Abstract
Sex-specific regulation of gene expression is the most plausible way for generating sexually differentiated phenotypes from an essentially shared genome. However, since genetic material is shared, sex-specific selection in one sex can have an indirect response in the other sex. From a gene expression perspective, this tethered response can move one sex away from their wild-type expression state and potentially impact many gene regulatory networks. Here, using experimental evolution in the model nematode Caenorhabditis elegans, we explore the coupling of direct sexual selection on males with the transcriptomic response in males and females over microevolutionary timescales to uncover the extent to which postinsemination reproductive traits share a genetic basis between the sexes. We find that differential gene expression evolved in a sex-specific manner in males, while in females, indirect selection causes an evolved response. Almost all differentially expressed genes were downregulated in both evolved males and females. Moreover, 97% of significantly differentially expressed genes in males and 69% of significantly differentially expressed genes in females have wild-type female-biased expression profile. Changes in gene expression profiles were likely driven through trans-acting pathways that are shared between the sexes. We found no evidence that the core dosage compensation machinery was impacted by experimental evolution. Together, these data suggest a defeminization of the male transcriptome and masculinization of the female transcriptome driven by direct selection on male sperm competitive ability. Our results indicate that on short evolutionary timescales, sexual selection can generate putative sexual conflict in expression space.
Keywords: sexual conflict, transcriptomics, C elegans
Significance.
The evolution of transcriptional sexual dimorphism is critical for generating sexual differentiation, and yet disentangling the effects of direct, sex-specific selection and indirect selection remains challenging. Here, we use expression analyses of females and males from experimental evolution populations evolved under strong postmating sexual selection to show that direct selection on males creates a male-like transcriptome in both sexes due to correlated effects. This work highlights how sexual selection can lead to sexual conflict in expression space over short timescales.
Introduction
Sexual dimorphism, ubiquitous across multicellular organisms, is rooted in the seeming conundrum that the sexes can be dramatically different yet share largely the same genome (Wyman et al. 2013; Kasimatis et al. 2017). On a functional level, this means that gene expression must be tightly regulated in a sex-specific manner (Grath and Parsch 2016; Mank 2017). On an evolutionary scale, it also means that selection acting differentially on one sex can have an immediate correlated response to selection on attributes of the other sex (Lande and Arnold 1983, 1985). This correlation underpins the classic Fisherian model of runaway sexual selection, as direct selection acting on a male display trait generates self-reinforcing indirect selection on female preference if they share some form of joint inheritance, either through pleiotropy or linkage (Fisher 1930; Lande 1981; Kirkpatrick 1982). Indirect selection can in principle generate a correlated evolutionary response on any number of female traits, however, particularly on polygenic traits for which alleles are more likely to be pleiotropic (Turelli 1985). For example, males of Pteridophora alberti, the King of Saxony bird-of-paradise, have highly elongated feathers equal to several body lengths on either side of their heads. Curiously, females display similar, but much smaller, feathers on their heads, possibly as residual responses to sexual selection in males (Darwin 1871; Cooper and Forshaw 1977). Much like these birds, opposite sex perturbation would be expected to be fairly short-lived on an evolutionary timescale, as natural selection against misexpression in the wrong sex leads to heightened sexual dimorphism (Lande 1980). The degree to which mating traits have additive genetic covariance, then, determines the opportunity for sexual conflict (Gavrilets et al. 2001; Arnqvist and Rowe 2005). While the evolution of sexual dimorphism has been extensively studied for macrolevel secondary sexual characteristics, such as plumage, territorial behavior, and body size, much less is known at the molecular level, particularly about the coupling between the primary domains of dimorphism: the response to sexual selection at the evolutionary level and the evolution sex-specific gene regulation at the functional level. We address that coupling here with an experimental evolution framework using the nematode Caenorhabditis elegans.
In internally fertilizing species, there is an additional layer to the reproductive process to those classically studied under sexual selection, namely, postinsemination reproductive interactions. Here, the male “trait” of interest can be any cell or protein that affects male fertilization success, while female “preference” is realized in any tissue, cell, or protein that interacts with the male ejaculate to affect female fertilization success. Sexual selection on male reproductive proteins has been inferred through molecular evolution analyses (Begun et al. 2000; Swanson and Vacquier 2002; Clark et al. 2006; Chapman 2011). These studies indicate that many seminal fluid proteins evolve rapidly due to sexually antagonistic coevolution, which in turn implies that direct selection is acting on both female and male postinsemination traits. However, identifying the specific female traits with which these male reproductive proteins interact has proved challenging. In this case, transcriptomic analysis can be useful for identifying widespread changes in gene expression after mating to uncover polygenic responses to selection within and across generations.
Few studies have measured direct selection on male postinsemination traits and the correlated response in females in a similar manner to male display and female preference traits. We previously used experimental evolution to select for increased sperm competitive ability in C. elegans males (Kasimatis et al. 2022). Experimental evolution provides an opportunity to reproducibly isolate directional selection on male characteristics from the potentially correlated responses in females using timescales when the opportunity for sexual conflict—the displacement of the sexes from their sex-specific fitness optima—is likely to be highest. We found that after 30 generations of evolution (i.e. on a microevolutionary timescale), males showed a strong, rapid response to postinsemination selection at both the phenotypic and genomic levels (Kasimatis et al. 2022). Specifically, postinsemination reproductive success increased by greater than 4-fold in the evolved populations. This reproductive fitness increase was underlain by a polygenic response of 82 significance peaks represented by 57 genes, 10 intergenic regions, and 15 putative pseudogenes. Together, we found that when the androdioecious C. elegans mating system is genetically engineered to reflect the ancestral dioecious mating state, there is overwhelming selection to improve maleness which likely reflects a response to the relaxed selection on males during the macroevolutionary transition from dioecy to androdioecy.
Here, we test if this strong, direct selection acting on male sperm competitive ability had an indirect impact on females. Specifically, we quantify the impact of direct postinsemination sexual selection on the transcriptomes of mated ancestral and evolved males and compare how selection impacts the transcriptome relative to the genome. Additionally, we examine the transcriptomes of mated females using a fully factorial crossing design of ancestral and evolved individuals to determine (i) whether strong, direct selection on sperm competition generates a response in females, and if so, (ii) is the female response evolved or plastic, and (iii) is the female response indicative of sexual conflict.
Results
A Strong Inference Framework for Expression Evolution
We selected 4 experimental evolution replicates that evolved under direct selection on sperm competitive ability for 30 generations to test for changes in the transcriptional profile of mated males and mated females. Ancestral and evolved males were crossed with the corresponding generation of ancestral or evolved females to assess how direct selection on males altered the male transcriptome. For females, we used a full factorial crossing design of ancestral and evolved females mated with ancestral and evolved males to determine if the female transcriptome changes and whether those changes were a result of selection (direct or indirect) or female plasticity. Populations of individuals for each cross were mated for 48 h after which individuals of the focal sex were isolated for transcriptomic analysis. The ancestorF-ancestorM cross represents the baseline transcriptomic profile of both males and females after mating before experimental evolution occurred. The evolvedF-evolvedM cross represents the transcriptomic profile of males and females after 30 generations of evolution. If direct sexual selection on males had no impact on a gene's expression level, then there would not be differential expression between the ancestorF-ancestorM and evolvedF-evolvedM cross for that gene. In males, expression of such genes would not contribute to the phenotype under selection, namely, sperm competitive ability. In females, expression of such genes would have no genetic covariance between females and males.
If instead, in females, a gene is differentially expressed between the ancestorF-ancestorM and evolvedF-evolvedM crosses, then females are responding to the sexual selection exerted on males. If the female transcriptional changes are a plastic response to increased sperm competitiveness, then male generation should be a major predictor; that is, females of the evolvedF-ancestorM cross should have expression profiles similar to the ancestorF-ancestorM cross and females of the ancestorF-evolvedM should have expression profiles similar to the evolvedF-evolvedM cross. Alternatively, if female transcriptional changes are a result of either direct selection in females induced by sperm competitiveness or indirect selection generated by genetic covariation between the sexes, then the expectation is that ancestral females will have similar expression profiles and evolved females will have similar expression profiles, regardless of male generation.
Direct Selection on Males Repeatably Downregulates the Evolved Male Transcriptome
We analyzed changes in male gene expression for each experimental evolution replicate separately (supplementary figs. S1 and S2, Supplementary Material online). Principal component analysis (PCA) was performed to compare samples based on male generation (i.e. G0 ancestor or G31 evolved). In each replicate, the first principal component partitioned samples by male generation and explained 68% to 82% of the total variance in gene expression (supplementary fig. S1, Supplementary Material online). Thus, there was a strong effect of direct selection on the male transcriptome. Differential gene expression analysis showed an overall signature of downregulation with 50% (Replicate B2) to 91% (Replicate A3) of genes being downregulated in evolved males. However, the degree of overlap between significantly differentially expressed genes (DEs) varied greatly among replicates (supplementary fig. S3a, Supplementary Material online). Ninety-eight percent of DEs in Replicate A3 were significant in at least one other replicate, while only 60% of DEs in Replicate A2 were shared with at least one other replicate. Replicate A2 also had the most DEs (n = 2,181) and showed the strongest increase in postmating reproductive success (Kasimatis et al. 2022), indicating replicate-specific variation in the transcriptional response.
Differential evolution between experimental evolution replicates is not surprising and could be caused by multiple processes, including selection acting on different segregating genetic variants and genetic drift. To focus on the consistent and repeatable gene expression changes that are most likely linked with the phenotype under selection, we combined all experimental evolution replicates. The final combined data set in males consisted of 3 independent ancestorF-ancestorM crosses and 12 independent evolvedF-evolvedM crosses.
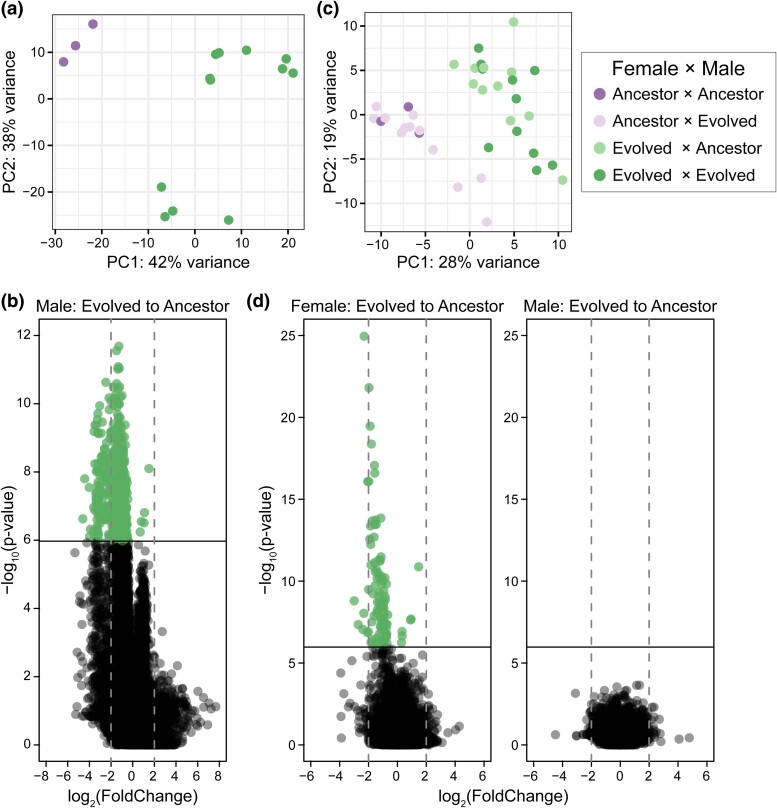
PCA again clustered samples by male generation, such that all ancestral males clustered and all evolved males clustered (Fig. 1a). The first principal component explained 42% of the variation in total gene expression and separated males by generation. The combined analysis, therefore, supports the individual replicate analysis and shows a strong effect of direct selection on the male transcriptome. We analyzed differential gene expression using a generalized linear model framework using male generation and correcting for average differences among evolutionary replicates. Differential expression analysis showed 589 genes to be significantly differentially expressed based on male generation of which 125 had at least a 2-fold change in expression (Fig. 1b; supplementary file S1, Supplementary Material online). All but 18 DEs were represented in individual replicate analyses, which indicates that the transcriptional changes identified are likely due to selection (supplementary fig. S3b, Supplementary Material online). Replicate did not contribute to significant differential gene expression (supplementary file S1, Supplementary Material online).
Fig. 1.
Differential gene expression is partitioned by ancestral (purple) and evolved (green) individuals. a) PCA of the variance stabilized data set partitions male generation along PC1. b) Differential gene expression was analyzed based on male generation and experimental evolution replicate. Only the male term contributed to gene expression changes (supplementary file S1, Supplementary Material online). Genes with a significant change in expression from ancestral to evolved males (shown in green) were determined using a Bonferroni cutoff. Positive expression changes correspond to upregulation in evolved males, while negative expression changes correspond to downregulation in evolved males. c) PCA of the variance stabilized data set partitions female generation along PC1. d) Differential gene expression was analyzed based on female generation, male generation, and experimental evolution replicate. Only the female term contributed to gene expression changes. Genes with a significant change in expression from ancestral to evolved females (shown in green) were determined using a Bonferroni cutoff. Positive expression changes correspond to upregulation in evolved females, while negative expression changes correspond to downregulation in evolved females.
Only 5 genes were significantly upregulated (mean fold increase = 1.0) in evolved males relative to ancestral males. They are located on autosomes and do not have ontology terms that clearly connect them to reproduction. The other 584 genes (99%) were downregulated (mean fold decrease = 1.5) in evolved males relative to ancestral males. The widespread signature of downregulation is not an artifact of biased count distribution (supplementary fig. S4a, Supplementary Material online). Rather, the genes which are highly upregulated in evolved males tended to occur in specific experimental evolution replicates, highlighting the importance of multiple levels of replication when identifying consistent patterns of selection. To determine if single nucleotide polymorphisms (SNPs) in the coding region or the cis-regulatory region of these genes were responsible for the changes in expression, we examined allele frequencies in our previously published genomic mapping data for these populations (Kasimatis et al. 2022). None of the DEs had significant allele frequency changes in the coding region. Two genes (apn-1 and apa-2) had SNPs in the cis-coding region; however, none of these SNPs showed a significant allele frequency change over the course of experimental evolution (supplementary file S2, Supplementary Material online). Together, these data indicate that direct selection on males is downregulating the male transcriptome through trans-acting pathways.
To assess the functional pathways in which these downregulated genes were involved, we examined their associated Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. GO enrichment analysis identified 20 ontology terms that were significantly enriched, many of which were related to some form of binding: DNA binding (n = 4), RNA binding (n = 2), and protein binding (n = 1) (supplementary file S3, Supplementary Material online). Eight KEGG pathways were significantly enriched, including 4 in the replication and repair class, 3 in the metabolism class, and 1 in the endocrine system class (supplementary file S3, Supplementary Material online).
Female Gene Expression Changes Indicate an Evolved Response to Selection Rather than Plasticity
First, to assess variation in the evolutionary response, we analyzed changes in female expression for each experimental evolution replicate separately. PCA clustered females by generation in each replicate (supplementary fig. S5, Supplementary Material online). The first principal component explained 30% to 46% of the total variance in gene expressed based on female generation alone. Male generation did not contribute to variance partitioning in the first 2 principal components. Replicated B3 had 2 outlier transcriptome replicates, which were censored in the downstream analyses (supplementary fig. S5, Supplementary Material online). Differential expression analysis supported that female generation alone determined expression changes as the male term did not identify DEs (supplementary fig. S6, Supplementary Material online). Female DEs recapitulated the male signature of downregulation with 92% to 96% of DEs being downregulated in evolved females. The majority of DEs overlapped with at least one other experimental evolution replicate, suggesting less replicate-specific evolution in females than in males (supplementary fig. S3c, Supplementary Material online).
Although the separate analysis of each replicate helps to highlight variance in evolution response, to more formally address evolutionary changes in the transcriptome that are repeatable across replicates, we used a combined analysis after correcting for among-replicate variance. The final censored, combined data set in females therefore consisted of 3 independent ancestorF-ancestorM crosses, 12 independent ancestorF-evolvedM crosses, 11 independent evolvedF-ancestorM crosses, and 11 independent evolvedF-evolvedM crosses. PCA of the full data set again clustered samples by female generation along the first principal component, which explained 28% of the total variance in gene expression levels (Fig. 1c). Male generation did not contribute to variance partitioning in the first 2 principal components, which captured 46% of the total variance. Thus, the combined PCA supports the individual replicate analyses and indicates that the female response is evolved rather than a plastic response based on male generation.
We formally examined the potential differences of ancestral and evolved males on the female transcriptional response, as well as the influence of evolutionary response to heightened sperm competition of the females themselves. Differential expression for individual genes was analyzed using a generalized linear model using female and male generation, correcting for average differences among evolutionary replicates. Neither male generation nor replicate contributed to significant changes in gene expression (Fig. 1d; supplementary file S1, Supplementary Material online). Rather, female generation determined differential gene expression, again demonstrating an evolved expression response (Fig. 1d). One hundred genes showed significant differential gene expression of which 8 genes had at least a 2-fold change in expression (Fig. 1d). Only 6 genes were significantly upregulated in evolved (mean fold increase = 0.7) females relative to ancestral females. Five of these genes are uncharacterized in function. The other 94 DEs were downregulated (mean fold decrease = 1.3) in evolved females relative to ancestral females (Fig. 1d). Again, widespread downregulation is not an artifact of biased count distribution (supplementary fig. S4b, Supplementary Material online). None of the significantly differentially expressed genes had significant allele frequency changes in the coding region or the cis-regulatory region (supplementary file S1, Supplementary Material online). Thus, changes in gene expression were again likely regulated through trans-acting pathways. No GO terms or KEGG pathways were significantly enriched in female DEs.
Indirect Selection on Females Drives Transcriptome Evolution
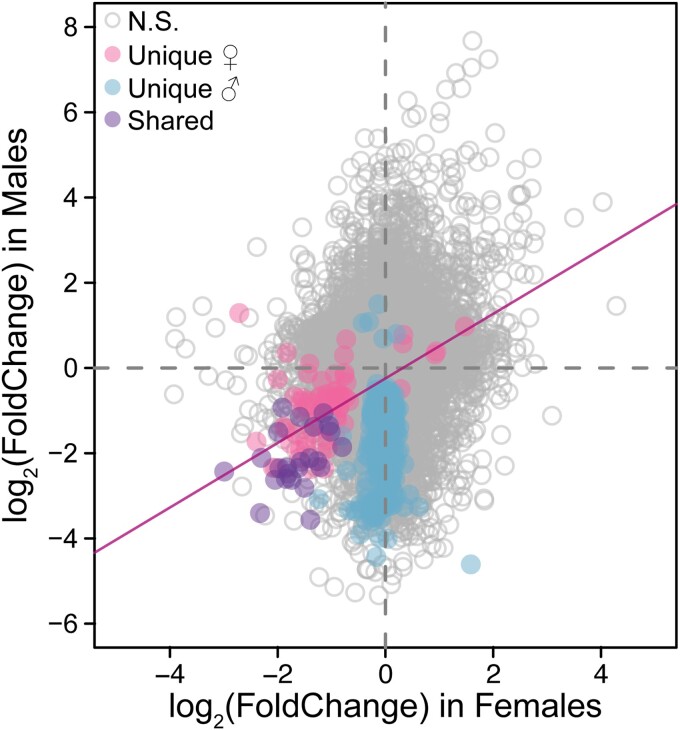
Since the female transcriptome response is evolved, we next examined whether the changes in female expression were due to direct selection in response to increased sperm competitiveness or indirect selection as a correlated response to the direct selection acting on males. To this end, we examined at the correlation in differential gene expression changes between the sexes. Genes that were only significantly differentially expressed in males (i.e. unique male DEs) were not correlated with female changes in gene expressed (n = 565, P = 0.27, adjusted R2 = 0.0004; Fig. 2). Rather, these genes changed expression in males alone. This widespread male-specific transcriptomic response indicates that direct selection on males predominantly changed the male transcriptome in a sex-specific manner.
Fig. 2.
A comparison of changes in gene expression levels between females and males. Positive expression changes correspond to upregulation in evolved females/males, while negative expression changes correspond to downregulation in evolved females/males. Genes that are not significantly differentially expressed in either sex are shown in gray. Genes that are significantly differentially expressed in males but not in females (i.e. unique male) are shown in blue. Genes that are significantly differentially expressed in females but not in males (i.e. unique female) are shown in pink. Genes that are significantly differentially expressed in both sexes (i.e. shared) are shown in purple. There is a significant relationship between the change in gene expression in females and males for all female DEs (F1,98 = 42.16, P < 0.001, adjusted R2 = 0.29).
Genes that were only significantly differentially expressed in females (i.e. unique female DEs) were, however, significantly correlated with corresponding changes in male expression (n = 76, F4,74 = 21.3, P < 0.001, adjusted R2 = 0.21). Therefore, these genes are largely being downregulated in females and males, though not significantly so in males (Fig. 2). This correlation holds when all female DEs—unique and shared between the sexes (n = 100)—are included (F1,98 = 42.16, P < 0.001, adjusted R2 = 0.29). Together, these data support that indirect selection on females was driving changes in the female transcriptome as female expression changes were not independent from those in male, while male expression changes occurred in a sex-specific manner.
Correlated Selection Defeminizes the Transcriptome of Males and Females
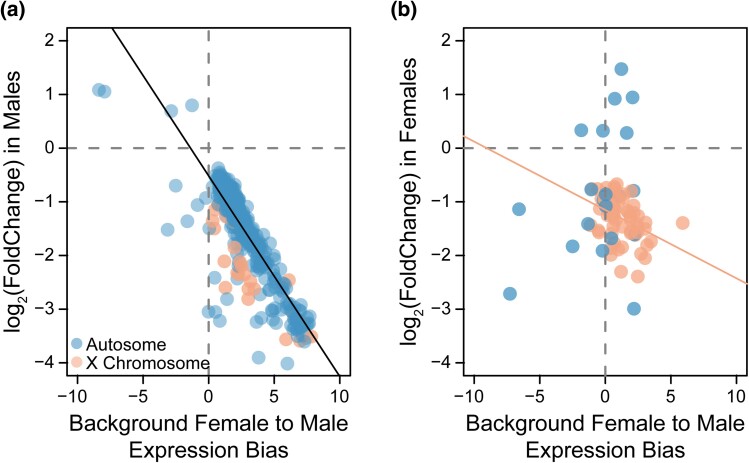
We hypothesized that selection was driving this widespread downregulation via either female-biased or nonsex-biased genes. In either case, the transcriptomic effect would be a less female-like and a more male-like transcriptome to support the increase in male reproductive fitness. To test this hypothesis, we examined the relationship between changes in gene expression after experimental evolution and sex-biased expression in wild-type females and males (Albritton et al. 2014). The majority (n = 563) of male DEs showed some degree of female-biased expression in wild-type individuals (Fig. 3a; χ2 = 837.3, df = 1, P < 0.001). A strong negative correlation exists between wild-type sex-bias and gene expression changes in males, such that more female-biased genes in wild type were more strongly downregulated after experimental evolution regardless of whether the gene was located on an autosome or the X chromosome (F1,568 = 1,562, P < 0.0001, adjusted R2 = 0.73). The reduction in female-bias indicates a defeminization of the male transcriptome.
Fig. 3.
Downregulated genes are female-biased. a) There is a significant negative correlation between wild-type sex-bias in expression and changes in expression after experimental evolution in males. This relationship holds regardless of genomic location. Positive wild-type gene expression values indicate female-biased expression and negative values indicate male-biased expression. Both sex-biased expression (female to male) and experimental evolution fold change are plotted on a log2 scale. b) There is a negative correlation between significant differentially expressed genes located on the X chromosome (shown in coral) and sex-biased expression in females. Only 22 genes were located on autosomes (shown in blue).
Similarly, the majority (n = 69) of female DEs also showed some degree of female-biased expression in wild-type individuals (Fig. 3b). Interestingly, 67 of the downregulated genes (81%; χ2 = 262.7, df = 1, P < 0.001) were located on the X chromosome, of which 57 were female-biased (Fig. 3b; χ2 = 89.8, df = 1, P < 0.001). There is a negative relationship between sex-biased expression and differential gene expression on the X chromosome, such that more female-biased genes had a greater fold decrease in expression (F1,65 = 12.4, P < 0.001, adjusted R2 = 0.15). This relationship suggests a pleiotropic effect in a trans-regulatory pathway impacting regulation of X-linked genes. No relationship exists between autosomal genes and differential gene expression (P = 0.14), though the low number of autosomal genes likely makes this comparison underpowered. Together, these data support a correlated expression response in females and indicate a skew toward a less female-like transcriptome in evolved females.
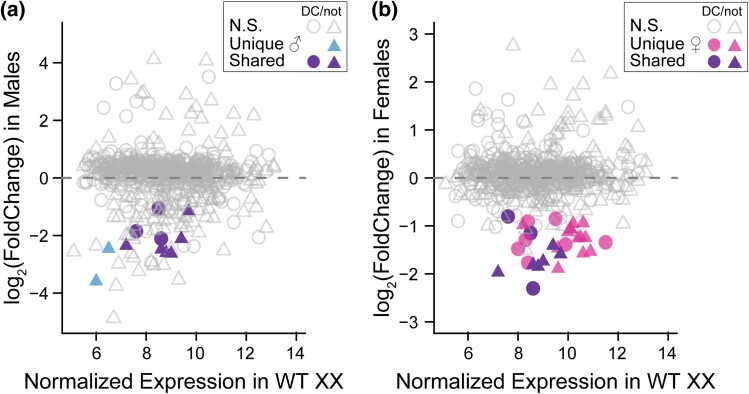
Significantly more DEs are located on the X chromosome in females than in males (χ2 = 319.5, df = 1, P < 0.0001) (supplementary fig. S7, Supplementary Material online). One major trans-regulatory pathway that could explain the correlation between downregulated genes and the X chromosome is dosage compensation. In Caenorhabditis nematodes, hermaphrodites/females have 2 copies of the X chromosome, each of which is partially downregulated to match the dosage of the single X chromosome in males (Meyer 2022). To determine if whole-scale changes in dosage compensation are responsible for the downregulated pattern in evolved females, we correlated differential gene expression with gene expression levels for dosage compensated and nondosage compensated X-linked genes (Jans et al. 2009). Only 31 (nmale = 2, nfemale = 20, nshared = 9) of the significant genes located on the X chromosome were present in both data sets (Fig. 4). Of these, 7 were dosage compensated in wild-type hermaphrodites/females (XX individuals) and 3 were dosage compensated in both sexes. These genes were further downregulated in evolved individuals, particularly in females, potentially suggesting an expression level lower than males. Nineteen genes in total were not dosage compensated in wild-type hermaphrodites/females, but were downregulated in evolved females by on average 1.4-fold. Similarly, the mean fold decrease in wild-type expression in males (XO individuals) for these same genes is 1.9-fold. Eight genes in total were not dosage compensated in wild-type males and were downregulated in evolved males by an average 2.4-fold. Thus, these results suggest that even genes that appear to be targets of dosage compensation can evolve away from their basal dosage compensated state.
Fig. 4.
Downregulation of X-linked genes is impacting dosage compensation. a) The relationship between normalized expression of dosage compensated (circles) and not dosage compensated (triangles) X-linked genes in wild-type hermaphrodites/females (XX individuals) versus differential gene expression in males. Genes with significant differential gene expression in males only are shown in blue, and shared genes are shown in purple. Both axes are plotted on a log2 scale. b) The relationship between normalized expression of dosage compensated (circles) and not dosage compensated (triangles) X-linked genes in wild-type hermaphrodites/females (XX individuals) versus differential gene expression in females. Genes with significant differential gene expression in females only are shown in pink, and shared genes are shown in purple.
The core dosage compensation genes sdc-2, sdc-3, dpy-27, and dpy-30 did not have significant changes in gene expression after experimental evolution (supplementary file S1, Supplementary Material online) nor were there significant allele frequency changes at or near these genes (Kasimatis et al. 2022). Additionally, there were no significant allele frequency changes at the rex or dox binding sites, indicating that the dosage compensation machinery itself is likely not the cause of widespread downregulation in evolved females. Therefore, alternative pathways must be involved in this defeminization of the male and female transcriptomes resulting from direct sexual selection acting on males.
Discussion
Sexual selection acts in a sex-specific manner yet can have an indirect evolutionary response in the other sex due to genetic associations created by their sharing the same genome (apart from heterogametic sex chromosomes, when present). When sexual selection acts on one sex, the other sex can be pulled away from their sex-specific optimum as a correlated response to selection, generating conflict between the sexes. Here, we explored how the transcriptome of newly evolved males responded to direct sexual selection on sperm competitive ability (Kasimatis et al. 2022) and whether this male-specific selection generated indirect selection on females by quantifying the ancestral and evolved transcriptomes of males and females mated with different experimental evolution generations.
Genes with significant expression changes between ancestral and evolved males were almost exclusively downregulated. Moreover, most of these genes are female-biased in wild-type individuals. These results suggest that the evolved male transcriptome moved away from a female-like state. This transcriptomic response supports our previous fitness data (Kasimatis et al. 2022) and indicates that over the course of experimental evolution these newly derived males improved their reproductive ability by becoming more like males under obligate male-female (dioecious) mating. This highlights the first, and fairly ancient, timescale for sexual conflict revealed by our data. The transition from the ancestral dioecious mating system within C. elegans has led to patterns of gene expression that favor hermaphroditic function—which is essentially very female-like—over male-specific function. Selection for increased male function generated in the populations analyzed here (Kasimatis et al. 2022), as well as that observed in other studies (LaMunyon and Ward 1998; Carvalho et al. 2014; Palopoli et al. 2015), leads to a rapid improvement in male reproductive traits such as sperm size and competitive ability. Our results illustrate that these macrophenotypic effects are reflected directly in the transcriptome via a shift in the balance of expression-level conflict away from female-biased function.
Further, as a byproduct of selection for improved male fitness, females displayed widespread concomitant signature of downregulation in the evolved female transcriptome. This represents sexual conflict on the second timescale. Selection on regulatory elements that allow enhanced male performance leads to a short-term correlated response to selection on the female transcriptome as well, making it more male-like. Whether we might expect sex-specific transcriptional regulation to rebalance itself over time depends on the specific fitness impacts of these transcriptional changes on females, which are difficult to assess here. Overall, the results indicate a direct defeminization of the male transcriptome and an indirect masculinization of the female transcriptome generated by direct selection on male fertilization success. Since gene expression levels impact trait function and, in turn, fitness, short-term sexual conflict was likely generated as a byproduct of direct sexual selection in males as female expression was pushed off its global optimum. While not definitive, a more female-like transcriptome is more likely to correlate with benefits on female fitness than a more male-like transcriptome. This give and take is likely to be an important feature structuring transcriptional regulation in many male-female species.
The factorial female mating design used here allows for changes in gene expression to be categorized as a plastic response to increased sperm competitiveness in males or an evolved response in the females themselves. If females were predisposed to increased sperm interactions, then male generation should have been a major contributor to gene expression changes. Such a scenario is analogous to a preexisting bias model of sexual selection (Ryan et al. 1990) and can lead to direct selection on female preference/resistance (Gavrilets et al. 2001; Arnqvist and Rowe 2005). However, we found no evidence that male evolutionary history impacted differential gene expression. Rather, all expression changes were determined based on whether a female was from the ancestral or evolved populations, indicating that differential gene expression was a result of indirect selection on females during experimental evolution. Under this scenario, optimal gene expression differs between the sexes, such that direct selection optimizing the male transcriptome generates an indirect response in females due to a between-sex genetic covariance in expression (Lande 1980; Lande and Arnold 1985; Barker et al. 2010). For our postinsemination model, selection was directed on sperm competitive ability. Since selection on males generated a polygenic response and the expression changes observed were likely due to trans-acting pathways, it is possible that multiple reproductive traits may be impacted.
A comparison between our previous study (Kasimatis et al. 2022) and the expression changes characterized here identified a nearly complete lack of overlap between allele frequency changes and changes in gene expression patterns. Interestingly, none of the expression changes in X-linked genes can be attributed to changes in their coding or cis-regulatory sequence. One potential reason for this discrepancy is that the genic changes were altering expression of upstream transcription factors that are expressed earlier in development. Kasimatis et al. (2022) identified genomic changes in the coding sequence of or nearby intergenic region of 4 transcription factors. Of particular note is the autosomal transcription factor sea-1, which is involved in sex determination and dosage compensation through hypoactivation of the X chromosome (Harris et al. 2020). Genomic analysis identified 7 SNPs with a significant allele frequency change located within a 30 base pair region of intron 2 (Kasimatis et al. 2022). It is possible that this region changes regulation of sea-1 itself or alters splicing in a way that increases the hypoactivation function of sea-1 and thus is responsible for the widespread downregulation of X-linked genes.
The majority of downregulated genes in females were located on the X chromosome. Because males are the heterogametic sex in C. elegans, alleles affecting gene expression on the X chromosome are directly exposed to selection without any potential shielding of an alternative allele due to dominance, as would occur in heterozygous females. Thus, selection can be more effective on X-bearing genes, especially when there is sex-specific selection (Charlesworth et al. 1987; Mank 2009; Vicoso and Charlesworth 2009). The response we observe here is thus consistent with the dominance theory of the rapid evolution of sex chromosomes and their role in speciation (Charlesworth et al. 1987; Charlesworth 1991).
Despite the preponderance of signal on the X chromosome, we found no evidence that mutations in the core dosage compensation pathway were the cause of this widespread downregulation. Downregulation of both dosage compensated and nondosage compensated genes suggests a potential dysregulation of dosage compensation. These changes further support a less wild-type hermaphrodite/female-like state. Wild-type dosage compensation levels may not represent an expression level that is optimal for both sexes (Mank et al. 2011; Wright and Mank 2012; Gu and Walters 2017). Sexual conflict over dosage compensation creates an obvious additional, but perhaps underappreciated, layer to understanding the evolution of sexual dimorphism across the genotype-phenotype-fitness landscape.
Previous experimental evolution studies suggest that male-limited evolution over tens of generations can decrease female fitness (Rice 1996; Prasad et al. 2007; Pischedda and Chippindale 2017). Conversely, female-limited or reduced sexual selection evolution tends to increase female fitness with little to no effect on male fitness (Holland and Rice 1999; Pitnick et al. 2001a, 2001b). These previous studies therefore suggest that sex-specific selection acting through males may have a higher potential to generate sexual conflict. However, this previous work has been blind to the traits under selection and therefore the genetic architecture underlying the sexual conflict response. Our work to bridge these layers of the genomic, transcriptomic, and fitness responses suggests a Red Queen scenario (Valen 1973; Brockhurst et al. 2014) relating genome evolution and transcriptional dimorphism during postinsemination interactions. Specifically, on microevolutionary timescales, strong sexual selection on males can rapidly improve “maleness” at the genomic and phenotypic levels (Kasimatis et al. 2022) as well as the transcriptomic level, as shown here. In turn, this selection on males pushes females away from their fitness optimum through indirect selection (Rice and Holland 1997; Rice and Chippindale 2001). Females may then respond through direct selection, which creates a macroevolutionary picture of a stable female-like state. Thus, examining the functional layers through which sexual dimorphism arises is critical for understanding the context in which sexual conflict can occur.
Sexual selection drives the evolution of some of the most elaborate phenotypic variation and complex behaviors. We show that sexual selection is just as powerful an evolutionary force on postinsemination traits. In particular, on microevolutionary timescales, females may not have the opportunity to counter the effects of indirect selection. As a consequence, females are being pulled away from their optimal transcriptional state. Given the seemingly high degree of pleiotropy between female and male reproductive networks, sexual conflict may evolve very rapidly in populations experiencing strong sexual selection.
Materials and Methods
Worm Culture and Mating Design
We previously evolved 6 replicated populations of C. elegans pseudofemales (fog-2) and males under enhanced postinsemination competition for 30 generations (Kasimatis et al. 2022). The complete experimental evolution details for the between-strain postinsemination selection only (BS-PO) regime can be found in Kasimatis et al. (2022). Briefly, evolving males mated with females for 24 h after which male sterility was induced to prevent further sperm transfer. Fully fertile competitor males were then added to the population to generation sperm competition. After a 24 h sperm competition period, progeny coming from the evolving males only was collected and propagated to the next generation. Hence, selection was acting on sperm defensive capability and sperm longevity. Here, we chose 4 evolved experimental evolution replicates (A2, A3, B2, B3), which spanned the range of reproductive increase, for transcriptomic analysis.
To generate mated male transcriptomes, we crossed ancestral (strain PX632) females to ancestral (strain PX632) males and evolved (G31 replicates A2, A3, B2, B3) females to evolved (G31 replicates A2, A3, B2, B3) males. The evolved crosses were performed for each experimental evolution replicate. To start a generation, age synchronized larval stage 1 (L1) worms were plated onto a 10 cm NGM-agar plates seeded with OP50 Escherichia coli at 20 °C with a density of 1,000 worms per plate (Brenner 1974; Kenyon 1988). Late larval stage 4 (L4) females (n = 40) and males (n = 40) were isolated onto medium NGM-agar plates (60 mm diameter) seeded with 100 μL OP50 E. coli. Three mating plates were set up for each cross. Mating plates were kept at 20 °C for 48 h. After the 48 h mating period, males were collected and pooled (n = 120) from all 3 mating plates per cross for bulk RNA isolation. Three biological replicate mating assays were conducted for each cross, each coming from an independent age synchronization event (n = 15 total male samples).
To generate mated female transcriptomes, we used a factorial design of ancestral (strain PX632) and evolved (G31 replicates A2, A3, B2, B3) females mated to ancestral (strain PX632) and evolved (G31 replicates A2, A3, B2, B3) males. Crosses were performed for each experimental evolution replicate following the same experimental procedure as males. After the 48 h mating period, females were collected and pooled (n = 120) from all 3 mating plates per cross for bulk RNA isolation. Three biological replicate mating assays were conducted for each cross, each coming from an independent age synchronization event (n = 39 total female samples).
Transcriptome Sequencing, Mapping, and Gene Calling
We performed bulk mRNA sequencing of mated males and females, separately, on day 3 of adulthood. Worms were isolated into 150 µL of S-Basal buffer and then pelleted to ∼20 µL. They were preserved with 250 µL Tri-reagent and flash frozen in liquid nitrogen. The samples went through 10 freeze-thaw cycle before RNA was isolated. RNA was isolated using the KAPA mRNA HyperPrep Kit (KK8580) with Illumina-compatible adapters. Libraries were prepared using the TrueSeq RNA Library Prep kit (Illumina) starting from 100 ng of RNA. 100 bp single reads for males and 100 bp paired-end reads were sequenced on an Illumina HiSeq 4000 at the University of Oregon Genomics and Cell Characterization Core Facility (Eugene, OR).
Reads were trimmed using skewer v0.2.2 (Jiang et al. 2014) to remove low-quality bases and TrueSeq adapters (parameters: -x AGATCGGAAGAGCACACGTCTGAACTCCAGTCA -y AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT -t 12 -l 30 -r 0.01 -d 0.01 -q 10). The trimmed reads were mapped to the C. elegans N2 reference genome (PRJNA13758-WS274) (Harris et al. 2020) through 2 passes using STAR v2.5.3a (Dobin et al. 2013). A table of gene counts was made using HTSeq v0.9.1 (Putri et al. 2022) (parameters: -s no -r pos -i ID -t gene -f) based on C. elegans N2 reference genome “longest isoform” annotations.
Differential Gene Expression Analysis
To compare-and-contrast changes in gene expression over time based on the generation (i.e. G0 or G31) of individuals at the time of mating, we tested for differential gene expression using the package DESeq2 v1.34.0 (Love et al. 2014) in R (R Core Team 2024). DESeq2 uses the generalized linear model framework to estimate the degree to which genes are differentially expressed. We first analyzed each experimental evolution replicate independently. For males, we fit the model: gene counts ∼ maleAnc,Evo. Since we had a full factorial design for mated females, we fit the model: gene counts ∼ femaleAnc,Evo + maleAnc,Evo. In females, experimental evolution replicate B3 had 2 outlier mating replicates (20_S36_L005 and 21_S37_L005), which were subsequently censored from the data set (supplementary fig. S5, Supplementary Material online). We then combined all experimental evolution replicates to capture only the expression changes that were repeatable across experimental evolution and reanalyzed the differential gene expression model. Normalized count data and gene expression fold change values were extracted from the DESeq2 linear models. Principal components were visualized using the DESeq2 function “plotPCA” of the variance stabilized data. Briefly, this function first calculates and identifies the 500 genes with the greatest variance in expression and then calculates a singular value decomposition of this matrix.
Significance values were determined by fitting a generalized linear model in R with generation and replicate as fixed effects. This approach is more robust to variation between experimental evolution replicates and therefore provides a more conservative estimate of significantly differentially expressed genes. Significance was based on a Bonferroni cutoff. These censored, combined data were used for subsequent analyses (supplementary file S1, Supplementary Material online).
We crossed referenced the genes with significant differential expression with the genomic significance peaks identified in Kasimatis et al. (2022). Additionally, we examined whether the cis-regulatory regions of significant differentially expressed genes—defined as 2 kb upstream of the start codon—had any SNPs with a significant change in allele frequency, but none of these met the significance peak threshold defined in Kasimatis et al. (2022) (supplementary file S2, Supplementary Material online).
We tested for enrichment of GO terms and KEGG pathways in significantly differentially expressed genes using the package goseq (Young et al. 2010) in R (R Core Team 2024). goseq performs GO analysis taking transcript length biases into account. Median transcript lengths were calculated using from TxDb.Celegans.UCSC.ce11.ensGene (Team and Maintainer 2019) based on the UCSC Genome Browser C. elegans genome version “ce11”. GO enrichment was calculated using the “Wallenius” method with a 0.05 false-discovery rate cutoff (Benjamini and Hochberg 1995) (supplementary file S3, Supplementary Material online).
Sex-Biased Gene Expression and Dosage Compensation
We compared the change in expression of significant genes with their wild-type expression profiles in C. elegans fog-2 females and males (Albritton et al. 2014) to examine any potential changes in sex-biased expression profiles. The relationship between sex-biased expression and differentiation gene expression in each sex was analyzed by fitting a linear model in R.
The dosage compensation profiles for C. elegans hermaphrodites (XX individuals) and males (XO individuals) were taken from Jans et al. (2009), as were the positions of rex and dox binding sites. We then correlated dosage compensation profiles with all genes in our transcriptomic data set. The positions of rex and dox sites were compared with genomic SNP data from Kasimatis et al. (2022) to determine if any significant allele frequency changes occurred in these regions.
Supplementary Material
Acknowledgments
We thank Ruben Lancaster and Alex Smith for the experimental assistance and Anastasia Teterina for the transcriptome analysis advice. We thank Locke Rowe, Stephen Wright, and 2 anonymous reviewers for their insightful discussion.
Contributor Information
Katja R Kasimatis, Institute of Ecology and Evolution, University of Oregon, Eugene, OR, USA.
John H Willis, Institute of Ecology and Evolution, University of Oregon, Eugene, OR, USA.
Christine A Sedore, Institute of Ecology and Evolution, University of Oregon, Eugene, OR, USA.
Patrick C Phillips, Institute of Ecology and Evolution, University of Oregon, Eugene, OR, USA.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
K.R.K. and P.C.P. devised the project. K.R.K. and C.A.S. performed the crosses. J.H.W. prepared the transcriptomic libraries. K.R.K. analyzed the data. K.R.K. wrote the manuscript with the support of the other authors.
Funding
This research was supported by National Institute for General Medicine grant R35GM131838 to P.C.P.
Data Availability
Raw transcriptomic reads are available at NCBI SRA under accession number PRJNA1013082. Differential expression analysis results for the combined data are in supplementary file S1, Supplementary Material online. Cross-referencing of cis-regulatory SNPs is available in supplementary file S2, Supplementary Material online. GO and KEGG pathway enrichment results are available in supplementary file S3, Supplementary Material online. All scripts are publicly available on the GitHub repository: https://github.com/katjakasimatis/expevol_femaletranscriptomics. The experimental evolution ancestral strain (PX632) and evolved replicates are available from the Phillips Lab upon request.
Literature Cited
- Albritton SE, Kranz AL, Rao P, Kramer M, Dieterich C, Ercan S. Sex-biased gene expression and evolution of the x chromosome in nematodes. Genetics. 2014:197(3):865–883. 10.1534/genetics.114.163311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G, Rowe L. Sexual conflict. New Jersey: Princeton University Press; 2005. [Google Scholar]
- Barker BS, Phillips PC, Arnold SJ. A test of the conjecture that g-matrices are more stable than b-matrices. Evolution. 2010:64(9):2601–2613. 10.1111/j.1558-5646.2010.01023.x. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P, Todd BL, Waldrip-Dail HM, Clark AG. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics. 2000:156(4):1879–1888. 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995:57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974:77(1):71–94. 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GD. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci. 2014:281(1797):20141382. 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Phillips PC, Teotónio H. Hermaphrodite life history and the maintenance of partial selfing in experimental populations of Caenorhabditis elegans. BMC Evol Biol. 2014:14:117. 10.1186/1471-2148-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T. Seminal fluid-mediated fitness traits in Drosophila. Heredity (Edinb). 2011:87(Pt 5):511–521. 10.1046/j.1365-2540.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991:251(4997):1030–1033. 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987:130(1):113–146. 10.1086/284701. [DOI] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006:131(1):11–22. 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Cooper W, Forshaw J. The birds of paradise and bower birds. Massachusetts: Collins; 1977. [Google Scholar]
- Darwin C. The descent of man and selection in relation to sex. London: Murray; 1871. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. New York: Oxford University Press; 1930. [Google Scholar]
- Gavrilets S, Arnqvist G, Friberg U. The evolution of female mate choice by sexual conflict. Proc Biol Sci. 2001:268(1466):531–539. 10.1098/rspb.2000.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grath S, Parsch J. Sex-biased gene expression. Annu Rev Genet. 2016:50(1):29–44. 10.1146/annurev-genet-120215-035429. [DOI] [PubMed] [Google Scholar]
- Gu L, Walters JR. Evolution of sex chromosome dosage compensation in animals: a beautiful theory, undermined by facts and bedeviled by details. Genome Biol Evol. 2017:9(9):2461–2476. 10.1093/gbe/evx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, et al. WormBase: a modern model organism information resource. Nucleic Acids Res. 2020:48(D1):D762–D767. 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland B, Rice WR. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc Natl Acad Sci U S A. 1999:96(9):5083–5088. 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009:23(5):602–618. 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014:15:182. 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis KR, Moerdyk-Schauwecker MJ, Lancaster R, Smith A, Willis JH, Phillips PC. Post-insemination selection dominates pre-insemination selection in driving rapid evolution of male competitive ability. PLoS Genet. 2022:18(2):e1010063. 10.1371/journal.pgen.1010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis KR, Nelson TC, Phillips PC. Genomic signatures of sexual conflict. J Hered. 2017:108(7):780–790. 10.1093/jhered/esx080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The nematode Caenorhabditis elegans. Science. 1988:240(4858):1448–1453. 10.1126/science.3287621. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. Sexual selection and the evolution of female choice. Evolution. 1982:36(1):1–12. 10.2307/2407961. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc Biol Sci. 1998:265(1409):1997–2002. 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980:34(2):292–305. 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci U S A. 1981:78(6):3721–3725. 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983:37(6):1210–1226. 10.2307/2408842. [DOI] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. Evolution of mating preference and sexual dimorphism. J Theor Biol. 1985:117(4):651–664. 10.1016/S0022-5193(85)80245-9. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am Nat. 2009:173(2):141–150. 10.1086/595754. [DOI] [PubMed] [Google Scholar]
- Mank JE. The transcriptional architecture of phenotypic dimorphism. Nat Ecol Evol. 2017:1(1):16. 10.1038/s41559-016-0006. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011:65(8):2133–2144. 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. The X chromosome in C. elegans sex determination and dosage compensation. Curr Opin Genet Dev. 2022:74:101912. 10.1016/j.gde.2022.101912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli MF, Peden C, Woo C, Akiha K, Ary M, Cruze L, Anderson JL, Phillips PC. Natural and experimental evolution of sexual conflict within Caenorhabditis nematodes. BMC Evol Biol. 2015:15:93. 10.1186/s12862-015-0377-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda A, Chippindale AK. Direct benefits of choosing a high-fitness mate can offset the indirect costs associated with intralocus sexual conflict. Evolution. 2017:71(6):1710–1718. 10.1111/evo.13240. [DOI] [PubMed] [Google Scholar]
- Pitnick S, Brown WD, Miller GT. Evolution of female remating behaviour following experimental removal of sexual selection. Proc Biol Sci. 2001a:268(1467):557–563. 10.1098/rspb.2000.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S, Miller GT, Reagan J, Holland B. Males’ evolutionary responses to experimental removal of sexual selection. Proc Biol Sci. 2001b:268(1471):1071–1080. 10.1098/rspb.2001.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad NG, Bedhomme S, Day T, Chippindale AK. An evolutionary cost of separate genders revealed by male-limited evolution. Am Nat. 2007:169(1):29–37. 10.1086/509941. [DOI] [PubMed] [Google Scholar]
- Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022:38(10):2943–2945. 10.1093/bioinformatics/btac166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. Foundation for Statistical Computing. 2024. [accessed 2024 Jun 15]. https://www.R-project.org/.
- Rice WR. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature. 1996:381(6579):232–234. 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- Rice WR, Chippindale AK. Intersexual ontogenetic conflict. J Evol Biol. 2001:14(5):685–693. 10.1046/j.1420-9101.2001.00319.x. [DOI] [Google Scholar]
- Rice WR, Holland B. The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol. 1997:41(1):1–10. 10.1007/s002650050357. [DOI] [Google Scholar]
- Ryan MJ, Fox JH, Wilczynski W, Rand AS. Sexual selection for sensory exploitation in the frog Physalaemus pustulosus. Nature. 1990:343(6253):66–67. 10.1038/343066a0. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Reproductive protein evolution. Annu Rev Ecol Syst. 2002:33(1):161–179. 10.1146/annurev.ecolsys.33.010802.150439. [DOI] [Google Scholar]
- Team B, Maintainer B. TxDb.Celegans.UCSC.ce11.ensGene: Annotation package for TxDb objects. R package version 3.4.6. 2019 [accessed 2024 Mar 1]. 10.18129/b9.bioc.txdb.celegans.ucsc.ce11.ensgene. [DOI]
- Turelli M. Effects of pleiotropy on predictions concerning mutation-selection balance for polygenic traits. Genetics. 1985:111(1):165–195. 10.1093/genetics/111.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen LV. A new evolutionary law. Evol Theory. 1973:1:1–30. [Google Scholar]
- Vicoso B, Charlesworth B. Effective population size and the faster-X effect: an extended model. Evolution. 2009:63(9):2413–2426. 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- Wright AE, Mank JE. Battle of the sexes: conflict over dosage-sensitive genes and the origin of X chromosome inactivation. Proc Natl Acad Sci U S A. 2012:109(14):5144–5145. 10.1073/pnas.1202905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman MJ, Stinchcombe JR, Rowe L. A multivariate view of the evolution of sexual dimorphism. J Evol Biol. 2013:26(10):2070–2080. 10.1111/jeb.12188. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-Seq: accounting for selection bias. Genome Biol. 2010:11(2):R14. 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw transcriptomic reads are available at NCBI SRA under accession number PRJNA1013082. Differential expression analysis results for the combined data are in supplementary file S1, Supplementary Material online. Cross-referencing of cis-regulatory SNPs is available in supplementary file S2, Supplementary Material online. GO and KEGG pathway enrichment results are available in supplementary file S3, Supplementary Material online. All scripts are publicly available on the GitHub repository: https://github.com/katjakasimatis/expevol_femaletranscriptomics. The experimental evolution ancestral strain (PX632) and evolved replicates are available from the Phillips Lab upon request.