Abstract
Thymidine analog mutations (TAMs) in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) confer resistance to zidovudine (AZT) by increasing the rate of ATP-dependent phosphorolysis of the terminal nucleotide monophosphate (primer unblocking). By contrast, the L74V mutation, which confers resistance to didanosine, sensitizes HIV-1 to AZT and partially restores AZT susceptibility when present together with one or more TAMs. To compare rates of primer unblocking in RTs carrying different clusters of TAMs and to explore the biochemical mechanism by which L74V affects AZT susceptibility, ATP-mediated rescue of AZT-blocked DNA synthesis was assayed using a series of purified recombinant RTs. Rates of primer unblocking were higher in the 67N/70R/219Q RT than in the 41L/210W/215Y enzyme and were similar to rates observed with an RT carrying six TAMs (41L/67N/70R/210W/215Y/219Q). The presence of 74V in an otherwise wild-type RT reduced the rate of primer unblocking to a degree similar to that observed with the M184V mutation for lamivudine resistance, which also sensitizes HIV-1 to AZT. Introduction of 74V into RTs carrying TAMs partially counteracted the effect of TAMs on the rate of primer unblocking. The effect of 74V was less marked than that of the 184V mutation in the 67N/70R/219Q and 41L/210W/215Y RTs but similar in the RT carrying six TAMs. These results demonstrate that L74V enhances AZT susceptibility by reducing the extent of its removal by ATP-dependent phosphorolysis and provides further evidence for a common mechanism by which mutations conferring resistance to didanosine and lamivudine sensitize HIV-1 to AZT.
Nucleoside analogs are prodrugs that inhibit the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1). Once phosphorylated by cellular kinases to their active triphosphate forms, these drugs compete with the natural deoxynucleoside triphosphates for incorporation by RT into the nascent reverse transcript. Because these inhibitors lack a 3′-OH they function as chain terminators, blocking further DNA polymerization. Nucleoside reverse transcriptase inhibitors (NRTIs) are a cornerstone of antiretroviral therapy and have contributed importantly to the dramatic reduction in HIV-related morbidity and mortality in the developed world (34, 50). However, the high prevalence of drug resistance limits the clinical benefits of NRTIs in many patients.
In the case of zidovudine (AZT), resistance emerges in a stepwise manner by accumulation of thymidine analog resistance mutations (TAMs) at RT codons 41, 67, 70, 210, 215, and 219 (3, 19, 23). The combined presence of three to six TAMs results in high-level (>500-fold) AZT resistance and contributes significantly to cross-resistance to other nucleoside RT inhibitors (46). Data from several studies suggest that these mutations usually are found in two distinct clusters: those linked to a T215Y mutation, and those linked to the K70R mutation (18, 28). When present together with other TAMs, the 215Y mutation most often occurs as a 41L/215Y, 41L/210W/215Y, or 41L/67N/210W/215Y combination. By contrast, the 70R mutation is most often found as 67N/70R, 70R/219Q (or E or N), 67N/70R/219Q (E,N), or 67N/70R/215F/219Q (E,N) combinations (48, 49).
Biochemical studies show that TAMs confer resistance to AZT and other NRTIs by accelerating the rate at which the terminal AZT monophosphate (or other dideoxynucleoside monophosphate) is removed through phosphorolytic cleavage. In this reaction, AZT monophosphate (AZT-MP) is transferred from the blocked DNA to pyrophosphate (PPi) or a PPi donor by RT, which in turn resumes polymerization (1, 16, 25, 30, 31). Current data suggest that ATP is the physiologically relevant pyrophosphate donor for nucleotide excision intracellularly (25, 30, 32). The degree of nucleotide excision correlates with the number of TAMs in RT (32). Most of these data have been generated using RTs carrying the 215Y mutation in combination with various other TAMs, but to our knowledge the relative rates of nucleotide excision by RTs carrying the most common TAM patterns (41L/210W/215Y and 67N/70R/219Q) have not been reported.
By contrast, mutations selected by lamivudine (3TC) and didanosine (ddI) confer resistance by increasing selectivity of RT for the natural deoxynucleoside triphosphate. In the case of 3TC, the M184V substitution in the highly conserved YMDD domain increases the Ki for 3TC-TP by 80-fold (47) and confers high-level (≈1,000-fold) 3TC resistance to viruses carrying this mutation (14, 38, 44). Introduction of this mutation in the highly conserved YMDD domain decreases RT processivity and significantly decreases viral replication capacity relative to that of the wild type (2, 24, 26, 45). The 184V mutation also restores AZT susceptibility in viruses carrying AZT resistance mutations (44). Biochemical studies suggest that the 184V mutation increases AZT susceptibility by impairing the rescue of AZT-terminated reverse transcripts (16). The 184V mutation reduces primer unblocking in a mutant carrying five TAMs (41L/67N/70R/215Y/219Q) (6), but its effect in viruses carrying the 67N/70R/219Q or 41L/210W/215Y clusters of TAMs has not been reported.
The L74V mutation confers resistance to ddI by reducing the Ki for ddATP. As with M184V, the 74V mutation decreases RT processivity, impairs viral fitness relative to wild-type HIV-1, and restores AZT susceptibility in viruses carrying TAMs (13, 40-42). This effect may vary depending on the particular combination of TAMs present together with 74V (13). The biochemical mechanism by which 74V restores AZT susceptibility is not yet known. Because viruses carrying the 74V mutation share many properties in common with 184V mutants, we sought to determine whether 74V also impairs primer unblocking. Therefore, we evaluated the effect of the 74V and 184V mutations on ATP-mediated phosphorolysis of AZT-terminated primers in wild-type RT and in RTs carrying different combinations of TAMs.
MATERIALS AND METHODS
Reagents.
Tris base and FLAG peptide were purchased from Sigma-Aldrich Corp. (St. Louis, Missouri). Luria-Bertani (LB) broth, ampicillin, isopropyl-β-d-thiogalactopyranoside (IPTG), imidazole, and dithiothreitol were obtained from Fisher Scientific (Hampton, New Hampshire). All oligonucleotides were synthesized by IDT Inc. (Coralville, Iowa). High-fidelity Taq DNA polymerase was purchased from Invitrogen Corp. (Carlsbad, California) and restriction enzymes from New England Biolabs Inc. (Beverly, Massachusetts). 3′-Azido-3′-deoxythymidine-5′-triphosphate (AZT-TP) was purchased from TriLink Biotechnologies (San Diego, California). T4 polynucleotide kinase, dideoxynucleoside triphosphates, deoxynucleoside triphosphates, and ATP were obtained from MBI-Fermentas, Inc. (Hanover, Maryland). ATP was treated with inorganic pyrophosphatase (Roche Applied Science, Indianapolis, Indiana) at 37°C to remove the contaminating PPi. [γ32P]ATP was purchased from Amersham Biosciences Corp. (Piscataway, New Jersey). EDTA-free protease inhibitor cocktail (Complete) was obtained from Roche.
Plasmid construction.
Recombinant RT was purified using a double-tag strategy as described (27). All molecular biology procedures were performed using standard techniques. A PCR-based approach was used for the cloning of the HIV-1 HXB2 RT subunits. The primers used in the PCR included sense 5′-CCGGAATTCATTAAAGAGGAGAAATTAACT-3′, antisense (p51): 5′-CATGCCATGGTCACTAATGATGATGATGATGATGGGATCCACGCGGAACTAGGAAGGTTTCTGCTCCTAC-3′, and antisense (p66) 5′-ACGCAAGCTTTCACTA CTTGTCATCGTCAT-3′. In addition to the RT sequences, these oligonucleotides encoded the histidine (His) and FLAG tags at the carboxy terminus of the p51 and p66 subunits, respectively. The PCR products containing the open reading frames of the HXB2 p51 and p66 subunits were then isolated and cloned into the pET-duet vector (Novagen-EMD Biosciences Inc., San Diego, California).
Generation of mutant RTs.
The M41L, D67N, K70R, L74V, M184V, L210W, T215Y, and K219Q substitutions were introduced into wild-type HXB2 p51 and p66 RT subunits by site-directed mutagenesis using the GeneEditor in vitro site-directed mutagenesis kit (Promega Corp., Madison, Wisconsin), following the manufacturer's directions. Full-length sequencing of mutant RTs was performed to confirm the presence of the desired mutations and to exclude adventitious mutations that may have been introduced during the mutagenesis procedure.
Enzyme purification.
Reverse transcriptase was purified from Escherichia coli BL21 (Invitrogen) transformed with pET-duet plasmids encoding either the p51-His or p66-FLAG subunit. Bacteria were harvested by centrifugation, resuspended in lysis buffer [50 mM Tris-HCl (pH 8.3); 300 mM NaCl; 10 mM imidazole; and protease inhibitors], and lysed by passage through a microfluidizer processor model M-110EHI (Microfluidics Corp., Newton, Massachusetts). Cellular debris was removed by centrifugation, and the supernatant was mixed with nickel-nitrilotriacetic acid-agarose (QIAGEN Inc., Valencia, California) to allow binding of the His-tag to the beads and to allow RT dimerization (7). After washing with buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 20 mM imidazole, the p51-His monomer and p51-His/p66-FLAG heterodimer were eluted by the addition of elution buffer [50 mM Tris-HCl (pH 7.4); 150 mM NaCl; 250 mM imidazole]. The eluted proteins were then applied to a column packed with anti-FLAG(M2) agarose resin (Sigma-Aldrich), which was washed with buffer containing 50 mM Tris-HCl (pH 7.4) and 150 mM NaCl. The p51-His/p66-FLAG heterodimer was recovered with elution buffer [50 mM Tris-HCl (pH 7.4); 150 mM NaCl; and FLAG peptide (100 μg/ml)] and concentrated using an Amicon-Ultra (size cutoff, 50 kDa) centrifugal filter (Millipore Corp., Billerica, Massachusetts). Finally, dithiothreitol was added to a concentration of 1 mM.
Analyses by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining revealed that the p51 and p66 subunits were present at a 1:1 ratio and were ≥95% pure. Preparations generally yielded specific activities of approximately 40,000 units/mg of protein, calculated as described (43).
ATP-mediated primer unblocking.
32P labeling of the PPT-18 and PPT-21 primers, preparation of AZT-terminated DNAs, and the primer unblocking assay, were performed as described (16). The template and primer oligonucleotides used in the experiments were derived from the polypurine tract of the HIV-1 genome (17). They include PPT-57 (template), 5′-CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′, and PPT-18 (primer), 5′-TTAAAAGAAAAGGGGGGA-3′. Rescue of AZT-terminated DNA synthesis was performed in the presence of 3.5 mM ATP, 10 μM each of dCTP and dGTP, and 100 μM each of dTTP and ddATP. All reactions were performed at 37°C and stopped by adding 1 μl of formamide gel loading buffer (Ambion Inc., Austin, Texas) at 0, 1, 3, 10, 20, 30, 45, 60, and 90 min. Samples were boiled for 5 min and resolved on 8% polyacrylamide-7 M urea gels. Gels were exposed to a PhosphorImager screen, the data were acquired in a Storm 820 system, and radioactive bands were quantified with ImageQuant software (Amersham). All experiments were performed in triplicate.
RESULTS
Relative rates of ATP-mediated primer unblocking with RTs carrying different TAMs.
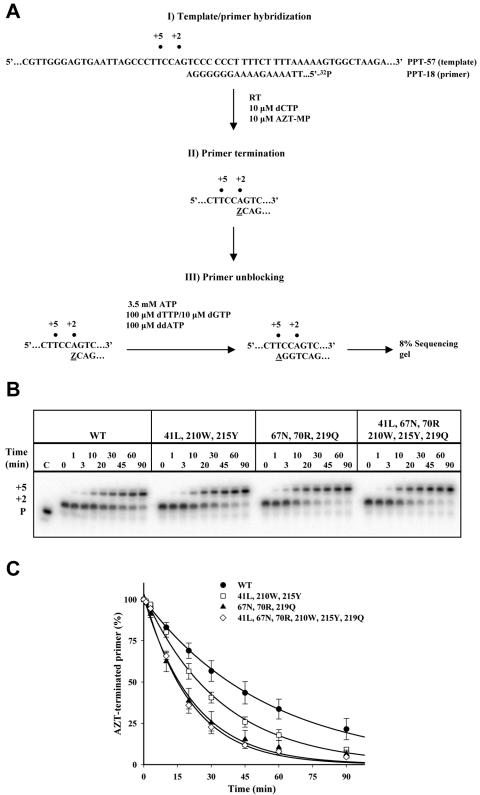
To compare the effect of TAMs characteristic of those found in clinical isolates on ATP-mediated primer unblocking, rescue of AZT-terminated DNA synthesis was assayed using RTs carrying M41L/L210W/T215Y (41L/210W/215Y RT), D67N/K70R/K219Q (67N/70R/219Q RT), and 41L/67N/70R/210W/215Y/219Q (6-TAM RT). As expected, the presence of TAMs increased the rate of ATP-mediated primer unblocking compared to the wild type (Fig. 1). Interestingly, the extent of primer unblocking was greater with the 67N/70R/219Q RT compared to the 41L/210W/215Y RT. Moreover, the extent of primer unblocking observed with 67N/70R/219Q RT was similar to that of the 6-TAM RT.
FIG. 1.
Effect of thymidine analog resistance mutations on ATP-mediated rescue of an AZT-blocked primer. A. Diagram depicting the three-step primer unblocking strategy used to monitor unblocking of an AZT-terminated primer. Z represents the incorporated AZT monophosphate at position +2 (terminated primer); +5 describes the position at which the unblocking reaction is terminated (rescued primer). In step I, the 32P-labeled primer PPT-18 hybridized to the template. In step II, the primer is extended by 2 nucleotides until AZT-MP is incorporated. In step III, addition of ATP, dGTP, dTTP, and ddATP at the indicated concentrations results in excision of AZT and resumption of DNA synthesis until incorporation of ddATP at position +5. B. Representative sequencing gels showing the rescue of the AZT-terminated primer for wild-type (WT), 41L/210W/215Y, 67N/70R/219Q, and 6-TAM RTs at the indicated times; P shows the position of migration of the primer. C. The percentages of terminated primer over time were determined for the wild-type (•), 41L/210W/215Y (□), 67N/70R/219Q (▴), and 6-TAM (⋄) RTs. Means and standard deviations from three experiments are shown; curves connecting the points represent the best fit for exponential decay.
Effect of the 74V mutation on primer unblocking.
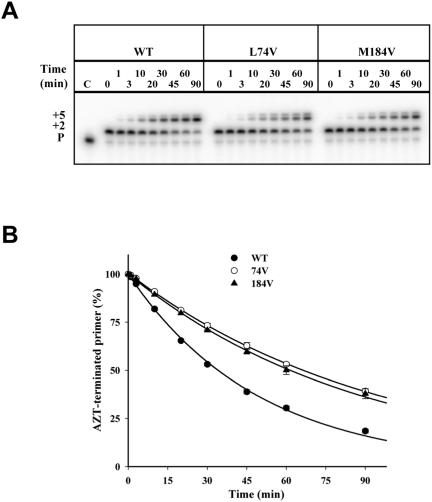
To determine the effect of the 74V mutation on primer unblocking, rescue of AZT-blocked DNA synthesis was compared in wild-type and 74V RTs. The rate of primer unblocking was significantly slower in the 74V RT compared to the wild-type enzyme (Fig. 2). The effect of 74V on reducing rescue of an AZT-blocked primer was comparable to the effect of the 184V mutation.
FIG. 2.
Effect of the L74V and M184V mutations on ATP-mediated rescue of an AZT-blocked primer. Reaction conditions were as in Fig. 1. A. Representative sequencing gels showing the rescue of the AZT-terminated primer for wild-type (WT), 74V, and 184V RTs at the indicated times. P shows the position for the migration of the primer. B. The percentages of terminated primer over time were determined for wild-type (•), 74V (○), and 184V (▴) RTs. Means and standard deviations from three experiments are shown; curves connecting the points represent the best fit for exponential decay. Some standard deviation bars are not visible due to small experimental variations.
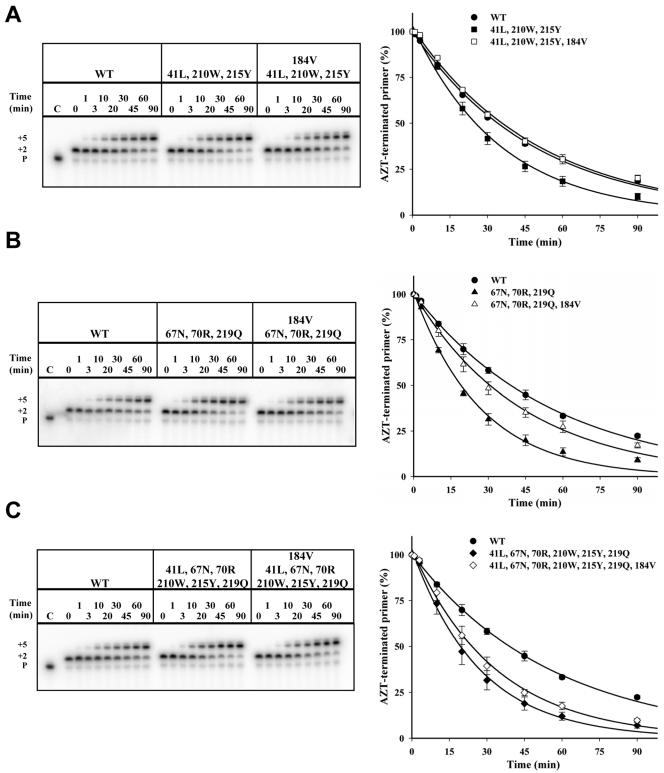
Effect of 184V and 74V mutations on primer unblocking in the presence of TAMs.
To determine the extent to which 184V altered the rate of primer unblocking in RTs carrying various combinations of TAMs, this substitution was introduced into the 41L/210W/215Y, 67N/70R/219Q, and 6-TAM RTs. When introduced into the 41L/210W/215Y RT, the 184V mutation reduced the rate of primer unblocking to nearly wild-type levels (Fig. 3A). When introduced into the 67N/70R/219Q RT, the 184V mutation resulted in intermediate rates of primer unblocking (Fig. 3B). Introduction of this mutation into the 6-TAM RT had a relatively modest though still significant effect on primer unblocking compared to the 6-TAM RT carrying the wild-type methionine at position 184 (Fig. 3C).
FIG. 3.
Effect of 184V on ATP-mediated primer rescue in the context of TAMs. Representative sequencing gels showing the rescue of the AZT-terminated primer for wild-type (WT) and mutant RTs at the indicated times (P shows the position for the migration of the primer). Means and standard deviations from three experiments are shown; curves connecting the points represent the best fit for exponential decay. A. Wild-type (•) versus 41L/210W/215Y (▪) versus 41L/210W/215Y/184V (□) RTs; B. wild-type (•) versus 67N/70R/219Q (▴) versus 67N/70R/219Q/184V (▵) RTs; C. wild-type (•) versus 6-TAM (♦) versus 6-TAM plus 184V (⋄) RTs.
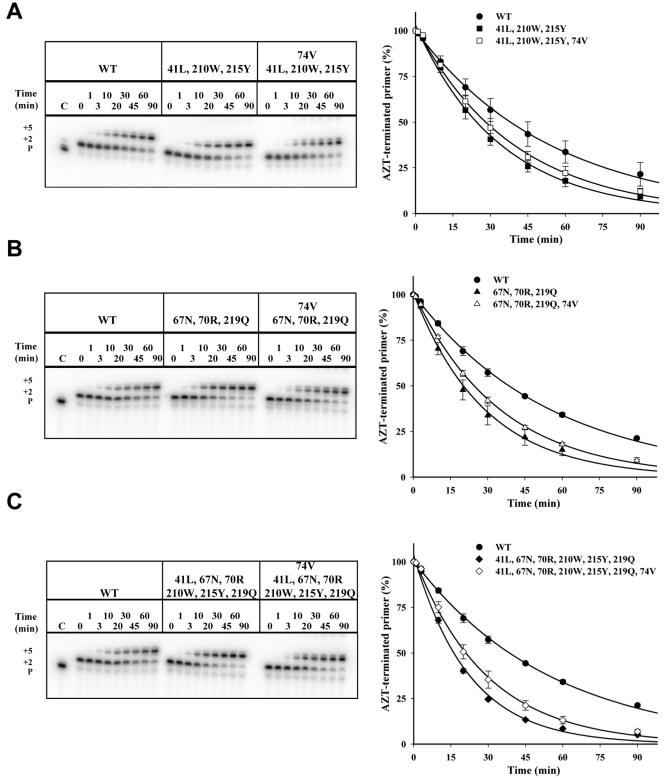
Similarly, the effect of the 74V mutation on primer unblocking in the presence of TAMs was determined by introducing this mutation into the 41L/210W/215Y, 67N/70R/219Q, and 6-TAM RTs. In contrast to the 184V mutation, introduction of 74V into the 41L/210W/215Y RT reduced the rate of primer unblocking only slightly (Fig. 4A); likewise, the effect of 74V on primer unblocking was relatively modest when introduced into the 67/70/219 RT or the 6-TAM RT (Fig. 4B and 4C, respectively). The magnitude of this effect was comparable to the effect of introducing the 184V mutation into the same 6-TAM background (Fig. 3C).
FIG. 4.
Effect of 74V on ATP-mediated primer rescue in the context of TAMs. Representative sequencing gels showing the rescue of the AZT-terminated primer for wild-type (WT) and mutant RTs at the indicated times (P shows the position for the migration of the primer). Means and standard deviations from three experiments are shown; curves connecting the points represent the best fit for exponential decay. A. Wild-type (•) versus 41L/210W/215Y (▪) versus 41L/210W/215Y/74V (□) RTs; B. wild-type (•) versus 67N/70R/219Q (▴) versus 67N/70R/219Q/74V (▵) RTs; C. wild-type (•) versus 6-TAM (♦) versus 6-TAM plus 74V (⋄) RTs.
DISCUSSION
In this study we showed that the L74V mutation in HIV-1 RT reduced ATP-mediated rescue of AZT-terminated DNA synthesis in wild-type and AZT-resistant enzymes. In otherwise wild-type RT, the magnitude of this effect was comparable to that observed with the M184V mutation. Both mutations counteracted the increased rate of primer unblocking observed in RTs carrying different combinations of TAMs representative of patterns commonly observed in clinical isolates. The 184V mutation reduced primer unblocking to wild-type levels in the 41L/210W/215Y RT and somewhat reduced the extent of primer unblocking in RTs with the 67N,70R and 219Q mutations or with all six TAMs. Introduction of 74V into these mutant enzymes partially counteracted the effect of TAMs on the rate of primer unblocking, but the effect was substantially less than that of the 184V mutation except in the context of the 6-TAM RT.
Our results demonstrate that the 74V mutation shares a common biochemical mechanism with other RT mutations that also sensitize HIV-1 RT to zidovudine, including the 184V mutation for 3TC resistance (6, 16), the K65R mutation for tenofovir resistance (K. L. White, N. A. Margot, J. M. Chen, A. S. Ray, R. Wang, M. Pavelko, T. Wrin, C. J. Petropoulos, M. McDermott, S. Swaminathan, and M. D. Miller, Abstr. 11th Conf. Retroviruses Opportunistic Infect., abstr. M-26, 2004), and the Y181C mutation for nevirapine and delavirdine resistance (39). Results similar to ours were reported by Frankel et al., who found a 50% reduction in the efficiency of ATP-dependent primer unblocking in RT carrying the 74V mutation (F. Frankel, D. Turner, B. Brenner, Y. Quan, and M. A. Wainberg, abstr. 27, Antiviral Ther. 9:S33, 2004), but that study did not examine the interaction of 74V with TAMs.
The results of both studies differ, however, from those of a previous report that found no effect of 74V on ATP-mediated extension of an AZT-blocked primer (5). This study examined the effect of ATP on the ability of different RTs to copy an entire oligonucleotide template. As noted by the authors of that study, the presence of only a few sites in the template at which AZT could be incorporated might have reduced the sensitivity of their assay to detect the effects of specific mutations on ATP-mediated primer unblocking (5). By contrast, the assay employed in our studies and those of Frankel et al. examined ATP-mediated primer unblocking at a single template position.
Previous studies of primer unblocking have used RTs with a wide variety of TAMs but have not directly compared the rates of ATP-mediated rescue of DNA synthesis using enzymes carrying combinations of TAMs commonly encountered in clinical isolates. For example, one report focused on enzymes carrying 67N/70R, 215F/219Q, or 67N/70R/215F/219Q, but did not study enzymes carrying mutations at 41L and 210W (1). Another report compared primer unblocking in RTs carrying 67N/70R/215F/219Q or 67N/70R/215Y/219Q (a relatively uncommon combination among clinical isolates), as well as enzymes carrying 41L and/or 215Y, but did not include enzymes carrying the 210W substitution in their studies (32). A third study examined primer unblocking in RT carrying 41L/67N/210W/215Y (with or without the 44D and 118I mutations), but did not include enzymes carrying the 67N/70R/219Q cluster of TAMs (15). We found higher rates of ATP-mediated rescue of an AZT-blocked primer with the 67N/70R/219Q RT compared to the 41L/210W/215Y enzyme. This observation is somewhat surprising, given that clinical studies have generally shown that the combined presence of the 41L, 210W, and/or 215Y mutations is associated with worse clinical or virologic response to AZT and other nucleoside RT inhibitors (21, 33).
Several factors could account for differences between our results and those of previous studies. It is possible that variation in the RT backbone between the molecular clone of HIV-1 used in our studies (HXB2) and clinical isolates may account for these differences. Differences in the absolute rates of primer unblocking and extension as a function of ATP and deoxynucleoside triphosphate concentrations have been reported, but in those reports the relative order of primer unblocking efficiency for different mutant enzymes remained similar at the different concentrations tested (5, 6). Therefore, we do not believe that the substance of our conclusions would be changed by use of different reagent concentrations. Alternatively, it is possible that phenotypic expression of AZT resistance in vivo results from a combination of primer unblocking and other, as yet unidentified factors. Our biochemical data should be extrapolated cautiously to the clinic, because the effects of TAMs and other nucleoside analog resistance mutations on clinical response to AZT-containing regimens result from a complex interplay of a variety of factors, including drug susceptibility, viral replication capacity, HIV-specific cellular immune responses, and the activity of other components of the antiretroviral regimen (9-11).
The rate of ATP-mediated rescue of an AZT-blocked DNA primer can be affected by a number of factors, including the rate of ternary complex formation (which requires positioning of AZT in the priming or P site of the enzyme and binding of the next incoming deoxynucleoside triphosphate at the nucleotide binding or N site), affinity of the enzyme for ATP, and positioning of the γ phosphate of ATP with respect to the phosphodiester bond between the terminal AZT-MP and the penultimate nucleoside monophosphate (4, 5, 30).
The aromatic side chains of 210W and 215Y or 215F are proposed on the basis of modeling studies to enhance ATP binding through stabilizing interactions with the adenine ring of ATP (5). This hypothesis is challenged by kinetic data as well as by mutagenesis studies that fail to show an effect of 215Y on RT affinity for ATP and suggest instead that this mutation optimizes the orientation of ATP for the primer excision reaction (29, 35). Our finding of higher rates of primer unblocking in the 67N/70R/219Q enzyme compared to the 41L/210W/215Y enzyme is consistent with the latter model that shows that the 67N/70R/219Q cluster in the absence of changes at the 215 position is also well positioned to make direct contact with ATP which facilitates a catalytically efficient alignment with the primer terminus (8).
In the case of the 184V mutation, it has been proposed that altered positioning of the template-primer allows the 3′ end of the AZT-MP-terminated primer to move more easily into the P position, thereby favoring formation of a closed (“dead end”) complex by binding of the incoming deoxynucleoside triphosphate (5, 37). The precise mechanism by which the 74V mutation reduces primer unblocking is not known. Crystal structures of RT complexed to the DNA substrate and the incoming deoxynucleoside triphosphate show that L74 is in proximity to the deoxynucleoside triphosphate binding site but does not make direct contact with the incoming nucleotide (20). Rather, this residue more likely makes contact with the +1 position of the DNA template and with the side chains of amino acids R72 and Q151, which are positioned at the base of the incoming deoxynucleoside triphosphate. In turn, Q151, together with D113, Y115, and F116, forms a small pocket that can accommodate the 3′-OH group of the incoming deoxynucleoside triphosphate or the 3′-azido group of AZT-TP.
We hypothesize that introduction of the L74V mutation disrupts proper interaction with Q151, altering optimal positioning of the 3′-azido group in the 3′ pocket, which in turn impairs ATP phosphorolysis of AZT. This model may also account for the reduced efficiency of incorporation of natural deoxynucleoside triphosphates in the L74V mutant (12). That the crystal structure of the L74V mutant RT shows no significant conformational changes in the structure of p66 compared to the wild-type enzyme argues for a subtle alteration of side chain interactions around the 3′ pocket (36). Alternatively, introduction of the β-branched amino acid valine at position 74 may alter primer positioning, which in turn may lead to a configuration that is unfavorable for nucleotide excision. This explanation seems less likely, as L74 does not appear to make direct contact with the 3′ end of the primer. Differences in the extent to which 74V and 184V reduce primer unblocking in various TAM backgrounds could be explained by their different locations in the RT structure. The 74V mutation is found at the base of the β2-β3 loop, whereas 184V is found several angstroms away in the catalytic loop (20).
In conclusion, we have shown that, like the 184V mutation, the 74V mutation reduces ATP-mediated rescue of an AZT-blocked primer. The finding that the 184V mutation had a greater effect on RTs carrying different 3-TAM combinations than did the 74V mutation corresponds to the clinical observation that 3TC resistance has a greater effect on ZDV susceptibility than does ddI resistance. The effect of both mutations on primer unblocking was blunted by the presence of a larger number of TAMs, which is consistent with the observation that the resensitizing effects of the 74V and 184V mutations can be overcome by accumulation of additional TAMs to yield viruses that are dually resistant to AZT and ddI or 3TC (22, 46). A more complete understanding of the precise mechanisms underlying these interactions may come from detailed kinetic analyses of primer unblocking, along with determination of the crystal structures of RTs carrying the combinations of mutations studied in this work. It would also be of interest to extend these findings by conducting biochemical studies with AZT-resistant RTs from other HIV-1 genetic backgrounds, including clinical isolates.
Acknowledgments
We are greatly indebted to B. Marchand (McGill University) for helpful advice and to Janet Steele for editorial assistance.
This research was supported by U.S. Public Health Service grants AI42567 and RR16482 (D.R.K.), the Harvard Medical School Center for AIDS Research Virology Core (AI060354), and the Canadian Institutes of Health Research (M.G.).
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Back, N. K. T., M. Nijhuis, W. Keulen, C. A. B. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher, C. A. B., E. O'Sullivan, J. W. Mulder, C. Ramautarsing, P. Kellam, G. Darby, J. M. A. Lange, J. Goudsmit, and B. A. Larder. 1992. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J. Infect. Dis. 165:105-110. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, P. L., H. Imamichi, S. G. Sarafinos, E. Arnold, and S. H. Hughes. 2004. Effects of the Δ67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. J. Virol. 78:9987-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., S. G. Sarafinos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, P. L., S. G. Sarafinos, E. Arnold, and S. H. Hughes. 2002. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 76:3248-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabodevilla, J. F., L. Odriozola, E. Santiago, and J. J. Martinez-Irujo. 2001. Factors affecting the dimerization of the p66 form of HIV-1 reverse transcriptase. Eur. J. Biochem. 268:1163-1172. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain, P. P., J. Ren, C. E. Nichols, L. Douglas, J. Lennerstrand, B. Larder, D. I. Stuart, and D. K. Stammers. 2002. Crystal structures of zidovudine- or lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215. J. Virol. 76:10015-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Aquila, R. T., V. A. Johnson, S. L. Welles, A. J. Japour, D. R. Kuritzkes, V. DeGruttola, P. S. Reichelderfer, R. W. Coombs, C. S. Crumpacker, J. O. Kahn, and D. D. Richman for the AIDS Clinical Trials Group 116B/117 Team and the Virology Committee Resistance Working Group. 1995. Zidovudine resistance and human immunodeficiency virus type 1 disease progression during antiretroviral therapy. Ann. Intern. Med. 122:401-408. [DOI] [PubMed] [Google Scholar]
- 10.Deeks, S. 2000. Determinants of virological response to antiretroviral therapy: implications for long-term strategies. Clin. Infect. Dis. 30(Suppl. 2):S177-184. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G., J. N. Martin, E. Sinclair, J. Harris, T. B. Neilands, H. T. Maecker, E. Hagos, T. Wrin, C. J. Petropoulos, B. Bredt, and J. M. McCune. 2004. Strong cell-mediated immune responses are associated with the maintenance of low-level viremia in antiretroviral-treated individuals with drug-resistant human immunodeficiency virus type 1. J. Infect. Dis. 189:312-321. [DOI] [PubMed] [Google Scholar]
- 12.Deval, J., J. M. Navarro, B. Selmi, J. Courcambeck, J. Boretto, P. Halfon, S. Garrido-Urbani, J. Sire, and B. Canard. 2004. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J. Biol. Chem. 279:25489-25496. [DOI] [PubMed] [Google Scholar]
- 13.Eron, J. J., Y. K. Chow, A. M. Caliendo, J. Videler, K. M. Devore, T. P. Cooley, H. A. Liebman, J. C. Kaplan, M. S. Hirsch, and R. T. D'Aquila. 1993. pol mutations conferring zidovudine and didanosine resistance with different effects in vitro yield multiply resistant human immunodeficiency virus type 1 isolates in vivo. Antimicrob. Agents Chemother 37:1480-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Q., Z. Gu, J. Hiscott, G. Dionne, and M. A. Wainberg. 1993. Generation of drug-resistant variants of human immunodeficiency virus type 1 by in vitro passage in increasing concentrations of 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girouard, M., K. Diallo, B. Marchand, S. McCormick, and M. Götte. 2003. Mutations of E44D and V1181 in the reverse transcriptase of HIV-1 play distinct mechanistic roles in dual resistance to AZT and 3TC. J. Biol. Chem. 278:34403-34410. [DOI] [PubMed] [Google Scholar]
- 16.Götte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götte, M., G. Maier, A. Mochi Onori, L. Cellai, M. A. Wainberg, and H. Heumann. 1999. Temporal coordination between initiation of HIV (+)-strand DNA synthesis and primer removal. J. Biol. Chem. 274:11159-11169. [DOI] [PubMed] [Google Scholar]
- 18.Hanna, G., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. Leigh-Brown, A. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181:904-911. [DOI] [PubMed] [Google Scholar]
- 19.Harrigan, P. R., I. Kinghorn, S. Bloor, S. D. Kemp, I. Najera, A. Kohli, and B. A. Larder. 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 70:5930-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catlytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 21.Japour, A. J., S. Welles, R. T. D'Aquila, V. A. Johnson, D. D. Richman, R. W. Coombs, P. S. Reichelderfer, J. O. Kahn, C. S. Crumpacker, and D. R. Kuritzkes for the AIDS Clinical Trials Group 116B/117 Study Team and the Virology Committee Resistance Working Group. 1995. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients following long-term zidovudine treatment. J. Infect. Dis. 171:1172-1179. [DOI] [PubMed] [Google Scholar]
- 22.Kuritzkes, D., D. Shugarts, M. Bakhtiari, D. Poticha, J. Johnson, T. Gingeras, M. Kennedy, and J. Eron. 2000. Emergence of dual resistance to zidovudine and lamivudine after long-term treatment of human immunodeficiency virus type 1 (HIV-1) infection with zidovudine plus lamivudine. J. Acquir. Immune. Defic. Syndr. 23:26-34. [DOI] [PubMed] [Google Scholar]
- 23.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 24.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 25.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, J., and D. R. Kuritzkes. 2001. A novel recombinant virus assay for comparing the relative fitness of HIV-1 reverse transcriptase variants. J. Acquir Immune Defic. Syndr. 27:7-13. [DOI] [PubMed] [Google Scholar]
- 27.Maier, G., U. Dietrich, B. Panhans, B. Schroder, H. Rubsamen-Waigmann, L. Cellai, T. Hermann, and H. Heumann. 1999. Mixed reconstitution of mutated subunits of HIV-1 reverse transcriptase coexpressed in Escherichia coli-two tags tie it up. Eur. J. Biochem. 261:10-18. [DOI] [PubMed] [Google Scholar]
- 28.Marcelin, A. G., C. Delaugerre, M. Wirden, P. Viegas, A. Simon, C. Katlama, and V. Calvez. 2004. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J. Med. Virol. 72:162-165. [DOI] [PubMed] [Google Scholar]
- 29.Matamoros, T., S. Franco, B. M. Vázquez-Álvarez, A. Mas, M. A. Martínez, and L. Menéndez-Arias. 2004. Molecular determinants of multi-nucleoside analogue resistance in HIV-1 reverse transcriptases containing a dipeptide insertion in the fingers subdomain. J. Biol. Chem. 279:24569-24577. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, P. R., S. E. Matsuura, A. Mohsin Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, M. D., N. Margot, B. Lu, L. Zhong, S. S. Chen, A. Cheng, and M. Wulfsohn. 2004. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J. Infect. Dis. 189:837-846. [DOI] [PubMed] [Google Scholar]
- 34.Palella, F. J., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV outpatient study investigators. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 35.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 36.Ren, J., R. Esnouf, A. L. Hopkins, E. Y. Jones, I. Kirby, J. Keeling, C. K. Ross, B. Larder, D. I. Stuart, and D. K. Stammers. 1998. 3′-azido-3′-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes. Proc. Natl. Acad. Sci. USA. 95:9518-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarafinos, S. G., K. Das, A. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 2004. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA. 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schinazi, R. F., R. M. Lloyd, M. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selmi, B., J. Deval, K. Alvarez, J. Boretto, S. Sarfati, C. Guerreiro, and B. Canard. 2003. The Y181C substitution in 3′-azido-3′-deoxythymidine-resistant human immunodeficiency virus, type 1, reverse transcriptase suppresses the ATP-mediated repair of the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 278:40464-40472. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, P. L., and C. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma, P. L., and C. Crumpacker. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St.Clair, M. H., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, and P. Kellam. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 43.Thimmig, R., and C. McHenry. 1993. HIV reverse transcriptase: Expression in E. coli, purification, and characterization of a functionally and structurally asymmetric dimeric polymerase. J. Biol. Chem. 268:16528-16536. [PubMed] [Google Scholar]
- 44.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA. 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakefield, J. K., S. A. Jablonski, and C. D. Morrow. 1992. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J. Virol. 66:6806-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitcomb, J. M., N. T. Parkin, C. Chappey, N. S. Hellman, and C. J. Petropoulos. 2003. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J. Infect. Dis. 188:992-1000. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, J. E., A. Aulabauch, B. Caligan, S. McPherson, J. K. Wakefield, S. Jablonski, C. D. Morrow, J. E. Reardon, and P. A. Furman. 1996. Human immunodeficiency virus type-1 reverse transcriptase. Contribution of Met-184 to binding of nucleoside 5′-triphosphate. J. Biol. Chem. 271:13656-13662. [DOI] [PubMed] [Google Scholar]
- 48.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, F. Ariasi, F. Volot, J. A. Gastaut, H. Gallais, J. Moreau, and J. Fantini. 1999. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J. Clin. Microbiol. 37:4099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, and J. Fantini. 2000. Mutation of L210W of HIV-1 reverse transcriptase in patients receiving combination therapy. Incidence, association with other mutations, and effects on the structure of mutated reverse transcriptase. J. Biomed. Sci. 7:507-513. [DOI] [PubMed] [Google Scholar]
- 50.Yeni, P. G., S. M. Hammer, M. S. Hirsch, M. S. Saag, M. Schechter, C. C. Carpenter, M. A. Fischl, J. M. Gatell, B. G. Gazzard, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, R. T. Schooley, M. A. Thompson, S. Vella, and P. A. Volberding. 2004. Treatment for adult HIV infection: 2004 recommendations of the International AIDS Society-USA Panel. JAMA 292:251-265. [DOI] [PubMed] [Google Scholar]