Abstract
Genetic diversity of viral isolates in human immunodeficiency virus (HIV)-infected individuals varies substantially. However, it remains unclear whether HIV-related disease progresses more rapidly in patients harboring virus swarms with low or high diversity and, in the same context, whether high or low diversity is required to induce potent humoral and cellular immune responses. To explore whether viral diversity predicts virologic control, we studied HIV-infected patients who received antiretroviral therapy (ART) for years before undergoing structured treatment interruptions (STI). Viral diversity before initiation of ART and the ability of the patients to contain viremia after STI and final cessation of treatment was evaluated. Seven out of 21 patients contained plasma viremia at low levels after the final treatment cessation. Clonal sequences encompassing the envelope C2V3C3 domain derived from plasma prior to treatment, exhibited significantly lower diversity in these patients compared to those derived from patients with poor control of viremia. Viral diversity pre-ART correlated with the viral replication capacity of rebounding virus isolates during STI. Neutralizing antibody activity against autologous virus was significantly higher in patients who controlled viremia and was associated with lower pretreatment diversity. No such association was found with binding antibodies directed to gp120. In summary, lower pretreatment viral diversity was associated with spontaneous control of viremia, reduced viral replication capacity and higher neutralizing antibody titers, suggesting a link between viral diversity, replication capacity, and neutralizing antibody activity.
Human immunodeficiency virus type 1 (HIV-1) infection is characterized by continuous viral replication at a high rate, which, combined with the error rate of the reverse transcriptase (14, 52), frequent recombination (19, 82), and host selection pressure, leads to a high genetic diversity in infected individuals (43, 66, 69, 80, 94). However, the level of diversity between individual patients can vary considerably. Various viral and host properties may contribute to the observed diversity: these include differences in virulence, subtype, immunogenicity and replication capacity of the transmitted viruses, the quasispecies composition of the infecting inoculum (transmission of single versus multiple quasispecies), host genetic factors such as chemokine receptor polymorphisms, HLA types, and gender differences (3, 12, 58, 70, 74-76, 83, 84, 88).
Whether or not HIV-related disease progresses more rapidly in patients harboring viruses with low or with high diversity levels is currently not known. While some have argued that higher viral diversity may induce broader HIV-specific immune responses, which subsequently could contain viral replication more efficiently (96), others have found that patients with limited genetic diversity showed delayed disease progression and mounted stronger immune responses than rapid progressors (49, 50, 80).
In the simian immunodeficiency virus (SIV) model viral properties were found to substantially impact disease progression (40). Likewise, in HIV infection, individuals with high viral diversity during primary HIV infection progressed more rapidly (45, 75). Taken together, these findings suggest that viral properties influence disease progression and are at least in part responsible for the high variability in viremia control between HIV-infected individuals.
We have recently shown that viral capacity is a driving factor in determining the magnitude of viral rebound and viral set point in chronic HIV-1 infection after cessation of therapy (90). Here, we investigated whether the diversity of the HIV-1 envelope (env) gene present in plasma before the initiation of antiretroviral therapy (ART) predicted the ability of individuals to spontaneously control viremia after cessation of longterm successful ART. We further investigated whether viral properties, immune correlates, or host genetic factors could be identified explaining differences in viremia control among patients. We found that low pretreatment diversity was associated with spontaneous control of plasma viremia after cessation of therapy, low viral replication capacity, and strong neutralizing activity.
(Presented in part at the 10th Conference on Retroviruses and Opportunistic Infections, 10 to 14 February 2003, Boston, Massachusetts, abstract 496.)
MATERIALS AND METHODS
Subject selection, samples, and study design.
From the 29 subjects enrolled at the University Hospital Zürich into the Swiss-Spanish Intermittent Treatment Trial (SSITT) (18), pretreatment plasma samples were available from 26 patients in the repository of the Swiss HIV Cohort Study. In 24/26 patients, PCR amplification of a 417-bp fragment spanning the C2 to C3 region of HIV-1 env was successful. Amplification failed in two patients infected with non-B subtypes (E/CRF1 and subtype C). Two patients were excluded because they did not complete the SSITT trial and one because treatment was initiated during primary HIV infection.
Patients underwent four consecutive STI cycles (2 weeks off and 8 weeks on treatment), followed by a fifth long treatment interruption (a minimum of 12 weeks off treatment if no adverse effects occurred) during SSITT. None of the patients experienced drug failure and all had undetectable viral loads (<50 RNA copies/ml) for >6 months before study entry. Results of the clinical trial and detailed patient characteristics have been reported (18, 21, 23, 61, 63). Written informed consent was obtained from all patients according to the guidelines of the Ethics Committee of the University Hospital Zurich. Twenty-one patients were eligible for the present analysis and the salient characteristics of the study subjects are shown in Table 1.
TABLE 1.
Patient characteristics
| Patient group and patient no. | Age (yr) | Gendera | HIV-1 infection (mo) | HIV subtype | Coreceptor usage of viral isolates (5th STI cycle) | ART received before STIb | HIV RNA before ART (copies/ml) | Viral diversity before ART (%) | Total HIV RNA copy input/RT-PCR | Duration of ART before STI (mo) | HIV RNA post-STI plateau (copies/ml) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control of viremia | |||||||||||
| 106 | 35 | m | >24 | B | R5 | AZT, 3TC | 12,000 | 0.71 | 10,000 | 37 | 5,128 |
| 107 | 42 | f | >24 | B | R5 | AZT, 3TC, NFV | 5,216 | 2.12 | 4,347 | 32 | 2,551 |
| 112 | 42 | m | 6-12 | B | R5 | AZT, 3TC, IDV | 27,100 | 1.61 | 22,583 | 32 | 6,866 |
| 117 | 30 | f | 12-24 | B | R5 | AZT, 3TC, RTV | 29,522 | 0.88 | 24,602 | 40 | 13,007 |
| 118 | 29 | m | 12-24 | B | R5 | AZT, 3TC, IDV | 16,927 | 0.94 | 14,106 | 34 | 3,099 |
| 123 | 36 | f | >24 | B | R5 | AZT, 3TC, RTV | 14,410 | 2.87 | 12,008 | 37 | 118 |
| 124 | 41 | m | 12-24 | B | R5 | ddI, d4T, NFV | 22,464 | 2.93 | 18,720 | 20 | 6,218 |
| No control of viremia | |||||||||||
| 101 | 39 | m | >24 | B | R5 | ddI, d4T, NFV | 10,752 | 5.51 | 8,960 | 20 | 32,607 |
| 103 | 41 | m | >24 | E/CRF1 | R5 | ddI, d4T, NFV | 9,758 | 2.81 | 8,132 | 16 | 102,953 |
| 109 | 34 | m | >24 | B | R5 | AZT, 3TC, RTV | 42,277 | 4.02 | 8,455 | 34 | 148,191 |
| 111 | 32 | m | >24 | B | R5 | ddI, d4T, NFV | 129,408 | 4.66 | 107,840 | 17 | 22,030 |
| 113 | 56 | m | 12-24 | B | R5 | AZT, 3TC, RTV | 107,303 | 2.27 | 89,419 | 35 | 34,110 |
| 114 | 29 | m | >24 | B | R5 | AZT, 3TC, RTV | 9,275 | 2.74 | 7,729 | 35 | 28,478 |
| 115 | 22 | m | 12-24 | B | R5 | D4T, 3TC, NFV | 15,118 | 1.60 | 12,598 | 26 | 9,368 |
| 119 | 34 | m | 12-24 | B | R5 | D4T, 3TC, SQV, RTV | 98,561 | 1.60 | 82,134 | 18 | 99,274 |
| 120 | 51 | m | 12-24 | B | R5 | AZT, 3TC, IDV | 100,618 | 2.97 | 83,848 | 30 | 38,252 |
| 121 | 35 | m | 3-6 | B | R5 | D4T, 3TC, NFV | 164,772 | 1.23 | 137,310 | 21 | 67,321 |
| 122 | 38 | m | >24 | B | R5 | AZT, 3TC, IDV | 12,451 | 3.34 | 10,376 | 33 | 24,982 |
| 126 | 46 | m | >24 | B | R5 | AZT, 3TC, RTV | 63,698 | 3.87 | 53,082 | 39 | 19,782 |
| 127 | 48 | f | >24 | B | R5 | D4T, 3TC, NFV | 25,417 | 2.95 | 21,264 | 26 | 106,923 |
| 128 | 39 | f | >24 | B | R5 | AZT, ddI, NFV | 9,404 | 4.90 | 7,837 | 29 | 20,236 |
| Median, control of viremia | 16,927 | 1.61 | 14,106 | 34 | 5,128 | ||||||
| Median, no control of viremia | 33,897 | 2.96 | 16,931 | 28 | 33,359 | ||||||
| P (Mann-Whitney) | 0.31 | 0.023 | 0.58 | 0.093 | 0.0004 |
m = male, f = female
AZT, zidovudine; 3TC, lamivudine; NFV, nelfinavir; IDV, indinavir; RTV, ritonavir; ddI, dideoxyinosine; d4T, stavudine; SQV, saquinavir.
RNA extraction and, HIV-1 quantification.
RNA extraction from plasma was performed as described (22). Plasma HIV-1 RNA was quantified using the Amplicor HIV Monitor test, version 1.5 (Roche Diagnostics, Rotkreuz, Switzerland) with modifications resulting in a detection limit of <20 HIV-1 RNA copies/ml (78).
Quantification of HIV-1 DNA.
Extraction of peripheral blood mononuclear cells (PBMC) and quantification of HIV-1 DNA was performed as previously described (13, 25). The results were normalized to HIV copies per 106 cells on the basis of total cellular DNA measurement.
Amplification of HIV-1 env.
Reverse transcription-PCR was a modification of the previously described method (31), which has been adapted to one-tube reactions in order to minimize the risk of contamination (24, 72, 86). Combined cDNA synthesis and PCR (RobusT RT-PCR System, Finnzymes, Espoo, Finland) was performed according to the manufacturer's recommendations. The outer primers V3Fout (5′-CAAAGGTATC CTTTGAGCCA AT-3′) and V3Bout (5′-ATTACAGTAG AAAAATTCCC CT-3′) were used at 10 pmol each per reaction. Duplicate 50-μl reactions using 25 μl starting template RNA were performed for each sample. This corresponds to the total yield from 800 to 900 μl of plasma. The thermocycling conditions were 45 min at 48°C and 2 min at 94°C, followed by 35 cycles of 30 seconds, 30 seconds, and 60 seconds at 94°C, 56°C, and 72°C, respectively, and 1 cycle of 10 min at 72°C.
For the nested PCR, 1 μl of the first-round product was amplified in a second-round PCR (HotStarTaq Master Mix, Qiagen, Basel, Switzerland) performed according to the manufacturer's recommendations in a 50-μl reaction volume with 20 pmol each of the modified inner primer V3Fin2 (5′-GAACAGGACC ATGTACAAAT GTCAGCACAG TACAAT-3′) and V3Bin (5′-GCGTTAAAGC TTCTGGGTCC CCTCCTGAG-3′) and the following cycling conditions: 15 min at 95°C, followed by 35 cycles of 94°C for 30 seconds, 56°C for 30 seconds and, 72°C for 1 min, with a final extension at 72°C for 10 min.
The average misincorporation rate under these nested PCR conditions was 0.18%, as tested by amplification of HIV-1 strain YU2 (GenBank M93258) and subsequent analysis of 16 clones. This error rate is in line with other reports (48, 74).
Cloning and sequencing.
One μl of the pooled and purified PCR products was ligated into the plasmid vector pCR 4 using the TOPO TA cloning system (Invitrogen, Groningen, Netherlands); 16 individual clones were picked after bacterial transformation and stored in a microwell plate at −20°C, each in 100 μl of water.; one μl of this mix was used in a PCR with 20-μl reaction volume, 10 pmol each of the inner primers, and otherwise identical conditions to the second-round PCR described above. This circumvents the need for overnight cultures and subsequent plasmid purification. The fidelity of the taq polymerase was tested by duplicate analysis of 32 clones. Identical sequences were found in the paired comparisons (error rate: 0/26,000 bp), which indicates that no artifacts are introduced by the additional PCR step.
One μl of each clonal PCR product containing approximately 20 to 30 ng DNA was sequenced in both directions with the nested primers described above using BigDye chain terminator chemistry and the automated sequencer ABI 3100 (Applied Biosystems, Rotkreuz, Switzerland).
Phylogenetic analyses.
Sequences were edited and aligned with Lasergene software version 5.06 (DNASTAR Inc., Madison, WI). The alignments were manually corrected to adjust sequence gaps with the reading frame. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 2.1 (41). Genetic distance estimates obtained from the Tamura-Nei 6-parameter model are reported throughout. Nevertheless, equivalent results were also obtained when simpler nucleotide substitution models like Jukes-Cantor or Kimura 2-parameter were used. Neighbor joining phylogenetic trees were constructed by MEGA using HXB2 as the reference sequence and bootstrapping (1,000 replications). Amino acid diversities were estimated by the Poisson Correction model assuming equal substitution rates and equal amino acid frequencies. Maximum likelihood phylogenetic trees were constructed by use of DNAML (J. Felsenstein, PHYLIP Phylogeny Inference Package version 3.6, distributed by the author, Department of Genetics, University of Washington, Seattle).
T-cell responses. (i) Assessment of HIV p24-specific CD4+ T-cell frequencies.
Peripheral blood lymphocytes (PBL) were depleted of CD8+ T cells prior to analysis using anti-CD8 monoclonal antibody-conjugated magnetic beads (Dynal United Kingdom Ltd., United Kingdom). Overlapping pooled peptides spanning full-length HIV p24 protein (5 μg/ml, NIBSC, United Kingdom) were used to determine HIV-specific CD4+ T-cell frequencies directly ex vivo by gamma interferon (IFN-γ) ELIspot analysis (61, 63). Nonspecific phytohemagglutinin (PHA) stimulation (5 μg/ml) was used as a positive control and medium alone was used as a negative control in all assays. Results are expressed as specific spot-forming cells (SFC) per 106 CD8-depleted PBL; background values were subtracted from the specific response before normalization to 106 CD8-depleted PBL. A positive response to a given antigen was defined as SFC/106 CD8-depleted PBL greater than 3 standard deviations above background. Spot quantification was automated and standardized using an ELISPOT plate reader (Autoimmun Diagnostika, Germany, software version 2.1).
(ii) Assessment of CD8+ T-cell responses.
Each patient was HLA-typed and tested repeatedly (mean number of times tested, 16; range, 3 to 26) to assess the frequency of responsive HIV-specific cytotoxic T lymphocytes (CTLs) in their peripheral blood lymphocytes. Patients were tested for CTL responses with synthetic peptides corresponding to previously described HLA class I-restricted optimal HIV CTL epitopes (61). The epitopes tested are described in the Los Alamos database (www.hiv.lanl.gov/content/immunology/tables/ctl_summary.html) in the context of HIV infection, and the exact list of epitopes tested has been published previously (76). We have evaluated in 58 HIV patients the degree of correlation between the CD8+ T-cell frequencies measured by IFN-γ ELIspot using either overlapping peptide pools spanning the entire HIV genome or our set of 82 previously described CTL epitopes (63) (data not shown). We found a good correlation (R = 0.52, P = 0.003) between these approaches, indicating that our optimal peptide-based analysis of CD8+ T-cell frequencies in the present manuscript is a valid approximation of HIV-specific CD8+ T-cell frequencies.
Viral replication kinetics in vitro. (i) Autologous patient viruses.
Autologous virus was isolated from patient PBMCs at the beginning of the fifth interruption cycle (weeks 42 to 50) by coculturing patient CD4+ T cells with stimulated PBMCs (98). The 50% tissue culture infectious dose and coreceptor usage of the obtained virus stocks were determined as described previously (90).
(ii) In vitro replicative capacity.
Virus inoculum (100 50% tissue culture infectious doses in 50 μl) was added to 12 replicate wells of a 96-well culture plate containing 2 × 105 stimulated PBMCs in 150 μl of culture medium. Culture supernatant was assayed for p24 antigen on days 4, 6, 10, and 14 postinfection by using an in-house p24 antigen enzyme-linked immunosorbent assay as described previously (57, 91). As the virus inoculum was not washed out at any stage of the experiment, the residual input p24 concentration was measured and subtracted from all test results. Cocultures were fed 100 μl of medium on days 6 and 10 postinfection To allow interisolate comparisons, infection experiments for all isolates were performed on the same day by using the same target cells, a mixture of three healthy donors, to eliminate distortion of the results by donor cell variability (90).
Plasma neutralization activity.
Autologous plasma neutralization activity was evaluated in a PBMC-based assay using replication-competent patient isolates as described (89). Virus inoculum was incubated with serial dilutions of heat inactivated patient or control plasma for 1 h at 37°C. Stimulated CD8-depleted PBMC were then infected with aliquots of this preincubation mixture. After 72 h, cultures were washed three times, then supplemented with medium and freshly stimulated PBMC. Cultures were incubated for 6 to 10 days and then assayed for p24 antigen.
Binding antibodies to gp120 antigen.
Plasma immunoglobulin G (IgG) titers to recombinant glycoprotein 120 (gp120) from the JR-FL strain (provided by W. Olson, Progenics Pharmaceuticals, Tarrytown, NY) were determined by enzyme-linked immunosorbent assay (ELISA) as described (7). Bound antibody was detected using alkaline phospatase-conjugated anti-human IgG (Sigma, St. Louis, MO) and the luminescence-generating CPD-Star system (Applied Biosystems, Rotkreuz, Switzerland) as described (89).
Genotypic analysis.
PBMC DNA from study subjects was genotyped at CCR2 (V64I), CCR5 (G-2455A), CCR5 (delta 32), CX3CR1 (T280 M), IL-10 (C-592A), RANTES (G-403A), RANTES (C-28G), macrophage inflammatory protein 1α (MIP-1α, T113C), and SDF-1 (3′A) genes by using TaqMan allelic discrimination techniques (Applied Biosystems). The contribution of the various alleles to HIV-1 susceptibility has been described elsewhere (12, 81, 87). Determination of HLA genotypes was performed as reported (63, 89).
Data analysis.
For each patient, the following viral life history parameters were calculated.
(i) HIV RNA before HAART.
The pretreatment VL corresponds to the plasma HIV RNA value recorded before highly active ART (HAART) when a plasma sample was stored and plasma env diversity could be generated at this timepoint.
(ii) Post-STI VL.
The post-STI VL value reflects the viral set point, i.e., the plateau of viremia post-STI, and was determined as the geometric mean of plasma RNA levels measured after week 40, when a steady state was reached (usually between weeks 46 and 64). For patient 107, the week 46 data point was part of a peak and was therefore omitted from the estimation of the plateau. Overall, 6 to 12 data points were used to calculate the plateau VLs.
(iii) Control of viremia.
Patients were classified into a controlling group and a noncontrolling group according to their abilities to control viremia in the absence of ART between weeks 40 and 76. Control of viremia was defined as maintenance of a VL of <5,000 RNA copies/ml for at least 8 weeks during this time period. The cutoff of 5,000 RNA copies/ml was set in the SSITT trial as clinical endpoint to segregate responders from nonresponders (18).
(iv) In vitro replication capacity (slope between values from days 0 and 6).
Slopes were calculated by performing linear regression analysis using the natural logarithm of p24 antigen values obtained on days 0, 4, and 6 postinfection.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism version 4 (GraphPad Software, San Diego, CA). Nonparametric tests were used for group comparisons (Mann-Whitney) and correlations (Spearman).
Nucleotide sequence accession number.
All reported sequences have been deposited in GenBank and assigned accession numbers DQ002026 to DQ002345 and AY656249 to AY656264.
RESULTS
Patient characteristics and cloning of the C2 to C3 region of HIV env.
In total, 336 clones (16 clones per patient, 21 patients) spanning the C2 to C3 region of the viral envelope were derived from pretreatment plasma HIV RNA. Patient characteristics are summarized in Table 1. The length of the sequenced region typically ranged from 343 to 349 bp. Major gaps exceeding 9 bp were found in six clones. However, elimination of these deleted sequences from distance estimation did not alter the diversity within the observed groups (<0.03% difference).
Viral diversity and phylogenetic reconstruction.
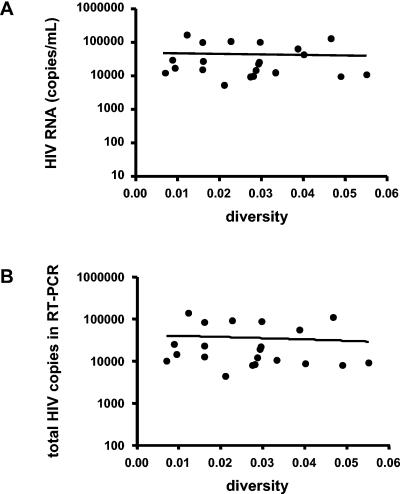
Pretreatment nucleotide sequence diversities ranged from 0.7 to 5.5%, and the amino acid diversities from 1.0 to 10.1%. To obtain representative samples of quasispecies, RT- and nested PCRs were performed in duplicate on HIV RNA extracts from 0.8 to 0.9 ml plasma. PCR products were pooled, cloned, and bidirectionally sequenced. All plasma samples tested revealed relatively high plasma virus concentrations (>5,000 copies/ml), making it likely that a range of viral strains rather than one dominant quasispecies was amplified. Furthermore, there was no correlation between viral diversity and plasma HIV RNA of all samples tested (r2 0.002, P = 0.84, Fig. 1). Thus, no evidence was found that low viral diversity was due to low input of HIV RNA for the samples tested. Taken together, our RT-PCR strategy and the relatively high plasma HIV RNA levels maximize the probability that the sequenced clones are representative of the actual viral population present in plasma in vivo.
FIG. 1.
Relationship between plasma virus concentrations and nucleotide sequence diversities observed in 16 clones isolated from each sample (A). The sequenced clones are representative of the actual viral populations present in vivo in 21 patients before antiretroviral therapy due to the high HIV copy number input into the RT-PCRs (total RNA extracted from 800 to 900 ul plasma was used) (B).
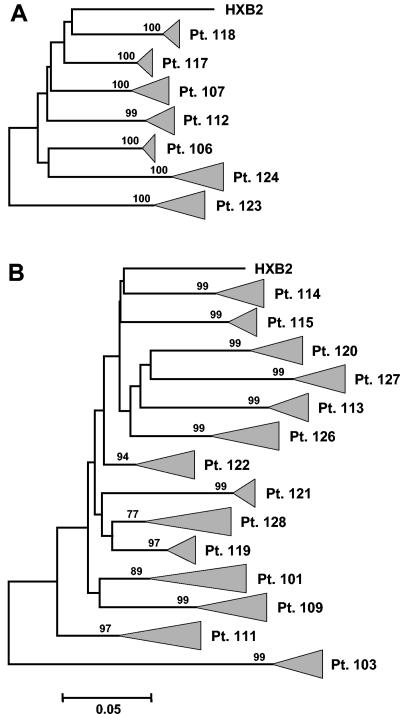
Sequences from each patient formed individual clusters with high bootstrap values (Fig. 2). Inferred neighbor joining phylogenetic trees with the 16 clones from each of the 21 subjects showed neither mixed interpatient clusters nor clustering with laboratory-adapted and “primary” HIV clones used in our laboratory (data not shown). Maximum likelihood trees also resulted in clear monophyletic clusters for clonal sequences from each patient (data not shown). We found diversity to be an intrinsic feature of a given host-virus interaction and not a simple function of time; the presumed duration of HIV infection did not correlate with diversity. However, time to infection could only be estimated within a certain range (Table 1).
FIG. 2.
Inferred neighbor-joining phylogenetic trees of clonal HIV-1 env C2-C3 sequences derived from seven patients in the controlling group (A) and 14 patients in the noncontrolling group (B). Triangles represent the compressed subtrees containing 16 clones isolated from plasma of each patient before antiretroviral therapy. The length of the triangle corresponds to the respective intrapatient diversity. Bootstrap values are indicated on each branch. The bar denotes 5% nucleotide divergence and diversity; HXB2 was included as external reference strain.
Relationship between pretreatment viral diversity and viremia after cessation of treatment.
To explore whether viral diversity had an impact on viral set points patients were classified into “controlling” and “noncontrolling” groups. Control of viremia was defined as spontaneous containment of the viral load to <5,000 RNA copies/ml for at least 8 weeks during the first 4 months after the last cessation of treatment (Table 1).
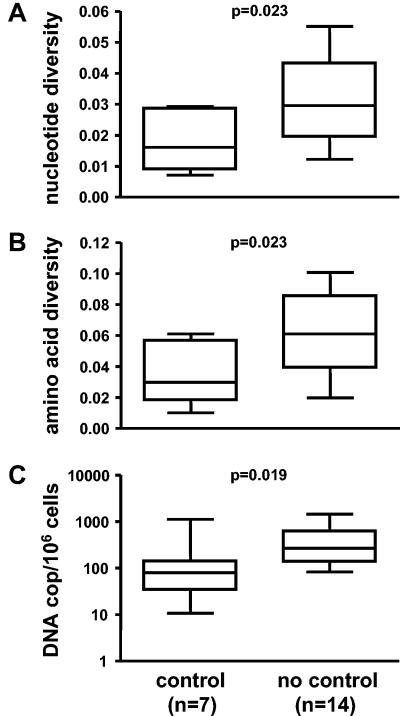
Pretreatment viral diversity was significantly lower in the seven patients who controlled viremia compared to that detected in the 14 noncontrollers (median nucleotide diversity: 1.61% for controllers [range, 0.7 to 2.9] and 2.96% for noncontrollers [range, 1.2 to 5.5], P = 0.02; median amino acid diversity: 2.97% for controllers [range, 1.0 to 6.1] and 6.11% for noncontrollers [range, 2.0 to 10.1], P = 0.02) (Fig. 3 A and 3 B). These findings are in agreement with our previous observation, which showed that containment of viremia in the controlling group was not induced by STIs but was rather the consequence of predefined viral and host properties (90). This was also supported by our finding that pretreatment viral load in the controlling group (median, 16,927; range, 5,216 to 29,522) tended to be lower than in the noncontrolling group (median, 33,897; range, 9275 to 164,772, Table 1). If the same pretreatment viral loads were taken as in the SSITT trial (geometric means of all preexisting pretreatment viral loads), then these pretreatment viral loads for the present study were significantly lower in the controllers (P = 0.019) compared to noncontrollers and these pretreatment viral load measurements did also significantly correlate with plateau viral load after STIs (P = 0.04) (data not shown).
FIG. 3.
Comparison between controlling and noncontrolling patients. Pretreatment nucleotide diversity (A) and amino acid diversity (B) within individual subjects (16 clones each). (C) HIV DNA levels in peripheral blood mononuclear cells during the first treatment interruption cycle following 1.6 to 3.9 years of continuous antiretroviral therapy. Box plots symbolize median value and interquartile range; whiskers extend over the entire range. P values were calculated by nonparametric Mann-Whitney test.
To define whether patients from the controlling group in general harbored fewer HIV infected cells than noncontrollers, we measured proviral DNA levels in peripheral blood mononuclear cells at the baseline of the first treatment interruption. HIV DNA levels were significantly lower in the controlling group (median, 80 [range, 11 to 1,127] copies per 106 cells) when compared to the noncontrolling group (median, 268 [83 to 1,446] copies per 106 cells, P = 0.02) (Fig. 3 C). Controllers were not treated longer with potent ART (Table 1). Therefore, a more pronounced decay of proviral DNA due to longer antiretroviral treatment can be excluded and lower proviral DNA levels in controllers are most likely due to an inherent limited viral replication capacity of viruses from controllers, when compared to noncontrollers, which is further supported by the same pattern found for pretreatment viral loads between groups (Table 1).
Relationship between in vitro replicative capacity and pretreatment env diversity.
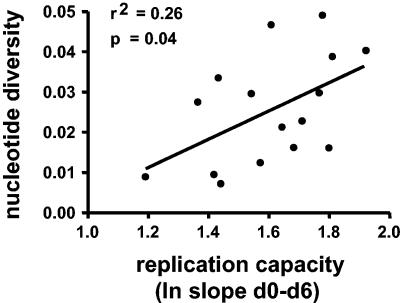
We have recently reported that the in vitro replicative capacity is a determinant of viral rebound and set point in chronic infection after cessation of therapy (90). Here, we explored the extent to which viral diversity is a defining characteristic of a virus population and whether, in turn, this predicts the fitness of a given virus swarm. To this end, we investigated the relationship between viral diversity, measured before the initiation of ART and the replicative capacity of virus isolates emerging several years later. The median time interval between determination of plasma env diversity and replication capacity during viral rebound of the last treatment interruption in SSITT was 41 months (range, 30 to 58 months). We found that the extent of pretreatment nucleotide env diversity was significantly correlated with the replication capacity determined years later (r2 = 0.26, P = 0.04, Fig. 4). Thus, we were able to demonstrate a link between virus swarms with a greater genetic diversity and virus isolates with higher replication capacity.
FIG. 4.
Correlation between pretreatment viral diversity and in vitro growth characteristics of virus isolated during the fifth treatment interruption. Replication capacity was derived from the profile of HIV p24 antigen production in cultures by linear regression (slope of natural logarithms of p24 levels over 6 days). Spearman correlation analysis: r2 = 0.26, P = 0.04, n = 16.
Relationship of pretreatment plasma env diversity, host genetic factors and adaptive immune responses.
In the analyses presented so far we have demonstrated that the replicative capacity of a patient's virus isolate, the genetic diversity of the virus and the patient's viral set point are significantly dependent on each other. However, thus far we have not gained insight into what determines whether a virus isolate manifests itself in a given host with high or low replicative capacity. Various innate and adaptive immune responses and genetic host factors have been demonstrated to affect viral fitness and disease progression.
In a first analysis we assessed if polymorphisms of genetic loci associated with faster or slower disease progression have an impact on viral diversity. To this end, patients were stratified according to the median diversity determined in our cohort and probed for polymorphisms associated with slow or rapid disease progression. We evaluated nine genetic polymorphisms: CCR2 (V64I), CCR5 (G-2455A), CCR5 (delta 32), CX3CR1 (T280 M), IL-10 (C-592A), RANTES (G-403A), RANTES (C-28G), MIP-1α (T113C), and SDF-1 (3′A) (33, 34, 56, 83, 92, 95), but none of these genes was found to be associated with either higher or lower genetic diversity.
A similar analysis was performed for HLA class I and II alleles that have previously been shown to be protective against disease progression (HLA-A*02, A*6802, A*11, B*27, B*51, B*57, B*58, Cw*08, and DRB1*01) or were found to be predictive for faster disease progression (HLA-A23, B*08, B*3501, B*45, B*53, and Cw*04) (11, 12, 47, 59, 76). Again, no correlation between HLA types associated with slower or faster disease progression and the degree of pretreatment viral diversity was found. Despite the fact that host genotypes tested in our patients did not explain differences in viral diversity, respectively in controlling viremia, we cannot rule out the possibility that host genotype exerts an influence. Indeed, effects of genetic polymorphisms on viral replication are relatively subtle and can generally only be demonstrated reliably in large patient cohorts, and multiple polymorphisms may act in concert. Thus, effects by genetic polymorphisms on viral replication in our small cohort could only be expected to be demonstrable, if there is an absolute association between particular alleles and viral replication.
Pretreatment viral diversity and cellular immune responses.
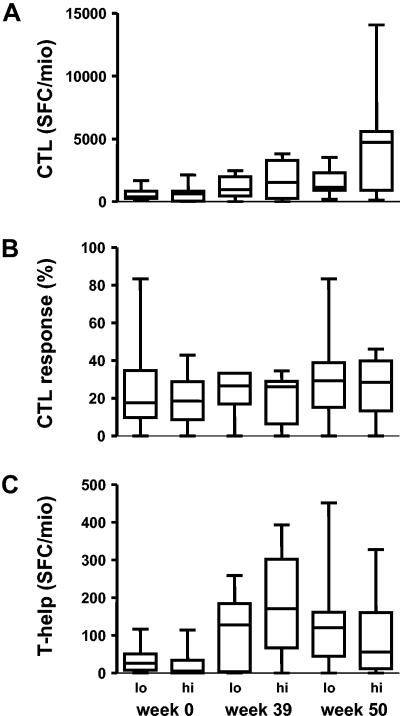
We previously studied CTL responses during the SSITT trial. Although there was an increase in the magnitude and breadth of cellular immune responses over the course of treatment interruptions, these variables were not associated with control of viremia (18, 61, 63). Here we specifically investigated whether controlling patients with low viral diversity in C2V3C3 would exhibit greater CTL responses compared to noncontrolling patients with higher C2V3C3 diversity. HIV-specific CTL responses were measured at week 0 (baseline), week 39 (before the fifth treatment interruption) and week 50 to 52 (during the final treatment interruption).
In line with the results of the larger patient group in the SSITT trial (63), no significant difference in magnitude (Fig. 5 A) or breadth (Fig. 5 B) of the CTL response was found between controlling and noncontrolling patients. During the STI trial, a transient increase in HIV Gag p24-specific CD4+ T-cell responses occurred (63), which was sustained in patients who had lower pretreatment viral loads. Here, we investigated whether patients with low pretreatment viral diversity were able to mount larger HIV-specific CD4+ T-cell responses when compared to patients with high viral diversity. However, no significant differences in CD4+ T helper responses between these groups were apparent (Fig. 5 C).
FIG. 5.
HIV-specific cellular immune responses in patients exhibiting low (lo = below median) and high (hi = above median) pretreatment viral diversity determined at week 0 (baseline of STI), week 39 (before the fifth treatment interruption) and week 50 (during the last treatment interruption). Magnitude of CD8+ cytotoxic T-cell response (CTL) expressed as total spot forming colonies (SFC) per 106 cells (A); 6-31 (median 18) different peptides were tested. The breadth of the CD8+ CTL response is indicated as the percentage of peptides tested that induced a response >50 SFC/106 cells (B). Magnitude of the HIV-specific CD4+ T-cell response expressed as SFC per 106 CD8-depleted PBMC (C).
Envelope diversity and neutralizing antibody response.
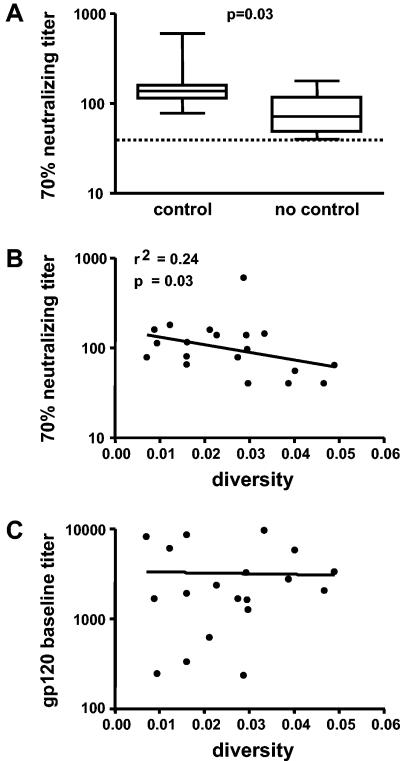
We next investigated if the neutralizing antibody response was associated with viral diversity. Neutralizing activity of plasma obtained at week 2 of the first treatment interruption was measured against the autologous virus isolate. We found that higher plasma neutralizing activity against autologous virus was associated with lower pretreatment nucleotide sequence diversity (r2 = 0.24, P = 0.034, Fig. 6 B) and amino acid diversity (r2 = 0.25, P = 0.028). In contrast, no association between diversity and titers of antibodies binding to monomeric anti-gp120 was found (Fig. 6 C). This finding underscores our previous observation that neutralizing antibody titers were higher in controlling than in noncontrolling patients (P = 0.031, Fig. 6 A) (89).
FIG. 6.
Comparison of 70% neutralizing titers found in 7 controlling and 12 noncontrolling patients (A). Neutralization activity of patient plasma derived from the first STI cycle was tested against autologous virus isolates derived during the fifth cycle. Dashed line represents the detection limit. Lower pretreatment diversity was correlated with higher neutralizing antibody activity (B, r2 = 0.24, P = 0.03, n = 19), whereas no correlation between diversity and titers of antibodies binding to monoclonal anti-gp120 was found (C).
DISCUSSION
It has been a longstanding debate as to whether HIV-related disease progresses more rapidly in patients harboring viruses with low or with high diversity and to what extent differences in diversity are related to host genetics and immune responses. To investigate this issue further, we performed a comprehensive virological, immunological, and genetic data analysis in chronically HIV-1-infected patients enrolled for years in the Swiss HIV Cohort Study: these patients were studied longitudinally prior to ART, during continuous ART, and while they subsequently participated in a recent structured treatment interruption trial (Swiss-Spanish Intermittent Therapy Trial) (18, 21, 23, 24, 55, 61 to 63, 76, 89, 90, 99, 100). Specifically, we wanted to determine whether envelope diversity from plasma HIV-1 RNA before initiation of antiretroviral treatment was predictive of an individual's ability to spontaneously control plasma viremia after cessation of ART. In addition, we probed viral properties and host characteristics, such as adaptive immune responses and host genetic factors, in order to elucidate the underlying mechanisms that steer viral diversity.
The key finding of these analyses was that envelope diversity before initiation of ART was significantly lower in patients who subsequently controlled plasma viremia during treatment interruption. This low pretreatment diversity observed in controllers implies that, in drug-naïve patients, viral strains had replicated to a lesser extent in controlling patients than in noncontrollers, who exhibited higher pretreatment diversities. Indeed, the fact that the degree of pretreatment env diversity not only predicted viremia upon cessation of ART but was also significantly correlated with the in vitro replication capacity of the viral isolates rebounding during this treatment interruption strongly suggests that the diminished replication properties of these viruses were most likely present before treatment and remained so. This view is supported further by the finding that PBMC from patients who controlled viremia and had low pretreatment diversity also harbored low levels of proviral DNA at the beginning of the treatment interruption. Thus, the viruses present in these patients not only replicated to a lesser extent, but also established a latent reservoir of reduced size (85, 101), which subsequently led to limited viral rebound and lower viral load plateaus during and after SSITT (18, 21, 25, 26, 100).
The questions of what causes lower diversity and how this relates to viral replication capacity then arise. HIV-specific cellular immune responses are thought to play a major role in controlling viremia (35, 44, 66, 77). However, although we have observed significant increases in HIV-specific CTL and T-helper responses in SSITT, those increases did not predict control of viremia but rather reflected antigen-driven expansions of T-cell responses due to viral recrudescence (18, 61, 63). Lacking correlation of HIV-specific immune responses, mostly assessed by antigen-induced IFN-γ production, with control of plasma viremia were also described in studies performed by other groups (1, 5, 10, 38), suggesting that there is not direct correlation between the magnitude of the HIV-specific T-cell response, as measured by current techniques, and the level of viral replication. Such a linear correlation would actually be unlikely to exist, since both antigen load (i.e., the level of viral replication) and HIV-specific T-cell immunity influence each other. Furthermore, it is likely that current techniques to measure overall HIV-specific immune responses with consensus or even with autologous HIV peptides may not adequately describe the in vivo functionality of those cells in the absence of knowledge about the contemporaneous full-length viral sequences. To this aim, longitudinal studies have to be performed starting at primary HIV infection, in which serial complete autologous viral sequencing is performed in conjunction with assessing T-cell responsiveness to contemporaneous T-cell antigens. Such studies have been performed and clearly indicated that CTL-driven epitope variation often occurs at various stages of infection (9, 36, 39, 43, 60, 64, 66).
In some of these studies it was shown that selection of CTL escape viruses was associated with reduced viral control and with loss of viral fitness (4, 27, 29).
Thus, CD8+ T-cell responses are clearly also involved in driving viral diversity. As such, the type of analysis of T-cell responses and viral diversity in this study does not address this question; however it indicates that there are no overall differences in the magnitude of T-cell frequencies between our patients with low or high env diversity.
Furthermore, it is conceivable that current techniques to measure HIV-specific immune responses may not adequately reflect the critical function of these cells that mediates control of HIV replication or that defense against HIV relies more on other adaptive or innate mechanisms (6, 71, 97). Interestingly, however, when exploring the humoral immune response we found a clear association between low pretreatment diversity and strong neutralizing antibody titers against autologous virus at baseline of the SSITT protocol.
Two possible scenarios can be envisioned: First, patients with low pretreatment diversity could have raised substantial neutralizing antibody responses early during infection and, subsequently were able to contain viral replication to a certain degree. In support of this, it has recently been shown that during and after primary infection, a neutralizing antibody response is generated that exerts a strong selection pressure on HIV env and leads to the rapid emergence of viral escape mutants (69, 94). Thus, it can be hypothesized that the containment of viremia seen in a subset of our patients might have been due to the continuous generation of neutralizing antibodies that lead to sustained suppression of viremia (32).
Second, patients with low pretreatment diversity may have been infected by viruses of low pathogenicity, which subsequently allowed a strong neutralizing antibody response to emerge. Over recent years, evidence has accumulated that properties of the infecting virus are important determinants of disease progression (8, 15, 16, 20, 30, 37, 42, 67, 79, 93). Kimata et al. showed that infection of macaques with SIV strains, derived from SIV-infected macaques at different time points after infection, exhibited differential pathogenicity in the new host; viruses derived from later stages being more pathogenic. This difference in pathogenicity was not due to the emergence of syncytium-inducing viruses but was attributed to the fact that animals infected with the early and intermediate phase viruses were able to raise significant neutralizing antibody responses, while the rapid progressors infected with late stage viruses failed to do so (40).
Further evidence that viral properties are important comes from the same group, which showed that patients who harbor viruses with higher diversity during primary infection, most likely representing infection events by multiple viruses, have higher viral set points and faster disease progression than patients with low diversity (45, 75). Taken together, one could hypothesize that our patients who harbored viruses with low level replication kinetics and low pretreatment diversity were primarily infected with viruses of reduced pathogenicity, which then allowed a more prominent neutralizing antibody response to develop that aided subsequent control of viremia. The fact that viruses with lower pretreatment diversity and reduced replication capacity also exhibited increased sensitivity to β-chemokines (90) supports the view that these viruses were intrinsically different from viruses derived from noncontrollers independently of the immunological response (data not shown).
Nevertheless, other factors that have been associated with better control of viral replication in large-cohort studies, such as host genetic polymorphisms and HLA types, must also be considered (12). In this study, neither genetic host polymorphisms in the genes for CCR2, CCR3, CCR5, RANTES, MIP-1α, SDF-1, or IL-10 nor HLA genotypes were associated with the degree of viral pretreatment diversity. In addition, no major variants harboring mutations associated with drug resistance were detected either in controllers or in noncontrollers (55, 99), thus excluding potential differences in replication capacity caused by resistance to antiretroviral drugs (51, 65, 68).
Our data are in agreement with Markham (50), Mani (49), and McNearney (54), who found that, among seroconverters, rapid progressors exhibited higher viral diversity than moderate or slow progressors. In contrast, several studies have found the opposite: rapid progression associated with low viral diversity (2, 17, 28, 46, 53, 96).
For example, Wolinsky et al. (96) described stronger HIV-specific CTL responses in patients with high env diversity and lower viral set points compared to rapid progressors, who exhibited low diversity and weak CTL responses. However, we observed no relationship between either HIV-specific CD8+ CTL responses or CD4+ T helper cell responses and viral diversity (Fig. 5). What could be the reasons for such conflicting results? In many of the earlier studies, only limited numbers of patients were studied. Furthermore, patients were mostly selected from large cohorts according to extremes of disease progression, e.g., rapid CD4 cell loss versus stable CD4 count. This strategy bears a high potential to also select for other factors associated with rapid or slow disease progression, such as certain chemokine receptor polymorphisms and HLA alleles, which now are known to influence HIV disease progression substantially in some cases. Host genetic factors and viral determinants were not described in most of these studies. Thus, strong bias due to unknown viral and host factors may have been introduced and may at least in part have been responsible for discrepant findings and therefore comparability of the conflicting studies is limited.
We acknowledge the potential limitations of our study. Our study was relatively small, limiting the power of the genetic analysis, with a risk of beta-type error (concluding that there was no difference, although in fact one did exist).
In addition, we could not follow patients from the time of seroconversion, and thus we cannot exclude completely that controlling patients were infected for shorter time periods. Nevertheless, the time between when infection occurred and when antiretroviral treatment was started could be estimated (Table 1). We found no significant difference when the numbers of patients with time to infection <24 months and >24 months were compared (P = 0.4) between controllers and noncontrollers. Moreover, a lack of influence of infection time is further supported by the observation that in the controllers we have found very low diversities (<1%) in three patients, which were at the most conservative estimate infected for at least 1 to 2 years when diversity was measured, whereas such low diversity was not seen in any of the noncontrollers. Recently, we have observed diversities of <1% (median, 0.42; range, 0.26 to 0.96) in 10 subjects with well-characterized primary HIV infection (73). In the noncontrollers, diversities of <1% were never detected despite the fact that in this group estimates of time from infection to determination of diversity could also be estimated to 12 to 24 months. Taken together the available data shows no suggestion that controllers were infected for a shorter period of time than the noncontrollers.
Moreover, the prospective nature of our study and the correlation found between pretreatment diversities, in vitro replication capacity and neutralizing antibodies (representing different assay types at different time points) make a bias based on patient selection even more unlikely. All viruses studied were R5 users, and during STIs no coreceptor switch was detected (90); also, all but one were subtype B, thus further reducing potential confounding factors (58).
In conclusion, we found that low pretreatment viral diversity was associated with better spontaneous control of plasma viremia after the last treatment interruption. The observation that viruses rebounding from patients with low pretreatment diversity years after successful ART demonstrated a reduced ability to replicate ex vivo, independent of the host cellular immune response, suggest that intrinsic viral properties, at least partly, were responsible for reduced viral replication, which resulted in lower viral diversity. Whether the higher neutralizing antibody titers found in controlling patients are cause or consequence of this reduced viral diversity and impaired replication capacity will need to be determined in longitudinal seroconverter studies. Nevertheless, our data suggest that viral characteristics in conjunction with neutralizing antibody activity are participating in shaping the course of HIV disease.
Acknowledgments
This study was financed within the framework of the Swiss HIV Cohort Study, supported by the Swiss National Science Foundation (grant no. 3345-062041). Support was provided by the Swiss National Science Foundation (grant no. 3345-65168 and 3100AO-103748/1 to H. F. Günthard and A. Trkola and grant no PP00B-102647 to A. Trkola), the Swiss HIV Cohort Study project 290, and a grant from the EMBDO Foundation (to H. F. Günthard), a subcontract to A.T. from NIH grant R37 AI36082 to J. P. Moore, and a grant from the Kanton of Zürich. D. Price is a Medical Research Council (United Kingdom) Clinician Scientist.
We thank our patients for their commitment, Christine Schneider and Roland Hafner for excellent patient care, Jean-Marie Tiercy for HLA typing, Gabi Bleiber for chemokine receptor polymorphism testing, Amalio Telenti for critical reading of the manuscript, Esther Beerli, Tuyeth Trinh Lu, Friederike Burgener, and Erika Schlaepfer for laboratory support, and Ingrid Nievergelt and Christine Vögtli for administrative assistance.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnarelli, P., F. Mazzola, S. Menzo, M. Montroni, L. Butini, and M. Clementi. 1999. Host-specific modulation of the selective constraints driving human immunodeficiency virus type 1 env gene evolution. J. Virol. 73:3764-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, J. D., F. M. Hecht, T. Wrin, M. R. Segal, C. A. Ramstead, T. J. Liegler, M. P. Busch, C. J. Petropoulos, N. S. Hellmann, J. O. Kahn, and R. M. Grant. 2004. Higher CD4+ T-cell counts associated with low viral pol replication capacity among treatment-naive adults in early HIV-1 infection. J. Infect. Dis. 190:251-256. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. R., J. M. Brenchley, D. A. Price, S. C. De Rosa, D. C. Douek, M. Roederer, and R. A. Koup. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281:65-78. [DOI] [PubMed] [Google Scholar]
- 7.Binley, J. M., P. J. Klasse, Y. Cao, I. Jones, M. Markowitz, D. D. Ho, and J. P. Moore. 1997. Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J. Virol. 71:2799-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. [DOI] [PubMed] [Google Scholar]
- 9.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat.Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 10.Cao, J., J. McNevin, S. Holte, L. Fink, L. Corey, and M. J. McElrath. 2003. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 77:6867-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 12.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med 54:535-551. [DOI] [PubMed] [Google Scholar]
- 13.Christopherson, C., Y. Kidane, B. Conway, J. Krowka, H. Sheppard, and S. Kwok. 2000. PCR-Based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 15.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deeks, S. G., R. M. Grant, T. Wrin, E. E. Paxinos, T. Liegler, R. Hoh, J. N. Martin, and C. J. Petropoulos. 2003. Persistence of drug-resistant HIV-1 after a structured treatment interruption and its impact on treatment response. AIDS 17:361-370. [DOI] [PubMed] [Google Scholar]
- 17.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagard, C., A. Oxenius, H. Gunthard, F. Garcia, M. Le Braz, G. Mestre, M. Battegay, H. Furrer, P. Vernazza, E. Bernasconi, A. Telenti, R. Weber, D. Leduc, S. Yerly, D. Price, S. J. Dawson, T. Klimkait, T. V. Perneger, A. McLean, B. Clotet, J. M. Gatell, L. Perrin, M. Plana, R. Phillips, and B. Hirschel. 2003. A prospective trial of structured treatment interruptions in human immunodeficiency virus infection. Arch. Intern. Med. 163:1220-1226. [DOI] [PubMed] [Google Scholar]
- 19.Fang, G., B. Weiser, C. Kuiken, S. M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, O. Anzala, and H. Burger. 2004. Recombination following superinfection by HIV-1. AIDS 18:153-159. [DOI] [PubMed] [Google Scholar]
- 20.Fenyo, E. M., L. Morfeldt-Manson, F. Chiodi, B. Lind, A. von Gegerfelt, J. Albert, E. Olausson, and B. Asjo. 1988. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J. Virol. 62:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, M., R. Hafner, C. Schneider, A. Trkola, B. Joos, H. Joller, B. Hirschel, R. Weber, and H. F. Günthard. 2003. HIV RNA in plasma rebounds within days during structured treatment interruptions. AIDS 17:195-199. [DOI] [PubMed] [Google Scholar]
- 22.Fischer, M., W. Huber, A. Kallivroussis, P. Ott, M. Opravil, R. Luthy, R. Weber, and R. W. Cone. 1999. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J. Clin. Microbiol. 37:1260-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer, M., B. Joos, B. Hirschel, G. Bleiber, R. Weber, H. F. Günthard, and the Swiss HIV Cohort Study. 2004. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J. Infect. Dis. 190:1979-1988. [DOI] [PubMed] [Google Scholar]
- 24.Fischer, M., B. Joos, J. K. Wong, P. Ott, M. Opravil, B. Hirschel, R. Weber, and H. F. Günthard. 2004. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J. Infect. Dis. 189:273-285. [DOI] [PubMed] [Google Scholar]
- 25.Fischer, M., A. Trkola, B. Joos, R. Hafner, H. Joller, M. A. Muesing, D. R. Kaufman, E. Berli, B. Hirschel, R. Weber, and H. F. Günthard. 2003. Shifts in cell-associated HIV-1 RNA but not in episomal HIV-1 DNA correlate with new cycles of HIV-1 infection in vivo. Antivir. Ther. 8:97-104. [PubMed] [Google Scholar]
- 26.Fischer, M., J. K. Wong, D. Russenberger, B. Joos, M. Opravil, B. Hirschel, A. Trkola, H. Kuster, R. Weber, and H. F. Günthard. 2002. Residual cell-associated unspliced HIV-1 RNA in peripheral blood of patients on potent antiretroviral therapy represents intracellular transcripts. Antivir. Ther. 7:91-103. [PubMed] [Google Scholar]
- 27.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med 10:275-281. [DOI] [PubMed] [Google Scholar]
- 28.Ganeshan, S., R. E. Dickover, B. T. Korber, Y. J. Bryson, and S. M. Wolinsky. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 30.Grovit-Ferbas, K., J. Ferbas, V. Gudeman, S. Sadeghi, M. B. Goetz, J. V. Giorgi, I. S. Chen, and W. A. O'Brien. 1998. Potential contributions of viral envelope and host genetic factors in a human immunodeficiency virus type 1-infected long-term survivor. J. Virol. 72:8650-8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Günthard, H. F., S. D. Frost, A. J. Leigh-Brown, C. C. Ignacio, K. Kee, A. S. Perelson, C. A. Spina, D. V. Havlir, M. Hezareh, D. J. Looney, D. D. Richman, and J. K. Wong. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haigwood, N. L., D. C. Montefiori, W. F. Sutton, J. McClure, A. J. Watson, G. Voss, V. M. Hirsch, B. A. Richardson, N. L. Letvin, S. L. Hu, and P. R. Johnson. 2004. Passive immunotherapy in simian immunodeficiency virus-infected macaques accelerates the development of neutralizing antibodies. J. Virol. 78:5983-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ioannidis, J. P., D. G. Contopoulos-Ioannidis, P. S. Rosenberg, J. J. Goedert, A. De Rossi, T. Espanol, L. Frenkel, M. J. Mayaux, M. L. Newell, S. G. Pahwa, C. Rousseau, G. Scarlatti, S. Sei, L. Sen, and T. R. O'Brien. 2003. Effects of CCR5-delta32 and CCR2-64I alleles on disease progression of perinatally HIV-1-infected children: an international meta-analysis. AIDS 17:1631-1638. [DOI] [PubMed] [Google Scholar]
- 34.Ioannidis, J. P., P. S. Rosenberg, J. J. Goedert, L. J. Ashton, T. L. Benfield, S. P. Buchbinder, R. A. Coutinho, J. Eugen-Olsen, T. Gallart, T. L. Katzenstein, L. G. Kostrikis, H. Kuipers, L. G. Louie, S. A. Mallal, J. B. Margolick, O. P. Martinez, L. Meyer, N. L. Michael, E. Operskalski, G. Pantaleo, G. P. Rizzardi, H. Schuitemaker, H. W. Sheppard, G. J. Stewart, I. D. Theodorou, H. Ullum, E. Vicenzi, D. Vlahov, D. Wilkinson, C. Workman, J. F. Zagury, and T. R. O'Brien. 2001. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann. Intern. Med. 135:782-795. [DOI] [PubMed] [Google Scholar]
- 35.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T-cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200:1243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann, D., M. Munoz, G. Bleiber, S. Fleury, B. Lotti, R. Martinez, W. Pichler, P. Meylan, and A. Telenti. 2000. Virological and immunological characteristics of HIV treatment failure. AIDS 14:1767-1774. [DOI] [PubMed] [Google Scholar]
- 38.Kaufmann, D. E., P. M. Bailey, J. Sidney, B. Wagner, P. J. Norris, M. N. Johnston, L. A. Cosimi, M. M. Addo, M. Lichterfeld, M. Altfeld, N. Frahm, C. Brander, A. Sette, B. D. Walker, and E. S. Rosenberg. 2004. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 78:4463-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 41.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 42.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 43.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 44.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long, E. M., H.-L. J. Martin, J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat.Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 46.Lukashov, V. V., C. L. Kuiken, and J. Goudsmit. 1995. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J. Virol. 69:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald, K. S., L. Matukas, J. E. Embree, K. Fowke, J. Kimani, N. J. Nagelkerke, J. Oyugi, P. Kiama, R. Kaul, M. A. Luscher, S. Rowland-Jones, J. Ndinya-Achola, E. Ngugi, J. J. Bwayo, and F. A. Plummer. 2001. Human leucocyte antigen supertypes and immune susceptibility to HIV-1, implications for vaccine design. Immunol. Lett. 79:151-157. [DOI] [PubMed] [Google Scholar]
- 48.Malet, I., M. Belnard, H. Agut, and A. Cahour. 2003. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J. Virol. Methods 109:161-170. [DOI] [PubMed] [Google Scholar]
- 49.Mani, I., P. Gilbert, J. L. Sankale, G. Eisen, S. Mboup, and P. J. Kanki. 2002. Intrapatient diversity and its correlation with viral setpoint in human immunodeficiency virus type 1 CRF02_A/G-IbNG infection. J. Virol. 76:10745-10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markham, R. B., W. C. Wang, A. E. Weisstein, Z. Wang, A. Munoz, A. Templeton, J. Margolick, D. Vlahov, T. Quinn, H. Farzadegan, and X. F. Yu. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T-cell decline. Proc. Natl. Acad. Sci. USA 95:12568-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 53.McDonald, R. A., D. L. Mayers, R. C. Chung, K. F. Wagner, S. Ratto-Kim, D. L. Birx, and N. L. Michael. 1997. Evolution of human immunodeficiency virus type 1 I sequence variation in patients with diverse rates of disease progression and T-cell function. J. Virol. 71:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNearney, T., Z. Hornickova, R. Markham, A. Birdwell, M. Arens, A. Saah, and L. Ratner. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA 89:10247-10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metzner, K. J., S. Bonhoeffer, M. Fischer, R. Karanicolas, K. Allers, B. Joos, R. Weber, B. Hirschel, G. Kostrikis, and H. F. Gunthard. 2003. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J. Infect. Dis. 188:1433-1443. [DOI] [PubMed] [Google Scholar]
- 56.Michael, N. L., L. G. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C. E. Wang, and H. W. Sheppard. 1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160-1162. [DOI] [PubMed] [Google Scholar]
- 57.Moore, J. P., Y. Cao, D. D. Ho, and R. A. Koup. 1994. Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J. Virol. 68:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neilson, J. R., G. C. John, J. K. Carr, P. Lewis, J. K. Kreiss, S. Jackson, R. W. Nduati, D. Mbori-Ngacha, D. D. Panteleeff, S. Bodrug, C. Giachetti, M. A. Bott, B. A. Richardson, J. Bwayo, J. Ndinya-Achola, and J. Overbaugh. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 61.Oxenius, A., A. R. Mclean, M. Fischer, D. A. Price, S. J. Dawson, R. Hafner, C. Schneider, H. Joller, B. Hirschel, R. E. Phillips, R. Weber, and H. F. Günthard. 2002. Human immunodeficiency virus-specific CD8(+) T-cell responses do not predict viral growth and clearance rates during structured intermittent antiretroviral therapy. J. Virol. 76:10169-10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oxenius, A., D. A. Price, S. J. Dawson, H. F. Günthard, M. Fischer, L. Perrin, E. Ramirez, C. Fagard, B. Hirschel, G. Scullard, J. N. Weber, A. R. Mclean, and R. E. Phillips. 2002. Residual HIV-specific CD4 and CD8 T-cell frequencies after prolonged antiretroviral therapy reflect pretreatment plasma virus load. AIDS 16:2317-2322. [DOI] [PubMed] [Google Scholar]
- 63.Oxenius, A., D. A. Price, H. F. Günthard, S. J. Dawson, C. Fagard, L. Perrin, M. Fischer, R. Weber, M. Plana, F. Garcia, B. Hirschel, A. McLean, and R. E. Phillips. 2002. Stimulation of HIV-specific cellular immunity by structured treatment interruption fails to enhance viral control in chronic HIV infection. Proc. Natl. Acad. Sci. USA 99:13747-13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oxenius, A., D. A. Price, A. Trkola, C. Edwards, E. Gostick, H. T. Zhang, P. J. Easterbrook, T. Tun, A. Johnson, A. Waters, E. C. Holmes, and R. E. Phillips. 2004. Loss of viral control in early HIV-1 infection is temporally associated with sequential escape from CD8+ T-cell responses and decrease in HIV-1-specific CD4+ and CD8+ T-cell frequencies. J. Infect. Dis. 190:713-721. [DOI] [PubMed] [Google Scholar]
- 65.Prado, J. G., S. Franco, T. Matamoros, L. Ruiz, B. Clotet, L. Menendez-Arias, M. A. Martinez, and J. Martinez-Picado. 2004. Relative replication fitness of multi-nucleoside analogue-resistant HIV-1 strains bearing a dipeptide insertion in the fingers subdomain of the reverse transcriptase and mutations at codons 67 and 215. Virology 326:103-112. [DOI] [PubMed] [Google Scholar]
- 66.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritola, K., C. D. Pilcher, S. A. Fiscus, N. G. Hoffman, J. A. Nelson, K. M. Kitrinos, C. B. Hicks, J. J. Eron, Jr., and R. Swanstrom. 2004. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J. Virol. 78:11208-11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roederer, M., J. M. Brenchley, M. R. Betts, and S. C. De Rosa. 2004. Flow cytometric analysis of vaccine responses: how many colors are enough? Clin Immunol. 110:199-205. [DOI] [PubMed] [Google Scholar]
- 72.Rusert, P., M. Fischer, B. Joos, C. Leemann, H. Kuster, M. Flepp, S. Bonhoeffer, H. F. Gunthard, and A. Trkola. 2004. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology 326:113-129. [DOI] [PubMed] [Google Scholar]
- 73.Rusert, P., H. Kuster, B. Joos, B. Misselwitz, C. Gujer, C. Leemann, M. Fischer, G. Stiegler, H. Katinger, W. C. Olson, R. Weber, L. Aceto, H. F. Günthard, and A. Trkola. Virus isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. J. Virol. 79:8454-8469. [DOI] [PMC free article] [PubMed]
- 74.Sagar, M., E. Kirkegaard, E. M. Long, C. Celum, S. Buchbinder, E. S. Daar, and J. Overbaugh. 2004. Human immunodeficiency virus type 1 (HIV-1) diversity at time of infection is not restricted to certain risk groups or specific HIV-1 subtypes. J. Virol. 78:7279-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sagar, M., L. Lavreys, J. M. Baeten, B. A. Richardson, K. Mandaliya, B. H. Chohan, J. K. Kreiss, and J. Overbaugh. 2003. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J. Virol. 77:12921-12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 101:12266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 78.Schockmel, G. A., S. Yerly, and L. Perrin. 1997. Detection of low HIV-1 RNA levels in plasma. J. Acquir. Immune. Defic. Syndr. Human Retrovirol. 14:179-183. [DOI] [PubMed] [Google Scholar]
- 79.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shin, H. D., C. Winkler, J. C. Stephens, J. Bream, H. Young, J. J. Goedert, T. R. O'Brien, D. Vlahov, S. Buchbinder, J. Giorgi, C. Rinaldo, S. Donfield, A. Willoughby, S. J. O'Brien, and M. W. Smith. 2000. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL-10. Proc. Natl. Acad. Sci. USA 97:14467-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shriner, D., A. G. Rodrigo, D. C. Nickle, and J. I. Mullins. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 167:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, and S. J. O'Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 84.Spira, S., M. A. Wainberg, H. Loemba, D. Turner, and B. G. Brenner. 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 51:229-240. [DOI] [PubMed] [Google Scholar]
- 85.Strain, M. C., H. F. Gunthard, D. V. Havlir, C. C. Ignacio, D. M. Smith, A. J. Leigh-Brown, T. R. Macaranas, R. Y. Lam, O. A. Daly, M. Fischer, M. Opravil, H. Levine, L. Bacheler, C. A. Spina, D. D. Richman, and J. K. Wong. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strain, M. C., S. Letendre, S. Pillai, T. Russell, C. C. Ignacio, H. F. Günthard, B. Good, D. M. Smith, C. Wiley, S. M. Wolinsky, M. Furtado, I. Grant, D. D. Richman, J. Marquies-Beck, J. Durelle, J. A. McCutchan, R. J. Ellis, and J. K. Wong. 2005. Genetic composition of human immunodeficiency virus type 1 in cerebrospinal fluid and blood without treatment and during failing antiretroviral therapy. J. Virol. 79:1772-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Telenti, A., V. Aubert, and F. Spertini. 2002. Individualising HIV treatment-pharmacogenetics and immunogenetics. Lancet 359:722-723. [DOI] [PubMed] [Google Scholar]
- 88.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 89.Trkola, A., H. Kuster, C. Leemann, A. Oxenius, C. Fagard, H. Furrer, M. Battegay, P. Vernazza, E. Bernasconi, R. Weber, B. Hirschel, S. Bonhoeffer, and H. F. Günthard. 2004. Humoral immunity to HIV-1: kinetics of antibody responses in chronic infection reflects capacity of immune system to improve viral set point. Blood 104:1784-1792. [DOI] [PubMed] [Google Scholar]
- 90.Trkola, A., H. Kuster, C. Leemann, C. Ruprecht, B. Joos, A. Telenti, B. Hirschel, R. Weber, S. Bonhoeffer, and H. F. Günthard. 2003. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J. Virol. 77:13146-13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Rij, R. P., S. Broersen, J. Goudsmit, R. A. Coutinho, and H. Schuitemaker. 1998. The role of a stromal cell-derived factor-1 chemokine gene variant in the clinical course of HIV-1 infection. AIDS 12:F85-90. [PubMed] [Google Scholar]
- 93.van 't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and nonsyncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 95.Winkler, C., W. Modi, M. W. Smith, G. W. Nelson, X. Wu, M. Carrington, M. Dean, T. Honjo, K. Tashiro, D. Yabe, S. Buchbinder, E. Vittinghoff, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Detels, S. Donfield, A. Willoughby, E. Gomperts, D. Vlahov, J. Phair, and S. J. O'Brien. 1998. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 279:389-393. [DOI] [PubMed] [Google Scholar]
- 96.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Z. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 97.Wolint, P., M. R. Betts, R. A. Koup, and A. Oxenius. 2004. Immediate cytotoxicity but not degranulation distinguishes effector and memory subsets of CD8+ T cells. J. Exp. Med. 199:925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong, J. K., M. Hezareh, H. F. Günthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 99.Yerly, S., C. Fagard, H. F. Günthard, B. Hirschel, and L. Perrin. 2003. Drug resistance mutations during structured treatment interruptions. Antivir. Ther. 8:411-415. [PubMed] [Google Scholar]
- 100.Yerly, S., H. F. Günthard, C. Fagard, B. Joos, T. V. Perneger, B. Hirschel, and L. Perrin. 2004. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18:1951-1953. [DOI] [PubMed] [Google Scholar]
- 101.Yerly, S., T. V. Perneger, S. Vora, B. Hirschel, and L. Perrin. 2000. Decay of cell-associated HIV-1 DNA correlates with residual replication in patients treated during acute HIV-1 infection. AIDS 14:2805-2812. [DOI] [PubMed] [Google Scholar]