Abstract
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) complex comprises three gp120 exterior glycoproteins each noncovalently linked to a gp41 transmembrane glycoprotein. Monomeric gp120 proteins can elicit antibodies capable of neutralizing atypically sensitive test viruses in vitro, but these antibodies are ineffective against representative primary isolates and the gp120 vaccines failed to provide protection against HIV-1 transmission in vivo. Alternative approaches to raising neutralizing antibodies are therefore being pursued. Here we report on the antibody responses generated in rabbits against a soluble, cleaved, trimeric form of HIV-1JR-FL Env. In this construct, the gp120 and gp41 moieties are covalently linked by an intermolecular disulfide bond (SOS gp140), and an I559P substitution has been added to stabilize gp41-gp41 interactions (SOSIP gp140). We investigated the value of DNA priming and compared the use of membrane-bound and soluble priming antigens and of repeat boosting with soluble and particulate protein antigen. Compared to monomeric gp120, SOSIP gp140 trimers elicited approximately threefold lower titers of anti-gp120 antibodies. Priming with DNA encoding a membrane-bound form of the SOS gp140 protein, followed by several immunizations with soluble SOSIP gp140 trimers, resulted in antibodies capable of neutralizing sensitive strains at high titers. A subset of these sera also neutralized, at lower titers, HIV-1JR-FL and some other primary isolates in pseudovirus and/or whole-virus assays. Neutralization of these viruses was immunoglobulin mediated and was predominantly caused by antibodies to gp120 epitopes, but not the V3 region.
The need for an effective vaccine to prevent the spread of human immunodeficiency virus type 1 (HIV-1) is well recognized, as is the likelihood that such a vaccine would require a component able to elicit broadly reactive and potent neutralizing antibodies (NAb) (reviewed in references 20, 26, 29, 35, and 40). The only relevant HIV-1 antigens for induction of NAb are the envelope glycoproteins encoded by the viral env gene. The functional Env complex comprises three gp120 surface glycoproteins each noncovalently linked to a gp41 transmembrane glycoprotein. The resulting trimeric structure is capable of receptor binding and the subsequent conformational changes that drive the fusion of the viral and cell membranes (3, 25, 55, 71). Various forms or subcomponents of the HIV-1 Env complex have been tested as vaccine immunogens in animals and humans over the past 20 years (26, 29, 40). The most studied proteins have been individual, monomeric gp120 subunits, expressed as recombinant proteins from mammalian cells. These gp120 monomers induce Ab capable of recognizing the immunizing antigen and of neutralizing atypically sensitive HIV-1 strains, including those that have been adapted to replication in vitro (51). However, Ab raised to gp120 proteins have not proven capable of neutralizing representative primary strains of HIV-1 at significant titers and they failed to elicit any protection against HIV-1 infection when they were tested in two efficacy trials (18). Hence, many research groups worldwide are now trying to develop Env-based immunogens that might perform better than gp120 monomers.
There are two major obstacles to the success of a NAb-based vaccine: HIV-1 sequence variation and the defenses that the Env complex has evolved to thwart the humoral immune system. Sequence variation may turn out to be the more difficult problem to solve, but it will only become relevant if we can first learn how to elicit Ab capable of successfully countering even a small fraction of circulating strains (13, 26, 46, 68). The defenses of Env against Ab have two principal but interlocking functions: to limit the induction of NAb in the first place and then reduce the probability that any elicited Ab can bind to the functional Env complex on the virion surface and thereby neutralize infectivity. Components of the defenses include the extensive glycosylation of both the gp120 and gp41 subunits, particularly gp120 since half its molecular weight is attributable to N-linked sugars; the structural flexibility of the CD4 and coreceptor binding sites on gp120; the limited availability of T-helper epitopes on Env; the creation of the coreceptor binding site on gp120 only after CD4 binding, combined with its inaccessibility after the Env complex has attached to cell surface CD4; and the limited exposure of conserved gp41 structural elements that are formed only during the fusion process itself (13, 30, 34, 36, 37, 70, 80, 81). The net effect is that only a few anti-Env Ab can bind to preexisting or new epitopes on the Env complex before or during the fusion process. The broadly reactive NAb that have been identified were isolated from HIV-1-infected humans. Although we can never be certain what forms of Env they were raised against in vivo, one possibility is that the stimulating antigen was a native structure present on virus particles or virus-infected cells (13, 50, 51, 55).
One approach to eliciting NAb more efficiently is to try to make and then modify proteins that mimic the native trimeric Env structures that exist on the virion surface (13). These trimers are, however, not easy to create in recombinant form, and several steps need to be taken to facilitate the process. The most common initial modification is to truncate the full-length gp160 protein immediately prior to the transmembrane domain of gp41 so that a soluble gp140 protein can be secreted and purified (5, 16, 33, 61, 73, 74, 79). However, the gp140 proteins are often unstable if they are proteolytically processed at the natural site that links gp120 with the gp41 ectodomain (gp41ECTO), and they dissociate spontaneously into their constituent subunits (5). One solution to this problem is to alter the cleavage site to prevent proteolysis, leading to the production of uncleaved gp140 proteins (gp140UNC) (15, 16, 61, 73, 74, 79). In some cases, additional trimer-stabilizing modifications have been made to gp140UNC proteins (15, 17, 62, 73, 74, 79).
We have taken a different approach by introducing an engineered disulfide bond between the gp120 and gp41ECTO subunits to stabilize their interaction, thus allowing the expression of a cleaved gp140 protein based on HIV-1JR-FL (5). The resulting proteins, designated SOS gp140, still have instability between the individual gp41ECTO subunits (59). An additional substitution (I559P) was therefore introduced within gp41ECTO to create a more stable protein known as SOSIP gp140 (58).
Uncleaved gp140 proteins and cleaved SOSIP gp140 proteins derived from many different HIV-1 isolates all form oligomers when expressed in vitro, with dimers, trimers, tetramers, and some higher-molecular-weight aggregates being commonly observed (15, 16, 58, 61, 73, 74, 79). Usually, but not always, the trimer fraction is purified prior to immunogenicity or other experiments because the other oligomeric forms presumably represent aberrant Env configurations caused, for example, by the formation of nonphysiological intersubunit disulfide bonds. Several studies have suggested that some versions of gp140UNC proteins do offer a modest but significant improvement for NAb induction, compared to monomeric gp120 proteins (2, 10, 67, 75). However, care must be taken when judging the magnitude and meaning of incremental improvements in Env immunogenicity, taking into account the now widespread use of new assays that offer a modest increase in sensitivity (41). Whether cleavage, or the lack of it, has enough influence on the conformation of gp140 trimers to significantly alter their immunogenicity has not been determined but is worth evaluating. As a first step, we report here on our initial assessments of the immunogenicity of cleaved trimeric JR-FL SOSIP gp140 proteins in rabbits using a DNA-priming, protein-boosting vaccination regimen.
MATERIALS AND METHODS
Env proteins and plasmids used for immunizations.
All plasmids and Env glycoproteins were derived from the genetic background of HIV-1JR-FL, a subtype B, R5 isolate (31). Except for monomeric gp120JR-FL, which was expressed from a standard env gene in stably transfected Chinese hamster ovary cells (64), all Env proteins were expressed using codon-optimized genes. Their design is summarized in Fig. 1A. The proteins were transiently expressed in 293T cells using the high-level mammalian expression vector pPPI4 as described elsewhere (5). A pPPI4 plasmid lacking an env insert served as an empty-vector control. The sequence integrity of all clones was confirmed prior to use.
FIG. 1.
Env constructs and SOSIP.R6 trimer purification. (A) Schematic of the env DNA constructs used for the priming immunization. Highlighted are the approximate size of the Env protein construct relative to full-length Wt gp160, the inclusion of a Wt (REKR) or enhanced (RRRRRR) cleavage site motif, the approximate position of the SOS cysteine bond (C = C), and the approximate position of the gp41-gp41 trimer-stabilizing mutation (I559P). The C1 to C5 domains of gp120 are also identified. Ecto, TM, and Cyt indicate the positions of the gp41 ectodomain, the membrane-spanning domain, and the cytoplasmic tail, respectively. (B) SDS-PAGE analysis of the purified HIV-1 gp120 and SOSIP.R6 gp140 proteins. The purified proteins, solubilized in Laemmli sample buffer with (reduced) or without (nonreduced) 20 mM dithiothreitol, were resolved on a 4 to 12% Bis-Tris polyacrylamide gradient gel and stained using Coomassie blue. (C) Blue-Native PAGE analysis of the purified HIV-1 gp120 and SOSIP.R6 gp140 proteins. The proteins were resolved on a 4 to 12% Bis-Tris polyacrylamide gradient gel and visualized using Coomassie blue. The values on the left are molecular sizes in kilodaltons.
The gp140 wild-type (Wt) vector expresses a soluble gp140 protein containing an unmodified cleavage site between gp120 and gp41ECTO. When expressed in cell culture, such proteins are only partially processed by cellular proteases, leading to mixtures of uncleaved and cleaved oligomeric gp140 proteins. The cleaved proteins readily dissociate into their gp120 and gp41ECTO components (5). The configurations adopted by gp140 Wt proteins after expression from DNA vectors in vivo are not known.
The SOSIP.R6 gp140 construct expresses a soluble gp140 protein containing an intermolecular disulfide bond (SOS) to stabilize the interaction between gp120 and gp41ECTO, together with an additional substitution (I559P) within gp41 that increases the association between the gp41ECTO components (58). The protein also contains a hexa-arginine (R6) motif in place of the original cleavage site between gp120 and gp41ECTO that is designed to increase the extent of gp140 cleavage into its constituents (6). When expressed in cell culture, the SOSIP.R6 gp140 protein forms a mixture of monomers, dimers, trimers, and higher-molecular-weight forms, with trimers usually predominating (reference 58 and data not shown). Again, how the protein appears upon in vivo expression is unknown.
The SOS.R6(T) construct expresses a membrane-bound version of SOS gp140 with a minimal cytoplasmic tail. It contains the SOS disulfide bond and the cleavage-enhancing R6 substitution but without the I559P mutation. SOS.R6(T) was made by first amplifying a Wt gp140Δct fragment from the full-length gp160 JR-FL template essentially as described elsewhere (32). The gp140Δct fragment contains the gp41 transmembrane domain with a short, three-amino-acid cytoplasmic tail. The additional SOS and R6 modifications were then made as described previously (5, 6). Truncation of the cytoplasmic domain of gp41, as in the SOS.R6(T) protein, circumvents natural sequences that promote down-regulation of Env, including the membrane-proximal Tyr-based endocytosis motif. Elimination of these sequences has been shown to increase the cell surface expression of Env in vitro and the magnitude of Env-induced immune responses in vivo (12, 76).
Env protein expression and purification.
The JR-FL SOSIP.R6 protein was expressed as a mixture of monomeric and oligomeric species from transiently transfected 293T cells (58). Briefly, the cells were cotransfected with the pPPI4-SOSIP.R6 and pcDNA3.1-Furin plasmids at a 2:1 ratio using polyethylenimine as the transfection reagent and cultured for 48 h (5, 9). Cell culture supernatants were collected, supplemented with protease inhibitors, concentrated, and stored at −80°C until further use.
The recombinant HIV-1JR-FL gp120 glycoprotein was expressed and purified as previously described (64). The SOSIP.R6 gp140 protein was purified from the concentrated supernatants by initial fractionation with 50% ammonium sulfate. The supernatant solution was subsequently processed using Galanthus nivalis lectin affinity chromatography (Vector Laboratories, Burlingame, CA). The trimeric form of SOSIP.R6 gp140 was separated from other monomeric and oligomeric Env proteins by gel filtration chromatography on Superose 6 columns (Amersham Biosciences, Piscataway, NJ) equilibrated with phosphate-buffered saline (PBS). The eluate fractions containing the trimeric SOSIP.R6 gp140 protein were identified by Blue-Native polyacrylamide gel electrophoresis (PAGE) analysis (59) and then combined and concentrated. The concentrations of purified Env proteins were measured by UV spectroscopy (64).
SOSIP.R6 gp140 trimers were also used for immunizations after capture onto 50-nm-diameter protein G paramagnetic beads (Miltenyi Biotec, Auburn, CA) coated with the human NAb 2G12 (66) to create a particulate antigen. The production and general properties of these bead-immobilized antigens will be described elsewhere (N.S. et al., unpublished data).
Immunization protocol.
The care, maintenance, and immunization of rabbits were carried out by Aldevron LLC, Fargo, ND, under contract. The facility operates in full compliance with the Animal Welfare Act, abides by the principles outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals, follows U.S. Department of Agriculture guidelines on the use of laboratory animals, and has Institutional Animal Care and Use Committee approval for the electroporation and immunization procedures.
One-year-old female New Zealand albino rabbits were anesthetized using an intramuscular (bicep) injection of a cocktail of ketamine, xylazine, and acepromazine prior to the injection of the pPPI4-based, codon-optimized Env expression plasmid into the hind limb skeletal muscle. A preimmunization blood sample (8 to 10 ml) was taken by venipuncture prior to injection of the sterile filtered plasmid (1 mg in PBS, administered as two 0.5-mg doses at two sites). A mild current was then applied by a caliper-type electrode, allowing the uptake of DNA to be enhanced by electroporation (39, 69). The rabbits were DNA immunized using this electroporation procedure at weeks 0 and 4, followed by several boosting immunizations with a JR-FL-based Env protein construct formulated in QS-21 adjuvant (Antigenics, Framingham, MA). For the protein immunizations in the later stages of the SOS.R6(T)-SOSIP.R6 study group (arm E), from week 37, Enhanzyn (Corixa Corp., Seattle, WA) was used as an alternative adjuvant to QS-21. For immunization, Env proteins were administered by injection into multiple anatomical sites (300 μl intramuscularly, into each hind leg, 50 μl intradermally at six sites, and 100 μl subcutaneously into the neck region; a total of 1 ml) as described elsewhere (67). The timing of each blood drawing or dose administration and the constructs used for each study animal are outlined in Fig. 2A and 3A.
FIG. 2.
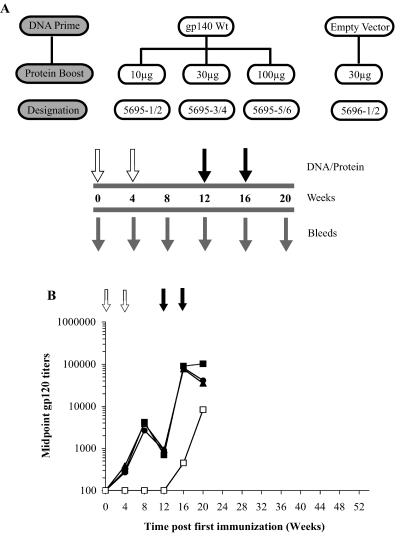
Anti-gp120 binding Ab elicited by immunization in the pilot study. (A) The schematic for the pilot study highlights the DNA construct (gp140 Wt, or empty vector) used for priming (open arrows), the dose of gp120 protein (10, 30, or 100 μg) used for each of the protein boosts (filled arrows), and the serum collection times (grey arrows). (B) Generation of anti-gp120 Ab in the pilot study. Rabbits were primed with 1 mg of pPPI4 expressing either codon-optimized soluble gp140 Wt Env (filled symbols) or the empty-vector control (open squares) and then boosted with 10 μg (filled triangles), 30 μg (filled squares), or 100 μg (filled circles) of soluble gp120 at the times indicated. The anti-gp120 Ab responses were measured by ELISA. Each datum point represents the mean (n = 2 animals) midpoint anti-gp120 binding titer for each arm.
FIG. 3.
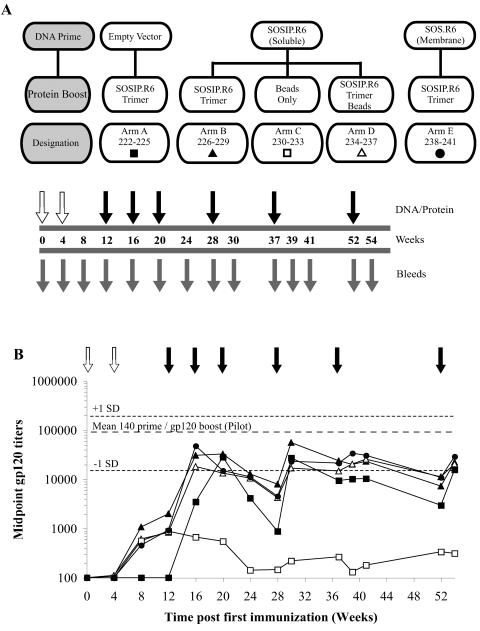
Anti-gp120 binding Ab elicited by SOSIP.R6 immunization. (A) The schematic for the second-stage study highlights the DNA construct [empty vector, SOSIP.R6, or SOS.R6(T)] used for priming (open arrows) the protein construct (SOSIP.R6 trimer, SOSIP.R6 trimer coupled to paramagnetic beads, or empty beads) used for each of the protein boosts (filled arrows), and the serum collection times (grey arrows). The designation numbers for the individual animals in each arm (A to E) are also listed. (B) Rabbits were primed with 1 mg pPPI4 expressing soluble gp140 SOSIP.R6 Env (▴, □, ▵; arms B, C, and D) or membrane-bound SOS.R6 Env (•; arm E) or primed with the empty-vector control (▪; arm A). The animals were then boosted with 30 μg of SOSIP.R6 trimer (▪, ▴, •; arms A, B, and E), 30 μg of SOSIP.R6 trimer coupled to beads (▵; arm D), or empty beads as a control (□; arm C) at the times indicated. The anti-gp120 Ab responses were measured by ELISA. Each datum point represents the mean (n = 4 animals) midpoint anti-gp120 binding titer for each arm. The mean (± standard deviation) anti-gp120 titer in the gp140 Wt DNA-primed and gp120-boosted animals (5695-1 to 5695-6) at week 20 of the pilot study is indicated by the dotted lines for comparison.
Binding of immune sera to monomeric gp120 and V3 peptides.
The cyclic peptides V3JR-FL (Ac-CTRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAHC-NH2) and V3HXB2 (Ac-CTRPNNNTRKRIRIQRGPGRAFVTIGKIGNMRQAHC-NH2) were synthesized by the American Peptide Company Inc. PA1 is a V3JR-FL-specific murine monoclonal Ab (MAb), as defined by its ability to bind the cyclic V3JR-FL peptide, but not the cyclic V3HXB2 peptide or a gp120JR-FL protein with V3 deleted, in immunoassays (N.S. et al., unpublished). The CD4-immunoglobulin G2 (CD4-IgG2) molecule has been described elsewhere (65). The binding of MAb, CD4-IgG2, and rabbit immune sera to monomeric gp120 was measured by enzyme-linked immunosorbent assay (ELISA) using the appropriate anti-species-alkaline phosphatase conjugate and the AMPAK colorimetric detection system (Dako Diagnostics) as previously described (43, 47). For assessment of Ab binding to V3 peptides, each peptide (10 μg/ml) was used directly to coat Immulon II plates overnight. The plates were then blocked with excess milk protein, and then the assay was continued as described above for the monomeric gp120 ELISA (42). Midpoint binding titers were estimated by interpolation.
HIV-1 neutralization assays.
Full-length gp160 proteins were derived from HIV-1 isolates JR-FL, ADA, YU2 (all obtained from T. Dragic, Albert Einstein College of Medicine, New York, NY), and SF162 (C. Cheng-Mayer, Aaron Diamond AIDS Research Center, New York, NY). Amphotropic murine leukemia virus Env (MuLV) was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH; contributed by N. Landau). Full-length gp160MN was made by PCR amplification of proviral DNA extracted from peripheral blood mononuclear cells (PBMC) acutely infected with HIV-1MN using a virus stock supplied by D. Montefiori, Duke University Medical School, Durham, NC. The procedure was similar to that described previously (19).
The generation of Env-pseudotyped virus stocks in 293T cells by calcium phosphate transfection and the use of the engineered HIV-1 coreceptor-bearing cell lines U87.CD4.CCR5 and U87.CD4.CXCR4 (3,000 cells per well) for Env-pseudotyped virus infection have been described previously (28, 32). The amount of input pseudovirus was normalized by infectivity (virus titer) rather than by p24 antigen content. Virus infection was measured by determining luciferase expression in relative light units (RLU). Various components of animal sera can potentially interfere with neutralization assays, particularly those using HIV-1 Env-pseudotyped viruses, by causing nonspecific inhibition or enhancement of virus infection and/or luciferase expression. In our experience, the magnitude of such interference can be both (pseudo)virus dependent and animal dependent. To correct for these interfering factors, a preimmune serum sample from the same animal was processed identically to the postimmune samples in each experiment in order to determine the percent neutralization at each dilution. Percent neutralization was defined as [1 − (RLUpostimmune/RLUpreimmune)] × 100. The effect of this adjustment was, in most cases, negligible; neutralization titers derived using the preimmune serum correction were usually very similar to those obtained using the standard control wells, containing only pseudovirus and cells, as a reference. Pseudoviruses expressing MuLV Env were also used to detect any nonspecific interference, since anti-HIV-1 Env Ab would not be expected to cross-neutralize them.
The neutralization properties of selected pre- and postimmune sera were also evaluated under contract, by ViroLogics Inc. (South San Francisco, CA), using their automated Phenosense HIV Entry neutralization assay (7, 56). This method also involves measuring the infectivity of Env-complemented, luciferase-encoding pseudoviruses in a single-cycle assay using CCR5/CXCR4-expressing U87.CD4 cells.
Neutralization assays were also performed using replication-competent HIV-1JR-FL and HIV-1MN. These studies examined virus replication on mitogen-activated PBMC using p24 antigen production as the readout. They were carried out essentially as previously described (32), except that a washout procedure was performed on day 1 to remove any potentially interfering components of the animal sera. Again, pre- and postimmune sera from each rabbit were tested in the same neutralization assay to permit the identification of any nonspecific interference. Simian immunodeficiency virus of macaques strain 239 (SIVmac239) was also included to provide an indication of any nonspecific effects of the rabbit sera against replication of a virus that should not be sensitive to any NAb present.
Ig purification.
Total Ig was purified from final blood drawing rabbit sera using the T-Gel Purification Kit (Pierce, Inc., Rockford, IL) according to the manufacturer's instructions, except that an azide-free buffer (50 mM NaH2PO4, pH 8) was used for Ab elution. The amount of recovered rabbit IgG was quantified using the Easy-Titer Rabbit IgG Assay Kit (Pierce, Inc.), the average recovery being 70% (n = 10). The extent of IgG recovery for each individual serum sample was always taken into account when determining neutralization or ELISA titers to allow comparisons between purified IgG and the corresponding unfractionated serum.
Ab depletion from sera by gp120 or V3 peptides.
A cyclic V3JR-FL peptide, gp120JR-FL, or bovine serum albumin (BSA) was coupled to cyanogen bromide-activated Sepharose 4B beads according to the manufacturer's instructions (Amersham Biosciences). Briefly, the beads (0.5 g of powder) were hydrated and washed extensively in 1 mM HCl, washed in ∼1.75 ml of coupling buffer (0.1 M NaHCO3, 0.5 M NaCl, pH 8.3), and then resuspended in the same buffer to form an ∼50% slurry. This slurry was divided into three equal portions and incubated overnight at 4°C with BSA (1 mg), cyclic V3JR-FL peptide (1 mg), or gp120JR-FL (0.5 mg). The beads were then washed extensively in coupling buffer, and the excess binding sites were blocked in 0.1 M Tris-HCl (pH 8) for 2 to 3 h at room temperature. Any noncovalently associated proteins were removed by washing first in 0.1 M sodium acetate-0.5 M NaCl (pH 4) and then in Tris-salt buffer (0.1 M Tris-HCl, 0.5 M NaCl, pH 8). Serum samples were diluted in Tris-salt buffer and then added to sterile 1.5-ml Eppendorf tubes containing ∼50 μl of 50% slurry for overnight incubation at 4°C. The treated serum was recovered by low-speed centrifugation (3,000 × g, 3 min). The beads were washed extensively in Tris-salt buffer, and then bound Ab were eluted in 0.2 M glycine (pH 2.5 to 3). The pH of the eluted solution was immediately neutralized using 1 M Tris buffer (pH 8.5). The eluted Ab were concentrated using sterile microcentrifuge-sized filter units (Millipore) and then resuspended in a volume of PBS corresponding to the starting volume of the original serum. All buffers had been passed through 0.2-μm-pore-size filters prior to use to help preserve the sterility of the Ab preparations.
RESULTS
Expression and characterization of trimeric SOSIP.R6 gp140 protein.
The designs of the Env proteins encoded within the various plasmids used for DNA and protein immunization are summarized in Fig. 1A. When 293T cells were transiently transfected in vitro, we found that approximately fivefold less of the codon-optimized JR-FL env gene was needed to express the same amount of Env protein than when the natural gene was used (data not shown). Hence, the codon-optimized gene was used to produce preparative amounts of protein for immunizations. The purified SOSIP.R6 gp140 protein was approximately 90% proteolytically processed, such that only a minor fraction (∼10%) of the 140-kDa band remained after treatment with the reducing agent dithiothreitol (Fig. 1B, lane 3). No detectable amounts of free gp120 were present in the SOSIP.R6 gp140 preparation, indicating that the intersubunit disulfide bond remained substantially intact during purification. Although the amount of disulfide-linked aggregates varied among production lots, the overall amount was consistently ≤20% (Fig. 1B, lane 4). The major protein contaminants seen on a reducing sodium dodecyl sulfate (SDS)-PAGE gel migrated as bands with molecular masses of ∼170 kDa and ∼25 kDa. The ∼170-kDa band was identified as bovine alpha 2 macroglobulin by its reactivity with specific Ab in a Western blot assay (data not shown). The identity of the ∼25-kDa band is not known.
Considering the amount of uncleaved protein and the presence of impurities and aggregates, the overall purity of the trimeric SOSIP.R6 gp140 protein purified from the supernatants of transiently transfected 293T cells was ∼75%. The trimeric nature of the purified protein was confirmed by Blue-Native PAGE analysis (Fig. 1C) and by analytical size exclusion chromatography (data not shown).
Pilot study on the immunogenicity of monomeric gp120JR-FL.
As a prelude to studies with SOSIP.R6 gp140, we first conducted a pilot experiment using a DNA prime, subunit protein boost format (Fig. 2A). In particular, we sought to identify the optimal concentration of the boosting antigen to use in future studies. In this first study, we used a soluble gp140 Wt-expressing construct in the DNA-priming phase, with monomeric gp120JR-FL as the boosting antigen. Each animal (two per experimental group) was primed with DNA at weeks 0 and 4 and then received two boosts of QS21-adjuvanted gp120 (10 μg, 30 μg, or 100 μg) at weeks 12 and 16. A fourth group of animals was primed with a plasmid that lacked an env insert (empty-vector control) before receiving two gp120 immunizations (30 μg) at weeks 12 and 16. All eight animals were bled every 4 weeks for 20 weeks.
The outcome of the immunizations was gauged initially by determining anti-gp120 binding Ab midpoint titers by ELISA (Fig. 2B). Endpoint titers, defined as the maximum dilution that yielded a signal twofold above the background, were typically greater than the midpoint titers, but we consider them to be less useful as comparators. As expected, DNA priming made a substantial difference in the speed and magnitude of the Ab responses to the subsequent protein boosts. Thus, at week 20 (final blood drawing), the anti-gp120 midpoint titers in the two animals that received only the empty-vector control plasmid were 12-fold lower (mean titer, 7 × 103), and developed more slowly, than in the animals that had been primed with the DNA plasmid expressing the gp140 Wt protein (mean titer, 9 × 104). In the latter animals, there was only a weak anti-gp120 response after the DNA-priming phase, but a substantial increase occurred after the first boost with soluble gp120 protein. The second boost had only a minor effect. The amount of gp120 given as the protein boost (10 to 100 μg) made little difference (± threefold) in the magnitude of the anti-gp120 response. Hence, 30 μg was selected as the amount to use in follow-up studies.
Second-stage study on the immunogenicity of SOSIP.R6 Env trimers.
Based upon the results of the pilot study, we initiated a second-stage experiment that had a similar format: DNA priming at weeks 0 and 4 and protein boosts at weeks 12 and 16, with 30 μg of protein used in the boosting immunizations (Fig. 3A). As in the pilot study, blood was drawn from the rabbits every 4 weeks for analysis. The initial aims of the experiment were fourfold: first, to obtain information on the absolute immunogenicity of SOSIP.R6 gp140 trimers; second, to compare the immunogenicity of these trimers with gp120 monomers using sera retained from the pilot study for this comparison; third, to determine whether bead-immobilized SOSIP.R6 trimers were superior to soluble forms of the same protein at eliciting an Ab response in the boosting phase (compare arms B and D); and fourth, to evaluate the relative merits of soluble (SOSIP.R6 gp140) or membrane-associated [SOS.R6(T)] forms of Env proteins in the DNA-priming phase (compare arms B and E). In one arm (arm A), DNA priming was with the control empty vector only; in another (arm C), beads lacking Env proteins were used as a control in the boosting phase.
Anti-gp120 binding Ab midpoint titers were again determined to provide an initial measurement of the overall immunogenicity of the test antigens (Fig. 3B). The limitations of DNA immunizations without the later use of an Env-based boosting antigen were clearly revealed by the outcome of arm C, in which the anti-gp120 titers never exceeded 1 × 103. Conversely, although an anti-gp120 response did develop when nonprimed animals were immunized with SOSIP.R6 gp140 trimers, the titers were much lower than seen in the DNA-primed animals, at least initially. However, after the second protein immunization, anti-gp120 titers were very similar in all the animals receiving SOSIP.R6 gp140 trimers, whether or not they had been primed earlier with DNA. Thus, at this time, the mean anti-gp120 titers in arms A and B were 2.8 × 104 and 3.2 × 104, respectively (Fig. 3B). The anti-gp120 responses induced by priming with membrane-bound Env [SOS.R6(T); arm E] or by boosting with the bead-based, particulate form of SOSIP.R6 gp140 trimers (arm D) were, on average, slightly (∼2.5-fold) lower than those in animals primed with SOSIP.R6 and boosted with SOSIP.R6 gp140 trimers (arm B). Thus, at week 20, the mean titers in arms B, D, and E were 3.2 × 104, 1.3 × 104, and 1.5 × 104, respectively.
When the anti-gp120 titers induced by Env trimers were compared with those raised in response to monomeric gp120 in the pilot study, the trimers were found to be less immunogenic. Thus, the SOSIP.R6 gp140 trimers elicited anti-gp120 titers typically approximately threefold lower than had been induced by monomeric gp120, i.e., ∼3 × 104 compared to ∼1 × 105 (Fig. 3B), though these differences were not significant (Mann-Whitney U test, P > 0.05).
We considered it possible that additional immunizations might increase the quality or quantity of the Ab raised to the Env trimers, so we decided to extend the second-stage study for another 34 weeks by further boosting the animals. Thus, the rabbits were reimmunized with the same forms of Env protein or with control beads at weeks 20, 28, 37, and 52 (Fig. 3A). We realized that this extension to the study meant that we would now lack a gp120 comparison arm for time points beyond week 20, but we felt that the additional information that we might obtain could still be useful in the design of future experiments.
The additional immunization at week 20 did not further boost the anti-gp120 titers in any of the rabbits from any arm, suggesting that the response to the first two boosts (weeks 12 and 16) was already maximal. This supposition was reinforced by the outcome of the protein boost at week 28. By that time, the anti-gp120 titers in the DNA-primed animals had decayed slightly (3- to 4-fold) from the week 20 level, whereas the mean titers in animals that had not received a DNA prime (arm A) had decayed by more than 30-fold. After the week 28 boost, the anti-gp120 titers rose to levels very similar to those achieved after the week 16 boosting. The further boosts at weeks 37 and 52 were also unable to further increase the maximum anti-gp120 titers; if anything, the responses to the later boosts were a little less than to the boosts at weeks 16 and 28 (Fig. 3B). Although the anti-gp120 titers in arms A and B were similar at week 20, the average titers were approximately threefold higher at weeks 24 and 28 in the animals that had received the SOSIP.R6 DNA prime (arm B) than in the corresponding non-DNA-primed animals (arm A) (P < 0.05; Mann Whitney U test). Thus, it is possible that DNA priming induced a slightly more robust immune response.
We also considered it worth evaluating whether switching adjuvants from QS-21 to Enhanzyn could increase the strength of the immune response to Env antigens. Hence, from week 37 onward, we boosted the SOS.R6(T)-primed and SOSIP.R6 gp140-immunized animals (arm E) with Env proteins formulated in Enhanzyn adjuvant. Prior to week 37, the gp120 binding titers for animals in arm B (SOSIP.R6 prime with SOSIP.R6 gp140 trimer boost) and arm E were comparable. This modification of the procedure did not, however, have any effect on the ensuing anti-gp120 titers; between weeks 39 and 54, the anti-gp120 titers were not significantly different for animals in arms E and B (Wilcoxon paired signed-rank test, P > 0.05).
Neutralization of HIV-1 Env-pseudotyped viruses.
Anti-gp120 binding Ab are not, of course, a measurement of functional anti-Env responses. We therefore measured neutralization titers at several time points in both the pilot and second-stage studies using HIV-1JR-FL and HIV-1MN Env-pseudotyped viruses in a reporter gene assay (Table 1). The former virus represents the autologous strain for our Env-based immunogens and is a primary isolate of average neutralization resistance. We used HIV-1MN as a second pseudovirus in the initial round of neutralization studies. HIV-1MN is a T-cell line-adapted (TCLA) virus that has been used extensively in vaccine-related research over the past decade (26) and is sensitive to neutralization by virtue of its in vitro passage history (21, 38, 44). The use of the corresponding HIV-1MN pseudovirus therefore allows us to detect and quantify low levels of NAb, although such Ab may have little relevance to the neutralization of more resistant primary viruses (45, 50). Each serum was tested at least twice.
TABLE 1.
Neutralization of primary and TCLA Env-pseudotyped HIV-1 by gp120- and SOSIP.R6-immunized rabbits
| Study | DNA/protein | Animal | Neutralization of Env-pseudotyped HIV-1 by rabbit sera froma:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wk 20
|
Wk 39
|
|||||||||||||
| JR-FL
|
MN
|
JR-FL
|
MN
|
|||||||||||
| 50% | 70% | 90% | 50% | 70% | 90% | 50% | 70% | 90% | 50% | 70% | 90% | |||
| Pilot | gp140 Wt/gp120 | 5695-1 | — | — | — | 14 | — | — | ||||||
| 5695-2 | — | — | — | >160 | >160 | 97 | ||||||||
| 5695-3 | — | — | — | >160 | 124 | 45 | ||||||||
| 5695-4 | — | — | — | >160 | 158 | 66 | ||||||||
| 5695-5 | — | — | — | 102 | 40 | 17 | ||||||||
| 5695-6 | — | — | — | 38 | 25 | 13 | ||||||||
| Second stage | ||||||||||||||
| Arm A | Empty vector/SOSIP.R6 | 222 | — | — | — | >160 | 98 | 33 | — | — | — | 148 | 92 | 35 |
| 223 | — | — | — | >160 | 141 | 45 | 21 | — | — | >160 | 101 | 27 | ||
| 224 | 12 | — | — | 22 | 11 | — | 22 | — | — | 57 | 28 | 12 | ||
| 225 | — | — | — | >160 | 103 | 31 | — | — | — | >160 | >160 | 81 | ||
| Arm B | SOSIP.R6/SOSIP.R6 | 226 | — | — | — | >160 | 120 | 44 | — | — | — | >160 | >160 | 113 |
| 227 | — | — | — | 154 | 84 | 25 | — | — | — | >160 | >160 | 50 | ||
| 228 | — | — | — | >160 | 125 | 33 | 29 | — | — | >160 | >160 | >160 | ||
| 229 | 21 | — | — | >160 | 138 | 39 | 20 | — | — | >160 | 129 | 33 | ||
| Arm C | SOSIP.R6/empty beads | 230 | — | — | — | — | — | — | — | — | — | — | — | — |
| 231 | — | — | — | — | — | — | — | — | — | 20 | — | — | ||
| 232 | — | — | — | — | — | — | — | — | — | 16 | — | — | ||
| 233 | — | — | — | — | — | — | — | — | — | — | — | — | ||
| Arm D | SOSIP.R6/SOSIP.R6 beads | 234 | — | — | — | 22 | — | — | — | — | — | >160 | 127 | 36 |
| 235 | — | — | — | 15 | — | — | — | — | — | >160 | 109 | 24 | ||
| 236 | — | — | — | >160 | >160 | >160 | 21 | — | — | >160 | >160 | >160 | ||
| 237 | — | — | — | — | — | — | — | — | — | >160 | 104 | 22 | ||
| Arm E | SOS.R6(T)/SOSIP.R6 | 238 | — | — | — | 91 | 28 | — | 34 | 14 | — | >160 | >160 | 81 |
| 239 | — | — | — | — | — | — | 22 | — | — | >160 | 153 | 30 | ||
| 240 | — | — | — | >160 | 111 | 45 | 25 | — | — | >160 | >160 | >160 | ||
| 241 | 16 | — | — | 39 | 19 | — | >160 | 85 | — | 150 | 76 | 22 | ||
The reciprocal of the dilution which resulted in 50%, 70%, or 90% neutralization of primary (JR-FL) or TCLA (MN) Env-pseudotyped HIV-1 infectivity on coreceptor-bearing U87.CD4 cells (mean of two to six tests). A dash indicates that <50% neutralization was observed at the 1:10 dilution. Note that the pilot study was terminated after 20 weeks, so no formal comparison of responses at week 39 was possible.
When the week 20 sera were tested against the more sensitive HIV-1MN pseudovirus, neutralization was detected frequently at both the 50% and 90% levels, and the responses could be titrated (Table 1). The strongest and most consistent neutralization of HIV-1MN was seen with sera from arms B (SOSIP.R6 gp140 prime and boost) and A (no prime, SOSIP.R6 gp140 boost). Boosting with bead-captured SOSIP.R6 gp140 was inferior to boosting with the same protein free in solution (compare arms D and B). However, the response to HIV-1MN in one bead-boosted animal, no. 236, was the strongest observed in the entire study. A comparison between the use of membrane-bound (arm E) and soluble (arm B) forms of Env in the DNA-priming phase suggested that expression of soluble proteins in vivo was the more effective strategy; thus, only three of the four animals in arm E generated any NAb against HIV-1MN, compared to four of four in arm B, and at lower titers. The animals receiving two DNA immunizations only (arm C) did not develop any NAb to HIV-1MN, implying that a protein boost in one form or another was necessary for NAb generation. Of note, however, is that the NAb responses to HIV-1MN in the animals receiving SOSIP.R6 gp140 boosts (arms A and B) were very similar to those elicited by monomeric gp120 boosting in the pilot study. This was particularly so in the two animals given 30 μg of gp120 (5695-3 and 5695-4), the same amount of Env protein used in the second-stage study. Thus, at 20 weeks, the NAb responses to SOSIP.R6 gp140 trimers (particularly arms A and B) and monomeric gp120 were similar, as judged by comparison of the 50%, 70%, and 90% endpoint titers against HIV-1MN (Table 1).
None of the week 20 sera raised to any of the gp120 or trimeric gp140 proteins could neutralize HIV-1JR-FL by >70% in the pseudovirus assay, and neutralization of this virus even at the 50% level was rarely observed (Table 1). Thus, sera from only 3/16 SOSIP.R6 gp140-immunized animals (arms A, B, D, and E) were positive at the 50% neutralization level, compared with 0/6 sera from animals primed with gp140 Wt DNA and boosted with gp120.
Overall, the NAb responses in the SOSIP.R6 gp140 and gp120 recipients were very similar after 20 weeks. The comparability of the NAb responses to the trimeric and monomeric proteins, despite the markedly lower titers of anti-gp120 binding Ab in the trimer recipients, suggests that the use of trimeric SOSIP.R6 gp140 Env proteins may reduce the development of non-NAb without penalizing NAb induction.
The second-stage study was continued on an ad hoc basis after week 20 by performing a further four immunizations. As noted above, the extension of the study beyond the originally planned 20 weeks eliminated any direct comparison between the responses to the trimeric SOSIP.R6 proteins and those elicited by gp120 monomers in the pilot study, which ended at week 20. A cross-sectional analysis at week 39 revealed that sera from 9/16 animals (arms A, B, D, and E) were now able to neutralize HIV-1JR-FL pseudoviruses by at least 50%, and the titers were increased compared to those observed at week 20 (Table 1). By week 39, animals within the SOS.R6(T)-primed group (no. 238 to 241) had developed the highest and most consistent response against HIV-1JR-FL among the groups of animals boosted with the SOSIP.R6 gp140 trimer. This difference was not apparent at week 20, when either HIV-1JR-FL or HIV-1MN neutralization titers were inspected. Indeed, serum from one rabbit (no. 241) had 50% and 70% neutralization titers to HIV-1JR-FL of >160 and 85, respectively, by week 39. Neutralization of HIV-1MN by week 39 sera was generally stronger than that seen at week 20 in the second-stage study, although when the response was already high at week 20, it increased little further, if at all, by week 39. Thus, the NAb response to the TCLA clone was maximal after only two protein immunizations, while NAb against the autologous primary isolate clone increased with time.
To examine the NAb responses to HIV-1MN and HIV-1JR-FL pseudoviruses in more detail, we selected a panel of sera from 10 rabbits in the second-stage study for further analysis, based on their neutralization potencies at week 39. The animals chosen were no. 222, 223, 224, 228, 229, 234, 236, 238, 240, and 241 (Fig. 4). A longitudinal analysis allowed us to examine separately the various influences of time, of the use of Enhanzyn adjuvant from week 37 in the SOS.R6(T)-primed, SOSIP.R6 gp140-immunized group, and of the additional immunizations themselves.
FIG. 4.
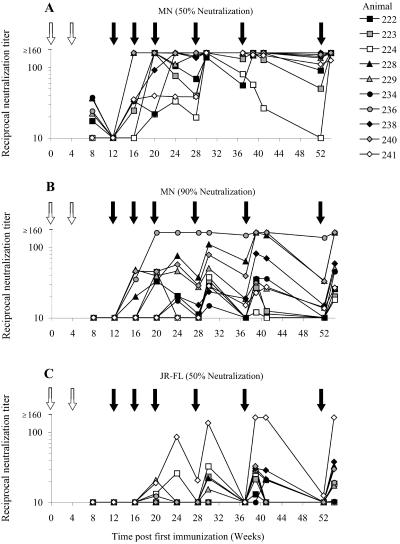
Neutralization of HIV-1JR-FL and HIV-1MN Env-pseudotyped viruses by selected sera. A panel of antisera taken from 10 rabbits immunized with trimeric SOSIP.R6 Env (second-stage study) was assembled to assess the generation of NAb over time. NAb titers against Env-pseudotyped HIV-1MN (A, B) and Env-pseudotyped HIV-1JR-FL (C) were determined at 12 time points over the 54-week time course, using a 50% (A, C) or 90% (B) endpoint. Symbols are used to represent the following groups: empty vector primed and SOSIP.R6 trimer boosted (squares), SOSIP.R6 primed and SOSIP.R6 trimer boosted (triangles), SOSIP.R6 primed and bead-SOSIP.R6 trimer boosted (circles), and membrane-bound SOS.R6(T) primed and SOSIP.R6 trimer boosted (diamonds). The timings of DNA priming (open arrows) and protein boosting (filled arrows) events are indicated. Note that the rabbit sera were not titrated beyond a dilution of 1:160; hence, differences between animals, or the heights of the peaks and depths of the troughs, cannot be determined for values beyond this level.
In 5 of the 10 animals, low levels of NAb against HIV-1MN could be detected at week 8, following the two DNA immunizations, although only at the 50% neutralization level (Fig. 4A). Ab capable of neutralizing HIV-1MN by 90% were detected from week 16, initially in only 4 of the 10 animals and then later in all animals. The average peak titer was 86, with a range of 28 to >160 (Fig. 4B). The magnitude of the NAb response to HIV-1MN generally followed the immunization schedule. During the 8- to 15-week periods between immunizations, the NAb titers in some sera decreased by as much as fivefold but were then restored, or in some cases increased, by the subsequent protein immunizations. There was also some variation between the animals in this substudy in the time taken to generate the peak 90% neutralization titer against HIV-1MN. The peak titers were observed at weeks 20 (no. 223 and 236), 30 (no. 224, 229, and 241), 39 (no. 222, 228, 238, and 240), and 54 (no. 234), and there was no obvious pattern related to the use of a DNA- or protein-based immunogen.
In contrast to what was seen with HIV-1MN, NAb against HIV-1JR-FL developed more slowly and at lower titers (Fig. 4C). Ab capable of neutralizing HIV-1JR-FL by 50% were detectable only after the two DNA and two protein immunizations, from week 20 onward. Although all 10 selected animals did generate NAb against HIV-1JR-FL over this extended study period (weeks 20 to 54), almost all responses were modest and transient (50% titers of <40). Sera from animal 241 [SOS.R6(T) prime, SOSIP.R6 gp140 boost] were capable of neutralizing HIV-1JR-FL by ≥90% at weeks 41 (titer, 10) and 54 (titer, 24), at which times the 50% titers were >160, but none of the other selected sera could do so. The relatively high-titer NAb response of animal 241 was unlikely to be attributable to the use of Enhanzyn adjuvant from week 37 onward, since atypically potent NAb titers were also observed in this animal between weeks 24 and 37. However, it is possible that Enhanzyn improved the NAb responses elicited in animals 238 and 240. Sera from both animals lacked neutralizing activity against HIV-1JR-FL prior to the switch to Enhanzyn at week 37, but by week 39, 50% titers of 34 and 25 had developed (Fig. 4C).
Neutralization of PBMC-grown, replication-competent virus.
Pseudovirus-based assays are typically more sensitive than those based on the use of PBMC-grown virus with PBMC as the target cells (7, 41). The latter assay has been the generally accepted standard for many years. Hence, we compared selected final blood drawing (week 54) sera for the ability to neutralize both replication-competent HIV-1JR-FL and HIV-1MN in a PBMC assay and the corresponding Env-pseudotyped viruses on the appropriate U87.CD4 coreceptor-bearing cells (Table 2). SIVmac239 and MuLV Env-pseudotyped viruses served as negative controls for the PBMC and pseudovirus assays, respectively. None of the rabbit sera neutralized these control viruses, suggesting that any neutralization of HIV-1JR-FL and HIV-1MN was likely to be a specific effect and not due to toxicity or other interfering factors (data not shown).
TABLE 2.
Comparative neutralization of Env-pseudotyped and PBMC-grown HIV-1
| Animal | Neutralization of Env-pseudotyped or PBMC-grown HIV-1 by sera from SOSIP.R6 gp140-immunized rabbitsa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Env pseudotyped HIV-1
|
PBMC-grown HIV-1
|
|||||||||||
| JR-FL
|
MN
|
JR-FL
|
MN
|
|||||||||
| 50% | 70% | 90% | 50% | 70% | 90% | 50% | 70% | 90% | 50% | 70% | 90% | |
| 222 | — | — | — | >160 | 111 | 26 | — | — | — | 14 | — | — |
| 223 | — | — | — | >160 | 74 | 20 | — | — | — | — | — | — |
| 224 | 18 | — | — | >160 | 65 | 18 | — | — | — | 18 | — | — |
| 228 | 33 | 10 | — | >160 | >160 | >160 | — | — | — | 37 | 26 | 14 |
| 229 | 17 | — | — | >160 | >160 | 50 | — | — | — | 34 | 22 | 11 |
| 234 | 10 | — | — | >160 | >160 | 45 | — | — | — | 24 | 12 | — |
| 236 | 19 | — | — | >160 | >160 | >160 | — | — | — | 154 | 100 | 45 |
| 238 | 38 | 15 | — | >160 | >160 | 59 | — | — | — | 35 | 21 | — |
| 240 | 29 | — | — | >160 | >160 | >160 | — | — | — | 103 | 67 | 29 |
| 241 | >160 | >160 | 24 | 125 | 75 | 26 | 101 | 56 | — | — | — | — |
The reciprocal of the dilution which resulted in 50%, 70%, or 90% neutralization of infectivity. Neutralization of Env-pseudotyped HIV-1 was determined on coreceptor-bearing U87.CD4 cells, while neutralization of PBMC-grown HIV-1 was determined on PBMC. A dash indicates that <50% neutralization was observed at the 1:10 dilution.
The infectious-virus-based PBMC assay and the Env pseudotype assay in U87.CD4 cells generated broadly similar patterns of neutralization data, although as expected the PBMC assay was substantially the less sensitive of the two (Table 2). The greater neutralization sensitivity of the Env-pseudotyped viruses is also shown by the use of CD4-IgG2 as a reference reagent. Thus, HIV-1JR-FL pseudovirus was approximately three times more sensitive to CD4-IgG2 at the 50% level (mean 50% inhibitory concentration [IC50], 0.11 ± 0.07 μg/ml; n = 29) compared to PBMC-grown HIV-1JR-FL (0.32 ± 0.24 μg/ml; n = 14). At the 90% neutralization level, the differential was approximately twofold (HIV-1JR-FL pseudovirus, 0.72 ± 0.44 μg/ml; replication-competent HIV-1JR-FL, 1.32 ± 0.74 μg/ml). A greater differential (7- to 10-fold) was seen between HIV-1MN pseudoviruses (IC50, 0.01 ± 0.01 μg/ml; IC90, 0.05 ± 0.06 μg/ml; n = 22) and PBMC-grown HIV-1MN (IC50, 0.10 ± 0.09 μg/ml; IC90, 0.38 ± 0.57 μg/ml; n = 6).
Only four of the test sera from SOSIP.R6 gp140-immunized animals (no. 228, 229, 236, and 240) could neutralize HIV-1MN by 90% (mean titer, 25) in the PBMC assay (Table 2). Three of these sera (no. 228, 236, and 240) were the most potent against HIV-1MN in the pseudovirus assay (90% titers of >160). However, serum from only 1 SOSIP.R6 gp140-immunized animal (no. 241) neutralized JR-FL by ≥50% in PBMC, whereas sera from 8/10 animals could neutralize HIV-1JR-FL at the 50% level in the Env pseudotype assay. When the longitudinal serum set from animal 241 was tested, the rise and fall of the neutralization titers against PBMC-grown HIV-1JR-FL and HIV-1JR-FL Env pseudotype virus followed a similar temporal pattern, although the absolute titers were lower in the PBMC assay (data not shown). Hence, the principal difference between the two assay systems is quantitative rather than qualitative.
Breadth of Ab reactivity elicited by Env subunit protein immunization.
While the ability of animal antisera to neutralize the homologous strain on which the subunit immunogen was based is a relevant parameter, their breadth of reactivity against a range of heterologous isolates is likely to be a more meaningful measurement of the potential of any candidate HIV-1 immunogen (13, 26).
We first assessed the cross-neutralization activities of the panel of 10 selected final blood drawing rabbit antisera (Table 2) against an extended virus test panel that included the Env-pseudotyped viruses HIV-1SF162, HIV-1ADA, and HIV-1YU2 and a panel of seven PBMC-grown primary isolates. Selected sera from gp120- and SOSIP.R6 gp140-immunized animals were also tested by an external laboratory, ViroLogics Inc.
Atypically sensitive Env-pseudotyped HIV-1SF162 was neutralized to a high titer by sera from gp120-immunized animals in the pilot study (mean 50% neutralization titer ± standard deviation, 108 ± 4) and by sera from the SOSIP.R6 gp140-immunized animals (titer, 112 ± 43). However, none of these antisera could neutralize more resistant Env-pseudotyped HIV-1ADA or HIV-1YU2, even at the 50% level, at a dilution of 1 in 10 (data not shown).
Final blood drawing sera from animals 228, 236, and 241 were tested against a panel of seven PBMC-grown R5 isolates, representing Env subtypes A (n = 2), B (n = 3), and C (n = 2), in a PBMC assay. No activity (50% neutralization titers of <10) was observed against these viruses with any of the test sera (data not shown).
Four sera from the second-stage study (weeks 20 and 54) and one from the pilot study (week 20) were also assessed for neutralizing activity using the Phenosense HIV Entry Env pseudotype neutralization assay (56). Seven HIV-1 strains were used, including well-characterized env clones with a range of sensitivities to NAb (Table 3). The pseudoviruses used in this assay were recently defined as ranging from highly sensitive to highly resistant as follows: HIV-1SF162, HIV-1NL4/3 > HIV-1BaL, HIV-11196 > HIV-1JR-FL, and HIV-1JR-CSF > HIV-11168 (7). A similar ranking was observed in the present study. Sera from SOSIP.R6 gp140- or gp120-immunized animals neutralized the four most sensitive isolates (HIV-1SF162, HIV-1NL4/3, HIV-1BaL, and HIV-11196). Neutralization of more resistant HIV-1JR-FL was, however, only observed using sera from SOSIP.R6 gp140-immunized animals 236 and 241. In contrast, the latter two sera were both inactive against HIV-1JR-CSF and HIV-11168. As was also observed in the in-house pseudovirus assays (Tables 1 and 2), serum from animal 241 was the most potent against HIV-1JR-FL and had modest activity against HIV-1MN, whereas serum 236 had more modest activity against HIV-1JR-FL but strongly inhibited HIV-1MN.
TABLE 3.
Assessment of cross-neutralization activity in the ViroLogics Inc. Phenosense assay
| Animal or control | Time serum sample obtained (wk) | Neutralization of Env-pseudotyped HIV-1 by rabbit seraa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SF162 | NL4/3 | BaL | 1196 | JR-FL | JR-CSF | 1168 | MLV | ||
| 5695-4 | 0 | — | — | — | — | — | — | — | — |
| 20 | 4,658 | 90 | 63 | 32 | — | — | — | — | |
| 228 | 0 | — | — | — | — | — | — | — | — |
| 20 | 2,233 | 154 | 55 | — | — | — | — | — | |
| 54 | 4,167 | 785 | 230 | 65 | — | — | — | — | |
| 229 | 0 | — | — | — | — | — | — | — | — |
| 20 | 810 | 40 | — | — | — | — | — | — | |
| 54 | 1,473 | 247 | 90 | 46 | — | — | — | — | |
| 236 | 0 | — | — | — | — | — | — | — | — |
| 20 | 1,499 | 1,327 | 29 | — | — | — | — | — | |
| 54 | 7,769 | 7,895 | 218 | 101 | 32 | — | — | — | |
| 241 | 0 | — | — | — | — | — | — | — | — |
| 20 | 3,379 | 36 | 46 | 40 | — | — | — | — | |
| 54 | 5,061 | 313 | 100 | 79 | 112 | — | — | — | |
| Reference HIV-1+ plasma N16 | Mean | 8,497 | 2,036 | 678 | 413 | 52 | 133 | 26 | — |
The reciprocal of the dilution which resulted in 50% neutralization of Env-pseudotyped HIV-1 infectivity on coreceptor-bearing U87.CD4 cells. A dash indicates that <50% neutralization was observed at the 1:25 dilution. Plasma (N16) was from an HIV-1-infected individual and was used as a reference standard to control for assay-to-assay variation.
Plasma sample N16, obtained from an HIV-1 subtype B-infected individual, was recently selected from a panel of 27 such plasma samples for its potent and broadly reactive NAb response (7). We used it to gauge the neutralizing activity of sera from the gp140 SOSIP.R6-immunized animals, no. 236 and 241, by performing a comparative analysis. The primary Env-pseudotyped viruses HIV-1 BaL, 1196, and JR-FL were neutralized to comparable extents by these two rabbit sera and by the N16 plasma (Table 3). Thus, immunization of rabbits 236 and 241 induced NAb of a potency similar to those generated during natural infection, although the breadth of neutralization by the rabbit sera was more restricted.
Overall, there was reasonable concordance between the pseudovirus neutralization data generated using the commercial Phenosense assay and the in-house tests. No obviously toxic effects of the test sera were apparent in any of the assays, and the control MuLV Env-pseudotyped virus was not inhibited.
Taken together, the above sets of data demonstrate that immunization with SOSIP.R6 gp140 elicited Ab capable of neutralizing the relatively resistant autologous primary isolate HIV-1JR-FL in both pseudovirus and whole-virus formats. A few heterologous Env subtype B Env-pseudotyped viruses that are known to be fairly sensitive to NAb were also neutralized by some of the test sera. However, the overall ability of the sera to cross-neutralize primary isolates was modest.
Qualitative analysis of NAb responses.
To gain some further understanding of what component was responsible for the neutralizing activity present in these rabbit antisera, we performed a series of control and/or analytical experiments with selected serum samples.
First, we purified Ig fractions from serum samples 5695-2, 5695-3, 5695-4, and 5695-6 from week 20 of the pilot study and from sera 228, 236, and 241 from week 54 of the second-stage study. We then compared the abilities of the Ig fractions and the unfractionated sera to neutralize HIV-1JR-FL and HIV-1MN in our standard in-house Env pseudotype assay. The neutralizing activity was retained in the purified Ig fractions, showing that Ab, and not other antiviral molecules such as chemokines or cytotoxic serum proteins, were responsible for the inhibition of viral infection caused by the corresponding unfractionated sera (data not shown). Only minor quantitative differences between the unfractionated sera and the purified Ig fractions were recorded.
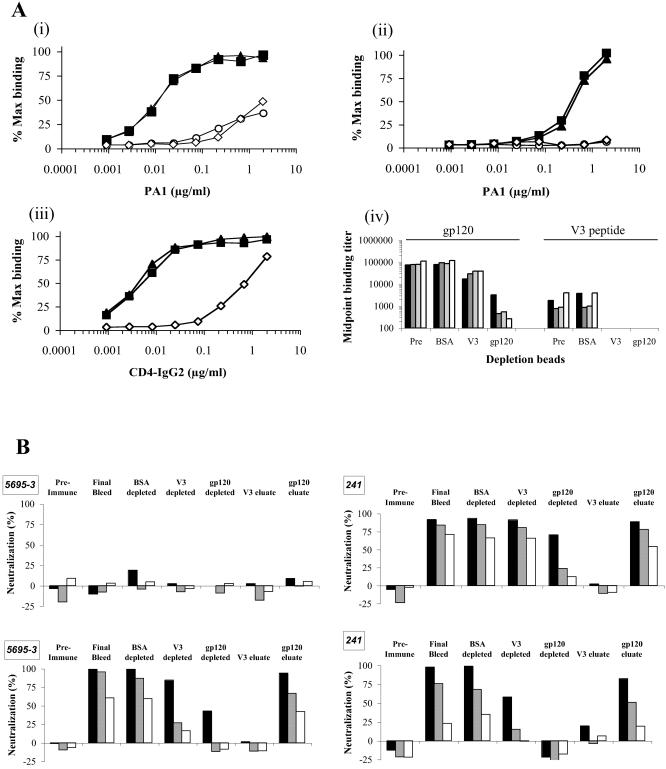
We next evaluated whether neutralization was attributable to Ab that recognized JR-FL gp120 and, more specifically, its V3 region. To deplete Ab directed against gp120 or the V3 loop, selected final blood drawing sera (those with the highest NAb titers) were incubated with bead-immobilized BSA (negative control), gp120JR-FL, or a cyclic V3JR-FL peptide. The depletion procedure was validated by spiking anti-V3JR-FL MAb PA1 or the CD4-IgG2 molecule into a rabbit pre-blood-drawing serum pool and then testing the ability of the gp120- and V3 peptide-containing beads to remove these agents from the spiked serum (Fig. 5A, parts i to iii). Both the gp120- and V3 peptide-containing beads depleted >99% of the added PA1 MAb from the test serum, and the gp120-beads removed a similar proportion of spiked CD4-IgG2, as judged by the shifts in the titration curves. As expected, the V3 peptide-beads did not remove CD4-IgG2 and neither PA1 nor CD4-IgG2 was removed by the control BSA-beads (Fig. 5A and data not shown).
FIG. 5.
Neutralization of HIV-1JR-FL pseudovirus by rabbit antisera is predominantly mediated by non-V3, gp120-directed antibodies. (A) BSA (filled triangles)-, V3 peptide (open circles)-, or gp120 (open diamonds)-coupled cyanogen bromide-Sepharose beads were used to deplete a rabbit preimmune serum pool spiked with V3-specific MAb PA1 (i, ii) or the CD4-IgG2 molecule (iii) before assay by gp120 binding ELISA (i, iii) or V3 peptide ELISA (ii). The nondepleted serum was also assayed (filled squares). (iv) Rabbit final blood drawing sera from the pilot study, 5695-3 (filled bars), and follow-up study, 228 (dark grey bars), 236 (light grey bars), and 241 (open bars), were left untreated (Pre) or depleted using BSA-, V3-, or gp120-coupled beads before determination of midpoint binding titers against gp120 or V3 peptide by ELISA as indicated. (B) Untreated and bead-treated sera 5695-3 and 241 were also used in pseudovirus neutralization assays against HIV-1JR-FL (upper parts) or HIV-1MN (lower parts). Bars represent serum dilutions of 1:10 (filled bars), 1:40 (grey bars), and 1:160 (open bars). Representative data were derived from two or three experiments.
When final blood drawing sera from four selected Env protein-immunized rabbits (pilot study animal 5695-3 and second-stage study animals 228, 236, and 241) were treated, the gp120-beads decreased the anti-gp120 titers, on average, by 33-, 166-, 199-, and 423-fold, respectively (Fig. 5A, part iv, and data not shown). Hence, the depletion procedure was effective at removing antigen-induced anti-gp120 Ab from sera. The same four sera were also incubated with the V3 peptide-beads; in this case, the anti-gp120 titer decreased only four-, three-, two- and threefold, respectively, implying that only a minor fraction of the total anti-gp120 Ab in the sera could recognize the V3 region of gp120. Both the gp120- and V3 peptide-beads were able to remove essentially all the V3 peptide-reactive Ab from all four animal sera. Similar reductions in gp120 binding titers were observed when three other pilot study sera, 5695-2, 5695-4, and 5695-6, were depleted using gp120-beads (data not shown).
Rabbit antisera 5695-3 (pilot study) and 241 [SOS.R6(T) DNA prime followed by SOSIP.R6 gp140 protein boost] were then tested for the ability to neutralize HIV-1JR-FL and HIV-1MN in the Env pseudotype assay before and after bead depletion. We selected serum 241 because of its relatively strong neutralizing activity compared to most of the other sera; we sought to determine what Ab specificities were responsible for this activity. Serum sample 5695-3 from the pilot study was chosen for comparison. The bead-treated and untreated sera and also preimmune sera from the same animals were tested in the neutralization assays at three different dilutions (Fig. 5B).
As expected, the serum from the gp120-immunized rabbit did not neutralize HIV-1JR-FL before or after treatment with the gp120- or V3 peptide-beads. Its ability to neutralize HIV-1MN was completely abolished by exposure to the gp120-beads (the dilution providing 50% inhibition [ID50] decreased from >160 to <10), and it was substantially depleted by the V3 peptide-beads (ID50, 28). We cannot discount the possibility that some Ab specific for V3MN were not depleted from these sera by the use of a V3JR-FL peptide. NAb activity could be recovered from the gp120-beads after glycine elution, but this procedure was unsuccessful at eluting NAb from the V3 peptide-beads, presumably because the affinity of the Ab-peptide interaction was very high. Results similar to those generated with serum from animal 5695-3 were obtained using final blood drawing sera from three other gp120-immunized animals (5695-2, 5695-4, and 5695-6) before and after gp120-bead depletion (data not shown).
The HIV-1JR-FL-neutralizing activity of antiserum 241 was not measurably affected by treatment with the V3 peptide-beads (the ID50 remained >160), but it was substantially depleted by the gp120-beads (the ID50 decreased to 23). In contrast, the activity of the same serum against HIV-1MN was dependent upon the presence of V3-directed Ab; their removal by the V3 peptide-beads substantially reduced the titer against HIV-1MN (the ID50 decrease from 99 to 16). The gp120-beads had a similar effect (ID50, <10) (Fig. 5B). Similar observations were made using final blood drawing sera from animals 228 and 236, albeit to a lower titer than no. 241 (data not shown).
We noted above that the most active rabbit sera could neutralize HIV-1JR-FL pseudovirus (and the neutralization-sensitive HIV-1SF162 pseudovirus) but not the HIV-1ADA or HIV-1YU2 pseudovirus. The V3 peptide depletion experiments suggested that the V3 region was unlikely to be the target for these NAb. However, we explored this issue further by altering the JR-FL V3 sequence to that of ADA by site-directed mutagenesis to create the single-residue change E322D. We then tested whether the resulting HIV-1JR-FL/V3-ADA pseudovirus was resistant to neutralization by serum 241. The serum was found to neutralize both HIV-1JR-FL and HIV-1JR-FL/V3-ADA pseudoviruses with identical titers (50% neutralization, >160). Because HIV-1ADA is resistant to serum 241, it seems unlikely that Ab to V3 were responsible for the neutralization of HIV-1JR-FL by this serum. However, the target(s) of the NAb present in serum 241 remains to be determined. The resistance of HIV-1ADA and HIV-1YU2 to neutralization by the same serum is probably due to more global differences in Env configuration, rather than in V3 alone. This conclusion is consistent with other studies on the global neutralization resistance of various viruses (43, 52, 53).
DISCUSSION
We describe here our first studies on the immunogenicity of disulfide-stabilized, cleaved forms of the trimeric HIV-1 Env complex (SOSIP.R6 gp140) in rabbits using a DNA prime, soluble-protein boost regimen. The principal aims of our experiments were threefold: first, to obtain information on the absolute immunogenicity of SOSIP.R6 gp140 trimers; second, to determine whether bead-immobilized SOSIP.R6 gp140 trimers are superior to soluble forms of the same protein at eliciting an Ab response in the boosting phase; and third, to evaluate the relative merits of soluble or membrane-associated forms of Env proteins in the DNA-priming phase. Further experiments are required to define the optimum regimen for immunizing animals with DNA and/or protein forms of SOSIP gp140 with the goal of eliciting NAb. However, we were able to identify conditions under which SOSIP.R6 gp140 trimers can elicit Ab that neutralize HIV-1JR-FL and certain other representative primary isolates in pseudovirus and/or whole-virus assays. The findings have been incorporated into the design of follow-on immunogenicity studies designed to directly compare SOSIP.R6 gp140 trimers and gp120 monomers.
We found that it was beneficial to use a DNA-priming procedure, compared to immunization with subunit proteins only. The advantages provided by DNA priming related more to the rate at which Ab responses were generated after subunit boosting and, to an extent, their longevity, than to the magnitude and breadth of the final NAb responses. In addition, Ab responses in primed animals were somewhat more robust and less subject to decline between protein boosts. Thus, NAb titers (or even anti-gp120 binding Ab titers) were a little higher in the DNA-primed animals after completion of all the protein boosts, compared to animals that received protein only. These findings are broadly consistent with other reports on the outcome of DNA immunizations in small animals (reviewed in references 23, 27, and 57).
We also compared the use of membrane-bound and soluble forms of gp140 proteins (i.e., cleaved, disulfide-stabilized proteins that contained or lacked the gp41 transmembrane domain) during the DNA-priming phase. The exact configuration(s) in which gp140 proteins are expressed in vivo after immunization with DNA is not known; indeed, to our knowledge, the issue has never been studied. It is, for example, uncertain whether the gp140 proteins are cleaved, wholly or in part, after DNA immunization. Env proteins derived from primary strains of HIV-1, when expressed on the surface of env-transfected cells in vitro, are rarely fully cleaved, and they differ from native, fusion-competent trimers in their exposure of various NAb and non-NAb epitopes (1, 22, 24, 32, 54). As our hypothesis is that Env cleavage is relevant to the design of the best antigenic mimics of the native trimer, we incorporated cleavage-enhancing substitutions into some of the gp140 proteins that were expressed in the DNA-priming phase (6). The R6 modification was included because Env proteins expressed on the cell surface in vitro are usually incompletely cleaved (22, 32, 60, 77). While this modification does improve the level of Env cleavage in transiently transfected 293T cells, assessed by surface biotinylation (data not shown), the impact that it has under in vivo conditions has not been determined. We did observe that expressing membrane-bound proteins in the DNA-priming phase offered a small but measurable advantage over soluble gp140 proteins in terms of the subsequent immune responses to the soluble-protein boosts. This advantage could be due to differences in the quality, or in the quantity, of the Env proteins expressed in the DNA-priming phase.
We used a membrane-bound protein that contains a minimal cytoplasmic domain. Truncation of the cytoplasmic domain of gp41 is a commonly used technique to increase Env cell surface expression for antigenicity studies by circumventing the natural down-regulatory sequences (8, 11, 12, 49, 72). Both the truncated form of Wt Env and the SOS mutant retain their fusion function and can be incorporated into infectious Env-pseudotyped particles generated by transfection of 293T cells (1, 4). Truncation of the cytoplasmic tail does, however, reduce Env incorporation into virions generated in T cells in vitro and hence decreases their infectivity (48). Recent cross-linking experiments suggest that full-length Env expressed on the surface of virions, and by implication the cell surface, is trimeric (14, 56). Taken together, we considered it unnecessary to further modify the SOS.R6(T) Env form by addition of the trimer-stabilizing mutation I559P.
When different forms of gp140 protein were compared in the protein-boosting phase, we found that the soluble proteins were slightly more immunogenic than the same proteins captured onto beads to make a particulate antigen. We evaluated the bead-captured antigens because, in principle, particulate antigens offer some theoretical immunogenicity advantages over small proteins and because the capture of Env proteins onto beads offers some benefits from the purification perspective. They may also be advantageous for the induction of cell-mediated immune responses. More details of the production and properties of the bead-immobilized Env proteins will be described elsewhere (N.S. et al., unpublished).
Following two DNA primings and two subunit protein boosts (week 20), the anti-gp120 binding Ab titers were approximately threefold lower in the gp140 trimer-immunized animals than in the gp120 recipients. Although we have not investigated the specificity of the Ab to the two forms of the Env protein, we reasoned that the titer difference could reflect an inherently reduced immunogenicity of the trimers. One possibility is that nonneutralizing epitopes are occluded within the trimer but exposed on the monomer; another is that the nonneutralizing face of gp120 is occluded in the trimer by the gp41ECTO domain (5, 59). Alternatively, neutralizing epitopes could be less well presented on the trimer than on the monomer because of epitope occlusion within the quaternary structure of the multimeric protein. This seems unlikely, however, because the reduced titer of binding Ab was not mirrored by a reduction in the titers of NAb in the same sera at the same time.
One goal of the study, as initially designed, was to determine whether SOSIP.R6 gp140 trimers are superior to monomeric gp120 proteins for NAb induction. This determination necessitated a comparison of the results of a pilot study (using gp120 monomers) and a second-stage study, which evaluated different versions of gp140 trimers. When the initial immunizations (two DNA primings, two soluble-protein boosts) were completed after 20 weeks, there was no indication that the trimers were superior to the monomers as immunogens. We therefore made an ad hoc decision to extend the immunization period in the second-stage study beyond the 20 weeks used in the pilot study to see if additional protein boosts could improve the Ab response to the trimers. Consequently, no formal comparison between gp120 monomers and gp140 trimers is possible after week 20. We will address this lacuna in follow-up studies that have been initiated, taking into account what we have learned from the experiments reported here.
In the later stages of the second-stage study, we were able to induce NAb to both neutralization-sensitive TCLA strain HIV-1MN and autologous primary strain HIV-1JR-FL with more consistency than in the early weeks of the experiment. In some animals, particularly no. 228, 238, 240, and 241, the titers of NAb against HIV-1JR-FL were respectable, particularly in the Env pseudotype assay. The neutralization titers elicited against HIVJR-FL toward the end of the study were substantially higher than those obtained at week 20, in the absence of any similar increase in anti-gp120 titers, suggesting that a prolonged immunization regimen is necessary to improve the quantity and/or quality or NAb elicited. It should be noted, however, that our general experience and those of others is that Env-pseudotyped viruses are 3- to 10-fold more sensitive to neutralization than the corresponding primary isolate, for reasons that have yet to be fully understood (7, 41). Thus, neutralization titers against pseudoviruses need to be interpreted cautiously; in the real world, vaccine-induced NAb must counter true, infectious primary viruses. These caveats notwithstanding, we could draw certain conclusions from the neutralization assays.
It is possible to elicit by immunization with a trimeric form of Env, SOSIP.R6 gp140, at least in a subset of rabbits, Ab capable of neutralizing a typically resistant HIV-1 clone, JR-FL, in a single-round Env pseudotype assay. In most of the animals, these NAb were only present at low titers, but in rabbit 241, the 50% and 90% neutralization titers against HIV-1JR-FL reached >160 and ∼20, respectively, by the final blood drawing. Ab from this animal were even capable of neutralizing PBMC-grown HIV-1JR-FL in a PBMC assay; we are not aware of other reports of the induction of NAb to JR-FL by Env-based vaccines. We have yet to understand what factors caused rabbit 241 to respond so strongly. An atypically robust response to Env was also observed in a single rabbit in a study of T-helper epitopes (30). Little, if any, of the NAb to JR-FL were directed against the V3 region, but essentially all of them were removed by incubation of the sera with monomeric gp120. Hence, their target is one or more gp120 epitopes outside the V3 region. Identifying this epitope(s) will be a priority for future studies, as knowledge like this could guide the development of new immunization strategies (13).
Even the most potent of the rabbit antisera had a very limited ability to cross-neutralize heterologous strains; the induction of a broadly reactive NAb response remains a daunting challenge. Moreover, the generation of a low level of NAb responses to the autologous strain did require an intensive and prolonged immunization regimen. NAb responses of the magnitude observed in this study are unlikely to provide protective immunity against HIV-1 infection. In particular, we need to improve the consistency with which the stronger responses are elicited. Overcoming the paucity of T-helper epitopes on Env may be important (30, 63, 78). Although there is a growing trend for Env trimers to offer some immunogenicity advantages over gp120 monomers (2, 10, 67, 75), more development work needs to be done to improve their immunogenicity. This applies both to the trimers we have studied here and to others whose basis is the deletion of the cleavage site between gp120 and gp41ECTO (15, 16, 61, 74, 79). It is also necessary that different env gene sequences be compared, in case some Env proteins are more immunogenic than others. Modifications designed to increase epitope exposure must also be evaluated, as should alternative ways to present Env proteins to the immune system. All of these issues are the subject of ongoing studies by us and others.
Acknowledgments
We are grateful to Anthony Villa, Trinity Zang, Irene Vishnefsky (Progenics), Maciej Paluch, Val Monrose, and Ashley Palmer (Weill Medical College) for excellent technical assistance. The earlier or current contributions of Mika Vesanen, James Binley, Min Lu, and Jason Gardner to the overall development of this project are appreciated. We thank David Montefiori (Duke University) for performing neutralization assays, Jessica Harms (Aldevron Inc.) for carrying out the rabbit immunization studies, and Terri Wrin (ViroLogic Inc.) for the Phenosense neutralization assays. Tanya Dragic and Cecilia Cheng-Mayer are thanked for some of the Env plasmids used in this study.
This work was supported in part by NIH grants AI 36082 and AI 45463 and NIH contract N01 AI 30030 (HIV Vaccine Design and Development Team) and by the International AIDS Vaccine Initiative. J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College, Cornell University, gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Abrahamyan, L. G., R. M. Markosyan, J. P. Moore, F. S. Cohen, and G. B. Melikyan. 2003. Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77:5829-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schulke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S. L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 9.Boussif, O., F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix, and J.-P. Behr. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 92:7297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bower, J. F., X. Yang, J. Sodroski, and T. M. Ross. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 78:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu, Z., L. Ye, A. Vzorov, D. Taylor, R. W. Compans, and C. Yang. 2004. Enhancement of immunogenicity of an HIV Env DNA vaccine by mutation of the Tyr-based endocytosis motif in the cytoplasmic domain. Virology 328:62-73. [DOI] [PubMed] [Google Scholar]
- 12.Bultmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 75:5263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 14.Center, R. J., R. D. Leapman, J. Lebowitz, L. O. Arthur, P. L. Earl, and B. Moss. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Center, R. J., J. Lebowitz, R. D. Leapman, and B. Moss. 2004. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J. Virol. 78:2265-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, B., Y. Cheng, L. Calder, S. C. Harrison, E. L. Reinherz, J. J. Skehel, and D. C. Wiley. 2004. A chimeric protein of simian immunodeficiency virus envelope glycoprotein gp140 and Escherichia coli aspartate transcarbamoylase. J. Virol. 78:4508-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, J. 2003. Public health. AIDS vaccine trial produces disappointment and confusion. Science 299:1290-1291. [DOI] [PubMed] [Google Scholar]
- 19.Connor, R. I., K. E. Sheridan, C. Lai, L. Zhang, and D. D. Ho. 1996. Characterization of the functional properties of env genes from long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 70:5306-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desrosiers, R. C. 2004. Prospects for an AIDS vaccine. Nat. Med. 10:221-223. [DOI] [PubMed] [Google Scholar]
- 21.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 22.Dubay, J. W., S. R. Dubay, H.-J. Shin, and E. Hunter. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J. Virol. 69:4675-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estcourt, M. J., A. J. McMichael, and T. Hanke. 2004. DNA vaccines against human immunodeficiency virus type 1. Immunol. Rev. 199:144-155. [DOI] [PubMed] [Google Scholar]
- 24.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 26.Garber, D. A., G. Silvestri, and M. B. Feinberg. 2004. Prospects for an AIDS vaccine: three big questions, no easy answers. Lancet Infect. Dis. 4:397-413. [DOI] [PubMed] [Google Scholar]
- 27.Giri, M., K. E. Ugen, and D. B. Weiner. 2004. DNA vaccines against human immunodeficiency virus type 1 in the past decade. Clin. Microbiol. Rev. 17:370-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon, C. J., M. A. Muesing, A. E. Proudfoot, C. A. Power, J. P. Moore, and A. Trkola. 1999. Enhancement of human immunodeficiency virus type 1 infection by the CC-chemokine RANTES is independent of the mechanism of virus-cell fusion. J. Virol. 73:684-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham, B. S. 2002. Clinical trials of HIV vaccines. Annu. Rev. Med. 53:207-221. [DOI] [PubMed] [Google Scholar]
- 30.Grundner, C., M. Pancera, J. M. Kang, M. Koch, J. Sodroski, and R. Wyatt. 2004. Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology 330:233-248. [DOI] [PubMed] [Google Scholar]
- 31.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 32.Herrera, C., C. Spenlehauer, M. S. Fung, D. R. Burton, S. Beddows, and J. P. Moore. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffs, S. A., S. Goriup, B. Kebble, D. Crane, B. Bolgiano, Q. Sattentau, S. Jones, and H. Holmes. 2004. Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine 22:1032-1046. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistence: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 35.Klausner, R. D., A. S. Fauci, L. Corey, G. J. Nabel, H. Gayle, S. Berkley, B. F. Haynes, D. Baltimore, C. Collins, R. G. Douglas, J. Esparza, D. P. Francis, N. K. Ganguly, J. L. Gerberding, M. I. Johnston, M. D. Kazatchkine, A. J. McMichael, M. W. Makgoba, G. Pantaleo, P. Piot, Y. Shao, E. Tramont, H. Varmus, and J. N. Wasserheit. 2003. The need for a global HIV vaccine enterprise. Science 300:2036-2039. [DOI] [PubMed] [Google Scholar]
- 36.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 37.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 39.Mathiesen, I. 1999. Electropermeabilization of skeletal muscle enhances gene transfer in vivo. Gene Ther. 6:508-514. [DOI] [PubMed] [Google Scholar]
- 40.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 41.Moore, J. P., and D. R. Burton. 2004. Urgently needed: a filter for the HIV-1 vaccine pipeline. Nat. Med. 10:769-771. [DOI] [PubMed] [Google Scholar]
- 42.Moore, J. P., Y. Cao, A. J. Conley, R. Wyatt, J. Robinson, M. K. Gorny, S. Zolla-Pazner, D. D. Ho, and R. A. Koup. 1994. Studies with monoclonal antibodies to the V3 region of HIV-1 gp120 reveal limitations to the utility of solid-phase peptide binding assays. J. Acquir. Immune Defic. Syndr. 7:332-339. [PubMed] [Google Scholar]
- 43.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 46.Moore, J. P., P. W. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 50.Parren, P. W., M. C. Gauduin, R. A. Koup, P. Poignard, Q. J. Sattentau, P. Fisicaro, and D. R. Burton. 1997. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol. Lett. 58:125-132. [DOI] [PubMed] [Google Scholar]
- 51.Parren, P. W., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 52.Parren, P. W., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: Biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 56.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson, H. L. 2002. New hope for an AIDS vaccine. Nat. Rev. Immunol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 58.Sanders, R. W., M. Vesanen, N. Schulke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Si, Z., N. Phan, E. Kiprilov, and J. Sodroski. 2003. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res. Hum. Retrovir. 19:217-226. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, A. Fong, S. R. Coates, L. Leung, M. Wininger, J. J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava, I. K., K. VanDorsten, L. Vojtech, S. W. Barnett, and L. Stamatatos. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Surman, S., T. D. Lockey, K. S. Slobod, B. Jones, J. M. Riberdy, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2001. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 98:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 65.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, D. R. Burton, and D. D. Ho. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-immunoglobulin G. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 69.Widera, G., M. Austin, D. Rabussay, C. Goldbeck, S. W. Barnett, M. Chen, L. Leung, G. R. Otten, K. Thudium, M. J. Selby, and J. B. Ulmer. 2000. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J. Immunol. 164:4635-4640. [DOI] [PubMed] [Google Scholar]
- 70.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 71.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 72.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye, L., Z. Bu, A. Vzorov, D. Taylor, R. W. Compans, and C. Yang. 2004. Surface stability and immunogenicity of the human immunodeficiency virus envelope glycoprotein: role of the cytoplasmic domain. J. Virol. 78:13409-13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhan, X., K. S. Slobod, S. Surman, S. A. Brown, T. D. Lockey, C. Coleclough, P. C. Doherty, and J. L. Hurwitz. 2003. Limited breadth of a T-helper cell response to a human immunodeficiency virus envelope protein. J. Virol. 77:4231-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, C. W., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 80.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zwick, M. B., E. O. Saphire, and D. R. Burton. 2004. gp41: HIV's shy protein. Nat. Med. 10:133-134. [DOI] [PMC free article] [PubMed] [Google Scholar]