MIC and peptide concentrations inducing toxicity in macrophages differ by at least 1 order of magnitude, these data suggest that the MAP therapeutic index for the treatment of bacterial infections could be favorable. In addition, the general lack of hemolysis of human erythrocytes, even at concentrations of 120 μg/ml and after prolonged incubation (24 h) is encouraging for in vivo use of these compounds.
Inner membrane permeabilization by M4, M5, and M6, as assayed by unmasking of cytoplasm β-galactosidase, occurred with minimal lag. Although the exact mechanism of action of the peptides on bacteria is unclear, the evidence reported here is consistent with the “carpeting” model of Shai (23, 24). We also presume that our dendrimeric peptides act by coating the membrane until they reach a trigger concentration, at which they collapse inward, disrupting the lipid bilayer and behaving like detergents.
Recent studies have investigated the effect of different basic residues on peptide antibacterial activity and cell selectivity (28, 31). The primary amine of Lys and the guanidinium group of Arg seem to interact differently with phospholipids (28). We found that replacement of Lys in position 3 by Arg (M28) improved antimicrobial activity against E. coli ATCC 25922, whereas replacement of Lys in position 2 (M29) and replacement of Lys in positions 2 and 3 (M30) decreased peptide activity. Peptides derived from the alanine scanning of M6 all had lower levels of antimicrobial activity. These findings suggest that the activity of M6 is determined by a combination of factors such as sequence, hydrophobicity, and position of cationic residue.
In conclusion, these studies demonstrate that peptides with antimicrobial activity can be selected from random phage display libraries. Synthesis of antimicrobial peptides in tetrabranched form increases peptide activity, both by decreasing peptide MIC and MBC and by dramatically increasing peptide stability to proteolysis by biological fluids.
The tetrabranched peptides described here (M4, M5, M6, and M28) are potent, protease-stable antimicrobial agents with in vitro activity against bacteria of clinical importance. Due to their low MICs, low cytotoxcity, and high stability, these novel antimicrobial peptides seem promising candidates for the clinical development of new antibacterial drugs.
Acknowledgments
This work was supported by the Italian Ministry of Education, University and Research (PRIN2002, 058925_005 to L.B.).
Special thanks go to Silvia Scali for peptide synthesis.
REFERENCES
- 1.Adessi, C., and C. Soto. 2002. Converting a peptide into a drug: strategies to improve stability and bioavailability. Curr. Med. Chem. 9:963-978. [DOI] [PubMed] [Google Scholar]
- 2.Blondelle, S. E., E. Perez-Paya, and R. A. Houghten. 1996. Synthetic combinatorial libraries: novel discovery strategy for identification of antimicrobial agents. Antimicrob. Agents Chemother. 40:1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondelle, S. E., E. Takahashi, R. A. Houghten, and E. Perez-Paya. 1996. Rapid identification of compounds having enhanced antimicrobial activity by using conformationally defined combinatorial libraries. Biochem. J. 313:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 5.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinisation in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracci, L., C. Falciani, B. Lelli, L. Lozzi, Y. Runci, A. Pini, M. G. De Montis, A. Tagliamonte, and P. Neri. 2003. Synthetic peptides in the form of dendrimers become resistant to protease activity. J. Biol. Chem. 278:46590-46595. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw, J. P. 2003. Cationic antimicrobial peptides: issues for potential clinical use. BioDrugs 17:233-240. [DOI] [PubMed] [Google Scholar]
- 8.Breitkreutz, D., P. Boukamp, C. M. Ryle, H. J. Stark, D. R. Roop, and N. E. Fusenig. 1991. Epidermal morphogenesis and keratin expression in c-Ha-ras-transfected tumorigenic clones of the human HaCaT cell line. Cancer Res. 51:4402-4409. [PubMed] [Google Scholar]
- 9.Epand, R. M., and H. J. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11-28. [DOI] [PubMed] [Google Scholar]
- 10.Goodman, M., C. Zapf, and Y. Rew. 2001. New reagents, reactions, and peptidomimetics for drug design. Biopolymers 60:229-245. [DOI] [PubMed] [Google Scholar]
- 11.Hancock, R. E. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E., and D. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E., and A. Patrzykat. 2002. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disord. 2:79-83. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, R. E., and A. Rozek. 2002. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206:143-149. [DOI] [PubMed] [Google Scholar]
- 15.Lozzi, L., B. Lelli, Y. Runci, S. Scali, A. Bernini, C. Falciani, A. Pini, N. Niccolai, P. Neri, and L. Bracci. 2003. Rational design and molecular diversity for the construction of anti-alpha-bungarotoxin antidotes with high affinity and in vivo efficiency. Chem. Biol. 10:411-417. [DOI] [PubMed] [Google Scholar]
- 16.Marcotte, I., K. L. Wegener, Y. H. Lam, B. C. Chia, M. R. de Planque, J. H. Bowie, M. Auger, and F. Separovic. 2003. Interaction of antimicrobial peptides from Australian amphibians with lipid membranes. Chem. Phys. Lipids 122:107-120. [DOI] [PubMed] [Google Scholar]
- 17.Mayo, K. H., J. Haseman, H. C. Young, and J. W. Mayo. 2000. Structure-function relationships in novel peptide dodecamers with broad-spectrum bactericidal and endotoxin-neutralizing activities. Biochem. J. 349: Pt. 3:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents. Approved guideline M26-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 19.Parpart, A. K., P. B. Lorenz, E. R. Parpart, J. R. Gregg, and A. M. Chase. 1947. The osmotic resistance (fragility) of human red cells. J. Clin. Investig. 26:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pini, A., F. Viti, A. Santucci, B. Carnemolla, L. Zardi, P. Neri, and D. Neri. 1998. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J. Biol. Chem. 273:21769-21776. [DOI] [PubMed] [Google Scholar]
- 21.Posnett, D. N., H. McGrath, and J. P. Tam. 1988. A novel method for producing anti-peptide antibodies. Production of site-specific antibodies to the T cell antigen receptor beta-chain. J. Biol. Chem. 263:1719-1725. [PubMed] [Google Scholar]
- 22.Scott, J. K., and G. P. Smith. 1990. Searching for peptide ligands with an epitope library. Science 249:386-390. [DOI] [PubMed] [Google Scholar]
- 23.Shai, Y. 1995. Molecular recognition between membrane-spanning polypeptides. Trends Biochem. Sci. 20:460-464. [DOI] [PubMed] [Google Scholar]
- 24.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 25.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 26.Tam, J. P. 1988. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam, J. P., Y. A. Lu, and J. L. Yang. 2002. Antimicrobial dendrimeric peptides. Eur. J. Biochem. 269:923-932. [DOI] [PubMed] [Google Scholar]
- 28.Tokunaga, Y., T. Niidome, T. Hatakeyama, and H. Aoyagi. 2001. Antibacterial activity of bactenecin 5 fragments and their interaction with phospholipid membranes. J. Pept. Sci. 7:297-304. [DOI] [PubMed] [Google Scholar]
- 29.Wenzel, R. P., and M. B. Edmond. 2000. Managing antibiotic resistance. N. Engl. J. Med. 343:1961-1963. [DOI] [PubMed] [Google Scholar]
- 30.Wu, M., E. Maier, R. Benz, and R. E. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235-7242. [DOI] [PubMed] [Google Scholar]
- 31.Yang, S. T., S. Y. Shin, C. W. Lee, Y. C. Kim, K. S. Hahm, and J. I. Kim. 2003. Selective cytotoxicity following Arg-to-Lys substitution in tritrpticin adopting a unique amphipathic turn structure. FEBS Lett. 540:229-233. [DOI] [PubMed] [Google Scholar]
- 32.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]