Abstract
Human cytomegalovirus (HCMV) attachment and entry stimulates the expression of cellular interferon-inducible genes, many of which target important cellular functions necessary for viral replication. Double-stranded RNA-dependent host protein kinase (PKR) is an interferon-inducible gene product that limits viral replication by inhibiting protein translation in the infected cell. It was anticipated that HCMV encodes gene products that facilitate the evasion of this PKR-mediated antiviral response. Using a Δγ134.5 herpes simplex virus type 1 (HSV-1) recombinant that triggers PKR-mediated protein synthesis shutoff, experiments identified an HCMV gene product expressed in the initial hours of infection that allows continued protein synthesis in the infected cell. Recombinant HSV-1 viruses expressing either the HCMV TRS1 or IRS1 protein demonstrate that either of these HCMV gene products allows the Δγ134.5 recombinant viruses to evade PKR-mediated protein shutoff and maintain late viral protein synthesis.
One of the mechanisms by which a cell responds to viral infection and replication is activation of double-stranded RNA activated protein kinase (PKR) (36). The activated enzyme phosphorylates the α subunit of translation initiation factor 2 (eIF-2α) and inhibits protein synthesis initiation in the infected cell (36, 42). This innate antiviral response to infection limits viral growth during the initial phases of viral infection prior to recruitment of the adaptive immune response (36). Viruses have evolved numerous ways to block the effect of activated PKR, and the genes in several herpesvirus genomes have been identified (2, 5, 6, 13, 14, 19, 26-28, 39).
The genetic basis or mechanism of viral resistance to the host protein shutoff response has been proposed in four of the eight human herpesviruses (herpes simplex virus type 1 [HSV-1], HSV-2, Epstein-Barr virus, and human herpesvirus 8 [HHV-8]) (5, 13, 19, 39). Herpesviruses encode gene products on both the sense and antisense strands of the genome and produce complementary transcripts forming double-stranded RNA (dsRNA), and they are therefore particularly susceptible to PKR-mediated host protein shutoff. In the case of HSV-1, studies using Δγ134.5 mutant viruses have revealed cryptic genetic mechanisms for evading the host protein shutoff response (7, 9, 29). These cryptic genetic loci were likely retained by HSV-1 for alternative functions, whereas in genetically related herpes viruses they represent the primary mechanism for evading the innate antiviral response (9). HCMV attachment and entry in human foreskin fibroblasts (HFF) activates an interferon response and alters gene expression in the cell (4, 47). Interferon stimulates the synthesis of proteins that target different aspects of viral infection in the cell, such as protein synthesis initiation (PKR), nucleocapsid transport/viral RNA synthesis (Mx proteins), and degradation of mRNA in infected cells (RNase L) (36). A prior study showed that human cytomegalovirus (HCMV) contains genes that code for continuing viral protein synthesis in the infected cell and preventing the PKR-mediated protein shutoff response (11).
The objective of this study was to define the HCMV gene or genes responsible for HCMV evasion of the host protein shutoff response. During revision of the manuscript, investigators reported that the HCMV TRS1 and IRS1 genes could complement a vaccinia recombinant lacking the PKR evasion gene E3L (10). Using an alternative approach, we demonstrate that an HCMV gene expressed in the initial hours of infection can complement Δγ134.5 HSV-1 virus infection, thus restoring wild-type viral protein synthesis in the infected cells. Recombinant Δγ134.5 viruses expressing either the HCMV TRS1 or IRS1 protein further demonstrate that these gene products are instrumental in the viral evasion of PKR-mediated protein shutoff, as eIF-2α is maintained in the unphosphorylated form and the virus exhibits late viral protein synthesis in the infected cell.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
Vero cells were obtained from the American Type Tissue Culture Collection and were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% newborn calf serum (8). Primary HFF cells were prepared as previously described and maintained for a maximum of 10 passages in 10% fetal bovine serum (FBS) (45).
The mammalian expression plasmids pHCMV 214, pHCMV 215, and pHCMV 231, encoding the polyhistidine-tagged IRS1, TRS1, and IRS263 protein-coding domains, respectively, were kindly provided by T. Shenk (Princeton University, Princeton, NJ) and have been described previously (35). The plasmid pCK1029 was created by inserting a 1.3-kb fragment, encoding two 8-base PacI restriction sites flanking the HCMV IE promoter and the coding domain of enhanced green fluorescent protein (EGFP) (Clontech, Palo Alto, CA), into the ApoI site in the UL3, UL4 intergenic region of plasmid pRB4841 (kindly provided by B. Roizman, University of Chicago, Chicago, IL). The plasmids pCK1114 and pCK1116 contain the SpeI fragment from the mammalian expression plasmids pHCMV215 and pHCMV214, respectively, inserted in the HSV-1 UL3, UL4 intergenic region. The plasmid pCK3008 was constructed by inserting the IRS1 gene in frame with the carboxyl-terminal epitope and polyhistidine tag of pcDNA3.1 V5/His. The IRS1 protein coding domain was amplified from AD-169 viral DNA using Pfu polymerase and the 5′ BamHI TRS1 (5′-GGATCCTCAATGGCCCAGCGCAAC-3′) and 3′ HindIII IRS263 (5′-AAGCTTATGATGAACGTGGTGAGGG-3′) oligonucleotides. The amplified DNA was then incubated with Taq polymerase, gel isolated, and cloned into pcDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA). The clone was verified by restriction digestion analysis and detection of immunoreactive protein of the expected mass, using IRS1 antisera in transient expression assays. A 4.4-kb BglII, AvrII fragment encoding the HCMV immediate early (IE) promoter and epitope-tagged IRS1 gene product from pCK3008 was then inserted into the UL3/4 intergenic region, thus creating the pCK1127 clone.
HSV-1(F) and AD169 are the prototypical HSV-1 and HCMV strains, respectively, used in these experiments (16, 33). The HSV-1 recombinant virus, R3616, described previously, lacks 1,000 bp in both copies of the γ134.5 gene (12). The γ134.5 gene product is the principal defense against PKR-mediated host protein shutoff, and the Δγ134.5 virus R3616 triggers the host protein shutoff response in human cells (13). The recombinant herpes viruses in this study were created by cotransfection and homologous recombination as previously described (32). C101 is a Δγ134.5 HSV-1 recombinant that expresses EGFP. It was isolated from among the progeny created by cotransfection of the plasmid pCK1029 and R3616 DNA and was purified with Vero cells by EGFP-positive plaque selection. The Δγ134.5 HSV-1 recombinant C130, which expresses the HCMV TRS1 gene product, was isolated in Vero cells on the basis of loss of EGFP expression after cotransfection of plasmid pCK1114 and the PacI-digested C101 DNA in rabbit skin cells. The C130 repair virus, C131, was created by cotransfection of the PacI-digested C130 DNA and the plasmid pCK1029 containing an EGFP expression cassette in the UL3, UL4 intergenic region and selection of EGFP-positive plaques. Recombinants C132 and C134 are Δγ134.5 HSV-1 viruses expressing the nonimmunoreactive and immunoreactive HCMV IRS1 gene product. They were selected by loss of GFP expression after cotransfection of PacI-digested C101 viral DNA and pCK1116 or pCK1127, respectively. The C134 repair virus, C135, was constructed by cotransfection of the PmeI-digested C134 DNA with plasmid pCK1029 and purified based on EGFP-positive plaque selection in Vero cells.
Western blotting.
The antibodies used in these studies and their sources are as follows. The rabbit polyclonal antibody against phospho-eIF-2α (p-serine-51) (44-728) and mouse monoclonal antibody against total eIF-2α (AHO0802) were purchased from Biosource International, Camarillo, CA. The polyclonal antibody against ICP0 was kindly provided by B. Roizman (University of Chicago, Chicago, IL); the mouse monoclonal antibodies against the immediate early 1 (IE1) protein (p63-27) and pp65 (65-8) were kindly provided by W. Britt (University of Alabama at Birmingham); and the TRS1 and IRS1 monoclonal antisera were kindly provided by T. Shenk (Princeton University, Princeton, NJ) (21, 31, 35).
Immunoblot experiments were performed as previously described, using equivalent protein mass (10 μg) loading (8). In summary, nitrocellulose sheets containing the electrophoretically separated proteins were incubated in blocking solution (5% bovine serum albumin in Tris-HCl-buffered saline [TBS] containing 0.01% Tween) for at least 1 h, reacted with antibody diluted in TBS for at least 4 h, and then washed five times with wash buffer (TBS containing 0.1% Tween). The nitrocellulose filter was next incubated with either an appropriate alkaline phosphatase or peroxidase-conjugated antibody diluted in wash buffer for a minimum of 90 min. The filter was then washed five times with wash buffer. The alkaline phosphatase-stained immunoblots were developed using 150 μg/ml 5-bromo-4-chloro-3-indolylphosphate (BCIP) and 300 μg/ml nitroblue tetrazolium in AP buffer (100 mM Tris-HCl [pH 9.5], 5 mM MgCl2, and 100 mM NaCl), whereas the peroxidase-stained immunoblots were developed using enhanced chemiluminescence as recommended by the manufacturer (Pierce, Rockford, IL).
DNA hybridization studies.
Purification, restriction digestion, electrophoretic separation, and transfer to Zeta probe membranes (Bio-Rad, Hercules, CA) by capillary transfer of viral DNA has been described previously (8). The separated and immobilized PstI-digested R3616, C101, C130, C131, and C132 DNAs were hybridized with a probe encoding the HSV-1 UL3 and UL4 sequences from the pRB4841 plasmid.
PCR and RT-PCR studies.
To demonstrate expression of HCMV IRS1 transcript in the C132-infected cells, reverse transcriptase (RT)-PCR was performed. Vero cells were infected with C130 and C132 at a multiplicity of infection (MOI) of 10, and total RNA was isolated from the infected cells at 18 hours postinfection (hpi) using an RNAqueous 4sure purification kit (Ambion, Austin, TX) per the manufacturer's instructions. The RNA was further digested with RNase-free DNase I (Ambion, Austin, TX) for 30 min at 37°C, followed by enzyme inactivation. First-strand synthesis was performed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and oligo(dT)18-21 primers on 1 μg (each) of C130 and C132 RNA. Subsequent PCR was performed on 1/10 of the first strand product using the following primers recognizing the 3′ unique domain of the IRS1 gene product: 5′ BamHI BspHI IRS263 (5′-GGATCCATCATGACAGAGCGTCAAAGTC-3′) and 3′ HindIII IRS263 stop (5′-AAGCTTCAATGATGAACGTGGTGAGG-3′).
UV inactivation of HCMV.
HCMV was UV inactivated by exposing 3 × 106 PFU of virus to 150 mJ of UV irradiation using a cross-linking chamber (Bio-Rad, Hercules, CA). Following UV irradiation, sodium pyruvate was added to a final concentration of 5 mM to neutralize any peroxide/superoxides produced during UV inactivation as described previously (17).
Immunofluorescence.
HFF cells were seeded on glass coverslips and mock infected or exposed to HCMV AD169 or UV-inactivated HCMV at an MOI of 3 in medium containing 10% FBS. After 2 h, the inoculum was replaced with 10% FBS. At 24 hpi, the cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde in PBS, and processed as previously described (38). Briefly the cells were permeabilized in 0.1% NP-40 in PBS for 10 min., washed five times with PBS, and blocked for 30 min with 10% goat serum in PBS at room temperature. After five washes with PBS, the cells were sequentially incubated for 1.5 h with pp65-specific monoclonal antibody (MAb) 65-8 ascites, washed with PBS, and incubated with IE1-specific (exon 4) MAb 63-27 ascites at 37°C. After being washed five times with PBS, the coverslips were incubated with goat anti-mouse immunoglobulin G1 (IgG1)-fluorescein isocyanate conjugated antibody (1:100 in PBS; Southern Biotechnology, Birmingham, AL) and Alexa Fluor 594 goat anti-mouse IgG2a antibody (1:400 in PBS; Molecular Probes, Eugene, OR) for 1 h at 37°C and Hoechst for 10 min. Washed coverslips were mounted onto slides using Slowfade antiphotobleaching reagent (Molecular Probes, Eugene, OR). Images were captured at ×400 magnification with an Olympus BX41 fluorescent microscope using Image-Pro Plus software (version 4.5) and processed using Adobe Photoshop 7.0.
In vivo protein synthesis.
For the HCMV/HSV-1 coinfection studies, duplicate cultures of HFF cells grown in 3.8-cm2-well plates were mock infected or infected with HCMV (AD169) or UV-inactivated HCMV at an MOI of 3 PFU/cell in DMEM-1% FBS. After 2 h, the inocula were replaced with DMEM containing 10% FBS. At 6 h postinfection, the mock- and HCMV-infected cells were either mock infected or infected with R3616 at an MOI of 10 PFU/cell for 2 h. At 23 h post-HCMV infection, the cultures were incubated with 199V medium lacking methionine [199V (-) MET] but supplemented with 50 μCi of l-[35S]methionine (>1,000 Ci/mmol; Amersham-Pharmacia)/ml of media. After 1 h of labeling, the cells were rinsed in ice-cold phosphate-buffered saline lacking Ca2+ and Mg2+ (PBS-A), scraped, resuspended in disruption buffer, boiled, and loaded on a 12% (vol/vol) polyacrylamide gel cross-linked with bis-acrylamide. The proteins were electrophoretically separated, transferred to nitrocellulose membranes, and subjected to autoradiography.
The protein labeling experiments for wild-type HSV-1 and the recombinant viruses R3616, C101, C130, C131, C132, C134, and C135 were performed as described previously (13). Briefly, HFF cells grown in 3.8-cm2-well plates were mock infected or infected with HSV-1(F) or recombinant virus at an MOI of 10. At 14 hpi, medium was removed and replaced with 199V (−) MET supplemented with l-[35S]methionine for 1 h. The cells were washed and lysed, and the proteins were electrophoretically separated and analyzed by autoradiography as described above.
RESULTS
In the initial hours of infection, CMV expresses a gene product that blocks the host protein shutoff response and the phosphorylation of eIF-2α.
To test the hypothesis that HCMV encodes a gene product that blocks the host protein shutoff response, coinfection experiments using either HCMV or UV-inactivated HCMV and a recombinant HSV-1 virus that triggers host protein shutoff were performed. Duplicate HFF cell cultures were either mock infected or infected with wild-type HCMV (AD169) or UV-inactivated HCMV at an MOI of 3 as described in Materials and Methods. At 6 hpi, the mock-infected and HCMV- and UV-inactivated HCMV-infected cells were either mock infected or superinfected with an HSV-1 Δγ134.5 recombinant virus (R3616) at an MOI of 10 PFU/cell. At 23 h post-initial infection (17 h after R3616 superinfection), the cultures were metabolically labeled for the final hour of infection, washed, lysed, and solubilized in loading buffer. Ten micrograms of total protein from each of the samples was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After the samples were transferred to nitrocellulose membranes, autoradiography and immunoblotting were performed.
The results of these experiments are presented in Fig. 1. Abundant radiolabeled proteins are detected in the mock, HCMV, and UV-inactivated HCMV singly infected cells (Fig. 1A, lanes 1, 3, and 5). In the R3616 superinfected mock- and UV-HCMV-infected HFF cells, there is reduced detection of radiolabeled proteins characteristic of PKR-mediated protein shutoff and the inhibition of late viral protein synthesis (Fig. 1A, lanes 2 and 6). In contrast, radiolabeled proteins in the HCMV/R3616-coinfected cells, characteristic of continued HSV-1 viral protein synthesis, are detected (Fig. 1A, lane 4). These data suggested that HCMV infection complemented the Δγ134.5 virus but that HCMV entry and delivery of virion-associated gene products were insufficient to restore wild-type protein synthesis in the R3616-infected cells.
FIG. 1.

Composite of autoradiograph, immunoblot, and immunofluorescence images. HCMV encodes a genetic mechanism expressed in the initial hours of infection to block phosphorylation of eIF-2α and maintain protein synthesis. (A) An autoradiographic image of the electrophoretically separated pulse-labeled proteins from mock-infected (Mock), HCMV-infected (HCMV), or UV-inactivated HCMV-infected (UV) HFF cells subjected to further mock or R3616 superinfection. (B) Immunoblot image shows abundant IE1 immunostaining in the cells infected with HCMV alone and reduced staining in the HCMV/R3616-infected cells. (C) Immunoblot showing equivalent ICP0 immunostaining in the R3616-infected cells. (D) Immunoblot image demonstrates abundant phosphorylated eIF-2α in both mock/R3616 and UV-inactivated HCMV/R3616-superinfected cells but minimal levels in the HCMV/R3616-superinfected cells. (E) Immunoblot demonstrating equivalent levels of total eIF-2α in the infected cell samples. (F) Indirect immunofluorescence of mock-infected (Mock), HCMV-infected (HCMV), and UV-inactivated HCMV-infected (UV) cells. The first row shows detection of virion-associated protein pp65 (pUL83) in both HCMV- and UV-HCMV-infected cells. The second row of images demonstrates the presence of IE1 staining only in HCMV-infected cells. The third row shows Hoechst nuclear staining in all samples.
The relative abundance of radiolabeled proteins differs dramatically between the HCMV- and the HCMV/R3616-infected cells (Fig. 1A, lanes 3 and 4). The presence of discrete metabolically labeled proteins is characteristic of HSV-1-infected cells and suggests that HSV-1 infection limits HCMV gene expression (Fig. 1A, lane 4). HSV-1 encodes several gene products (virion host shutoff, ICP27) that selectively enhance viral gene expression and limit cellular gene expression within 2 to 3 h of HSV-1 infection (18, 24, 41). Based upon the time course of the infection, the results suggested that HSV-1 infection limited HCMV gene expression as early as 8 hpi. To examine whether R3616 coinfection downregulated HCMV gene expression, immunostaining experiments were performed to examine the most abundant HCMV IE gene product (IE1). In comparison with the HCMV-infected cell lysate, there is reduced IE1 staining for the HCMV/HSV-1-coinfected cell lysate (Fig. 1B, lanes 3 and 4). As anticipated, IE1 was not detected by immunostaining in the mock-infected or UV-inactivated HCMV-infected cell samples (Fig. 1B, lanes 1, 2, 5, and 6). These data support the hypothesis that HSV-1 superinfection reduced subsequent HCMV gene expression and suggested that the complementing HCMV gene was likely expressed in the initial hours of infection, before the Δγ134.5 virus blocked further HCMV gene expression.
To verify that R3616 efficiently superinfected HCMV-infected cells and that the decreased IE1 production was not a global diminution in both HSV-1 and HCMV gene expression, immunoblotting with an antibody against the HSV-1 ICP0 protein was performed. The results show equivalent ICP0 immunostaining for all of the R3616-infected samples and demonstrate that R3616 infection and gene expression proceeds independent of prior HCMV infection (Fig. 1C, lanes 2, 4, and 6). As expected, ICP0 is not detected in the HSV-1-uninfected cells (Fig. 1C, lanes 1, 3, and 5). Taken together, these data indicated that Δγ134.5 HSV-1 infection and gene expression proceed in HCMV-infected cells and that HSV-1 gene expression likely reduced subsequent HCMV IE1 gene expression. These data suggested that the HCMV gene product that complements the Δγ134.5 virus was expressed during the initial hours of HCMV infection.
Experiments next examined if the late viral protein synthesis seen for HCMV/R3616-coinfected cells was from viral evasion of the PKR-mediated protein shutoff response. To test this hypothesis, immunostaining experiments to determine the relative abundance of phosphorylated eIF-2α in the infected cell samples were performed. In uninfected cells, PKR exists in a monomeric, unactivated form. In Δγ134.5-infected cells, viral dsRNA triggers PKR activation, characterized by PKR dimerization, autophosphorylation, and selective phosphorylation of eIF-2α, ultimately leading to the cessation of protein synthesis. As anticipated, minimal phosphorylated-eIF-2α in the mock-infected and UV-inactivated HCMV-exposed cell samples was detected (Fig. 1D, lanes 1 and 5). In contrast, phosphorylated eIF-2α was readily detected in both the R3616-infected mock-infected and R3616/UV-HCMV-coinfected cell samples (Fig. 1D, lanes 2 and 6). This is consistent with the autoradiograph results from the pulse-labeling experiment, demonstrating a reduction of protein synthesis in the infected cells (Fig. 1A, lanes 2 and 6). The level of phosphorylated eIF-2α detected in the HCMV singly infected cells is similar to that in the mock-infected cell sample (Fig. 1D, lanes 1 and 3). These data suggest that either HCMV does not trigger PKR-mediated protein shutoff in the initial 24 h of infection or the virus encodes a protein that precludes this antiviral response. In the HCMV/R3616-coinfected cell sample, phosphorylated eIF-2α is undetectable, indicating that HCMV prevents R3616-induced PKR-mediated protein shutoff (Fig. 1D, lane 4). To verify that eIF-2α existed in the unphosphorylated form in the HCMV singly infected and HCMV/R3616-coinfected cells, immunostaining with an antibody that detects both phosphorylated and unphosphorylated eIF-2α was performed. The results show that equivalent eIF-2α is detected in all cell samples (Fig. 1E, lanes 1 to 6). Taken together, these data suggested that a transcribed HCMV gene product, but not a virion-associated gene product, complemented the Δγ134.5 virus and precluded Δγ134.5-induced host-mediated protein shutoff.
To verify that the level of UV energy used to inactivate HCMV did not damage the capsid and prevent efficient viral attachment, entry, and delivery of the virion-associated gene products, immunofluorescence studies were performed. The HCMV virion-associated protein pp65 (pUL-83) is detectable in both the nuclei of HCMV- and UV-HCMV-infected cells but not in mock-infected cells at 24 hpi (Fig. 1F, top row). Consistent with the IE1 immunostaining studies shown in panel B, the synthesized gene product IE1 is present only in the HCMV-infected cells (Fig. 1F, middle row). Finally, Hoechst nuclear staining of the HCMV-, UV-HCMV- and mock-infected samples demonstrates similar sample size (Fig. 1F, bottom row). Taken together, these data indicate that an HCMV-transcribed gene product is required to abrogate phosphorylation of eIF-2α and the cessation of protein synthesis induced during Δγ134.5 infection.
Genotypes of the viruses used or derived for this study.
Experiments next focused on identifying the HCMV gene that complemented the Δγ134.5 recombinant virus and enabled late viral protein synthesis. An HCMV gene expressed in the initial hours of infection appeared likely based upon the HSV-1 and HCMV coinfection experiments. Furthermore, it was anticipated that a recombinant HCMV lacking the gene would be attenuated and replicate poorly, as has been observed with Δγ134.5 recombinants. In Δγ134.5-infected cells, the virus arrests at the onset of viral DNA replication and late viral gene expression. This results in decreased virion production and extracellular spread (37). Two recombinant HCMV viruses, an IE2 internal-deletion virus and a TRS1 deletion recombinant, had been recently described to exhibit a similar growth defect (3, 37). We hypothesized that these HCMV genes may be involved in viral evasion of the PKR-mediated protein shutoff response and that their absence leads to a delay in the transition to the late phase of HCMV infection. Initial studies suggested that the IE2 gene was not directly involved in PKR evasion but that the HCMV TRS1 gene could complement R3616 late viral protein synthesis (data not shown).
To test the hypothesis that the HCMV TRS1 gene enables viral evasion of host protein shutoff induced by HSV-1 coinfection, Δγ134.5 recombinants encoding either the HCMV TRS1 or HCMV IRS1 gene products were constructed. Schematics of the recombinant viruses and the anticipated hybridization patterns for hybridization studies are presented in Fig. 2. The locations of the HCMV IRS1, TRS1, IRS263, IE2 genes, the HSV-1 γ134.5, UL3, UL4, and the prototypical DNA sequence arrangements of HSV-1 and HCMV are also shown in Fig. 2 (35, 37).
FIG. 2.
Schematic representation of the DNA sequence arrangement of the wild-type HSV-1 and HCMV genome and the recombinant HSV-1 viruses used in this study. Both HCMV (line 2) and HSV-1 (line 6) have group E genomes characterized by two covalently linked components, L and S, each composed of unique sequences (UL and US) flanked by inverted repeat sequences. The locations of the HCMV IRS1, IRS263, and TRS1 genes in the wild-type HCMV genome are shown (line 1). The HCMV IE2 gene and the location of the in-frame deletion mutation in IE2 86 exon 5 (Δ) is also shown (line 3). Line 4 demonstrates the location of one of the two copies of the HSV-1 γ134.5 gene. In the Δγ134.5 parent virus, R3616, both copies of the γ134.5 gene have been deleted, as represented in line 5. Line 7 represents the UL3, UL4 genetic domain in the wild-type and R3616 genome. Line 9 shows the UL3, UL4 domain of the recombinant virus C101, derived from R3616, containing the CMV immediate early promoter-driven EGFP gene. The recombinant virus C130, represented in line 11, contains the HCMV TRS1 gene under control of the HCMV IE promoter in the UL3, UL4 intergenic region of a Δγ134.5 virus. The Δγ134.5 recombinant C132 (expressing IRS1 transcript but not IRS1 protein) and C134 are represented in line 13 and 15, respectively. They contain the CMV IE promoter and HCMV IRS1 gene in the UL3, UL4 intergenic region. Lines 8, 10, 12, 14, and 16 represent the predicted fragments produced by PstI restriction digestion of the viral DNAs. The repair viruses C131 and C135 are not included but are predicted to be identical schematically to the C101 virus (line 9). P, PstI.
HCMV and HSV-1 share a common genomic arrangement characteristic of class E genomes, consisting of two covalently linked long and short genetic domains, each composed of a unique domain flanked by inverted repeat domains (Fig. 2, lines 2 and 6) (43, 44). The recombinant viruses constructed for this study lack both copies of the γ134.5 gene, the principal HSV-1 gene involved in evasion of the PKR host protein shutoff response (Fig. 2, lines 4 and 5) (12, 13). The R3616 UL3, UL4 genetic domain is shown (Fig. 2, line 7). The Δγ134.5 recombinant virus C101 contains a 1,600-bp sequence encoding the CMV IE promoter-driven EGFP gene product flanked by the 8-bp sequence recognized by the PacI restriction enzyme in the UL3, UL4 intergenic region (Fig. 2, line 9). C101 was derived by cotransfection of R3616 viral DNA and pCK1029 plasmid DNA and sequential EGFP-positive plaque purification in Vero cells. To determine if the HCMV TRS1 or HCMV IRS1 gene precludes the shutoff of protein synthesis in Δγ134.5 HSV-1-infected cells, recombinant C130 (encoding the HCMV TRS1 gene product) and recombinant C132 (encoding the IRS1 gene product) were constructed initially (Fig. 2, lines 11 and 13). Subsequent RT-PCR and immunostaining studies showed that the IRS1 recombinant, C132, while expressing IRS1 transcript, did not make immunoreactive protein. Therefore, recombinant C134, which expresses immunoreactive IRS1 protein, was constructed. The C130-repair virus (C131) and C134-repair virus (C135) are not shown but have the same genetic organization as C101 (Fig. 2, line 9).
A novel technique, described in Materials and Methods, was used for constructing the virus; this technique enables selection of the virus on nonhuman cell lines and therefore reduces the selective pressure for secondary revertant mutations in the Δγ134.5 virus. To eliminate the selective advantage provided by second site mutations that restore wild-type protein synthesis, a technique initially described by Glorioso and colleagues was refined to facilitate the selection and repair of recombinant viruses rapidly without exposing the recombinant viruses to human cells (22). Recombinant C101 is a Δγ134.5 virus that encodes a colorimetric selection agent, destabilized EGFP, which flanked by PacI restriction enzyme sites in the UL3, UL4 intergenic region of the HSV-1 genome and is instrumental in this novel method (Fig. 2, line 9). The 8-bp sequence recognized by the PacI restriction enzyme does not occur naturally in the 152,260 bp of the HSV-1 genome, and its presence in the recombinant viral DNA enables selective digestion at the intergenic region. The PacI-digested DNA is not readily replicated after transfection in mammalian cells. However, new recombinants are synthesized efficiently and selected in nonhuman cells by cotransfecting the cut viral DNA with a “rescue” plasmid carrying the new genetic information flanked by homologous domains that bridge the digested viral DNA. Recombinant Δγ134.5 viruses do not elicit the host-mediated protein shutoff response in Vero cells. This allows the nonselective growth of either Δγ134.5 or wild-type HSV-1 viruses.
The DNA hybridization studies are shown in Fig. 3. Hybridization of the electrophoretically separated, immobilized PstI-digested viral DNA with a probe spanning the HSV-1 UL3 and UL4 open reading frames revealed a shift from a single 3.07-kb PstI fragment in R3616 (Fig. 4A, lane 1) to two PstI fragments of 2.09 and 1.29 kb in C101 (Fig. 3A, lane 2) created by two novel PstI sites in the EGFP cassette in C101. The 1.6-kb PstI fragment containing the EGFP open reading frame and part of the CMV promoter does not contain HSV-1 sequence and does not hybridize with the probe but is present when hybridized with an EGFP-containing probe (data not shown). The recombinant C130 encodes the HCMV TRS1 gene product inserted between the UL3 and UL4 genes of Δγ134.5 HSV-1. The HCMV TRS1 gene contains two PstI restriction sites (21 bp apart) in the 5′ sequence domain. These unique restriction sites create two detectable fragments, of 4.04 kb and 3.38 kb, in the PstI-digested C130 viral DNA (Fig. 3A, lane 3). The repair virus, C131, is predicted to have a similar genetic organization as C101, and the DNA hybridization studies confirm this (Fig. 3A, lanes 2 and 4). Recombinant C132 contains the HCMV IRS1 sequence in the UL3, UL4 intergenic region. The HCMV IRS1 and TRS1 genes share a common 5′ genetic domain but diverge in the 3′ region. Consequently, a similar hybridization pattern is seen with the C130 and C132 PstI-digested viral DNA. The C132 recombinant shares an identical 3.38-kb fragment (encoded by the UL3 and 5′ domain of the IRS1 gene) as C130; however, the unique 3′ sequence produces a slightly slower-migrating 4.19-kb fragment by Southern blotting (Fig. 3A, lane 5). Recombinant C134 (expressing immunoreactive IRS1 protein) also contains a CMV IRS1 gene in the UL3, UL4 intergenic region. This recombinant was derived from a different plasmid and the cloning strategy eliminated a 960-bp sequence upstream from the HCMV IE promoter and 510 bp downstream from the polyadenylation site. The probe, therefore, hybridizes with both a 2.42-kb and a 3.68-kb fragment in the C134 recombinant. The repair virus, C135, is genetically similar to C101 and C131 and also produces a 1.29-kb and a 2.09-kb restriction fragment (Fig. 3A, lane 7). Immunostaining studies, using pooled IRS1 and TRS1 antisera, demonstrate that the C130 and C134 recombinant viruses produce immunoreactive TRS1 and IRS1 protein, respectively, whereas the C132 recombinant does not (Fig. 3B, lanes 3, 5, and 6). Reverse transcription and PCR amplification further demonstrate that the C132 virus expresses IRS1 transcript, indicating that the recombinant contains an IRS1 gene with either a frameshift mutation or a premature stop codon (Fig. 3C, lane 3).
FIG. 3.
Composite of the Southern hybridization, immunoblot, and RT-PCR images. (A) Image of the electrophoretically separated DNA fragments from PstI digested viral DNAs hybridized with a specific probe encoding the HSV-1(F) UL3 and UL4 gene products. (B) Immunoblot image demonstrating TRS1 or IRS1 protein production in the C130- and C134-infected cell samples using pooled monoclonal antisera against TRS1 and IRS1. (C) IRS1 transcript is detectable by RT-PCR in the C132-infected cells and does not represent DNA contamination, as demonstrated by the absence of PCR product in the RNA-alone sample.
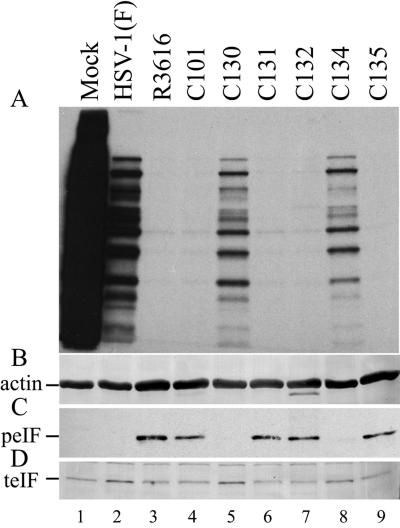
FIG. 4.
Composite of autoradiograph and immunoblot images. (A) Autoradiographic image of the pulse-labeled mock-infected and HSV-1(F), R3616, C101, C130 to C132, C134, and C135 virus-infected cell samples. (B) Immunoblot, using antibody against actin, showing evidence of equivalent mass loading. (C) An immunoblot, using antibody directed against phosphorylated eIF-2α (Ser-51), demonstrates phosphorylated eIF-2α in the R3616-, C101-, C131-, C132-, and C135-infected cell samples and its absence in the mock-, HSV-1(F)-, C130-, and C134-infected samples. (D) An immunoblot, using antibody against total eIF-2α, demonstrates that eIF-2α is detectable in both mock-infected and infected cells.
A Δγ134.5 recombinant encoding the HCMV TRS1 gene product exhibits the wild-type protein synthesis phenotype.
Transient expression studies suggested that the HCMV TRS1 complemented Δγ134.5 late viral protein synthesis. To further test this hypothesis, cells infected with Δγ134.5 recombinant viruses expressing the TRS1 and IRS1 gene products were examined for late HSV-1 viral protein synthesis by using pulse-labeling experiments. Replicate cultures of HFF were mock infected or infected with HSV-1(F), R3616, C101, C130-C132, C134, and C135 at an MOI of 10. The cultures were labeled with [35S]methionine at 15 hpi for 1 h and then processed for autoradiography as described in Materials and Methods.
Both mock- and HSV-1(F)-infected cell samples contain abundant radiolabeled protein (Fig. 4A, lanes 1 and 2). As discussed previously, the HSV-1(F)-infected cell sample contains distinct radiolabeled proteins characteristic of wild-type HSV-1 protein synthesis. In contrast, there is decreased detection of radiolabeled proteins in the R3616 and C101 infected cell samples, characteristic of the Δγ134.5 protein synthesis phenotype (Fig. 4A, lanes 3 and 4). Both of these recombinants lack the γ134.5 gene and are incapable of precluding PKR-mediated protein shutoff. In the cells infected with recombinant viruses expressing the TRS1 (C130) or IRS1 (C134) protein, radiolabeled proteins are readily detectable, indicative of continued protein synthesis in the infected cell (Fig. 4A, lanes 5 and 8). In contrast, there is decreased detection of radiolabeled proteins in the C131, C132, and C135 and infected cells (Fig. 4A, lane 6, 7, and 9). These data indicate that expression of either the HCMV TRS1 or IRS1 protein can confer the wild-type HSV-1 protein synthesis phenotype in Δγ134.5-infected cells.
To verify that inhibition of PKR-mediated protein shutoff was the basis for the late viral protein synthesis witnessed with the C130- and C134-infected cells, equivalent-mass samples (10 μg), as demonstrated by equivalent actin immunostaining (Fig. 4B, lanes 1 to 9), were electrophoretically separated and immunostained for phosphorylated eIF-2α. The results show that phosphorylated eIF-2α in the R3616, C101, C131, C132 and C135-infected cell samples, characteristic of PKR-mediated protein shutoff, is readily detected (Fig. 4C, lanes 3, 4, 6, 7, and 9). In the mock-, HSV-1(F)-, C130-, and C134-infected cell samples, phosphorylated eIF-2α is undetectable by immunostaining and correlates with continued protein synthesis in these cells (Fig. 4B, lanes 1, 2, 5, and 8). Total eIF-2α immunostaining verifies the presence of eIF-2α in the C130 and C134 samples (Fig. 4D, lanes 5 and 8). These data indicate that in cells infected with a Δγ134.5 recombinant virus expressing either the HCMV TRS1 or IRS1 protein, eIF-2α is maintained in the unphosphorylated state, thus allowing continued late viral protein synthesis similar to that observed with wild-type HSV-infected cells.
DISCUSSION
Eukaryotic cells contain biochemical systems that limit viral gene expression (36). Instrumental in these defense pathways are proteins which can be activated by viral induced changes in the cell. Once activated, these proteins inhibit critical cellular functions and limit the impact of viral gene expression in the cell. One of these pathways is the dsRNA-activated protein shutoff response, which inhibits protein synthesis initiation in the infected cell. This antiviral response is controlled by a cellular kinase, PKR, which remains inactive in unstressed cells, but is induced by interferon stimulation (36). Upon binding dsRNA of sufficient size to permit dimerization, the kinase activates, autophosphorylates, and then selectively phosphorylates eIF-2α inhibiting protein synthesis initiation in the cell (36). Cell stress responses (heat shock and growth factor withdrawal) can also trigger PKR-mediated protein shutoff and concomitant growth suppression (15, 30, 46). In addition to regulating protein synthesis in the cell, PKR also is involved in dsRNA-mediated NF-κB activation and interferon regulatory factor 1 (IRF1)-dependent gene transcription (23).
Viruses have evolved genes that counteract these host defense mechanisms to permit viral gene expression and replication in the infected cell. In the case of PKR-mediated protein shutoff, viral evasion strategies have been shown to include proteolytic degradation of PKR (poliovirus), inhibition of PKR activation (adenovirus VAiRNA, HHV-8 vIRF-2, influenza NS1, vaccinia E3L), prevention of eIF-2α phosphorylation by activated PKR (vaccinia virus K3L), or the dephosphorylation and regeneration of unphosphorylated eIF-2α (HSV-1 γ134.5 protein) (2, 5, 14, 19, 25, 27, 28, 39).
HCMV is predicted to produce complementary transcripts that anneal to form dsRNA. Moreover, HCMV infection triggers an interferon response in the infected cell, inducing the synthesis of interferon-regulated genes integral to the host antiviral response (4, 47). Based upon these findings, it was anticipated that HCMV encoded a protein that enabled evasion of the interferon-mediated antiviral response. Investigators using a vaccinia recombinant published studies showing that HCMV encodes a gene product that permits continued protein synthesis in the HCMV-infected cell (11). By using chemicals that block viral DNA polymerase activity, it was predicted that the gene was expressed prior to viral DNA replication (11). Investigators further reported, using a vaccinia recombinant lacking E3L, that the HCMV IRS1 and TRS1 genes could counteract the host antiviral response pathway. Our studies confirm this and show that either the IRS1 or TRS1 protein can also complement an HSV-1 recombinant incapable of recruiting protein phosphatase 1α and evading PKR-mediated protein shutoff.
The following findings are presented in this report: (i) HCMV infection complements HSV-1 Δγ134.5 infection and enables late viral protein synthesis. (ii) EIF-2α is maintained in an unphosphorylated form in the HCMV/Δγ134.5 coinfected cells, indicating that HCMV encodes a gene product that targets PKR-mediated protein shutoff. (iii) The HCMV gene product responsible is expressed in the initial hours of infection and virion-associated gene products are not sufficient to permit continued protein synthesis in the infected cells. (iv) Independent transfer of the HCMV TRS1 and IRS1 gene to an HSV-1 Δγ134.5 recombinant demonstrates that expression of either HCMV TRS1 or IRS1 protein changes the phenotype of the Δγ134.5 recombinant, resulting in continued late viral protein synthesis and the maintenance of eIF2α in the unphosphorylated state. (v) A recombinant virus that expresses HCMV IRS1 transcript but does not make IRS1 protein is unable to reverse PKR-mediated protein shutoff. This suggests that IRS1 or TRS1 protein is necessary for evasion of PKR-mediated host protein shutoff in the recombinant infected cells.
These data show that both the HCMV TRS1 or IRS1 gene products can independently complement Δγ134.5 recombinant viruses and reverse PKR-mediated protein shutoff response in the infected cell. Both the IRS1 and TRS1 genes share common promoters and a common sequence encoded in the repeated sequences (c and c′ domain) of the HCMV short genetic domain. The 3′ sequences of the IRS1 and TRS1 genes are encoded in the US domain. Consequently, the gene products diverge in the C-terminal region, while sharing the same N-terminal 549 amino acids. The HCMV IRS263 gene is expressed at immediate early and late times in infection, although accumulation peaks early in infection. The transcript is encoded in the short unique domain of the genome and encodes an approximately 30-kDa protein (35). HCMV TRS1 is delivered as a virion-associated protein and is also synthesized de novo as an immediate early gene product with nuclear and cytoplasmic localization. The greatest accumulation occurs late in HCMV infection, when TRS1 exhibits a predominant cytoplasmic distribution. TRS1 is required for oriLyt-dependent DNA replication and enhances reporter gene expression in conjunction with different HCMV gene products in transfection assays (20, 35, 40). The mechanism of TRS1 transactivation enhancement has not been elucidated. The TRS1 gene product can cooperate with ppUL69 and pp71 to enhance IE activity and requires the IE2 gene product to enhance ICP36 late promoter activity (34, 35). There is no evidence that TRS1 acts directly in oriLyt replication or in HCMV gene transactivation.
An HCMV TRS1 deletion recombinant that exhibited a growth arrest similar to that observed in Δγ134.5 recombinants was reported (3). The ΔTRS1 recombinant virus contained a substitution mutation in the central domain of the TRS1 coding domain (nucleotides 226,115 to 227,585) (ADsubTRS1) and replicated poorly in low-multiplicity infections, but was capable of late viral protein synthesis (3). Investigators found that the ADsubTRS1 virus was defective and required complementation, whereas a recombinant lacking the central domain of the IRS1 gene (ADsubIRS1) and the wild-type virus grew similarly (3). In some respects, the behavior of the ADsubTRS1 recombinant is similar to that of the Δγ134.5 mutants. They both arrest at late stages of infections (following the onset of viral DNA synthesis) with decreased virion assembly and reduced virus recovery. However, there are distinct differences between the behavior of the Δγ134.5 and the ΔTRS1 virus. The ADsubTRS1 synthesizes viral DNA and late HCMV viral proteins in infected cells, like that seen in wild-type infection. The principal defects observed with the AdsubTRS1 virus involved abnormal localization of pp65 and pp71 and the arrest of virion assembly (1, 3).
How do HCMV TRS1 and IRS1 preclude the host protein shutoff response and are these findings biologically significant? One possible explanation is that the phenotype elicited in this study does not represent the function of TRS1 or IRS1 in HCMV infection. This explanation would suggest that HCMV encodes a gene product capable of inhibiting a principal cellular antiviral defense but does not utilize the gene for this purpose during HCMV infection. A second possibility is that TRS1 is the primary defense against the host protein shutoff response during HCMV infection. In prior studies, a TRS1 deletion virus (ADsubTRS1) synthesized late HCMV proteins similarly to wild-type virus-infected cells and does not appear to elicit the host protein shutoff response (3) These findings do not exclude the possibility that HCMV TRS1 inhibits PKR-mediated protein shutoff in infected cells. In the AdsubIRS1 or AdsubTRS1 recombinant, the presence of TRS1 or IRS1, respectively, would preclude PKR-mediated host protein shutoff. The IRS1 TRS1 double deletion would address this possibility.
Another interesting paradox is that de novo synthesized HCMV TRS1 or IRS1 is required to protect against host protein shutoff, while virion-associated IRS1 or TRS1 protein is not sufficient to preclude the host protein shutoff response. This is not unprecedented. Viral gene products satisfy diverse functions during the lytic cycle. These functions, encoded by different domains of the gene, can be regulated based upon cellular location, kinetic expression, or the presence of associated viral gene products. In particular, studies of an HSV-1 second-site mutant expressing Us11 early in infection have shown that only the de novo synthesis of the US11 gene product, but not the virion-associated US11, inhibits PKR-mediated host protein shutoff (8).
HCMV TRS1 and IRS1 share a common 1,648-bp sequence encoded within the repeat short domain of the HCMV genome. The similar protein synthesis phenotype exhibited by C130 (TRS1 Δγ134.5) and C134 (IRS1 Δγ134.5) HSV-1 recombinant suggests that a shared sequence domain in HCMV TRS1 and IRS1 is important for evasion of PKR-mediated shutoff of protein synthesis. Definition of the essential gene product domain by fine mapping remains to be performed.
Acknowledgments
I thank Giang Tong for technical assistance, William Britt for scientific discussions and monoclonal antibodies used for immunofluorescence studies, Jackie Parker and Mark Pritchard for scientific discussions, Tom Shenk for the generous gift of the TRS1, IRS1, and IRS1263 expression clones, Veronica Sanchez and Deborah Spector for the IE2 internal deletion recombinant viruses, and Sameera Kasaam for reading and editing of the manuscript.
These studies were aided by grants from NIH (AI01680) and from the Research Institute of Alabama Children's Hospital Foundation.
REFERENCES
- 1.Adamo, J. E., J. Schroer, and T. Shenk. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 78:10221-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, T. L., G. N. Barber, and M. G. Katze. 1993. Degradation of the interferon-induced 68,000-Mr protein kinase by poliovirus requires RNA. J. Virol. 67:791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankenship, C. A., and T. Shenk. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J. Virol. 76:12290-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burysek, L., and P. M. Pitha. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 75:2345-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, K., O. Elroy-Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 268:12837-12842. [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, G. Y. Gillespie, and B. Roizman. 2002. Second-site mutation outside of the US10-12 domain of Δγ134.5 herpes simplex virus 1 recombinant blocks the shutoff of protein synthesis induced by activated protein kinase R and partially restores neurovirulence. J. Virol. 76:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassady, K. A., M. Gross, and B. Roizman. 1998. The second-site mutation in the herpes simplex virus recombinants lacking the γ134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J. Virol. 72:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Child, S. J., S. Jarrahian, V. M. Harper, and A. P. Geballe. 2002. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J. Virol. 76:4912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 13.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, M. V., H. W. Chang, B. L. Jacobs, and R. J. Kaufman. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67:1688-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan, R., and J. W. Hershey. 1984. Heat shock-induced translational alterations in HeLa cells. Initiation factor modifications and the inhibition of translation. J. Biol. Chem. 259:11882-11889. [PubMed] [Google Scholar]
- 16.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 17.Fortunato, E. A., M. L. Dell'Aquila, and D. H. Spector. 2000. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 97:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krisky, D. M., P. C. Marconi, T. J. Oligino, R. J. Rouse, D. J. Fink, J. B. Cohen, S. C. Watkins, and J. C. Glorioso. 1998. Development of herpes simplex virus replication-defective multigene vectors for combination gene therapy applications. Gene Ther. 5:1517-1530. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. B., S. R. Green, M. B. Mathews, and M. Esteban. 1994. Activation of the double-stranded RNA (dsRNA)-activated human protein kinase in vivo in the absence of its dsRNA binding domain. Proc. Natl. Acad. Sci. USA 91:10551-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, T. G., N. Tang, S. Thompson, J. Miller, and M. G. Katze. 1994. The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol. Cell. Biol. 14:2331-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 28.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 30.Montine, K. S., and E. C. Henshaw. 1989. Serum growth factors cause rapid stimulation of protein synthesis and dephosphorylation of eIF-2 in serum deprived Ehrlich cells. Biochim. Biophys. Acta 1014:282-288. [DOI] [PubMed] [Google Scholar]
- 31.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 32.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227-232. [DOI] [PubMed] [Google Scholar]
- 33.Pritchett, R. F. 1980. DNA nucleotide sequence heterogeneity between the Towne and AD169 strains of cytomegalovirus. J. Virol. 36:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp, T. V., M. Schwemmle, I. Jeffrey, K. Laing, H. Mellor, C. G. Proud, K. Hilse, and M. J. Clemens. 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 21:4483-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudhakar, A., A. Ramachandran, S. Ghosh, S. E. Hasnain, R. J. Kaufman, and K. V. Ramaiah. 2000. Phosphorylation of serine 51 in initiation factor 2α (eIF2α) promotes complex formation between eIF2α(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry 39:12929-12938. [DOI] [PubMed] [Google Scholar]
- 43.Tamashiro, J. C., and D. H. Spector. 1986. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J. Virol. 59:591-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wadsworth, S., R. J. Jacob, and B. Roizman. 1975. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J. Virol. 15:1487-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, S. L., C. B. Hartline, N. L. Kushner, E. A. Harden, D. J. Bidanset, J. C. Drach, L. B. Townsend, M. R. Underwood, K. K. Biron, and E. R. Kern. 2003. In vitro activities of benzimidazole d- and l-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan, Q., K. A. Lord, I. Alamo, Jr., M. C. Hollander, F. Carrier, D. Ron, K. W. Kohn, B. Hoffman, D. A. Liebermann, and A. J. Fornace, Jr. 1994. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol. Cell. Biol. 14:2361-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]