Abstract
The 3′ nontranslated region (NTR) of the pestivirus Bovine viral diarrhea virus (BVDV), a close relative of human Hepatitis C virus, consists of three stem-loops which are separated by two single-stranded regions. As in other positive-stranded RNA viruses, the 3′ NTR of pestiviruses is involved in crucial processes of the viral life cycle. While several studies characterized cis-acting elements within the 3′ NTR of a BVDV replicon, there are no studies addressing the significance of these elements in the context of a replicating virus. To examine the functional importance of 3′ NTR elements, a set of 4-base deletions and deletions of each of the three stem-loops were introduced into an infectious BVDV cDNA clone. Emerging mutant viruses were characterized with regard to plaque phenotype, growth kinetics, and synthesis of viral RNA. The results indicated that presence of stem-loop (SL) I and the 3′-terminal part of the single-stranded region between stem-loops I and II are indispensable for pestiviral replication. In contrast, deletions within SL II and SL III as well as absence of either SL II or SL III still allowed efficient viral replication; however, a mutant RNA lacking both SL II and SL III was not infectious. The results of this study provide a detailed map of the essential and nonessential elements within the 3′ NTR of BVDV and contribute to our understanding of sequence and structural elements important for efficient viral replication of pestiviruses in natural host cells.
Bovine viral diarrhea virus 1 (BVDV-1), BVDV-2, Classical swine fever virus, and Border disease virus are members of the genus Pestivirus, which, together with the genera Flavivirus and Hepacivirus, constitute the family Flaviviridae (6, 22). Pestiviruses are causative agents of economically important livestock diseases such as classical swine fever, bovine viral diarrhea, and mucosal disease. Pestiviruses have a positive-sense single-stranded RNA genome of about 12.3 kb in length with one large open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTRs) (see references 6 and 31 for reviews). The ORF encodes a polyprotein of approximately 3,900 amino acids which is co- and posttranslationally processed into the mature viral proteins by viral and cellular proteases. The first third of the ORF encodes an autoprotease and four structural proteins, while the 3′ part of the RNA genome encodes the other nonstructural (NS) proteins. Nonstructural protein NS3 possesses multiple enzymatic activities, serine protease (39, 43, 44), NTPase (38), and helicase activity (41). NS5B represents the RNA-dependent RNA polymerase (RdRp) (26, 47). According to the effects in tissue culture, two biotypes of pestiviruses are distinguished, cytopathogenic and noncytopathogenic viruses (21, 29).
Pestiviruses replicate in the cytoplasm of infected cells. For a complete RNA replication cycle, the genomic RNA is first copied into minus-strand RNA, which in turn serves as the template for plus-strand RNA synthesis. RNA replication of pestiviruses requires viral RNA templates, the viral nonstructural proteins 3, 4A, 4B, 5A, and 5B, and probably cellular proteins. The pestivirus 5′ NTR starts with 70 nucleotides which precede an internal ribosome entry site, which mediates cap-independent translation of the viral polyprotein (5, 30, 36, 37). Analyses of BVDV mutants revealed that apart from the highly conserved 5′-terminal sequence motif 5′-GUAU the remainder of the 70 nucleotides which form stem-loop (SL) Ia are not required for efficient pestivirus replication in cell culture or in vivo (5, 20, 32). Moreover, it has been suggested that the corresponding complementary AUAC-3′ sequence motif of the viral minus strand probably represents a minimal cis-acting element essential for synthesis of viral genomic RNA (5, 20).
According to computer predictions and biochemical analyses, the 3′ NTR of BVDV folds into three stem-loop structures separated by single-stranded regions (15, 23, 46). Analyses with replicating BVDV subgenomic RNAs encompassing the 5′ and 3′ NTRs as well as the genomic region encoding the nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B revealed that the highly conserved 3′-terminal SL I and the single-stranded region between SL I and SL II play essential roles during RNA replication (46). According to recently communicated data, the remaining variable part of the 3′ NTR contains a complex RNA motif which serves as the binding determinant for a set of cellular proteins and is believed to coordinate the switch between viral translation and replication (23, 24). While these studies were based on a BVDV replicon system, there are no reports addressing the significance of 3′ NTR elements in the context of a replicating virus.
In the present study, several mutations were introduced into the 3′ NTR of an infectious BVDV full-length cDNA clone. After transfection of in vitro-transcribed RNAs, essential and nonessential elements within the 3′ NTR were defined. The mutants were characterized with regard to (i) specific infectivity of the RNA, (ii) growth properties, and (iii) accumulation of viral genomic RNA and proteins. The data presented deepen our understanding of sequence and structural elements within the 3′ NTR that are important for efficient replication of pestiviruses in their natural host cells.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK) cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco′s modified Eagle′s medium supplemented with 10% horse serum. Cells were tested regularly for the absence of pestiviruses by reverse transcription-PCR and immunofluorescence (3). The cytopathogenic BVDV-1 strain CP7-5A has been described previously (5).
Reverse transcription-PCR, molecular cloning, and nucleotide sequencing.
Reverse transcription of heat-denatured RNA and PCR were done as described previously (3). For determination of the 5′- and 3′-terminal sequences of BVDV CP7 mutant viruses, an RNA ligation method was employed as described (4, 5). Computer analysis of sequence data was performed using Heidelberg Unix Sequence Analysis Resources (Deutsches Krebsforschungszentrum, Heidelberg, Germany), which provides the GCG (Genetics Computer Group Inc., Madison, Wisconsin) software package (16).
Construction of 3′ NTR mutant plasmids.
Construction of BVDV full-length cDNA clones carrying mutations in the 3′ NTR was based on plasmid pCP7-5A (5). Compared to the authentic sequence of the parental BVDV strain CP7, pCP7-5A contains two nucleotide differences within the 3′ NTR. The two nucleotides present in pCP7-5A were replaced by the authentic sequence of CP7 wild-type virus resulting in pCP7-388, which served as the parental construct for all mutants described here. Compared to the formerly used construct pCP7-5A, the specific infectivity of RNA transcribed from pCP7-388 was about fivefold increased and plaques produced by the resulting virus CP7-388 were twofold larger (data not shown).
All nucleotide numberings included here refer to pCP7-388. Mutants M3, M5, M9, M10, M11, Del Sl I, and Del Sl II were constructed by PCR with the antisense primer carrying the desired mutation and the sense primer OLB49 (nucleotides 10756 to 10773) using plasmid pCP7-388 as the template. The PCR products obtained were cloned in pCR2.1 (Invitrogen) or pDrive (Qiagen) vectors. After control of the nucleotide sequences, the fragments were digested with ClaI and AatII and subsequently cloned in pCP7-388 precut with ClaI (nucleotide 11076) and AatII (nucleotide 12259). Mutants M1, M2, M4, M6, M7, M8, and M12 to M15 were constructed by Quikchange PCR (Stratagene, Heidelberg, Germany) using a subclone of pCP7-388, which encompasses nucleotides 10756 to 12294 of the CP7 sequence, followed by cloning and sequencing of the altered ClaI/AatII fragments into pCP7-388 precut with ClaI (11076) and AatII (12259).
For mutants M1, M2, and Del SL I, the fragments carrying the mutations were introduced into pCP7-388 using the ClaI site and an FseI site which is located directly downstream of the SmaI site used for linearization of the full-length cDNA. For construction of Del SL III and Del SL II-III, two AgeI sites were introduced into the cDNA clone CP7-388 at positions 12116 and 12157 (Del SL III), and positions 12116 and 12217 (Del SL II-III). After digestion with AgeI followed by religation, the truncated ClaI/AatII fragments were cloned into CP7-388 precut with ClaI and AatII (see above).
To reconstruct the pseudorevertants of mutants M3 and Del SL II, the sequences of the pseudorevertants were incorporated by Quikchange PCR into the parental cDNA clones harboring the original deletion. After cloning and sequencing, the resulting clones were digested with ClaI and AatII and the purified fragments were introduced into pCP7-388 precut with ClaI and AatII. Further details of the cloning strategies as well as primer sequences are available upon request.
Computer-predicted RNA folding.
Modeling of the RNA secondary structures was performed with the programs RNAFOLD, MFOLD, and FOLDANALYZE (16). Computer-predicted models of the RNA secondary structure were used to assess the locations and putative changes of the RNA secondary structure for each mutant.
In vitro synthesis and transfection of RNA.
In vitro transcription of RNA and transfection of MDBK cells were performed as described (5). About 2 μg of RNA was used for each transfection. For comparative analyses, the transcription/transfection experiments using sets of 3′ NTR mutants together with the parent virus CP7-388 were performed in parallel and repeated several times; these analyses included an immunofluorescence assay, determination of viral yields after transfection, and a plaque assay (see below).
Plaque assays.
MDBK cells were transfected with 2 μg of each RNA, and 10-fold serial dilutions of transfected cells together with 2 × 106 naive MDBK cells were seeded into six-well dishes. After incubation at 37°C for 4 h, the attached cells were overlaid with semisolid medium containing 0.6% low-melting-point agarose (Gibco-BRL) and 5% horse serum. After 6 days of incubation at 37°C, 2% (wt/vol) paraformaldehyde was used for fixation of the cells. After removal of the agarose overlays, the cells were washed with phosphate-buffered saline, followed by air drying, and subjected to immunostaining as described (5). For each of the mutant viruses and CP7-388, the diameters of 10 to 37 plaques were measured and used for calculation of mean values (average plaque sizes) and standard deviations (see Table 1 and Fig. 5B).
TABLE 1.
Specific infectivities and plaque sizes of 4-base deletion constructs
| Construct | Bases deleteda | Infectivity (PFU/μg RNA) | Mean diam of plaques (mm) ± SD |
|---|---|---|---|
| M1 | 167-170 | ||
| M2 | 159-162 | ||
| M3b | 140-143 | 1.2 × 101 | |
| M4 | 129-132 | ||
| M5 | 123-126 | ||
| M6 | 120-123 | 2.7 × 105 | 3.0 ± 0.7 |
| M7 | 118-121 | 2.0 × 105 | 2.8 ± 0.5 |
| M8 | 116-119 | 3.6 × 105 | 3.1 ± 0.6 |
| M9 | 106-109 | 9.0 × 105 | 4.1 ± 0.5 |
| M10 | 92-95 | 2.1 × 105 | 2.9 ± 0.8 |
| M11 | 79-82 | 1.8 × 106 | 3.8 ± 0.6 |
| M12 | 57-60 | 1.3 × 105 | 4.4 ± 0.5 |
| M13 | 48-51 | 1.5 × 105 | 4.9 ± 0.6 |
| M14 | 33-36 | 1.9 × 105 | 4.3 ± 0.4 |
| M15 | 26-29 | 4.3 × 105 | 4.6 ± 0.9 |
| CP7-388 | 1.2 × 106 | 4.9 ± 0.8 |
Numbers correspond to those in Fig. 1.
Emergence of pseudorevertants.
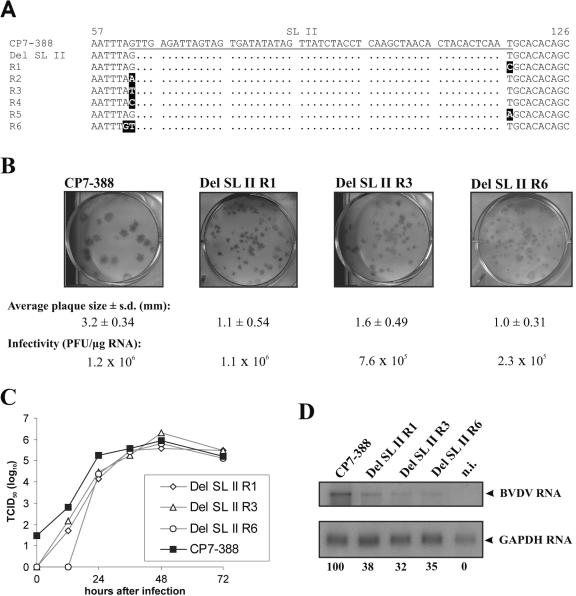
FIG. 5.
Analysis of pseudorevertants emerged after transfection of MDBK cells with RNA derived from construct Del SL II. (A) Sequence alignment of CP7-388 (top), Del SL II, and the six pseudorevertants R1 to R6. The underlined part of the CP7-388 sequence represents SL II. Nucleotide changes in the sequences of the pseudorevertants compared to the original deletion mutant Del SL II are highlighted. (B) Plaque assays and specific infectivities of CP7-388 and the genetically engineered mutants Del SL II R1, Del SL II R3, and Del SL II R6. For each virus, the average diameter of plaques, the standard deviation (s.d.), and the specific infectivity are indicated. (C) Growth kinetics of CP7-388, Del SL II R1, Del SL II R3, and Del SL II R6. MDBK cells were infected at an MOI of 0.1 with supernatants from the second tissue culture passage after transfection. The titers of released virus were determined over a 3-day period. (D) Northern blot analysis of total RNA from MDBK cells infected at an MOI of 0.1 with BVDV CP7-388 and the indicated BVDV mutants. RNAs were extracted at 24 h after infection. The relative amounts of viral genomic RNAs are indicated below the blot as a percentage of that of the parent CP7-388 (100%). For further details see the legend to Fig. 2B.
Determination of growth kinetics.
We infected 2 × 106 MDBK cells with transcript-derived virus at a multiplicity of infection (MOI) of 0.1. After adsorption for 1 hour at 37°C, the cells were washed six times with phosphate-buffered saline, overlaid with medium containing 10% horse serum, and incubated over a 3-day period. At the indicated time points aliquots (200 μl) of the supernatant were removed and used for titration on MDBK cells. The viral yields were determined as the titer of 50% tissue culture infectious doses (TCID50) per milliliter.
RNA preparation, gel electrophoresis, and Northern (RNA) hybridization.
For comparative analyses of viral RNA synthesis, 2 × 106 MDBK cells were infected with transcript-derived virus at an MOI of 0.1 and processed in parallel to cells used for determination of the growth kinetics. Preparation of total cellular RNA, gel electrophoresis, radioactive labeling of the probes, hybridization, and posthybridization washes were carried out as described previously (2). An EcoRI fragment (nucleotides 6278 to 7850) of the cDNA clone pCP7-388 was used as a BVDV-specific probe. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe was a kind gift of C. Grassmann (Institut für Virologie, Giessen, Germany) and has been described (28). The viral genomic RNA and the GAPDH RNA were detected by autoradiography, and the intensity of bands was determined by phosphorimaging (Typhoon 9200, Amersham Biosciences, Freiburg, Germany).
RESULTS
Mapping of essential and nonessential elements within the 3′ NTR by small deletions.
To study the significance of 3′ NTR elements for viral replication, we first constructed a set of 4-base deletion mutants. This approach was used to affect both sequence and structural elements. The 15 deletions introduced into the 3′ NTR of CP7-388 cover different paired and unpaired regions of the three stem-loops SL I, SL II, and SL III as well as the two single-stranded regions separating the stem-loops (Fig. 1 and Table 1). The cDNA clones carrying the mutations were used for in vitro transcription of full-length genomic RNAs.
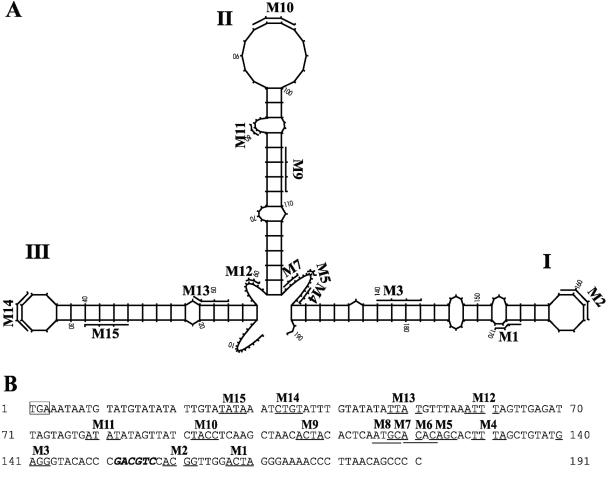
FIG. 1.
3′ NTR of BVDV strain CP7 and positions of 4-base deletions. (A) Predicted RNA secondary structure of the 3′ NTR of CP7, including locations of the engineered 4-base deletions (M1 to M15). The BVDV 3′ NTR stem-loops are designated according to the nomenclature proposed by Deng and Brock (15). The numbers correspond to those of the CP7 sequence shown in panel B. Mutants M6 and M8 are not displayed in this model. (B) Nucleotide sequence of the 3′ NTR of CP7. The translational stop codon is marked by a box. The positions of the 4-base deletions (M1 to M15) are underlined. The AatII restriction site is in italics.
After transfection of MDBK cells, the specific infectivities and virus yields were determined for each of the mutant RNAs and the RNA transcribed from the parent plasmid pCP7-388. Transfection of mutant RNAs M1, M2, M4, and M5 did not result in production of viral antigen, while all the other RNAs were infectious. The specific infectivities of M6 to M15 were between 1.3 × 105 PFU/μg and 1.8 × 106 PFU/μg and thus either not reduced or only moderately reduced compared to the specific infectivity of CP7-388 (1.2 × 106 PFU/μg) (Table 1). In contrast, the specific infectivity of the RNA from M3 (1.2 × 101 PFU/μg) was near the limit of detection. These differences correlated well with the percentage of immunofluorescence-positive cells determined 24 h posttransfection and the virus titers obtained at different time points after transfection (data not shown). To confirm the obtained results, the transcription/transfection experiments using the whole set of mutant RNAs were done in parallel and repeated twice. The separate experiments led to very similar results (data not shown).
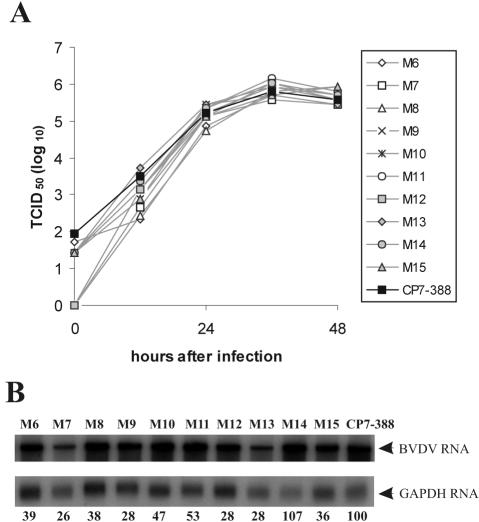
To characterize their growth properties, the growth rate and yield of the mutant viruses M6 to M15 were determined. Supernatants from cells transfected with the individual mutant RNAs were titrated and then used to infect MDBK cells at an MOI of 0.1. After incubation at 37°C, virus released into the medium was titrated over a 2-day period (Fig. 2A). The peak titer for CP7-388 was 8.5 × 105 TCID50/ml, achieved 36 h postinfection. All mutants reached very similar peak titers compared to CP7-388 (Fig. 2A). Furthermore, progeny virus recovered from the in vitro-transcribed RNAs of M6 to M15 and CP7-388 was characterized by plaque assay on MDBK cells using dilutions of cells directly after transfection (Table 1). The transcript-derived parent virus CP7-388 formed plaques with an average size of 4.9 mm. M6, M7, M8, and M10 produced plaques with an average size ranging from 2.8 to 3.1 mm, while plaques generated by M9 and M11 to M15 had an average size ranging from 3.8 to 4.9 mm. Taken together, both the specific infectivities and the growth properties of mutants M6 to M15 are similar to those of CP7-388.
FIG. 2.
(A) Growth curves of BVDV CP7-388 and mutant BVDV strains M6 to M15. Growth kinetics were determined on MDBK cells infected with supernatants of the second tissue culture passage of the indicated viruses at an MOI of 0.1. The titers of released virus were determined over a period of 48 h. (B) Northern blot analysis of total RNA from MDBK cells infected with BVDV CP7-388 and the indicated mutant BVDV strains at an MOI of 1. Mock-infected cells served as a negative control (not shown). RNAs were extracted at 18 h postinfection. The blot was hybridized with a BVDV CP7-specific cDNA fragment as well as a GAPDH-specific probe. The positions of the viral genomic RNA and the GAPDH RNA are indicated at the right. The amounts of viral genomic RNA were determined with a phosphorimager, and values were normalized by using the signals obtained with the GAPDH-specific probe. The relative amounts of viral genomic RNAs are indicated below the blot as a percentage of that of the parent CP7-388 (100%).
For analysis of viral RNA synthesis, cells were infected with the mutants M6 to M15 and CP7-388 at an MOI of 1. Total cellular RNA was prepared 18 h postinfection and used for Northern blot analysis (Fig. 2B). Viral RNA was visualized by hybridization with a BVDV-specific probe. The amounts of viral genomic RNA were quantified by phosphorimaging, and the values were normalized by using the signals obtained with a GAPDH-specific probe (Fig. 2B). The RNAs of CP7-388 and M14 were slightly more abundant than those of the other mutant viruses (M6 to M13 and M15) that produced 26 to 53% of the amount of RNA detected after infection with CP7-388. Northern blot analysis of RNAs prepared 24 h postinfection led to very similar results (data not shown).
Taken together, the results of our analyses demonstrated that the deletions within SL I and the 3′-terminal part of the single-stranded region directly upstream of SL I resulted in complete (M1, M2, M4, and M5) or almost complete (M3) loss of infectivity. In contrast, for all mutants with deletions affecting SL II, SL III, the single-stranded region between these stem-loops, and the 5′-terminal part of the single-stranded region between SL I and SL II, specific infectivity, plaque size, virus yield, and viral RNA synthesis were not reduced or only moderately reduced (Table 1 and Fig. 2). It can be concluded that SL I and the 3′-terminal part of the preceding single-stranded region of the BVDV genomic RNA represent essential elements for efficient replication of BVDV, while our analyses of 4-base deletions did not provide evidence for the presence of additional essential cis-acting elements in the remaining part of the 3′ NTR.
Search for secondary mutations within the 3′ NTR of the 4-base deletion mutants.
To investigate whether secondary mutations occurred within the 3′ NTR, each deletion mutant was passaged twice in MDBK cells. Using RNAs from the second tissue culture passage, the 3′-terminal sequences were amplified by reverse transcription-PCR and cloned in a bacterial plasmid; at least three cDNA clones were characterized for each mutant. For mutants M6 to M15, sequence analysis indicated the absence of any secondary mutations within the genomic region analyzed.
The specific infectivity of M3 was only 12 PFU/μg RNA (Table 1). The plaques that emerged after transfection with the RNA of M3 were <0.6 mm in size and thus significantly smaller than the plaques produced by CP7-388 (data not shown). The viruses obtained from five independently emerged plaques were biologically cloned and further characterized. Determination of the 3′-terminal sequences of these five viruses actually demonstrated the presence of mutations in the direct vicinity of the deletion site (Fig. 3A). Interestingly, the 3′ NTR sequences of pseudorevertants M3-2, M3-3, and M3-4 were identical.
FIG. 3.
Pseudorevertants recovered after transfection of RNA derived from construct M3. (A) 3′-terminal sequences of CP7-388 (top), deletion mutant M3, and the independently emerged pseudorevertants M3-1, M3-2, M3-3, M3-4, and M3-5. The underlined part of the CP7-388 sequence represents SL I. Nucleotide changes in the sequences of the pseudorevertants are highlighted. (B) Growth kinetics of the genetically engineered mutants M3R2 and M3R5 determined on MDBK cells infected at an MOI of 0.1 with supernatants of the second tissue culture passage after transfection with the respective RNAs. The titers of released virus were determined over a 3-day period. (C) Northern blot analysis of total cellular RNAs prepared 24 h after infection of MDBK cells with CP7-388, M3R2, and M3R5 at an MOI of 0.1. Mock-infected cells (n.i.) served as a negative control. The relative amounts of viral genomic RNAs are indicated below the blot as a percentage of that of the parent CP7-388 (100%). For further details see the legend to Fig. 2B. (D) Predicted RNA secondary structures of SL I and specific infectivities of CP7-388, M3, M3R2, and M3R5.
For further analyses, the mutations identified in the genomes of M3-2 and M3-5 were introduced into pCP7-388; the resulting mutant viruses were named M3R2 and M3R5, respectively. After transfection of the in vitro-synthesized RNA, infectious virus was recovered and propagated. The second tissue culture passage was used for analysis of growth kinetics, plaque sizes, and viral RNA synthesis. For both engineered mutants, the infectious virus titers obtained during the first 48 h after infection were reduced by about one (M3R2) or two (M3R5) orders of magnitude compared to the parent virus CP7-388 (Fig. 3B). Plaque assays revealed a small-plaque phenotype for both mutant viruses (data not shown); the average plaque sizes of M3R2 and M3R5 were reduced at least sevenfold compared to the plaques produced after infection with CP7-388 (data not shown).
For analysis of viral RNA synthesis, a Northern blot analysis of total cellular RNAs prepared 24 h after infection of MDBK cells with M3R2, M3R5, and CP7-388 at an MOI of 0.1 was performed and the relative amounts of viral genomic RNA were determined by phosphorimager analysis. As expected from its replication kinetics, the amounts of accumulated RNAs of M3R2 and M3R5 were only 7% and 2%, respectively, of the amount of CP7-388 RNA (Fig. 3C). Taken together, the replication efficiency of the mutants M3R2 and M3R5 is significantly reduced. According to a comparison of predicted RNA secondary structures, it can be assumed that the severely affected stability of SL I of the original mutant M3 was partly restored by the secondary mutations which led to the emergence of pseudorevertants such as M3R2 and M3R5 (Fig. 3D).
Deletion of entire stem-loops.
The results obtained prompted us to construct additional mutants, each lacking one of the three stem-loops SL I, SL II, and SL III as well as a mutant lacking both SL II and SL III (Del SL II-III). The RNA of mutant Del SL I, which lacks the 3′-terminal stem-loop SL I, and the RNA of mutant Del SL II-III were not infectious (Fig. 4A). The lack of infectivity of Del SL I is in agreement with the results obtained with mutant RNAs carrying 4-base deletions within SL I (see above). In contrast, removal of SL III did not lead to a significant reduction of the specific infectivity of the resulting mutant RNA (Fig. 4A). Further studies, including determination of growth kinetics (Fig. 4B) and plaque size (Fig. 4C) as well as analysis of viral RNA synthesis (Fig. 4D), revealed no significant differences between Del SL III and the parental CP7-388. These results demonstrate that SL III is not required for efficient replication of BVDV.
FIG. 4.
Analysis of BVDV mutants lacking entire stem-loops of the 3′ NTR. (A) Schematic drawing of the 3′ NTR of the parental BVDV strain CP7-388 and the engineered deletion mutants Del SL I, Del SL II, Del SL III, and Del SL II-III, lacking SL I, SL II, SL III, and SL II together with SL III, respectively. For each mutant RNA, the specific infectivity (PFU/μg RNA) is indicated. For Del SL I and Del SL II-III, transfection of in vitro-transcribed RNA into MDBK cells did not result in generation of infectious viruses. For Del SL II, the specific infectivity was slightly above the limit of detection. Note that the infectivity of Del SL III was not significantly reduced compared to that of the parental construct CP7-388. (B) Growth kinetics of Del SL III and CP7-388 determined on MDBK cells infected at an MOI of 0.1 with supernatants of the first tissue culture passage after transfection. The titers of released virus were determined over a 2-day period. (C) Plaques produced by BVDV CP7-388 and Del SL III at day 6 posttransfection. Plaques were visualized by immunostaining. (D) Northern blot analysis of total RNA from MDBK cells infected with Del SL III and BVDV CP7-388 at an MOI of 0.1. RNAs were extracted at 24 h postinfection. The relative amounts of viral genomic RNAs are indicated below the blot as a percentage of that of the parent CP7-388 (100%). For further details see the legend to Fig. 2B.
With respect to the mutant RNA lacking SL II (Del SL II), the specific infectivity was slightly above the detection limit (Fig. 4A). After two tissue culture passages of the supernatant from cells transfected with this RNA, titers of >4 × 105/ml infectious virus were obtained, suggesting the emergence of secondary mutations. Determination of the 3′-terminal sequences of the viruses obtained actually demonstrated the presence of a set of single or double point mutations located in the direct vicinity of the deletion site of Del SL II (Fig. 5A). All pseudorevertants which emerged after transfection of Del SL II lacked the entire SL II.
To study the effects of the absence of SL II further, the sequences of three of the identified pseudorevertants (R1, R3, and R6) (Fig. 5A) were introduced into the 3′ NTR of CP7-388. Transcription/transfection experiments indicated that the specific infectivities of the resulting RNAs (Del SL II R1, Del SL II R3, and Del SL II R6) were between 2.3 × 105 and 1.1 × 106 PFU/μg and thus not significantly or only slightly lower than that of CP7-388 (1.2 × 106 PFU/μg) (Fig. 5B). All engineered mutants lacking the entire SL II produced smaller plaques than CP7-388 (Fig. 5B). For further characterization, the growth kinetics of the three mutants and CP7-388 were determined using an MOI of 0.1. MDBK cells were infected with supernatants from cells transfected with the various RNAs. After incubation at 37°C, virus released into the medium was titrated over a 3-day period. With respect to the first 24 h postinfection, the titers produced by the viruses lacking SL II were about one order of magnitude lower than the titers of CP7-388, while at later time points all viruses including CP7-388 grew to very similar titers (Fig. 5C). Each of the viruses reached titers of >5 × 105 TCID50/ml in the cell-free tissue culture supernatant.
For comparative analysis of viral RNA synthesis, cells were infected at an MOI of 0.1 and processed in parallel to those used to determine the growth rates. Total cellular RNA was prepared 24 h postinfection and used for Northern blot analysis. Viral RNA was visualized by hybridization, and the intensity of bands was determined with a phosphorimager (Fig. 5D). The RNA of the parental virus CP7-388 was more abundant than that of any of the mutant viruses lacking SL II. Accordingly, there was a reduction of both the amounts of accumulated viral RNAs and the virus titers obtained at 24 h postinfection
Taken together, the results of our analyses of BVDV mutant RNAs lacking entire stem-loop structures within the 3′ NTR demonstrated that the presence of SL I is indispensable for pestiviral replication, while deletion of either SL II or SL III allowed efficient viral replication. However, a mutant lacking both SL II and SL III was not infectious. Accordingly, the replication ability of BVDV depends on the presence of SL I and the 3′-terminal part of the single-stranded region directly upstream of SL I together with either SL II or SL III.
DISCUSSION
The nontranslated regions of positive-strand RNA viruses contain cis-acting signals important for replication and translation of viral genomes (7, 8, 10, 12, 27, 33, 34, 42). Pestivirus genomic RNAs encompass 5′ and 3′ NTRs with lengths of approximately 380 to 400 and 190 to 270 nucleotides, respectively (1, 4). With regard to the 3′ NTR of pestiviral genomes, the 3′-most 70 nucleotides represent a highly conserved element, of which the last 56 to 60 nucleotides form a stable stem-loop structure, termed SL I (4, 15, 46). The remaining variable portion of the 3′ NTR has been reported to form a complex RNA motif encompassing two less-stable stem-loop structures, termed SL II and SL III (23).
Using a BVDV replicon, cis-acting elements in the 3′ NTR have been characterized previously (23, 24, 46). Such replicons usually lack the genomic region encoding the structural proteins, p7, and NS2; they are suited to dissect the role of RNA signals for RNA replication and translation. However, the importance of sequence and structural elements for other steps of the viral life cycle such as packaging and particle formation cannot be addressed.
The experiments described here were designed to study the significance of sequence and structural elements of the 3′ NTR of BVDV in the context of a replicating virus. Several deletions were introduced into the 3′ NTR of an infectious BVDV cDNA clone and subsequently analyzed. The results of our analyses show that SL I and the 3′-terminal part of the single-stranded region between SL I and SL II represent essential elements for viral replication, while transfection of mutant RNAs with small deletions within SL II and SL III and also mutant RNAs lacking either the entire SL II or SL III structure still resulted in recovery of efficiently replicating viruses; the latter were not or only slightly reduced in their ability to grow to high viral titers. Accordingly, lack of either SL II or SL III as well as deletion of the 5′-terminal part of the single-stranded region preceding SL I had no major negative effect on RNA replication, translation, packaging, or particle formation.
Our observation that all 4-base deletions within SL I (affecting both stem and loop regions) and the 3′-terminal part of the single-stranded region between SL I and SL II as well as complete removal of SL I abrogate viral replication highlights the crucial role of these elements for viral replication. This is in agreement with a previous study based on a BVDV replicon system (46). According to that study, mutations within SL I and the preceding single-stranded region lead to partial or complete inhibition of RNA replication.
The single-stranded region between SL I and SL II is highly conserved among various pestivirus species (1, 4), and it has been suggested that this region, similar to SL I, is important for viral RNA replication of a BVDV replicon (46). Our analysis revealed that 4-base deletions affecting the 3′ portion of this region completely inhibited viral replication, while deletions within the 5′-terminal portion of this single-stranded region had only minor negative effects on the replication capacity of the respective mutants (Fig. 2 and Table 1). Similar to our results obtained for the 3′ NTR of BVDV, it has been shown for other members of the Flaviviridae that 3′-terminal sequences are absolutely essential for viral RNA replication. For the related human hepatitis C virus, it is known that the highly conserved 3′X region and the poly(U/UC) tract represent indispensable elements implicated in viral RNA replication (19, 45). In addition, the essential nature of 3′-terminal sequences has also been demonstrated for GB virus-B as well as for members of the genus Flavivirus such as tick-borne encephalitis virus and dengue virus (11, 33, 34). Moreover, genetic analyses of 3′ NTR sequences of other plus-strand RNA viruses revealed the existence of cis-acting elements crucial for viral replication (9, 14, 17, 18).
For flaviviruses and hepatitis C virus, viral and cellular factors have been found to bind to the terminal regions of viral genomes and were proposed to play important roles in viral replication (7, 8, 12, 13, 25, 40). Recently, it has been reported that a group of cellular host factors, the so-called NFAR proteins, are implicated in RNA replication of a BVDV replicon, and it was shown that these proteins bind to SL II and SL III (23, 24). It will be interesting to determine whether some of the mutants described here differ with respect to binding of such cellular proteins.
In contrast to mutations within SL I and the 3′-terminal portion of the preceding single-stranded region, all 4-base deletions located in base-paired and loop regions of SL II and SL III were well tolerated. Interestingly, deletion of the entire SL II resulted in recovery of efficiently replicating pseudorevertants. Analysis of reconstructed mutants proved that single or double point mutations in the vicinity of the SL II deletion site reconstituted the specific infectivity to almost the wild-type level. It can be speculated that the point mutations repair either an essential sequence element or a structural conformation of the 3′ NTR required for efficient replication. Notably, the specific infectivity, viral replication efficiency, and amounts of accumulated viral RNA of the reconstructed mutants lacking SL II were not significantly reduced or only slightly reduced (Fig. 5). Using a different approach based on a BVDV replicon, it has been shown that point mutations affecting the sequence and structure of SL II significantly decreased the RNA replication efficiency but did not result in complete loss of replication capability (24).
With regard to SL III, the results of the present study provide clear evidence that a virus mutant lacking that stem-loop did not differ from the parental virus with respect to specific infectivity of viral RNA, growth kinetics, plaque size, or yield of viral RNA and proteins (Fig. 4 and data not shown). This observation is in conflict with the above-mentioned study, which concluded that proper formation of SL II and SL III is crucial for correct translation termination and efficient RNA replication of a BVDV replicon (24). While in the replicon system entire stem-loops were not deleted, RNA replication of replicons with mutations affecting the structure of SL III was severely inhibited. Accordingly, the significance of cis-acting elements determined in the context of a replicon lacking substantial parts of the viral genomic RNA may differ from their importance in the context of an infectious full-length viral genome.
Alternatively, the differences could be due to the use of different virus strains or viral sequences. The BVDV replicon DI9c used by Isken et al. (23, 24) was obtained by fusion of cDNA sequences derived from the genomes of BVDV strains CP7, CP9, and NADL and harbors chimeric 5′ and 3′ NTRs (35). In contrast to DI9c, our infectious BVDV cDNA clone used for the experiments described in this study represents an authentic copy of the genome of BVDV strain CP7.
Finally, the differences observed concerning the importance of SL II and SL III might be due to the use of different cell lines. In contrast to our study, where bovine kidney cells were used in all experiments, the experiments with the BVDV replicons were performed in baby hamster kidney 21 (BHK-21) cells, which are not susceptible to infection with pestiviruses. In this context, it is important to mention that BHK-21 cells, which have been used extensively to study RNA replication of BVDV subgenomic replicons, do not support efficient RNA replication of in vitro-transcribed BVDV full-length genomic RNAs (P. Becher, unpublished). Accordingly, RNA replication of BVDV replicons in BHK-21 cells is probably influenced by factors different from those important for replication of pestiviruses in natural host cells.
Acknowledgments
We thank M. Orlich and S. Widauer for excellent technical assistance.
This study was supported by Sonderforschungsbereich 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft and Intervet International BV.
REFERENCES
- 1.Avalos-Ramirez, R., M. Orlich, H.-J. Thiel, and P. Becher. 2001. Evidence for the presence of two novel pestivirus species. Virology 286:456-465. [DOI] [PubMed] [Google Scholar]
- 2.Becher, P., G. Meyers, A. D. Shannon, and H.-J. Thiel. 1996. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J. Virol. 70:2992-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher, P., M. Orlich, A. D. Shannon, G. Horner, M. König, and H.-J. Thiel. 1997. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 78:1357-1366. [DOI] [PubMed] [Google Scholar]
- 4.Becher, P., M. Orlich, and H.-J. Thiel. 1998. Complete genomic sequence of border disease virus, a pestivirus from sheep. J. Virol. 72:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becher, P., M. Orlich, and H.-J. Thiel. 2000. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J. Virol. 74:7884-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becher, P., and H.-J. Thiel. 2002. Genus Pestivirus (Flaviviridae), p. 327-331. In C. A. Tidona and G. Darai (ed.), The Springer index of viruses. Springer-Verlag, Heidelberg, Germany.
- 7.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 69:5650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, E. A., H. Zhang, L. H. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukh, J., C. L. Apgar, and M. Yanagi. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470-478. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C.-J., M.-D. Kuo, L.-J. Chien, S.-L. Hsu, Y.-M. Wang, and J.-H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, T., R. J. Sugrue, Q. Xu, K. W. Lee, Y.-C. Chan, and J. Fu. 1998. Recombinant dengue virus type1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology 246:409-417. [DOI] [PubMed] [Google Scholar]
- 14.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. Brown, and D. Cavanagh. 2001. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, R., and K. V. Brock. 1993. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 21:1949-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. A. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreher, T. W. 1999. Functions of the 3′ untranslated region regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 18.Duque, H., and A. C. Palmenberg. 2001. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3′ untranslated region. J. Virol. 75:3111-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frolov, I., M. S. McBride, and C. M. Rice. 1998. Cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillespie, J. H., J. A. Baker, and K. McEntee. 1960. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 50:73-79. [PubMed] [Google Scholar]
- 22.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H.-J. Thiel. 2000. Family Flaviviridae, p. 859-878. In C. M. Fauquet, M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 23.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isken, O., C. W. Grassmann, H. Yu, and S. E. Behrens. 2004. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., and M. M. C. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′ untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn, R. J., D. E. Griffin, H. Zhang, H. G. M. Niesters, and J. H. Strauss. 1992. Attenuation of Sindbis virus neurovirulence by using defined mutations in nontranslated regions of the genome RNA. J. Virol. 66:7121-7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lackner, T., A. Müller, A. Pankraz, P. Becher, H.-J. Thiel, A. E. Gorbalenya, and N. Tautz. 2004. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J. Virol. 78:10765-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, K. M., and J. H. Gillespie. 1957. Propagation of virus diarrhea virus of cattle in tissue culture. Am. J. Vet. Res. 18:953. [PubMed] [Google Scholar]
- 30.Lemon, S. M., and M. Honda. 1997. Internal ribosome entry site within the RNA genomes of hepatitis C virus and other flaviviruses. Semin. Virol. 8:274-288. [Google Scholar]
- 31.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 32.Makoschey, B., P. Becher, M. G. Janssen, M. Orlich, H.-J. Thiel, and D. Lütticken. 2004. Bovine viral diarrhea virus with deletions in the 5′-nontranslated region: reduction of replication in calves and induction of protective immunity. Vaccine 22:3285-3294. [DOI] [PubMed] [Google Scholar]
- 33.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Men, R., M. Bray, D. Clark, C. M. Chanock, and C.-J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers, G., N. Tautz, P. Becher, H.-J. Thiel, and B. Kümmerer. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol. 70:8606-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole, T. L., C. Wang, R. A. Popp, L. N. D. Potgieter, A. Siddiqui, and M. S. Collett. 1995. Pestivirus translation occurs by internal ribosome entry. Virology 206:750-754. [DOI] [PubMed] [Google Scholar]
- 38.Tamura, J. K., P. Warrener, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology 193:1-10. [DOI] [PubMed] [Google Scholar]
- 39.Tautz, N., K. Elbers, D. Stoll, G. Meyers, and H.-J. Thiel. 1997. Serine protease of pestiviruses: determination of cleavage sites. J. Virol. 71:5415-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warrener, P., and M. S. Collett. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 69:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 43.Wiskerchen, M. A., and M. S. Collett. 1991. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 184:341-350. [DOI] [PubMed] [Google Scholar]
- 44.Xu, J., E. Mendez, P. R. Caron, C. Lin, M. A. Murcko, M. S. Collett, and C. M. Rice. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71:5312-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi, M., and S. L. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, H., C. W. Grassmann, and S.-E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, W., L. L. Gutshall, and A. M. Del Vecchio. 1998. Identification and characterization of an RNA-dependant RNA polymerase activity within the nonstructural 5B region of bovine viral diarrhea virus. J. Virol. 72:9365-9369. [DOI] [PMC free article] [PubMed] [Google Scholar]