Abstract
The L74V and M184V mutations in the reverse transcriptase (RT) gene of human immunodeficiency virus type 1 (HIV-1) are frequently associated with resistance to the nucleoside reverse transcriptase inhibitors abacavir, didanosine, and lamivudine. Yet viruses containing any of these mutations often display hypersusceptibility to zidovudine (ZDV). Two distinct mechanisms have been described to explain HIV-1 drug resistance. One of these involves diminished rates of incorporation of the nucleotide analogue by mutated RT, while the other mechanism involves increased rates of phosphorolytic excision of the drug-terminated primer. To understand the biochemical mechanisms responsible for the hypersensitization of L74V-containing viruses to ZDV, we studied the efficiency of excision of ZDV-monophosphate (ZDV-MP)-terminated primers by recombinant wild-type and mutated HIV-1 RTs in cell-free assays. We observed that the L74V mutation in RT caused reductions in ATP-dependent removal of ZDV-MP from newly synthesized viral DNA. In addition, we determined that the L74V and M184V mutations did not affect the ratio between the populations of RT-DNA/DNA complexes found at pre- and posttranslocational stages; however, they might have affected proper alignment between incorporated chain terminator and pyrophosphate donor, substrate orientation, affinity for ATP, and/or primer-template substrate. Finally, we confirmed previous findings that L74V-containing viruses display diminished replication capacity and that this is associated with reduced levels of synthesis of early reverse-transcribed viral DNA molecules.
The development of resistance to antiretroviral drugs used in the treatment of infections caused by human immunodeficiency virus type I (HIV-1) is of major worldwide concern (31). Drug resistance is attributable to both the high replication rate of HIV-1 and the error-prone nature of HIV-1 reverse transcriptase (RT) (16, 33). In addition, the persistence of cellular reservoirs of virus and anatomic sanctuary sites ensures the continuing replication of viruses even in patients with suppressed plasma viremia. As a consequence, eradication of HIV-1 is not considered possible with currently available treatment regimens (37, 41).
Nucleoside analog reverse transcriptase inhibitors (NRTIs) of HIV-1 replication are a class of drugs that act as chain terminators of viral DNA synthesis by virtue of the fact that they lack a 3′-OH group. Combinations of NRTIs have been successfully used in the treatment of HIV-1-infected patients (6, 22). However, resistance has been reported in regard to all currently approved members of this class (17).
Two distinct mechanisms have been described to explain HIV-1 drug resistance to nucleoside analogues. One of these involves the diminished acceptance by mutated, resistant RT enzymes of NRTIs, such that these molecules are discriminated against and are no longer incorporated into the nascent viral DNA chain (19). The other mechanism involves increased phosphorolytic excision of the incorporated drug (1, 26).
The M184V mutation in the RT of HIV-1 is associated with high-level resistance to lamivudine and low- to intermediate-level resistance to didanosine (ddI) and abacavir (ABC) (2, 9, 36), while the K65R mutation is principally associated with a low to intermediate level of resistance to tenofovir both in vitro and in vivo (28, 45). The L74V mutation also confers low- to intermediate-level resistance to ddI and ABC (29).
Combinations of M184V and L74V are frequently found in patients treated with ABC-containing regimens. Both mutations share a number of other characteristics: all involve similar discriminatory mechanisms in regard to incorporation of relevant NRTIs that would ordinarily inhibit reverse transcription (12); all result in diminished RT processivity and viral replication capacity (40); and L74V- and M184V-containing mutant enzymes are severely impaired in regard to efficiency of RNA primer usage (8) and have been associated with zidovudine-hypersusceptibility and suppression of resistance (25, 43).
We have recently demonstrated that M184V-containing mutant enzymes compromise the phosphorolytic removal of incorporated ZDV and the ensuing rescue of DNA synthesis, which provides a plausible mechanism for drug hypersusceptibility (13). Other suppressor mutations that are implicated in ZDV hypersusceptibility, i.e., Y181C, which confers resistance to non-NRTIs and various mutations that reduce susceptibility to foscarnet (38, 42) show similar effects, although the location of these residues in the three-dimensional structure of HIV-1 RT differs substantially. In this study, we investigated molecular mechanisms involved in ZDV hypersusceptibility in the context of the L74V mutation and the combination of L74V and M184V.
(This work was performed by F. A. Frankel in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada.)
MATERIALS AND METHODS
Enzymes and nucleic acids.
Recombinant wild-type and mutated HIV-1 RTs (L74V, M184V and L74V/M184V) were prepared and purified as previously described (20). To study the rescue of chain-terminated DNA synthesis, both a DNA primer (PPT-18) and DNA template (PPT-57) were derived from the polypurine tract of the HIV-1 genome (14). Oligonucleotides were chemically synthesized and purchased from Invitrogen (Burlington, Ontario, Canada).
Cells and viruses.
H9 cells and the human lymphoblastoid T-cell lines MT-2 and MT-4 were grown in RPMI 1640 medium supplemented with 2 mM l-glutamine, 10% fetal calf serum, and penicillin/streptomycin as described (11). pNL4-3-derived wild-type viruses and viruses containing the L74V, M184V, and L74V/184V mutations were amplified in MT-4 cells as previously described (8) and stored at −80°C until use.
Drugs.
ZDV was purchased from Sigma-Aldrich Inc. (Oakville, Ontario, Canada). ZDV triphosphate (ZDV-TP) was purchased from Trilink Biotechnologies (San Diego, California).
Phenotypic resistance assay.
To determine the sensitivity of wild-type and L74V-containing viruses to ZDV, 2 × 106 MT-2 cells were infected as previously described with 105 TCID50 virus stock and incubated in 1 ml RPMI 1640 complete medium at 37°C for 2 h (8). The cells were then washed and plated in increasing concentrations of ZDV. The 50% inhibitory concentration (IC50) values were determined based on RT activity, which was measured in culture supernatants after 4 days as previously described (10).
Rescue of chain-terminated DNA synthesis with PPi or ATP.
The PPT-18 primer was radiolabeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase at 37°C for 30 min and purified on an 8% polyacrylamide-7 M urea gel. ZDV-terminated primer strands were detected as previously described (13). In brief, 0.425 μM PPT-18 primer was mixed with 1.275 μM PPT-57 DNA template and 32P-radiolabeled PPT-18 in a buffer containing 50 mM Tris-HCl, pH 7.8, and 50 mM NaCl. Next, the mixture was denatured at 95°C for 2 min, incubated at 70°C for 15 min, and cooled down at room temperature for 30 min. Then, 1.275 μM of wild-type or mutated RT, 10 μM dCTP, and 10 μM ZDV-TP were added to the prehybridized primer-template complex. Reactions were initiated by the addition of 6 mM MgCl2 and allowed to proceed for 30 min at 37°C.
The excision of the ZDV-terminated primer was initiated by adding a mix containing 100 μM dTTP, 10 μM dGTP, 100 μM ddATP, and 150 μM PPi or 3.5 mM ATP. A 35 mM solution of ATP was pretreated with 1 U of inorganic pyrophosphatase to control for putative contaminating PPi. DNA synthesis was monitored in time course experiments. Samples were resolved in an 8% polyacrylamide-7 M urea gel followed by overnight exposure on X-ray films (Kodak BioMax MR Film). Band intensities were analyzed by densitometry or by molecular imaging.
Site-specific footprinting.
Footprinting experiments were performed using a stable solution of potassium peroxynitrite (KOONO) as recently described (23).
Real-time PCR.
We established a real-time PCR assay to detect early versus late products in reverse transcription. Reverse-transcribed DNAs were detected using the Light Cycler Fast Start DNA SYBR Green I kit (Roche Diagnostics Corporation, Indianapolis, IN) according to the manufacturer's instructions. Briefly, 50 to 100 ng DNA was added to a master mix containing 3 to 4 mM MgCl2, 0.4 μM of each primer, and appropriate concentrations of deoxynucleoside triphosphates and Taq DNA polymerase in a 20 μl final volume. In all cases, an initial denaturing step of 95°C for 10 min and a total of 35 cycles were performed.
To determine the earliest product of reverse transcription, i.e., minus strong-stop viral DNA [(−)ssDNA], PCR conditions were as follows: 95°C for 10 seconds, 68°C for 5 seconds, 72°C for 6 seconds, and acquisition temperature 80°C for 1 second. To determine the levels of HIV-1 DNA representing intermediate products after the first strand transfer, PCR conditions were as follows: 95°C for 10 seconds, 68°C for 5 seconds, and 72°C for 12 seconds, including acquisition. To determine full-length HIV-1 DNA, PCR conditions were as follows: 95°C for 10 seconds, 64°C for 5 seconds, 72°C for 5 seconds, and acquisition temperature 80°C for 1 second. PCR specificity was assessed by melting curve analysis and further confirmed by agarose gel electrophoresis.
The primer sequences used to determine levels of HIV-1 (−)ssDNA, DNA representing intermediate products after the first strand transfer and full-length DNA have been previously described (21, 34). Briefly, primers PS and A55 bind to the region spanning R-U5 and were used to determine (−)ssDNA; primers pUT and A55 bind to the region spanning U3 and R and were used to determine intermediate products after the first strand transfer; primers pgag and Pst-A bind to the region spanning the gag gene and were used to determine full-length HIV-1 DNA. HIV-1 DNA copy numbers were normalized based on the amount of the β-globin gene, which was determined using the LC Control kit (Roche Diagnostics Corporation, Indianapolis, IN), according to the manufacturer's instructions. Standard curves were constructed by measuring triplicate dilutions of an AgeI-linearized pNL4-3 proviral clone over a range of 102 to 106 copies/μl.
Viral replication assays.
We infected 106 H9 cells with DNase I-treated wild-type or mutated HIV-1 at a multiplicity of infection (MOI) of 0.0001 in a 24-well plate as previously described (44). At 2, 4, 8, 12, and 24 h after infection, cells were washed twice with phosphate-buffered saline and resuspended in 200 μl phosphate-buffered saline. DNA was extracted using a QIAamp DNA minikit (QIAGEN Inc., Mississauga, Ontario, Canada) according to the manufacturer's instructions and quantified by optical density at 260 nm. Reverse-transcribed DNAs were quantified by real-time PCR as described above.
RESULTS
L74V-containing viruses show increased susceptibility to ZDV.
Previous tissue culture data have shown that viruses harboring the L74V mutation together with classical thymidine-associated mutations (TAMs) display increased susceptibility to ZDV compared to viruses containing TAMs only (43). Our tissue culture susceptibility assays using MT-2 cells showed that L74V-containing viruses increased susceptibility to ZDV by approximately 40% compared to wild-type virus. Furthermore, viruses containing the M184V mutation displayed similar levels of hypersusceptibility to ZDV (Table 1).
TABLE 1.
IC50s and resistance to ZDV in cell culture assaysa
| Virus | IC50 (nM) | Resistance (fold) |
|---|---|---|
| Wild type | 29.1 ± 5.0 | |
| L74V | 15.2 ± 3.3 | 0.5 |
| M184V | 17.4 ± 4.2 | 0.6 |
| L74V/M184V | ND | ND |
Values are means of at least three independent experiments ± standard deviation. ND, not determined.
L74V mutation in RT causes an approximate 50% reduction in the efficiency of ZDV-monophosphate excision from newly synthesized viral DNA.
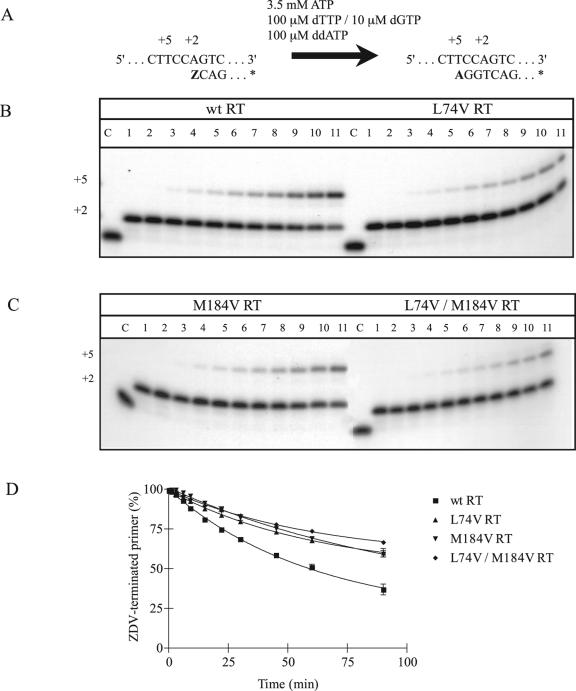
Several laboratories have described the role of ATP as a pyrophosphate donor in the excision of ZDV-MP from viral DNA (1, 4, 26). To study the mechanisms potentially associated with the observed increase in susceptibility to ZDV by L74V-containing RT and viruses, we evaluated the efficiency of excision of ZDV-MP from viral DNA in gel-based assays (Fig. 1A). We found that L74V RT displayed impaired rescue of ZDV-MP chain-terminated primers compared to wild-type RT in the presence of ATP (Fig. 1B). Whereas wild-type RT was able to unblock 50% of the ZDV-terminated primer after 59 min in this reaction, L74V RT and the doubly mutated RT L74V/184V required more than 90 min to achieve the same degree of excision (Fig. 1C and 1D). The M184V single mutant RT was also inefficient in regard to excision in the presence of ATP, as previously described (13).
FIG. 1.
Efficiency of excision of incorporated ZDV-MP in the presence of 3.5 mM ATP. The efficiencies of the reactions with wild-type and mutant RTs were compared in time course experiments. (A) Graphic representation of the cell-free system (PPT-57/PPT-18) used to monitor the excision of ZDV-MP from newly synthesized HIV-1 DNA. (B) Comparison between wild-type RT and L74V RT. (C) Comparison between M184V RT and the double mutant L74V/M184V RT. Lane C corresponds to control labeled primer. Lanes 1 to 11 show reaction products at 0, 1, 3, 6, 10, 15, 22, 30, 45, 60, and 90 min after addition of the excision mix, respectively. (D) Graphic representation of gel-based assays shown in B and C. Values are means of at least three independent experiments ± standard error.
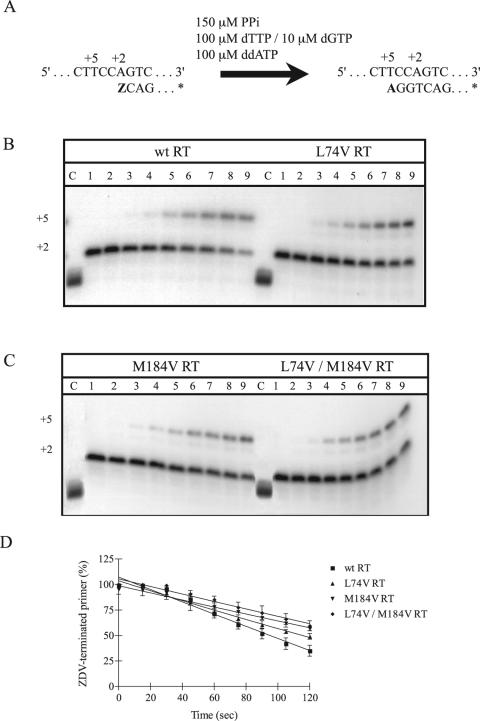
To study whether these diminished rates of excision depend on the chemical nature of the pyrophosphate donor we next evaluated the efficiency of excision of ZDV-MP in the presence of PPi in reactions extending between 15 seconds and 2 min (Fig. 2A). The results in Fig. 2B and 2D show that differences were observed between wild-type and L74V RTs, although they were not as pronounced as seen with ATP. Whereas wild-type RT unblocked 50% of the ZDV-terminated primer after 95 seconds, L74V RT required 114 seconds to achieve the same degree of excision. In addition, M184V RT and L74V/M184V RT displayed significantly impaired rescue of ZDV-terminated primer in the presence of PPi (Fig. 2C and 2D).
FIG. 2.
Efficiency of excision of incorporated ZDV-MP in the presence of 150 μM PPi. The efficiencies of the reactions with wild-type and mutant RTs were compared in time course experiments. (A) Graphic representation of the cell-free system (PPT-57/PPT-18) used to monitor the excision of ZDV-MP from newly synthesized HIV-1 DNA. (B) Comparison between wild-type RT and L74V RT. (C) Comparison between M184V RT and the double mutant L74V/M184V RT. Lane C corresponds to control labeled primer. Lanes 1 to 9 show reaction products at 0, 15, 30, 45, 60, 75, 90, 105, and 120 seconds after addition of the excision mix, respectively. (D) Graphic representation of gel-based assays shown in B and C. Values are means of at least three independent experiments ± standard error.
L74V mutation does not appear to affect the relative positioning of RT and its nucleic acid substrate.
Despite the fact that the various mutations that have been associated with ZDV hypersusceptibility are located in different regions of the RT enzyme, each of them appears to exert its effects through decreases in the rates of excision of the incorporated ZDV-MP. Previous reports have shown that the Y181C mutation can decrease the affinity of RT for ATP (36). K65R and M184V may likewise affect affinity and/or the orientation of the pyrophosphate donor, given the close proximity of these residues to the incoming nucleotide. However, L74 is also in contact with the template overhang, which suggests that the L74V mutation may influence the positioning of RT on its primer/template substrate (5).
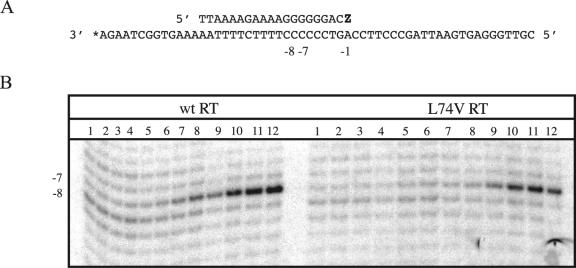
To test this hypothesis, we conducted site-specific footprinting experiments as recently described (23). The results in Fig. 3B show that footprinting of RT-DNA/DNA complexes with potassium peroxynitrite (KOONO), in the presence of increasing concentrations of the next templated deoxynucleoside triphosphate, resulted in a gradual shift of cleavage from template position −8 to −7. This one-nucleotide shift represents the difference between the pre- and posttranslocational configurations of RT. The phosphorolytic excision of incorporated nucleotides is only possible in the pretranslocation configuration with cleavage specificity at position −8, while the appearance of a band at −7 points to a dead-end complex that is present after translocation takes place. Therefore, the L74V mutation may be expected to facilitate the formation of the latter complex, which would help to explain the diminished rates of excision associated with this mutation.
FIG. 3.
Site-specific footprinting of wild-type RT and L74V RT with KOONO. (A) Sequence of the DNA/DNA primer/template substrate. The primer was terminated with ZDV-MP (Z) and subsequently incubated with increasing concentrations of the next complementary nucleotide. (B) Lanes 1 to 12 show footprints on 3′-end-labeled template strands in the presence of 0, 1, 3.125, 6.25, 12.5, 25, 50, 100, 250, 500, 1,000, and 2,000 μM dGTP, respectively.
However, this does not appear to be the case. Notably, footprints with complexes that contained a ZDV-terminated primer strand showed that approximately 20 μM of the next complementary nucleotide, dGTP, was required to obtain 50% of the posttranslocational configuration with both wild-type RT and RT containing L74V. Similar results were obtained with an enzyme that contained the M184V mutation (data not shown).
Viruses harboring L74V display decreased synthesis of (−)ssDNA, DNA produced after the first strand transfer, and full-length DNA in reverse transcription.
Recently, L74V- and M184V-containing enzymes have been shown to display diminished efficiency of initiation of minus- and plus-strand DNA synthesis (8). To study possible correlations between the decreased levels of minus- and plus-strand DNA synthesis by L74V and M184V RTs and the observed diminished replication capacity associated with L74V- and M184V-containing viruses, we evaluated the efficiency of the reverse transcription reaction by real-time PCR in a single round of infection.
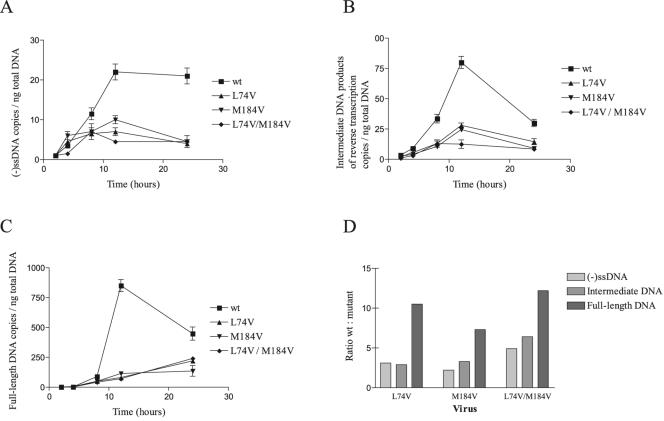
Quantification of the data showed that both wild-type and mutant viruses produced peak levels of (−)ssDNA, DNA produced after the first strand transfer, and full-length DNA at 12 h after infection, as previously documented (18). Moreover, the wild-type viruses produced 2.2 to 4.9 times more (−)ssDNA than did viruses containing either the L74V or M184V mutation or both L74V and M184V during this time (Fig. 4A). These differences were even more striking in the case of intermediate DNA products after the first strand transfer (Fig. 4B) and full-length DNA (Fig. 4C), for which 6.4-fold and 12.2-fold differences existed between the wild-type and doubly mutated L74V/M184V viruses, respectively (Fig. 4D).
FIG. 4.
Viral replication kinetics of different reverse-transcribed HIV-1 DNAs. We infected 106 H9 cells with wild-type and mutant HIV-1 viruses at an MOI of 0.0001. After 2, 4, 8, 12, and 24 h of infection, reverse-transcribed DNAs were quantified by real-time PCR as described in Materials and Methods. (A) (−)ssDNA. (B) Intermediate products of reverse transcription. (C) Full-length DNA. (D) Ratio between newly synthesized wild-type HIV-1 DNA and mutant HIV-1 DNA for the different reverse-transcribed products at 12 h of infection. Values are means of at least three independent experiments ± standard error.
DISCUSSION
Although the L74V mutation in HIV-1 RT is clinically associated with resistance to ddI and ABC (15, 24, 29), HIV-1 viruses harboring this mutation have been shown to display hypersusceptibility to ZDV (43). Even though the L74V mutation is known to result in diminished incorporation of both ddI and ABC in cell-free assays (36), the mechanism by which it can restore susceptibility to ZDV or establish hypersensitivity to ZDV has not been elucidated.
Both the M184V and K65R mutations in HIV-1 RT impair the rescue of chain-terminated DNA synthesis (3, 13, 27). Now we have shown that L74V-containing RTs also display impaired rescue of ZDV-MP chain-terminated primers compared with wild-type RT in the presence of ATP as a pyrophosphate donor. We postulate that this mechanism is probably responsible for the hypersensitivity to ZDV seen in tissue culture when viruses containing L74V are studied. In addition, we found slight differences in regard to unblocking of ZDV-terminated primers in the presence of PPi between wild-type and L74V RTs. Furthermore, the doubly mutated L74V/M184V RT displayed similar levels of ZDV-MP unblocking to those of M184V RT, suggesting that the effect of the M184V mutation exceeds that of the L74V mutation.
The presence of the L74V mutation in HIV-1 has been shown to suppress the appearance of TAMs (4). Recently, it has also been shown that HIV-1 RTs containing the L74V mutation in a TAM background significantly reduced ATP-mediated primer unblocking, thus partially reversing the effect of TAMs (L. R. Miranda, M. Götte, and D. R. Kuritzkes, submitted for publication) (30). This finding is consistent with the observations in this paper on the hypersensitization to ZDV of viruses containing L74V.
Several laboratories have proposed that ATP is the most important pyrophosphate donor under physiological conditions and excision can only occur at a pretranslocational stage (32, 35). Our footprinting experiments suggest that the L74V and M184V mutations do not affect the ratio between the populations of RT-DNA and DNA complexes found at the pre- and posttranslocational stages. Rather, these mutations may affect the precise alignment between the enzyme, the pyrophosphate donor ATP, and the chain-terminated primer-template such that the catalytic step is compromised. Alternatively, the enzyme may dissociate more frequently from its nucleic acid substrate before excision can occur. This would be consistent with reduced processivity of the mutant enzymes that contain L74V and/or M184V. These mutations may also affect affinity for ATP, in the pretranslocation position, which would likewise affect the excision reaction. Finally, the L74V and M184V mutations may affect the proper alignment between the incorporated chain terminator and the pyrophosphate donor.
Viruses harboring the L74V mutation have been shown to possess diminished replication capacity compared to wild-type viruses (39). Previous studies have also suggested that diminished RT processivity as well as diminished RNA primer usage may be factors associated with the decreased fitness of L74V or M184V-containing viruses (8). Here we have shown that L74V-, M184V-, and L74V/M184V-containing viruses display diminished synthesis of (−)ssDNA, DNA produced after the first strand transfer, and full-length DNA compared to wild-type viruses during a single round of infection. Moreover, the doubly mutated L74V/M184V virus showed the greatest reduction in this regard, consistent with the results of previously published viral replication assays (7).
The fact that L74V-containing viruses display diminished replication capacity as well as hypersensitivity to ZDV suggests that interpretation algorithms for genotypic resistance may need to be modified, although only randomized trials can validate a potential change in therapy.
These findings add to the evidence that the L74V and M184V mutations should be regarded as a group with regard to shared mechanisms of resistance to NRTIs and the consequences of these mutations on both RT enzymatic function and viral fitness. We now wish to study whether other drug resistance-related mutations in association with L74V and/or M184V will also lead to decreased efficiency of excision of chain-terminated primers.
Acknowledgments
We thank Anna Derjuga and Maureen Oliveira for technical assistance in real-time PCR and tissue culture assays, respectively.
Fernando A. Frankel is the recipient of a Canadian Institutes of Health Research (CIHR) doctoral fellowship award. This work was supported by grants from CIHR.
Footnotes
Dedicated to the memory of Silvio Frankel.
REFERENCES
- 1.Arion, D., and M. A. Parniak. 1999. HIV resistance to zidovudine: the role of pyrophosphorolysis. Drug. Resist. Updates 2:91-95. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, C. A., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−)enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, P. L., H. Q. Gao, and S. H. Hughes. 1998. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob. Agents Chemother. 42:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., C. Tantillo, A. Jacobo-Molina, R. G. Nanni, J. Ding, E. Arnold, and S. H. Hughes. 1994. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc. Natl. Acad. Sci. USA 91:4882-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P. K., B. Wynhoven, and P. R. Harrigan. 2004. 2004: which HIV-1 drug resistance mutations are common in clinical practice? AIDS Rev. 6:107-116. [PubMed] [Google Scholar]
- 7.Deval, J., J. M. Navarro, B. Selmi, J. Courcambeck, J. Boretto, P. Halfon, S. Garrido-Urbani, J. Sire, and B. Canard. 2004. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J. Biol. Chem. 279:25489-25496. [DOI] [PubMed] [Google Scholar]
- 8.Diallo, K., B. Marchand, X. Wei, L. Cellai, M. Gotte, and M. A. Wainberg. 2003. Diminished RNA primer usage associated with the L74V and M184V mutations in the reverse transcriptase of human immunodeficiency virus type 1 provides a possible mechanism for diminished viral replication capacity. J. Virol. 77:8621-8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, Q., Z. Gu, J. Hiscott, G. Dionne, and M. A. Wainberg. 1993. Generation of drug-resistant variants of human immunodeficiency virus type 1 by in vitro passage in increasing concentrations of 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, Q., Z. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, Q., M. A. Parniak, Z. Gu, and M. A. Wainberg. 1992. Generation of nucleoside-resistant variants of HIV-1 by in vitro selection in the presence of AZT or DDI but no by combinations. Leukemia 6(Suppl. 3):192S-195S. [PubMed] [Google Scholar]
- 12.Goldschmidt, V., and R. Marquet. 2004. Primer unblocking by HIV-1 reverse transcriptase and resistance to nucleoside RT inhibitors (NRTIs). Int. J. Biochem. Cell Biol. 36:1687-1705. [DOI] [PubMed] [Google Scholar]
- 13.Gotte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotte, M., M. Kameoka, N. McLellan, L. Cellai, and M. A. Wainberg. 2001. Analysis of efficiency and fidelity of HIV-1 (+)-strand DNA synthesis reveals a novel rate-limiting step during retroviral reverse transcription. J. Biol. Chem. 276:6711-6719. [DOI] [PubMed] [Google Scholar]
- 15.Harrigan, P. R., C. Stone, P. Griffin, I. Najera, S. Bloor, S. Kemp, M. Tisdale, and B. Larder. 2000. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J. Infect. Dis. 181:912-920. [DOI] [PubMed] [Google Scholar]
- 16.Hubner, A., M. Kruhoffer, F. Grosse, and G. Krauss. 1992. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural RNA. J. Mol. Biol. 223:595-600. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, R. T. D'Aquila, L. M. Demeter, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, A. Telenti, and D. D. Richman. 2004. Update of the drug resistance mutations in HIV-1: 2004. Top. HIV Med. 12:119-124. [PubMed] [Google Scholar]
- 18.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krebs, R., U. Immendorfer, S. H. Thrall, B. M. Wohrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 20.Le Grice, S. F., and F. Gruninger-Leitch. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 21.Liang, C., L. Rong, M. Laughrea, L. Kleiman, and M. A. Wainberg. 1998. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J. Virol. 72:6629-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locatelli, G. A., R. Cancio, S. Spadari, and G. Maga. 2004. HIV-1 reverse transcriptase inhibitors: current issues and future perspectives. Curr. Drug. Metab. 5:283-290. [DOI] [PubMed] [Google Scholar]
- 23.Marchand, B., and M. Gotte. 2003. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J. Biol. Chem. 278:35362-35372. [DOI] [PubMed] [Google Scholar]
- 24.Martin, J. L., J. E. Wilson, R. L. Haynes, and P. A. Furman. 1993. Mechanism of resistance of human immunodeficiency virus type 1 to 2′,3′-dideoxyinosine. Proc. Natl. Acad. Sci. USA 90:6135-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masquelier, B., D. Descamps, I. Carriere, F. Ferchal, G. Collin, M. Denayrolles, A. Ruffault, B. Chanzy, J. Izopet, C. Buffet-Janvresse, M. P. Schmitt, E. Race, H. J. Fleury, J. P. Aboulker, P. Yeni, and F. Brun-Vezinet. 1999. Zidovudine resensitization and dual HIV-1 resistance to zidovudine and lamivudine in the delta lamivudine roll-over study. Antivir. Ther. 4:69-77. [PubMed] [Google Scholar]
- 26.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 27.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Hum. immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 28.Miller, M. D., N. A. Margot, K. Hertogs, B. Larder, and V. Miller. 2001. Antiviral activity of tenofovir (PMPA) against nucleoside-resistant clinical HIV samples. Nucleosides Nucleotides Nucleic Acids 20:1025-1028. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V., M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce, and M. Tisdale. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:163-171. [DOI] [PubMed] [Google Scholar]
- 30.Miranda, L. R., and M. Gotte. 2005. Presented at the 12th Conference on Retroviruses and Opportunistic Infections, Boston, Mass., 23 February 2005, abstr. 699.
- 31.Morens, D. M., G. K. Folkers, and A. S. Fauci. 2004. The challenge of emerging and re-emerging infectious diseases. Nature 430:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naeger, L. K., N. A. Margot, and M. D. Miller. 2001. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir. Ther. 6:115-126. [PubMed] [Google Scholar]
- 33.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 34.Quan, Y., L. Rong, C. Liang, and M. A. Wainberg. 1999. Reverse transcriptase inhibitors can selectively block the synthesis of differently sized viral DNA transcripts in cells acutely infected with human immunodeficiency virus type 1. J. Virol. 73:6700-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 36.Ray, A. S., Z. Yang, J. Shi, A. Hobbs, R. F. Schinazi, C. K. Chu, and K. S. Anderson. 2002. Insights into the molecular mechanism of inhibition and drug resistance for HIV-1 RT with carbovir triphosphate. Biochemistry 41:5150-5162. [DOI] [PubMed] [Google Scholar]
- 37.Saksena, N. K., and S. J. Potter. 2003. Reservoirs of HIV-1 in vivo: implications for antiretroviral therapy. AIDS Rev. 5:3-18. [PubMed] [Google Scholar]
- 38.Selmi, B., J. Deval, K. Alvarez, J. Boretto, S. Sarfati, C. Guerreiro, and B. Canard. 2003. The Y181C substitution in 3′-azido-3′-deoxythymidine-resistant human immunodeficiency virus, type 1, reverse transcriptase suppresses the ATP-mediated repair of the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 278:40464-40472. [DOI] [PubMed] [Google Scholar]
- 39.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, P. L., and C. S. Crumpacker. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siliciano, J. D., and R. F. Siliciano. 2004. A long-term latent reservoir for HIV-1: discovery and clinical implications. J. Antimicrob. Chemother. 54:6-9. [DOI] [PubMed] [Google Scholar]
- 42.Spence, R. A., K. S. Anderson, and K. A. Johnson. 1996. HIV-1 reverse transcriptase resistance to nonnucleoside inhibitors. Biochemistry 35:1054-1063. [DOI] [PubMed] [Google Scholar]
- 43.St Clair, M. H., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 44.Victoria, J. G., D. J. Lee, B. R. McDougall, and W. E. Robinson, Jr. 2003. Replication kinetics for divergent type 1 human immunodeficiency viruses using quantitative SYBR green I real-time polymerase chain reaction. AIDS Res. Hum. Retroviruses 19:865-874. [DOI] [PubMed] [Google Scholar]
- 45.Wainberg, M. A., M. D. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]