ABSTRACT
While fungi can make positive contributions to ecosystems and agro-ecosystems, for example, in mycorrhizal associations, they can also have devastating impacts as pathogens of plants and animals. In undisturbed ecosystems, most such negative interactions will be limited through the coevolution of fungi with their hosts. In this article, we explore what happens when pathogenic fungi spread beyond their natural ecological range and become invasive on naïve hosts in new ecosystems. We will see that such invasive pathogens have been problematic to humans and their domesticated plant and animal species throughout history, and we will discuss some of the most pressing fungal threats of today.
INTRODUCTION
While fungi can make positive contributions to ecosystems and agro-ecosystems, for example, in mycorrhizal associations, they can also have devastating impacts as pathogens of plants and animals. In undisturbed ecosystems, most such negative interactions will be limited through the coevolution of fungi with their hosts. In this article, we explore what happens when pathogenic fungi spread beyond their natural ecological range and become invasive on naïve hosts in new ecosystems. We will see that such invasive pathogens have been problematic to humans and their domesticated plant and animal species throughout history, and we will discuss some of the most pressing fungal threats of today.
Terms Used in This Article
Accessory chromosome: A chromosome additional to the basic karyotype complement, that is, the complete set of chromosomes, which is not necessarily present in all strains of a given species.
Adaptive potential: The capacity of an organism to adapt in response to changes in its environment.
Agro-ecosystem: The organisms coexisting on agriculturally managed land, along with the agricultural environment, as it affects those organisms.
Clonal spread: Movement of a group of genetically identical individuals.
Ecosystem services: Tangible and intangible benefits or consequences of ecosystem function.
Elite germplasm: Seeds (or equivalent) of an organism, specifically optimized to a particular environment, usually through commercial breeding.
Emerging pathogen: A pathogen responsible for a disease that is appearing in a new host or in a new place or increasing its occurrence in an existing host.
Endophyte: Used in this context to mean a fungus that lives within plant tissues for at least part of its life cycle without causing apparent disease.
Evolutionary potential: Capacity of an organism to acquire and retain new alleles, or to undergo shifts in allele frequency, in response to selection pressure.
Extinction/extirpation: Extinction is generally considered to be the death of the last individual of the species, while extirpation is the loss of a species in the chosen geographic area of study, though it still exists elsewhere.
Generalist pathogen: Generalists thrive in a wide variety of environmental conditions and can make use of a variety of different resources (for example, different host plants).
Heterothallic: Species which have sexes that reside in physically separate individuals.
Horizontal gene transfer (HGT): Acquisition of genetic material by one organism from a second, often nonrelated, organism. May be direct or via a vector such as a virus.
Hypervirulence: Unusually high virulence.
Hypovirulence: Decreased virulence.
Innate immunity: Nonspecific defense mechanisms against pathogen attack of animals or plants, usually most relevant during the initial phase of that attack.
Invasive species: An organism outside its native range, spreading rapidly and causing ecological and/or economic damage.
Long-distance dispersal: Spread of an organism to areas well beyond its current range, for example in international air currents.
Mycorrhiza: A symbiotic association between a fungus and the roots of a vascular plant.
Pandemic: An outbreak of infectious disease that spreads exceptionally widely: through populations, across countries, continents, or even globally.
Pathogenicity: The ability of an organism to cause disease in a particular host.
Obligate pathogen: Pathogens that must infect a host to survive, in contrast to other pathogens that are capable of survival independent of their host.
Saprophyte: A microbe that lives on dead or decaying organic matter.
Two-speed genome: A genome in which certain parts, which may range from specific, fairly small regions such as pathogenicity islands or entire (often accessory) chromosomes, are seen to evolve at a faster rate than the core genetic material.
Vertical transmission: “Inheritance” between generations; may refer to transmission of a pathogen or of genetic material (contrast to HGT).
Virulence: The severity of symptoms caused by a pathogen on a susceptible host.
Zoonoses: Disease of humans originating in animals and transmitted either directly or via a vector.
THE HISTORICAL IMPORTANCE OF FUNGAL DISEASE
Devastating fungal diseases of plants and animals can be identified throughout history. The decimation of harvests and ensuing famines, together with various efforts to prevent such outbreaks, are frequently described in early records. For example, wheat stem rust, caused by the fungus Puccinia graminis (still a problematic and invasive pathogen today; see “Current Examples of Emerging Infectious Fungal Diseases”), has been much feared for centuries (1). In the late 4th century BCE, in ancient Greece, Aristotle noted that this disease was favored by warmth and moisture (2). Meanwhile, the Romans held a festival, Robigalia, offering sacrifices to placate the god Robigo and protect cereal crops from such diseases (1). Around this time—circa 400 BCE—Hippocrates coined the words “endemic” and “epidemic” to distinguish between “normal” diseases such as coughs and colds and “abnormal” diseases such as plagues, in his Hippocratic corpus Of the Epidemics. References to plagues and pestilences of plants and peoples can also be found in the Bible as being heaven-sent curses or punishments.
Ergotism and Witchcraft
Fungal diseases of crops have, on occasion, led to some surprising social and political outcomes. A fascinating example is that of the ergot fungus, Claviceps purpurea. This pathogen of rye and other outbreeding cereal crops has a unique life cycle. Its sexual spores germinate on the stigmatic surface of its hosts’ flowers, and its hyphae mimic pollen tube growth by extending downward to infect the ovary (3). The fungus then appropriates nutrients from the host to form a fruiting body, or sclerotium, in place of the developing seed. While fungal infection causes relatively little loss in yield, the grain quality is reduced due to the production of the fungal toxins ergotamine, ergine, and lysergic acid in the fruiting body (3). Ingestion of foodstuffs made from contaminated grain causes gastrointestinal symptoms followed by fatigue, depression, and painful muscular contractions. In more severe cases, vasoconstriction gives rise to “formication” (the feeling that the patient is covered in ants) and burning sensations and gangrene of the extremities, and psychoses may also occur (4). This suite of symptoms has long been recognized, with ergotism being dubbed “St. Anthony’s fire” in the 11th century (5), but their source eluded mankind for centuries. The risk posed by ergot toxicity has diminished considerably since the identification of its fungal cause, with screening for contaminated seeds now in place. Nevertheless, there have still been isolated outbreaks, for example, in France in 1951 and in Ethiopia as recently as 2001 (6, 7). The most notorious case of ergot poisoning is perhaps that which triggered the 17th century witch trials in Salem, Massachusetts.
In a 17th century society dominated by political upheaval, puritanical Christianity, and social bickering, a group of young girls (aged 9 to 17) developed gastrointestinal symptoms, tingling and crawling sensations in the fingers, muscular contractions, mania, depression, and psychosis (4, 8). Unable to reach a diagnosis, a doctor suggested that the girls were bewitched (9). The patients then laid accusations of witchcraft against, initially, women on the fringes of the society (4, 8, 9). Fear and hysteria swept through the community and did not loosen its grip until 20 “witches” had been executed. At this time, similar symptoms were seen in Finnmark, Norway. Here, however, it was the patients themselves who were accused of witchcraft. A witch hunt with even more tragic consequences than in Salem saw 137 people sent to trial, of whom around two-thirds were executed (10). In both Salem and Finnmark, it is thought that the illness which sparked the cycle of accusations, trials, and executions was due to ergotism (8, 9). Seasonal factors favoring the development of the fungus C. purpurea, including a cold winter followed by a humid spring, have been noted (4). Although other forces no doubt played a role in the extent of the accusations, it seems likely that C. purpurea triggered these ugly episodes, illustrating that fungi can influence human history in many ways.
Changing Landscapes: Chestnut Blight and Dutch Elm Disease
Emerging pathogens are those where the occurrence of the disease that they cause is entirely novel, or greatly increases, within a particular ecosystem. This may be due to the introduction of their host, either by man, due to a changed and newly permissive climate, or naturally; pathogen acquisition of new virulence genes or other traits allowing pathogenicity on a new host; or the appearance of a new genotype of a pathogen through hybridization, horizontal gene transfer, or other evolutionary events (discussed below in “Drivers for the Emergence of New Fungal Disease”). In the 20th century, two forest diseases emerged which destroyed large numbers of trees in Europe and North America. These serve as good examples of the damage done by emerging fungal pathogens in the recent past.
Chestnut blight is a disease of Castanea species, including the American chestnut Castanea dentata, the European Castanea sativa, and the Asian Castanea crenata. The causative fungus, Cryphonectria parasitica, is native to East Asia (11). Imports of C. crenata seedlings from Japan to North America appear to have been responsible for spreading C. parasitica to the United States (12). Once in North America, the fungus attacked native C. dentata trees. Within 50 years of the initial introduction of C. parasitica, mature C. dentata chestnut trees had been obliterated from the landscape. While uninfected rootstocks remain able to form new sprouts, these quickly become blighted, and no fruits are produced (11). C. dentata trees were previously the dominant canopy tree throughout the Appalachian forests (11), where their seas of white blossom held cultural importance. The loss of C. dentata trees also had profound ecological consequences, affecting both forest-floor and nearby aquatic habitats: chestnut leaves decompose easily and are of high nutritional value to invertebrates, compared to the leaves of the oak trees which largely replaced them (13). Impacts on mammal and bird species varied, with anecdotal accounts indicating a sharp drop in squirrel numbers due to the loss of mast production (14), while woodpeckers benefited from the increase in deadwood and insects therein (15). Meanwhile, seven moth species that lived or fed exclusively on chestnut became extinct (16). Thus, one fungus, C. parasitica, changed the forest landscape of the United States.
The spread of chestnut blight worldwide has been tracked by population genetic studies. In North America, genotypic data indicate that the source of the pathogen was Honshu, Japan, confirming the historical evidence of C. crenata seedling imports from this area in 1904. The American C. parasitica populations show considerable genetic divergence from the Asian populations; however, this is probably due to a small founder population, leading to random fixation of particular alleles in the emergent population (17). Overall, the data support a single or small number of introductions, followed by a rapid geographical spread. By contrast, C. parasitica spread slowly through Europe and without as marked destruction of the host C. sativa trees (17), which are less susceptible than American C. dentata. Additionally, European strains of C. parasitica themselves became infected with a virus, CHV-1, which caused fungal hypovirulence (17). Variation in the European fungal strains of C. parasitica is due to repeated introductions from both Japan and China. Despite the differences in spread pattern, American and European populations of the chestnut blight fungus show no genetic differentiation, indicating that there has been import and export of infected hosts between the two continents (17). Given the level of devastation the chestnut blight fungus unleashed in America—over 3 billion chestnut trees lost to disease—the fact that intercontinental trade in chestnuts was ongoing during this pandemic is, in retrospect, of grave concern.
A second fatal forest pathogen, which caused Dutch elm disease, spread across multiple continents in the 20th century, changing landscapes where grand elms once dominated the skyline. In contrast to chestnut blight, multiple pandemics of Dutch elm disease occurred. The final pandemic, in the 1970s, was the most damaging. Like the loss of American chestnuts, the loss of hundreds of millions of elm trees caused profound ecological changes, including the exposure of neighboring trees to storm damage and death and the creation of open woodland habitat that supported altered bird and mammal assemblages (11). The first pandemic, in the 1920s, was caused by the fungus Ophiostoma ulmi. This was a relatively weak pathogen, and it declined in disease severity naturally by the 1940s (18). However, a second, more aggressive pathogen, Ophiostoma novo-ulmi, appeared during the 1970s (11, 19). This species has limited ability to interbreed with O. ulmi, and hybrid forms show reduced fitness. Nevertheless, transient hybrids occurred, and these provided a “genetic bridge” across which genes flowed between the species. These genes included a particular mating type allele, MAT-1, which increased the capacity of O. novo-ulmi to undergo sexual recombination, increasing its evolutionary potential (20). As we will see in “Natural Factors Promoting the Emergence of New Invasive Fungal Diseases,” a high evolutionary potential is often important in the emergence and establishment of new infectious diseases.
Two subspecies of O. novo-ulmi exist, and both have invaded Europe: O. novo-ulmi subsp. novo-ulmi from the East and O. novo-ulmi subsp. americana from the West. These now hybridize freely (20). However, O. ulmi cannot coexist with the more aggressive O. novo-ulmi, so this pathogen is now locally extinct in many areas (20). As with chestnut blight, trade and transport are implicated in the spread of Dutch elm disease, with O. novo-ulmi subsp. americana reaching Europe in a consignment of logs sent from the United States to the United Kingdom in 1974 (20). In “Anthropogenic Factors Promoting the Emergence of New Invasive Fungal Diseases” we will discuss the importance of trade in plants and plant derivatives in the spread of agricultural and forest pathogens.
CURRENT EXAMPLES OF EMERGING INFECTIOUS FUNGAL DISEASES
Fungal diseases of plants have changed our landscapes and devastated our crops over the centuries. Since the Green Revolution, agricultural practices have favored intensive farming and the large-scale growing of hectare upon hectare of genetically uniform crops. Such plants are guarded from disease by one or two disease resistance genes or by the widespread spraying of fungicides. This plenitude of identical hosts has provoked widespread outbreaks of many known fungal foes on our major calorie crops. In this section, we focus on these emerging infectious diseases of domesticated crop plants and wild animals.
Diseases of Crop Plants
Soybean rust: Phakopsora pachyrhizi (Basidiomycete) (21–25)
What it infects
Soybean (Glycine max) and 95 other known legume species.
Becoming global
Sometimes called Asian soybean rust, this disease spread to Africa in the 1990s and then to South America and finally North America by 2004. Three main factors underlie its invasiveness: first, its asexual spores, which are produced in prolific numbers, can spread in global air currents. This includes cyclones and hurricanes, which cause unpredictability in terms of long-distance spread into new niches. These spores are then “rained out” of the atmosphere and deposited on wet leaves—ideal conditions for infection. Second, although P. pachyrhizi is a true obligate pathogen, it is a generalist: it can infect many different legume hosts. As well as soybean, it infects forage legumes such as vetch and lupin, as well as widely distributed weeds such as kudzu (Pueraria lobata), which is itself invasive. Third, P. pachyrhizi can rapidly generate new asexual spores by passing through its life cycle in as few as 9 days. Together, these factors ensure that the fungus spreads widely and finds a host readily. If soybean plants are absent, or only present seasonally, the pathogen sporulates on alternative hosts, repeatedly reinfecting newly planted soybean crops.
Why it matters
P. pachyrhizi causes an up to 80% reduction of soybean yields. This can lead to huge economic losses and social consequences, which have particularly affected parts of South America. An outbreak in 2001 in Paraguay caused severe losses and led to an outbreak in Brazil, in which up to 60% yield losses were reported. In 2002, over half a million metric tons of soybean were lost in Brazil alone. These losses, combined with the use of extra fungicides, cost Brazilian farmers 2 billion dollars during 2001 to 2003. The disease spread to Argentina in 2002 and Bolivia in 2003. In the United States, economic studies indicate that soybean rust could cause serious yield losses and increase production costs. In resource-poor farming areas, the resultant loss of livelihood leads to human suffering.
Mitigation
P. pachyrhizi is currently susceptible to treatment with azole and strobulurin fungicides. These target the fungal plasma membrane and interfere with mitochondrial electron transport, respectively. Prudent use of such chemicals, with careful monitoring, will hopefully lessen the risk of emergence of new, fungicide-resistant P. pachyrhizi strains. Moreover, meteorological monitoring and predictive modeling of global air currents and rainfall will aim to determine which crops are vulnerable and inform control measures.
Wheat stem rust: Puccinia graminis (Basidiomycete) (1, 26–32)
What it infects
Primarily a disease of wheat (Triticum aestivum), P. graminis can infect oats (Avena sativa), rye (Secale cereale), and barley (Hordeum vulgare), as well as the wild grasses Festuca and Lolium spp. The wheat stem rust fungus undergoes sexual reproduction on an alternative host, barberry (Berberis vulgaris).
Becoming global
Evidence suggests that the telial stage jumped onto grasses during their range expansion in the tertiary period, and the fungus was then further spread by agriculture. Barberry was identified as the alternative host in the 1780s. The fungus forms five spore types as it cycles between wheat and barberry. First, the teliospores, which have overwintered on wheat straw debris, germinate to form basidiospores which infect barberry, forming spermatia on the upper leaf surface. This triggers aeciospore formation on the lower leaf surface. These aeciospores are lifted into the boundary layer of the atmosphere and can travel thousands of kilometers before being deposited on wheat leaves, where they then form uredospores. These rust-red asexual uredospores are generated in prolific numbers and herald the start of epidemic disease levels. As the weather conditions deteriorate in autumn and the wheat harvests are gathered, the uredospore population declines in favor of the overwintering teliospores. Despite this complex dual-host life cycle, P. graminis is able to skip its telial stage to spread clonally in the absence of Berberis. Epidemics of wheat stem rust occurred in all wheat growing regions of the world during the 20th century; 60% of the U.S. wheat crop was lost in 1916, and eight more epidemics followed between 1920 and 1960. The popularity of Barberry as an ornamental in gardens and parks was implicated in these outbreaks. Meanwhile, Europe suffered epidemics in the 1930s, China in the 1950s, and Australia in the 1970s.
Why it matters
In addition to historical epidemics causing major yield losses worldwide, a new strain of P. graminis, known as Ug99, was discovered in Uganda in 1999. This new strain had spread to many African wheat growing countries, as well as Iran and Yemen, by 2005, and by 2014 had also been detected in Russia. There are now 11 known races of Ug99, each of which has broken the resistance offered by a different R gene bred into wheat. Ug99 is aggressive, causing significant yield losses, particularly to wheat varieties without effective disease resistance, and has shown itself to have high adaptive potential. It is feared that Ug99 will eventually reach other major wheat growing areas in Europe, Asia, and the Americas, causing catastrophic losses in global wheat yield.
Mitigation
Twentieth century epidemics were overcome by a combination of breeding for disease resistance in wheat and, in the United States, by the eradication of barberry plants. The former has mostly relied on gene-for-gene resistance, with durable protection derived from pyramiding of R genes. Destruction of over 50 million Berberis plants followed a major public education campaign (“Barberry or bread”), public participation, and legislation to make eradication compulsory.
Charcoal rot: Macrophomina phaseolina (Ascomycete) (21, 33–37)
What it infects
Over 500 host plant species are known. Individual strains show some degree of host specificity, but this is often incomplete, allowing the fungus to spread to infect other hosts. Crops infected by the charcoal rot fungus include soybean, sunflower, jute, cotton, cowpea, maize, sorghum, chickpea, peanut, canola, strawberry, guava, cassava, and melon. The fungus can also infect wild prairie grasses, as well as being an opportunistic human pathogen.
Becoming global
M. phaseolina was reported as the causal agent of guava wilt in India in 1990. It was identified as charcoal rot of strawberries in France in 1993 and of canola in the United States in 1994. In 1997, M. phaseolina was found causing stem rot of cassava in West Africa. In the 1990s, charcoal rot of soybean appeared in the United States. Charcoal rot of canola was found in Argentina in 2006 and Western Australia in 2009. Population genetics suggest this fungus spreads clonally, with its ability to infect wild species being important for its invasiveness.
Why it matters
M. phaseolina is a difficult fungus to identify. As such, it may have existed in many areas before it was first reported. However, charcoal rot began, more recently, to have to a serious impact on soybean yield in the United States. From 1996 to 2004, yield losses were estimated at around three quarters of a million metric tons per year. Thus, M. phaseolina was the second most economically important disease of soybean at this time. In Africa’s Sahel region over the same period, yield losses averaged around 10%; 100% yield loss is possible. The disease is most problematic in arid and semi-arid regions; the fungus thrives where soil nutrient content is low and temperatures are high. M. phaseolina is expected to become more geographically widespread under climate change models, and some host plants may be threatened with extinction. Increased occurrence of M. phaseolina is of further concern because it can act as an opportunistic mammalian pathogen.
Mitigation
M. phaseolina is hard to control, because its microsclerotia (hardened masses of fungal mycelium) persist in soil for up to 15 years. Soil treatment to mitigate the spread of fungal disease is problematic, not least because M. phaseolina shows resistance to many current fungicides. Moreover, antifungal prophylaxis does not prevent M. phaseolina infection in immunocompromised human patients, and the fungus can only be removed by wide surgical excision. Further research into this pathogen is needed, because its population genetic structure, life cycle, and manner of spread need to be better defined.
Leaf spot of barley: Ramularia collo-cygni, (Ascomycete) (38–41)
What it infects
This fungus primarily causes leaf spot disease on barley but is emerging as a pathogen of oats and wheat. It infects wild grasses including Triticum, Festuca, Lolium, Elymus, Agropyron, and Brachypodium spp.
Becoming global
R. collo-cygni was first described as a pathogen in 1893 in Italy, but rigorous scientific study of the fungus began in the 1980s. It was found in the United Kingdom in the late 1990s and detected in Europe, South and North America, the Middle East, Russia, Iceland, and Africa in the 2000s. This rapid emergence begs the question of whether it truly emerged or, instead, if improved recognition and detection led to its apparently sudden worldwide appearance. The latter seems a plausible explanation, because plants infected with R. collo-cygni remain symptomless while the fungus migrates to the awns (bristles on the glumes) and ears. The subsequent infection of the grains transmits the fungus from generation to generation and fuels further dispersal as infected germplasm is traded across the world.
Why it matters
This disease is largely symptomless in the early stages of infection, with symptoms appearing around the time of flowering and during grain set. This leads to low yield and low grain quality from a crop that appears uninfected until 3 weeks before the harvest is gathered. Recently, infections proved problematic in South America; high incidence and disease severity in Argentina in 2011 led to 70% grain yield losses, while in 2012, the fungus spread to six other South American barley growing areas, where it has now become a limiting factor in barley production. In 2002, widespread reports of barley leaf spot disease due to R. collo-cygni infection were reported in the United Kingdom. Hitherto, the fungus had been kept at bay by the use of strobilurin fungicides, but at this time, a point mutation known from related fungi rendered the fungus resistant to the quinone outside inhibitor fungicides. While the South American R. collo-cygni strain(s) has not yet acquired this mutation, if—or when—it does, grain costs and yield losses will rise.
Mitigation
Strobilurin fungicides are still in use in South America, but a limited lifetime should be expected and fungicide mixtures encouraged, given that fungicide resistance has arisen in R. collo-cygni elsewhere in the world. Trials have been conducted on seed treatments but without success, because the fungus behaves as a true endophyte, infecting the endosperm and embryo; treatments which kill the fungus are therefore detrimental to the seed itself.
Diseases of Animals
Amphibian chytridiomycosis: Batrachochytrium dendrobatidis and Batrachochytrium salamandrivorans (Chytridiomycota) (42–51)
What they infect
B. dendrobatidis has a broad host range, infecting all three classes of amphibian (anurans, caudates, and Gymnophiona). The fungus has a worldwide distribution occurring in 56 of 82 (68%) countries and infecting 42% of species. B. salamandrivorans has a more restricted range both in terms of its hosts, being found only in caudates (newts and salamanders), and geographic locations. Infection by these two chytrid fungi causes a cutaneous disease known as chytridiomycosis (Fig. 1).
FIGURE 1.

Mass mortalities of midwife toads, Alytes obstetricans, caused by Batrachochytrium dendrobatidis (photo: Matthew C. Fisher).
Becoming global
Following the discovery in 1998 that B. dendrobatidis was a key driver of declines in amphibian species in Australia and the Americas, attention has focused on determining where outbreaks of chytridiomycosis have occurred and whether there are any underlying spatio-temporal patterns that indicate the original sources of infection, as well as pathways of spread. Phylogenomic analyses have shown that the worldwide emergence of chytridiomycosis is most likely explained by the rapid transmission of a hypervirulent lineage, called the global panzootic lineage (BdGPL). This occurred on a global scale: this lineage has now been found in all continents alongside endemic lineages. Indeed, all mass-mortality and extinction events thus far attributed to chytridiomycosis caused by B. dendrobatidis are associated with the presence of BdGPL. In vivo laboratory assessments of the virulence of BdGPL have shown that this is indeed a hypervirulent lineage—a feature that confers “superbug” status on BdGPL. However, the questions surrounding the origin of BdGPL and the historical timing of this lineage’s emergence across different continents remain elusive. In contrast, B. salamandrivorans is known to have emerged much more recently, having been most likely transported from Southeast Asia to Europe via the pet trade in infected amphibians.
Why it matters
Amphibians have long been recognized as being the most endangered vertebrate class on the planet, and chytridiomycosis is now recognized as a proximal driver of many amphibian die-offs worldwide. Indeed, chytridiomycosis has been described as “the worst infectious disease ever recorded amongst vertebrates, in terms of the numbers of species impacted and its propensity to drive them to extinction” (47). Thus, chytridiomycosis can result in widespread population decline, even extinction, in a subset of species and has contributed to the threatened status of almost 400 species of amphibian.
Mitigation
Countering disease-driven amphibian declines should consist of a multifaceted approach adapted to the stages of pathogen emergence. These include tackling the prearrival stage, the advancing invasion front, epidemic outbreaks, and established disease. Current approaches include prevention through restricting global trade in infected amphibians because it is clear that international movement of traded species is key to the spread of chytrids into naïve, disease-free ecosystems. Other short-term solutions include ex situ breeding programs known as “amphibian arks,” alongside the cryopreservation of reproductive material. However, long-term, in situ, sustainable solutions are required if the goal of amphibian conservation is to be attained. This implies neutralizing the disease threat in wild populations using, for instance, antifungal compounds.
Bat white nose syndrome (WNS): Pseudogymnoascus destructans (Ascomycete) (52–55)
What it infects
First recognized in North America, P. destructans is now known to infect and cause clinical disease in at least six North American bat species. The fungus has also been described as infecting other species of bats in Eurasia and China.
Becoming global
WNS is a disease caused through infection of hibernating bats by P. destructans. First described following a cave die-off of little brown bats (Myotis lucifugus) in Schoharie County, New York, in the winter of 2006, the infection has since spread rapidly across eastern North America (Fig. 2). It was recently detected in Washington State, some 1,700 km from the index site. The mycosis is hypothesized to have been vectored into North America from Europe, where the fungus widely infects European bat species but where it does not cause observable mortality. Genetic analyses have confirmed that the North American variant of P. destructans is a single clone carrying a single mating type. This contrasts with the European genotypes, which are highly diverse and carry either one of the two heterothallic mating types.
FIGURE 2.

Mass mortalities of little brown bats, Myotis lucifugus (photo: Alan Hicks).
Why it matters
P. destructans causes mass mortalities in North American hibernacula because the fungus destroys cutaneous membranes, including the wings, of hibernating bats. By 2015, four notable North American bat species’ distributions had been entirely encompassed within the range of spread of P. destructans. As a result, species extirpations are predicted to occur in the near future.
Mitigation
Our current understanding suggests that the temperature of the bat hibernacula plays a role in WNS disease severity. Cooler and drier hibernacula appear to serve as refugia from disease impacts for some populations, because they carry lower fungal loads and thereby increase chances of bat survival. Indeed, it has been proposed that manipulating hibernacula entrances to create cooler and drier sites or restricting access to reduce bats’ use of the warmer and wetter portions of hibernacula has potential as a single intervention in the management of WNS. Other approaches proposed include antifungal or biological (probiotic) interventions that could be used to reduce inoculum loads of P. destructans in the short term, thus allowing the bats time to adapt and evolve defensive/protective immunological responses to this pathogen.
Human cryptococcosis: Cryptococcus neoformans and Cryptococcus gattii (Basidiomycete) (56–60)
What it infects
Cryptococcus is a genus within the Tremellales, an order of fungi within the Basidiomycetes called “jelly fungi,” due to their gelatinous fruiting bodies. They are commonly found growing on rotting wood as saprophytes. There are over 30 known species of Cryptococcus, of which two, C. neoformans and C. gattii, cause the majority of opportunistic human infections. Both species are readily recovered from the environment, where they can be isolated from the bark of a wide variety of tree species alongside other organic matter, most notably bird feces.
Becoming global
Genetic analysis has shown that C. neoformans and C. gattii have evolved independently from each other over the last approximately 30 to 40 million years. This separation has resulted in subtle variations in their virulence patterns. C. neoformans is the predominant species causing infection in immunocompromised individuals with HIV/AIDS. C. neoformans thus emerged in concert with the AIDS pandemic. C. gattii, however, causes rarer infections in putatively immunocompetent individuals. An unusual cluster of cryptococcal disease in otherwise healthy individuals was seen on Vancouver Island, British Columbia, in 1999. This infection, caused by C. gattii, has now expanded into mainland Canada and the northwestern United States and is known as the Pacific Northwest outbreak. It was likely seeded by infectious inocula from South America.
Why it matters
The virulence attributes of Cryptococcus stem from various adaptations it has undergone that allow it to survive in the environment. An array of recognized “dual-use” survival/virulence factors is known. The most notable of these is the thick enveloping polysaccharide capsule, which in the environment defends against attack by parasitic amoebae but which also allows the fungus to survive macrophage assault and to disseminate as an intracellular parasite within the human body. Such adaptations have led to widespread human infection where the prevalence of HIV is high. In 2009, the CDC estimated that there are close to one million cases per year, with at least 100,000, and perhaps 500,000, deaths per year in sub-Saharan Africa alone, where cryptococcal meningitis accounts for around 17% of AIDS mortality.
Mitigation
The most important method of controlling cryptococcal meningitis is to control HIV. In Europe and North America, the numbers of cryptococcal patient cases fell dramatically after introduction of effective antiretroviral therapy. However, despite the widespread use of antiretroviral therapy in sub-Saharan Africa, there is no evidence of a reduction of cryptococcal infections. This may be due to the emergence of antiretroviral therapy resistance in target populations provoked by nonadherence to treatment regimes. The recent introduction of a cheap and easily used antibody-based disease diagnostic “dipstick” test that detects cryptococcal antigen holds much promise. This test has the potential to allow earlier treatment of the disease with potent antifungal drugs. It has triggered the widespread advocacy calling for the affected populations to have access to the most advanced antifungal drugs at an affordable cost.
Azole-resistant aspergillosis: Aspergillus fumigatus (Ascomycete) (61–63)
What it infects
The genus Aspergillus comprises a few hundred fungal species found in various climates worldwide. Certain Aspergillus species cause a variety of clinical diseases in humans and animals. The disease spectrum ranges from allergic conditions such as allergic bronchopulmonary aspergillosis through to invasive tissue disease in immunosuppressed tissue-transplant patients. Allergic bronchopulmonary aspergillosis is primarily seen in patients with chronic Aspergillus colonization suffering from cystic fibrosis and chronic granulomatous disease. Patients with lung diseases such as chronic obstructive pulmonary disease can develop chronic pulmonary aspergillosis when the fungus grows and becomes locally invasive, thus causing tissue damage.
Becoming global
The triazole antifungals, including itraconazole, voriconazole, isavuconazole, and posaconazole, are widely used to treat diseases caused by Aspergillus infection. However, azole-resistant strains have recently emerged across six continents and pose a significant new therapeutic challenge. Although de novo azole resistance is known to have arisen occasionally in patients during azole therapy, the main burden is now known to have been caused by the acquisition of resistance from environmental isolates. Here, the evolution of azole resistance in A. fumigatus is attributable to the widespread use of azole-based fungicides, which are widely deployed to control fungal disease in agriculture. The resistance phenotype is associated with particular key mutations in the fungal cyp51A gene. The occurrence of rapid selective sweeps associated with such new alleles has been documented.
Why it matters
cyp51A-mediated resistance is increasing rapidly; in some settings up to 10% of patients with Aspergillus-related disease do not respond to azole treatment. The prognosis for these patients is poor, with up to 80% mortality.
Mitigation
Azole fungicides are widely used in agriculture; they rank as the most important antifungal from a global perspective. Worldwide measures to curb their use should now be considered due to their attendant selection for antifungal resistance on nontarget pathogenic fungi such as A. fumigatus. But intervention is problematic, because azole use is so widespread in agriculture. A ban on azole use in crop disease prevention would alter reliance on other classes of fungicide, such as strobilurins and fast-forward emergence of new fungicide-resistant genotypes. However, this highlights the need either to reserve the antifungal azoles for the treatment of human fungal disease or to develop new antifungal drugs for human use.
DRIVERS FOR THE EMERGENCE OF NEW FUNGAL DISEASE
Natural Factors Promoting the Emergence of New Invasive Fungal Diseases
Most organisms are able to resist infection by most potential pathogens through innate immunity (e.g., 64). Pathogens, in turn, constantly evolve to overcome host defenses, creating selection pressure for hosts to withstand such pathogens. There is thus a classic evolutionary arms race between host and pathogen (65). While it is relatively easy to understand this process, it is considerably harder to predict when, where, and how new diseases will emerge.
The key factor determining the likelihood that a microbe will become pathogenic for the first time, on a new host or in a new place, is its “evolutionary potential.” This is the organism’s potential for genetic adaptability, and it is underpinned by a combination of the organism’s biology, genomics, and population genetics. First, variability is a prerequisite for selection. A linked factor is effective population size, because more reproducing individuals generate more mutations on which selection can act. Second, reproductive mode affects evolutionary potential, because recombination creates new mixtures of genes (66, 67). Third, certain features of pathogen genomes may add to their evolutionary potential, increasing recombination, hybridization, variability, or rates of horizontal gene transfer (67, 68) (Fig. 3).
FIGURE 3.
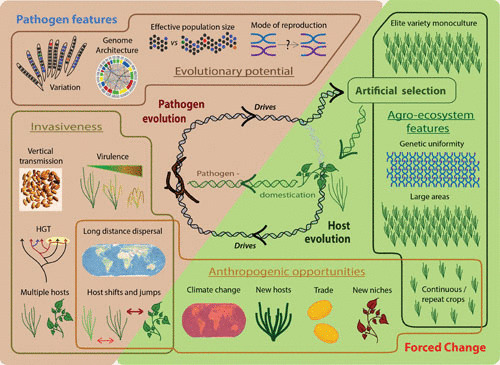
Factors influencing the emergence of infectious diseases of crop plants. These include features of the pathogens themselves as well as changes forced by the opportunities given to and pressures placed upon pathogens externally, as a result of human activity. Pathogen features prompting the emergence of new diseases include specialized genomes, sexual reproduction, large populations, and plentiful variation, which contribute to evolutionary potential. Invasiveness is key to the appearance of new emerging infectious diseases. This pathogen trait has natural components including high virulence and the capacity to infect multiple hosts and to transmit vertically, as well as such traits as long-distance dispersal and a propensity to undergo host shifts and jumps. Such traits may be natural or appear as a result of opportunities provided by anthropogenic changes to the environment. Such anthropogenic opportunities include climate change and trade and transport, introducing pathogens to new places, niches, and hosts. They may also take the form of pressures placed on pathogens by the continuous cultivation of a single crop year-round leading to genetic uniformity of available hosts over large areas. Such elite varieties are the product of artificial selection, which is the main driver of host evolution when the host is a domesticated crop. Host evolution can then drive pathogen evolution, leading to the appearance of new emerging infectious diseases and subsequent selection of new, resistant elite varieties. In addition, during the domestication of a crop, pathogens may be directly domesticated, short-cutting their adaptation to a particular host.
The biology and life cycle of the wheat pathogen Zymoseptoria tritici (Septoria leaf blotch disease) exemplifies all of these factors (Fig. 4). This fungus shows plentiful genetic variation, with evidence of gene flow between populations (69). It causes a polycyclic disease, with many cycles of asexual reproduction occurring within a wheat growing season, which allows selection for virulence (70). However, Z. tritici also reproduces sexually (71). The fungal genome consists of a core set of chromosomes and a suite of up to eight accessory chromosomes of unknown function, carrying genes which are largely silenced (72). The complement of these accessory chromosomes varies with fungal strain and can also change between generations. Further, there is evidence of multiple insertion and deletion events occurring in the accessory chromosomes. The changes in accessory chromosome number probably originate through nondisjunction at meiosis, followed by the fusion of sister chromatids. This leads to chromosomal degeneration and is probably the origin of the accessory chromosomes. The rapid evolution that takes place on accessory chromosomes (73) contributes to the pathogen’s adaptive potential (74) and helps deleterious changes to be lost while advantageous ones are preserved. Other hallmarks attesting to the plasticity of this fungus are seen in its genome, which contains 17% repetitive DNA, 70% of which is enriched in class 1 transposable elements, which are associated with high mutation rates (75). Z. tritici poses a major threat to wheat production throughout the temperate world (71, 76; see “Threats to Agriculture from Emerging Fungi”). Its ability to evolve rapidly not only challenges our ability to breed for durable disease-resistant wheat cultivars but also promotes the emergence of fungicide resistance.
FIGURE 4.
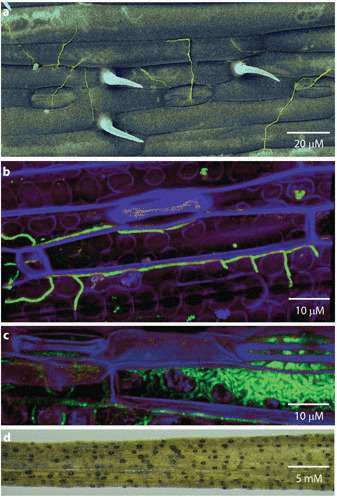
The wheat pathogen, Zymoseptoria tritici. (a) Scanning electron micrograph showing Z. tritici (green) on the surface of a wheat leaf. False color image. (b) Confocal fluorescence micrograph showing green-fluorescent protein-tagged Z. tritici (turquoise) beginning to colonize mesophyll tissues of a wheat leaf (purple). False-color image. (c) Confocal fluorescence micrograph showing Z. tritici (turquoise) proliferating with a wheat leaf at a late stage of infection. The fungal mass on the right of the image is a nascent pycnidium, the structure in which sporulation takes place. (d) Severely diseased wheat leaf showing Z. tritici symptoms, including mature pycnidia (black). Each black spot can release hundreds of spores. (Photos: Helen Fones; GFP-tagged Z. tritici: reference 127.)
Genomes like that of Z. tritici, where accessory chromosomes are associated with high evolutionary potential, are often termed “two-speed” genomes. This refers to the separation of their core set of slowly evolving, essential genes from their accessory genes, genomic regions, or chromosomes, which are enriched in repetitive DNA, transposable elements, or other features which make them hot spots for mutation, recombination, and even for the introgression of genes from other species (77). These rapidly evolving genomic regions are frequently associated with pathogenicity and may be lineage-specific, determining host range (74). Examples can be seen in the genus Fusarium, a collection of saprophytic and pathogenic fungi which can infect both plants and animals, including humans. Fusarium solani, also called Nectria haematococca, carries up to three accessory chromosomes, which include genes more similar to Aspergillus genes than to the rest of the F. solani genome (76). One such accessory chromosome encodes pathogenicity factors that allow the strain which carries it to infect pea plants, having degraded a host defense compound (a phytoalexin) (76, 78). The related generalist Fusarium oxysporum (Fig. 5; see section on Panama disease) also carries accessory chromosomes containing many transposable elements and virulence genes which define its host range. These accessory chromosomes can be readily exchanged, even between fungal strains which are not sexually compatible and between strains of different species in the genus Fusarium (79). Not all Fusarium species carry accessory chromosomes, but even those lacking this feature have rapidly evolving genomic regions associated with pathogenicity and virulence determinants (80). The wheat pathogen F. graminearum, which has recently re-emerged to cause worldwide problems (69), has four chromosomes that show high single nucleotide polymorphisms and rapid recombination rates in the telomeric regions at the ends of chromosomes, reflecting ancient chromosome fusions (81).
FIGURE 5.
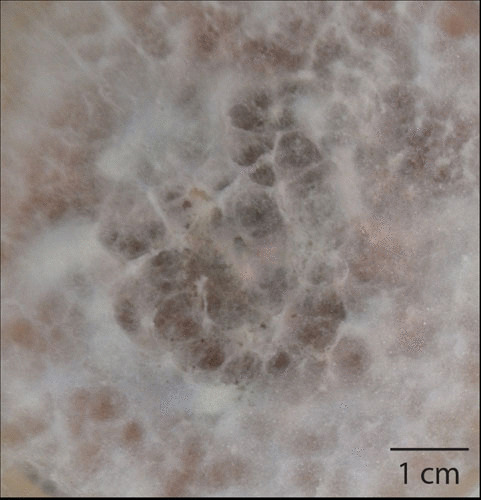
Fusarium oxysporum f. sp. cubense race 4 (Panama disease). Culture on potato-dextrose agar in a laboratory, showing characteristic purple coloring. (Photo: Helen Fones.)
The exchange of accessory chromosomes between Fusarium strains indicates that they undergo some form of HGT. HGT is common among bacteria and frequently allows the transfer of pathogenicity, virulence, or antibiotic resistance genes, creating “pathogenicity islands” in bacterial genomes. HGT had been assumed to be rare in eukaryotes, but new evidence concerning exchange of pathogenicity genes in particular suggests that it may be much more commonplace in filamentous fungi than realized. Such evidence usually comes from comparative genome data, the availability of which has recently exploded with the advent of rapid DNA sequencing technologies and analytical tools. Nucleotide sequences gained through HGT, including bacterial pathogenicity islands, can usually be recognized by a GC content different from the surrounding region of the genome and by a nearby abundance of transposase genes (encoding enzymes that move transposons). The F. solani chromosome containing the pea pathogenicity gene conforms to this pattern (76). There is a similar HGT footprint in the toxA gene of the notable wheat pathogen Pyrenophora tritici-repentis, which appears to have transferred from Phaesosphaeria nodorum, and in a cluster of pathogenicity-related genes in the maize smut pathogen Ustilago maydis. In the generalist plant pathogenic fungus Alternaria alternata, which provokes human allergies, such clusters are known to define host range (78).
So what are the endogenous attributes which favor the emergence of a new pathogen? Pathogens with large populations, high mutation rates, high rates of outcrossing and genetic recombination, and a high level of genomic plasticity, including the ability to exchange genetic material in a nonsexual manner such as HGT, are most likely to emerge as new pathogens: they have the evolutionary potential to overcome host defenses in a naïve plant or animal (79). Various routes have been identified for this emergence. Chief among these are host jumps and host shifts (82). It is relatively easy to see that the acquisition or loss of an accessory chromosome or a single or set of pathogenicity genes could lead to a host jump, by allowing a pathogen of one host suddenly to infect another. Historical examples of host jumps include the rice blast pathogen, Magnaporthe oryzae, which jumped from Setaria millet onto rice 7,000 years ago (78), and the ergot fungus, C. purpurea (see “Ergotism and Witchcraft”), whose closest relatives are symbionts of wild grasses and pathogens of arthropods (83, 84). More recently, the rice blast fungus jumped onto wheat in Brazil and, in 2016, onto wheat in Bangladesh (85, 86). Host shifts are smaller, between closely related hosts. Another route for the emergence of a new pathogen is through hybridization between two or more related fungal strains. An example of this mechanism was seen in Dutch elm disease (see “Changing Landscapes: Chestnut Blight and Dutch Elm Disease”), where hybridization between O. ulmi and the new, more aggressive O. novo-ulmi allowed the transfer of pathogenicity genes from the older species into the emergent, more devastating pathogen (20, 78). Other hybrid emerging fungal pathogens include poplar rust, Melampsora columbiana, and Botrytis allii, a postharvest disease of onions (20).
In addition to these factors, which promote the emergence of a new pathogen, there are also natural factors which play a role in determining pathogen invasiveness. Earlier in the article we saw that certain invasive plant pathogenic fungi share the ability to travel internationally in air currents (22, 23, 30). This allows worldwide spread of spores to infect any susceptible host population. In addition, it allows obligate pathogens to repeatedly reinvade areas where the host is not available year-round (87). Airflows are largely predictable, in both a local and global sense, allowing models to identify major risk areas based on pathogen source populations, spore release times, and prevailing air currents. But there are also unpredictable, extreme weather events such as cyclones or hurricanes (12, 87, 88) which can carry pathogens to new areas faster than expected. Interestingly, it now appears that for foliar crop pathogens, this long-distance dispersal is not a special adaptation, but rather an extension of the processes involved in short-distance spread among hosts within a field, combined with turbulent airflow at small spatial scales (87). Fungal foliar crop pathogens appear to be uniquely successful at long-distance dispersal, which may be an exaptation derived from their usual modes of transmission (primarily rain splash and wind dispersal) (87). Similar processes are, however, also probably contributing to the aerosolization and contemporary dispersal of azole-resistant A. fumigatus, leading to the occurrence of the resistant cyp51A alleles in regions where there is little drug pressure (61).
An alternative—or additional—method of spread involves passage from one host generation to the next. For crop pathogens, this means transmission via the seed. This is particularly effective when both infected seeds and young plants appear healthy. Infected seeds allow the pathogen to use the host’s own dispersal mechanisms while ensuring a susceptible host on arrival. International seed trade amplifies the effectiveness of this strategy (see “Anthropogenic Factors Promoting the Emergence of New Invasive Fungal Diseases”). An example of this vertical transmission is R. collo-cygni, which as we saw above, lives endophytically, causing no symptoms, until the plant begins to flower (40). There is a trade-off in virulence between intra- and interhost replication (78); endophytic growth is a solution which precludes any risk of host death before transmission.
Another feature of fungal pathogens which promotes their invasiveness is the ability to establish a year-round infection in a new area, often taking advantage of volunteer or wild plants. Such continuous infection means that there is a continuous inoculum source in the new location, even if the crop is not grown year-round. Alternatively, establishment of a year-round infection on crop plants in an area where they are grown year-round can provide year-round inoculum for invasion of nearby areas where the same crop is seasonal. Again, this ability is shared by many of the fungi discussed in “Current Examples of Emerging Infectious Fungal Diseases.” There are a number of ways fungi achieve this. Broad host range, as seen in M. phaseolina and Fusarium spp., is one strategy; generalist pathogens dispersing to a new area have a range of potential hosts available and are thus likely to be able to become established. The same effect can be achieved, however, by more specialist fungi with narrower ranges of alternative hosts if those hosts are themselves widespread. The fungi R. collo-cygni and P. graminis exemplify this strategy, because they infect a number of wild grass genera whose species are found throughout temperate regions (1, 40, 41). Similarly, P. pachyrhizi, while primarily studied as a soybean pathogen, is able to infect many legumes, including the invasive weed kudzu, whose large, widespread populations greatly increase host availability (23, 24). Alternatively, fungi may form resting spores (such as the microsclerotia formed by M. phaseolina), which can survive cold winters or persist through hot or dry summer periods. Such a strategy increases the chances of infecting any hosts that become available in a new environment as the seasons change.
Anthropogenic Factors Promoting the Emergence of New Invasive Fungal Diseases
In addition to the factors outlined above, some human activities affect the likelihood of new invasive fungal diseases emerging. The most important of these is trade and transport (11), followed closely by agricultural practices that lead to wide distribution and high-density cultivation of genetically uniform plants or livestock, susceptible to the same diseases (66).
In “Changing Landscapes: Chestnut Blight and Dutch Elm Disease,” we saw that the international trade in chestnut and elm continued during both the chestnut blight and Dutch elm disease epidemics. In agriculture and sylviculture, trade in products such as food or wood is commonplace, as is the worldwide trade in germplasm—including seeds, rhizomes, or cuttings of elite crop varieties. In “Natural Factors Promoting the Emergence of New Invasive Fungal Diseases,” we saw that seed-borne fungi can exploit both natural and anthropogenic seed dispersal mechanisms. Live plants and animals are also traded and may carry a range of pests and pathogens undetected into new areas. Such trade is common in asexually propagated crops such as banana (see section on Panama disease). Trade in live plants is implicated in the spread of the ash die-back fungus (Hymenoscyphus fraxineus; see “Threats to Natural Ecosystems from Emerging Fungi”) to the United Kingdom (89) and white pine blister rust fungus (Cronartium ribicola) to the United States, where the host, Pinus albicaulis, is now threatened, and Canada, where it is endangered (90, 91). Similarly, it is clear now that the international trade in Asian salamander species is enormous, with, for instance, over 2.3 million individuals of Cynops orientalis being imported into the United States from 2001 to 2009 (43). There has been much focus on how the transcontinental vectoring of Bsal in amphibians may have occurred. Molecular surveillance data have demonstrated that Bsal is relatively widespread in European captive collections (45) and is associated with Asian species as well as with outbreaks of disease in captive European species of caudates. A clear case has therefore been made that the international movement of traded species of amphibians is a key mechanism that is contributing to the spread of Bsal and its invasion of naïve, disease-free ecosystems. These potent arguments have recently resulted in a ban on the importation of 201 salamander species into the United States (92).
When potential hosts are introduced to a new area, there are a number of associated risks. First, the imported organisms may be infected with a disease not occurring in the new location. Such introductions of deleterious organisms are sometimes termed “pathogen pollution” (11). Second, the introduced organism can carry a pathogen or commensal organism that has previously caused little or no disease but which may, in the new location, acquire new virulence or pathogenicity genes via HGT or hybridization, creating a new pathogen. Third, such fungi might possess or obtain genes for virulence in hosts endemic to the new area. These risks are compounded by the fact that closely related species of fungi which have diverged allopatrically (while occupying separate, nonoverlapping geographic areas) will not have experienced selection pressure toward perfect reproductive isolation. Thus, reproductive barriers may be leaky, allowing the formation of “hopeful monsters” (93)—hybrids which, while usually having lower fitness than the parents, occasionally bring together pathogenicity and virulence determinants in devastating new combinations. Backcrossing to either parent can then fix these genes in otherwise well-adapted local pathogen populations. Fourth, climatic factors might favor an imported pathogen or potential pathogen such that it is no longer limited by hot summers, cold winters, or drought. Finally, biotic factors may favor an introduced organism, because by invading new territories it may leave behind competitors, predators, or parasites, thus undergoing “enemy release.” This allows the invader population to explode in numbers, further increasing the likelihood of favorable HGT, hybridization, recombination, or mutation events. Anthropogenic transport and mixing of pathogens can thus have serious consequences (11, 69, 87, 89–94).
Another anthropogenic factor in the emergence of fungal pathogens, particularly those of crop plants and, to a lesser extent, of livestock, is the unique cradle for pathogen evolution provided by agriculture. Agro-ecosystems are characterized by huge populations of genetically uniform individuals. A few varieties of one crop may constitute almost all of the host variability throughout particular climatic regions, or even worldwide (11, 78, 95). In contrast to the patchy distribution of hosts in natural ecosystems, large cropping areas mean that pathogen populations rarely experience bottlenecks or loss of diversity (87). Pathogen populations are enormous, and coinfection by multiple strains is the norm, increasing the rates of mutation, HGT, and hybridization (67). Similarly, intensive farming practices, with multiple crops per year, reduce the temporal diversity of host availability that is a feature of natural ecosystems. Moreover, fungal pathogens have no need for resting spores if the host is cultivated year-round and instead undergo extra cycles of asexual propagation and clonal spread, bolstering population sizes still further (76, 95). For example, the crop-destroying rice blast fungus M. oryzae is almost entirely clonal, while its wild relatives rely on sexual reproduction to produce the resting spores needed for persistence in natural ecosystems (11, 69). Similar processes are argued to have led to the extremely rapid emergence of P. destructans and WNS across North America, because bats disperse asexual conidia along their migratory routes (96).
Examples of the importance of agro-ecosystem uniformity in shaping the evolution of agricultural pathogens can be seen today in many economically important fungal crop pathogens. Z. tritici, for example, diverged from its nearest relatives around the time that wheat was domesticated and has its center of diversity in the same location—the Fertile Crescent—as wheat itself (78). The wild relatives of Z. tritici have a wider host range, which was lost in the genetic bottleneck that followed the initial jump by the ancestors of Z. tritici onto wheat. However, as wheat cultivation spread around the world, the pathogen went global too; there are signatures in its genome indicating rapid population growth and diversification (78, 95).
Similarly, many of the pathogens discussed in “Current Examples of Emerging Infectious Fungal Diseases” benefit from these features of agro-ecosystems. As seen in “Natural Factors Promoting the Emergence of New Invasive Fungal Diseases,” these invasive, pathogenic fungi share the ability to establish year-round populations through infection of a range of hosts including wild species (21, 40, 81, 91, 97, 98), allowing spread through agricultural and unmanaged ecosystems. Such pathogens often have long-distance dispersal ability, bolstering genetic diversity during establishment (12, 21, 66, 87). Further, many are accidentally propagated on volunteer host plants, which appear between growing seasons, along roadsides or in other anthropogenic environments (1, 23, 24, 41, 66) (see Fig. 3).
Apart from crop plants, there is only one organism whose pathogens show such rapid evolution and propensity for host shifts, host jumps, and HGT events. This is because the density and uniformity of the agro-ecosystem, and high levels of pathogen pollution within it, are matched by only one species: the species supported by the agro-ecosystems—ourselves. With a huge, dense population, Homo sapiens has provided its pathogens with a similar evolutionary cradle. This means that lessons may be learned in interdisciplinary research into the evolution and epidemiology of crop and human pathogens (78).
A final anthropogenic factor influencing the emergence of invasive pathogens is climate change. This has both direct and indirect effects on the spread of fungi. One study found that pests and pathogens of crops are moving poleward as global temperatures increase, with fungi leading the charge (99, 100). Limitations imposed by cold winter temperatures, both on fungi themselves and on their hosts, are gradually lifting at higher latitudes, while perhaps, higher summer temperatures impose restrictions closer to the equator. This process is exemplified by the increasing severity of outbreaks of chytridiomycosis across amphibian populations in the high Pyrenees as the onset of spring arrives earlier in the year (101).
In addition to these direct effects, climate change influences agriculture, with crops increasingly grown outside of their natural ranges. With the spread of crop species comes pathogen pollution and all its attendant risks, including enemy release for pathogens which are noncompetitive in the hosts’ home range. In addition, hosts will be exposed to pathogens native to the new location, which may adapt to them and eventually then spread into their original range (11, 78).
Panama disease as a case study concerning anthropogenic disease spread
Panama disease, also called Fusarium wilt, is an aggressive disease of banana first recorded in Australia in 1876. By 1950, it had spread throughout banana growing regions worldwide (102). Its spread was facilitated by the global monoculture of Gros Michel banana, the only variety grown for export at the time (102). Banana, a seedless triploid (three copies of every genome) resulting from hybridization of two Musa species, is propagated via rhizomes (103). Like infected seed (see “Anthropogenic Factors Promoting the Emergence of New Invasive Fungal Diseases”), symptomless but infected rhizomes can be transported around the world, carrying the disease with them (102). The disease can be soilborne, with microconidia entering roots and traveling through the vascular system, followed by further microconidation, allowing spread throughout the plant (104). Chlamydospores (thick-walled spores) survive in soil for up to 30 years. Gros Michel banana was first infected with this disease in Panama, providing its common name; the causal agent was identified in Cuba in 1910 and named F. oxysporum f. sp. cubense (Foc) (103). F. oxysporum causes vascular wilt disease in a wide range of plants, although individual formae speciales often show a restricted host range (see “Natural Factors Promoting the Emergence of New Invasive Fungal Diseases”). Within Foc are four races. Race 1 caused exceptional losses in Gros Michel plantations. Race 2 infects various varieties of cooking banana, whose fruits are not grown for export, and race 3 infects a different host species altogether (104). Abandoning Gros Michel, commercial growers turned to a new variety, Cavendish, which existed as a curiosity in collections in the United Kingdom and Honduras. Cavendish has appropriate qualities for export and resists Foc race 1 (103). Thus, trade in the world’s favorite fruit was saved—for a time. In the late 1960s, reports appeared of a similar wilt disease infecting Cavendish banana, which in 1994, was confirmed as a new race of Foc (104). Race 4 also infects plantains, so it threatens two important crops. Initially, Foc Race 4 was confined to the Eastern hemisphere; its discovery in Jordan in 2014 (104) means that it is now likely to spread through Africa and, in time, will probably reach the banana plantations of the Americas. Treatment is problematic, because Foc is highly resistant to fungicide. A new, resistant banana variety will be needed if drastic losses in production are to be prevented.
THREATS TO NATURAL ECOSYSTEMS FROM EMERGING FUNGI
We have already seen that fungal diseases can have huge impacts on tree species, for example, in Dutch elm disease and chestnut blight. “Changing Landscapes: Chestnut Blight and Dutch Elm Disease” described the effect of the devastation of the Appalachian chestnut on biodiversity, as well as changes to landscapes and the resulting cultural impact. A similar example, which has received extensive attention, is the case of ash dieback in the United Kingdom and Europe. Ash dieback is a fatal disease of ash (Fraxinus excelsior). It is caused by H. fraxineus (previously Chalara fraxinea), an ascomycete native to Asia which is thought to have spread into and around Europe on infected but asymptomatic nursery stock (76, 105) (Fig. 6). H. fraxineus has devastated ash woodland throughout Europe, with up to 100% of trees affected and with ash now considered a threatened species in several countries (106). This disease attracted public and scientific interest in the United Kingdom, where ash is common in woodlands and hedgerows. Studies indicate that around 45 species are dependent on ash, among a total of around 450 species associated with this tree (107). Ash is a pioneer of and keystone species in European riparian forests, as well as being a hedgerow plant and an urban amenity species (106).
FIGURE 6.

Hymenoscyphus fraxineus on ash. Left, hyphae (green; stained with fluorescein isothiocyanate-labeled wheat-germ agglutinin) of H. fraxineus, visualized by confocal microscopy, growing over and out of the vascular tissue of an ash leaf (red; stained with propidium iodide). Right, ash die-back symptoms on an ash seedling under laboratory conditions. (Photos: Helen Fones.)
H. fraxineus displays very high levels of outcrossing, resulting in rapid gene flow between populations (108–110). This means that even among emerging fungal diseases, H. fraxineus has high evolutionary potential. New genotypes with increased virulence or pathogenicity, whether arising by mutation, introgression, or recombination, quickly spread throughout European populations of this fungus. H. fraxineus shows much greater genetic diversity in Asia than in Europe, and Asian strains show higher virulence and pathogenicity to ash (111). With global trade and transport, it is only a matter of time until this destructive potential reaches Europe. Further, there are known to be various sympatric (species inhabiting the same geographic region) Hymenoscyphus species (112, 113) with which hybridization may ensue. As well as possible evolution of increased virulence, the possibility of host shifts onto additional Fraxinus or Oleaceae species, or even host jumps onto other trees entirely, is therefore high for this pathogen (113, 114, 128).
Another keystone species affected by an emerging fungal pathogen is the Hawaiian ‘Ōhi’a tree (Metrosideros polymorpha). This tree is threatened by the new disease rapid ‘Ōhi’a death, caused by the fungus Ceratocystis fimbriata. In this disease, the fungus infects the sapwood, causing death within weeks. Mortality rates are high: about 50% of infected trees die. Transmission may be by infected soil, cuttings, insect vectors, or windblown insect frass (powder produced by boring insects) (115, 116). The disease is of particular concern, because ‘Ōhi’a is a dominant species in the Hawaiian rainforest, determining succession and canopy structure and providing a habitat for many endangered species (115). Moreover, it is a key pioneer in the colonization of lava flows because it has a root system that is particularly able to penetrate cracks and is adapted to withstand toxic volcanic gases (117, 118).
Another ecosystem service provided by large tree species is carbon sequestration. American chestnut was an efficient carbon sink in its native range (119), capable of absorbing up to 35 metric megatons of CO2 had the trees not been destroyed by C. parasitica. Similarly, Dutch elm disease is calculated to have prevented the absorption of a metric megaton of CO2. These values do not include the vast potential carbon release from decaying wood of the dead trees (93).
Vertebrate biodiversity is an important component of food webs, and the losses of neotropical amphibian species have caused demonstrable changes in ecosystem functioning. This is compounded by the fact that amphibians are both important grazers in their aquatic tadpole forms as well as insect-predators in their terrestrial adult morphs. In Central America, chytridiomycosis has changed patterns of amphibian biodiversity. Here, community composition across the region was severely disrupted by epidemics at multiple sites, and many of the unique species of these communities were eliminated, with disproportionate effects on endemic species. This resulted in a homogenization of the regional fauna and ecological homogenization of reproductive mode and habitat (120). Alongside the losses of biodiversity, economic consequences are starting to be attributed to emerging fungal diseases of vertebrates. While correctly attributing the economic cost of ecosystem functions is still in its infancy, valuations have estimated the losses to U.S. agriculture that are the result of declines in bat populations at more than $3.7 billion per year (121). Indeed, there are many ecosystem services which are hard to quantify but may be threatened or diminished when species are decimated by emerging fungal diseases. These include prevention of flooding, provision of clean air and water, pollination of crops and other plants, as well as the cultural, religious, and recreational significance of host species to humans. Where biodiversity is threatened, we are also at risk of losing potential pharmaceuticals and various other genetic resources which could be used to improve domestic species.
THREATS TO AGRICULTURE FROM EMERGING FUNGI
As well as their devastating effects on natural ecosystems and wild species, emerging fungal pathogens pose serious threats to agricultural production and food security. This is evident from “Current Examples of Emerging Infectious Fungal Diseases,” where we saw the huge losses that can result from particular fungal pathogens. A recent, detailed study estimated that in the three major wheat growing countries of the European Union (United Kingdom, France, and Germany) losses caused by the fungus Z. tritici (Septoria leaf blotch of wheat) surpass €1,400 million yearly. This is despite deployment of the most resistant wheat varieties and application of fungicides at a further cost of over €900 million per growing season (70). These losses are due to just one disease, in conditions optimized for its control. It is clear that losses worldwide to fungal pathogens impact significantly on productivity and costs. Affordable food security will therefore depend on our ability to mitigate this threat.
Unfortunately, that ability is constantly challenged by the pathogens themselves. As we have discussed, emerging fungal pathogens are exceptionally adaptable. As well as increasing the risk that they will spread to new hosts and new places, this adaptability allows pathogens to repeatedly break the resistance provided by elite crop varieties. At least one wheat variety previously resistant to Z. tritici has failed due to new virulence acquisition by the pathogen (122); resistance breeding is complicated by linkage between Z. tritici resistance and low crop yield (123).
Not only do fungi have the ability to out-evolve and overcome the defenses of resistant hosts, but they can evolve or acquire genes for fungicide resistance, as we saw in “Azole-Resistant Aspergillosis.” As such, these strains overcome our main mechanisms for protecting crops from disease and thus ensuring food security. The very traits that promote their becoming invasive pathogens also increase the risk of their developing fungicide resistance. For example, few curative fungicides exist for Z. tritici in wheat; there is reliance on azoles and strobilurins, to which the fungus is beginning to show resistance (70). Indeed, azole-resistant fungi are becoming increasingly common, a problem partly attributed to the persistence of azoles in soil (124). This increases incidental contact with fungi, creating selection pressure for resistance, which may be conferred by a single gene such as Aspergillus nidulans atrB, encoding a multidrug transporter conferring resistance to azoles and other fungicides (125). This is concerning not just because of the implications for agriculture, but also because many of these fungi or their close relatives cause mycoses in humans. Examples include A. fumigatus, the cause of invasive aspergillosis, a potentially fatal disease generally treated with triazole fungicides (see “Azole-Resistant Aspergillosis”). A single amino acid change and a 34-bp tandem repeat in the promoter region of the Cyp51A gene encoding the target protein are sufficient to provide resistance to various frontline triazoles that are used to treat severe human infection (61, 124).
Similarly, the generalist fungus M. phaseolina (see “Current Examples of Emerging Infectious Fungal Diseases”), which attacks immunocompromised patients, is treated with azole fungicides, but azole-resistant fungal strains are appearing (33). Given the environmental persistence of azoles and the high evolutionary potential of crossover pathogens such as M. phaseolina (126), it may be necessary to curb the agricultural use of azoles to maintain their medical efficacy, as discussed in “Azole-Resistant Aspergillosis.” However, there are not yet effective replacements for azoles in agriculture.
CONCLUSIONS
Fungal infections have wreaked havoc with our harvests through the centuries. Fungi continue to cause widespread damage in crops. Recently, however, we have witnessed their growing impact on ecosystem resilience and on human health: there has been a dramatic decline in populations of amphibians and bats and an increased incidence of fungal infections in humans. Human activity and global climate change are intensifying fungal dispersal by modifying natural ecosystems and creating new opportunities for evolution. We must take steps to mitigate the risk of these infectious diseases spreading globally, for without action, fungal infections will lead to reduced crop yields, damaging food security. Increased agricultural reliance on fungicides will promote the evolution of fungicide resistance, which trait may then spread into pathogens of wild species and of ourselves. Biodiversity and human health will then suffer alongside agriculture, increasing the risk to ecosystems and ecosystem services at a time when pressures to exploit land to feed humans will be heightened. We must therefore ensure that research on the various host, pathogen, and environment interactions is fueled by adequate research funding and that disease detection is improved and then used to impose stricter global biosecurity protocols in the international trade of plant and animal species.
Contributor Information
Helen N. Fones, Department of Biosciences, University of Exeter, Exeter, EX4 4QD, United Kingdom
Matthew C. Fisher, Department of Infectious Disease Epidemiology, School of Public Health, Imperial College, London, St Mary’s Hospital, London W2 1PG, United Kingdom
Sarah J. Gurr, Department of Biosciences, University of Exeter, Exeter, EX4 4QD, United Kingdom University of Utrecht, 3584 CH, Utrecht, The Netherlands; Rothamsted Research, North Wyke, Okehampton, EX20 2SB, United Kingdom.
Joseph Heitman, Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC 27710.
Barbara J. Howlett, School of Biosciences, The University of Melbourne, Victoria, NSW 3010, Australia
REFERENCES
- 1.Leonard KJ, Szabo LJ. 2005. Stem rust of small grains and grasses caused by Puccinia graminis. Mol Plant Pathol 6:99–111. 10.1111/j.1364-3703.2005.00273.x [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Chester KS. 1946. The Nature and Prevention of the Cereal Rusts as Exemplified in the Leaf Rust of Wheat. Chronica Botanica Company, Waltham, MA. [PubMed] [Google Scholar]
- 3.Tudzynski P, Scheffer J. 2004. Claviceps purpurea: molecular aspects of a unique pathogenic lifestyle. Mol Plant Pathol 5:377–388. 10.1111/j.1364-3703.2004.00237.x [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Woolf A. 2000. Witchcraft or mycotoxin? The Salem witch trials. J Toxicol Clin Toxicol 38:457–460. 10.1081/CLT-100100958 [DOI] [PubMed] [Google Scholar]
- 5.Rudge FM. 1907. Orders of St. Anthony. The Catholic Encyclopedia. Robert Appleton Company, New York, NY. http://www.newadvent.org/cathen/01555a.htm. [Google Scholar]
- 6.Fuller J. 1969. The Day of St Anthony’s Fire. Hutchison, London, United Kingdom. [Google Scholar]
- 7.Urga K, Debella A, Agata N, Bayu A, Zewdie W. 2002. Laboratory studies on the outbreak of gangrenous ergotism associated with consumption of contaminated barley in Arsi, Ethiopia. Ethiop J Health Dev 16:317–323. 10.4314/ejhd.v16i3.9800 [DOI] [Google Scholar]
- 8.Matossian MK. 1982. Ergot and the Salem witchcraft affair. Am Sci 70:355–357. [PubMed] [PubMed] [Google Scholar]
- 9.Caporael LR. 1976. Ergotism: the satan loosed in Salem? Science 192:21–26. 10.1126/science.769159 [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Alm T. 2003. The witch trials of Finnmark, Northern Norway, during the 17th century: evidence for ergotism as a contributing factor. Econ Bot 57:403–416. 10.1663/0013-0001(2003)057[0403:TWTOFN]2.0.CO;2 [DOI] [Google Scholar]
- 11.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. 2004. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 19:535–544. 10.1016/j.tree.2004.07.021 [DOI] [PubMed] [Google Scholar]
- 12.Isard SA, Gage SH, Comtois P, Russo JM. 2005. Principles of the atmospheric pathway for invasive species applied to soybean rust. Biosci 55:851–861. 10.1641/0006-3568(2005)055[0851:POTAPF]2.0.CO;2 [DOI] [Google Scholar]
- 13.Ellison AM, Bank MS, Clinton BD, Colburn EA, Elliott K, Ford CR, Foster DR, Kloeppel BD, Knoepp JD, Lovett GM, Mohan J, Orwig DA, Rodenhouse NL, Sobczak WV, Stinson KA, Stone JK, Swan CM, Thompson J, Von Holle B, Webster JR. 2005. Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front Ecol Environ 3:479–486. 10.1890/1540-9295(2005)003[0479:LOFSCF]2.0.CO;2 [DOI] [Google Scholar]
- 14.Orwig DA. 2002. Ecosystem to regional impacts of introduced pests and pathogens: historical context, questions and issues. J Biogeogr 29:1471–1474. 10.1046/j.1365-2699.2002.00787.x [DOI] [Google Scholar]
- 15.Smith KG, Rodewald PG, Withgott J. 2000. Red-headed woodpecker (Melanerpes erythrocephalus), p 518. In Poole A, Gill F (ed), The Birds of North America. Academy of Natural Sciences, Philadelphia, PA. [Google Scholar]
- 16.Opler PA. 1978. Insects of American chestnut: possible importance and conservation concern. The American Chestnut Symposium, West Virginia University Press, Morgantown, WV. [Google Scholar]
- 17.Dutech C, Barrès B, Bridier J, Robin C, Milgroom MG, Ravigné V. 2012. The chestnut blight fungus world tour: successive introduction events from diverse origins in an invasive plant fungal pathogen. Mol Ecol 21:3931–3946. 10.1111/j.1365-294X.2012.05575.x [DOI] [PubMed] [Google Scholar]
- 18.Gibbs J, Brasier CM, Webber J. 1994. Dutch Elm Disease in Britain. Great Britain, Forestry Authority, Research Division, Farnham, United Kingdom. [Google Scholar]
- 19.Brasier CM. 1991. Ophiostoma novo-ulmi sp. nov., causative agent of current Dutch elm disease pandemics. Mycopathologia 115:151–161. 10.1007/BF00462219 [DOI] [Google Scholar]
- 20.Brasier CM, Kirk SA. 2010. Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol 59:186–199. 10.1111/j.1365-3059.2009.02157.x [DOI] [Google Scholar]
- 21.Sutrave S, Scoglio C, Isard SA, Hutchinson JM, Garrett KA. 2012. Identifying highly connected counties compensates for resource limitations when evaluating national spread of an invasive pathogen. PLoS One 7:e37793. 10.1371/journal.pone.0037793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isard SA, Gage SH, Comtois P, Russo JM. 2005. Principles of the atmospheric pathway for invasive species applied to soybean rust. Biosci 55:851–861. 10.1641/0006-3568(2005)055[0851:POTAPF]2.0.CO;2 [DOI] [Google Scholar]
- 23.Yorinori JT, Paiva WM, Frederick RD, Costamilan LM, Bertagnolli PF, Hartman GE, Godoy CV, Nunes J Jr. 2005. Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Dis 89:675–677. 10.1094/PD-89-0675 [DOI] [PubMed] [Google Scholar]
- 24.Pivonia S, Yang XB. 2004. Assessment of the potential year-round establishment of soybean rust throughout the world. Plant Dis 88:523–529. 10.1094/PDIS.2004.88.5.523 [DOI] [PubMed] [Google Scholar]
- 25.Del Ponte EM, Godoy CV, Li X, Yang XB. 2006. Predicting severity of Asian soybean rust epidemics with empirical rainfall models. Phytopathology 96:797–803. 10.1094/PHYTO-96-0797 [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Zadoks JC, van den Bosch F. 1994. On the spread of plant disease: a theory on foci. Annu Rev Phytopathol 32:503–521. 10.1146/annurev.py.32.090194.002443 [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Peterson PD. 2013. “The Barberry or Bread”: The Public Campaign to Eradicate Common Barberry in the United States in the Early 20th Century. APS Features. 10.1094/APSFeature-2013-08. 10.1094/APSFeature-2013-08. [DOI] [Google Scholar]
- 28.Visser B, Herselman L, Park RF, Karaoglu H, Bender CM, Pretorius ZA. 2011. Characterization of two new Puccinia graminis f. sp. tritici races within the Ug99 lineage in South Africa. Euphytica 179:119–127. 10.1007/s10681-010-0269-x [DOI] [Google Scholar]
- 29.Peterson PD, Leonard KJ, Miller JD, Laudon RJ, Sutton TB. 2005. Prevalence and distribution of common barberry, the alternate host of Puccinia graminis, in Minnesota. Plant Dis 89:159–163. 10.1094/PD-89-0159 [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan S, Singh H, Joshi LM, Saari EE. 1976. Meteorological conditions associated with long distance dissemination and deposition of Puccinia graminis tritici uredospores in India. Phytopathology 66:198–203. 10.1094/Phyto-66-198 [DOI] [Google Scholar]
- 31.Sibikeev SN, Markelova TS, Baukenova EA, Druzhin AE. 2016. Likely threat of the spread of race UG99 of Puccinia graminis f. sp. tritici on wheat in southeastern Russia. Russ Agric Sci 42:145–148. 10.3103/S1068367416020154 [DOI] [Google Scholar]
- 32.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V. 2011. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481. 10.1146/annurev-phyto-072910-095423 [DOI] [PubMed] [Google Scholar]
- 33.Sarr MP, Ndiaye M, Groenewald JZ, Crous PW. 2014. Genetic diversity in Macrophomina phaseolina, the causal agent of charcoal rot. Phytopathol Mediterr 53:163–173. [Google Scholar]
- 34.Arora P, Dilbaghi N, Chaudhury A. 2012. Opportunistic invasive fungal pathogen Macrophomina phaseolina prognosis from immunocompromised humans to potential mitogenic RBL with an exceptional and novel antitumor and cytotoxic effect. Eur J Clin Microbiol Infect Dis 31:101–107. 10.1007/s10096-011-1275-1 [DOI] [PubMed] [Google Scholar]
- 35.Islam MS, Haque MS, Islam MM, Emdad EM, Halim A, Hossen QM, Hossain MZ, Ahmed B, Rahim S, Rahman MS, Alam MM, Hou S, Wan X, Saito JA, Alam M. 2012. Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13:493. 10.1186/1471-2164-13-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur S, Dhillon GS, Brar SK, Vallad GE, Chand R, Chauhan VB. 2012. Emerging phytopathogen Macrophomina phaseolina: biology, economic importance and current diagnostic trends. Crit Rev Microbiol 38:136–151. 10.3109/1040841X.2011.640977 [DOI] [PubMed] [Google Scholar]
- 37.Su G, Suh SO, Schneider RW, Russin JS. 2001. Host specialization in the charcoal rot fungus, Macrophomina phaseolina. Phytopathology 91:120–126. 10.1094/PHYTO.2001.91.2.120 [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Walters DR, Havis ND, Oxley SJ. 2008. Ramularia collo-cygni: the biology of an emerging pathogen of barley. FEMS Microbiol Lett 279:1–7. 10.1111/j.1574-6968.2007.00986.x [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Fountaine JM, Fraaije BA. 2009. Development of QoI resistant alleles in populations of Ramularia collo-cygni. Asp Appl Biol 92:123–126. [Google Scholar]
- 40.Havis ND, Nyman M, Oxley SJ. 2014. Evidence for seed transmission and symptomless growth of Ramularia collo-cygni in barley (Hordeum vulgare). Plant Pathol 63:929–936. 10.1111/ppa.12162 [DOI] [Google Scholar]
- 41.Havis ND, Brown JK, Clemente G, Frei P, Jedryczka M, Kaczmarek J, Kaczmarek M, Matusinsky P, McGrann GR, Pereyra S, Piotrowska M, Sghyer H, Tellier A, Hess M. 2015. Ramularia collo-cygni: an emerging pathogen of barley crops. Phytopathology 105:895–904. 10.1094/PHYTO-11-14-0337-FI [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Fisher MC, Garner TWJ, Walker SF. 2009. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol 63:291–310. 10.1146/annurev.micro.091208.073435 [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Garner TWJ, Schmidt BR, Martel A, Pasmans F, Muths E, Cunningham AC, Weldon C, Fisher MC, Bosch J. 2016. Mitigating amphibian chytridiomycoses in nature. Philos Trans R Soc B 371: 20160207. 10.1098/rstb.2016.0207. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson DH, Aanensen DM, Ronnenberg KL, Powell CI, Walker SF, Bielby J, Garner TW, Weaver G, Fisher MC, Bd Mapping Group. 2013. Mapping the global emergence of Batrachochytrium dendrobatidis, the amphibian chytrid fungus. PLoS One 8:e56802. 10.1371/journal.pone.0056802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martel A, Blooi M, Adriaensen C, Van Rooij P, Beukema W, Fisher MC, Farrer RA, Schmidt BR, Tobler U, Goka K, Lips KR, Muletz C, Zamudio KR, Bosch J, Lötters S, Wombwell E, Garner TWJ, Cunningham AA, Spitzen-van der Sluijs A, Salvidio S, Ducatelle R, Nishikawa K, Nguyen TT, Kolby JE, Van Bocxlaer I, Bossuyt F, Pasmans F. 2014. Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346:630–631. 10.1126/science.1258268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, Clare F, Bosch J, Cunningham AA, Weldon C, du Preez LH, Anderson L, Pond SL, Shahar-Golan R, Henk DA, Fisher MC. 2011. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci USA 108:18732–18736. 10.1073/pnas.1111915108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gascon C, Collins JP, Moore RD, Church DR, McKay JE, Mendelson JR III. 2007. Amphibian conservation action plan. IUCN/SSC Amphibian Specialist Group, The World Conservation Union (IUCN), Gland, Switzerland.
- 48.Lips K. 2016. Overview of chytrid emergence and impacts on amphibians. Philos Trans R Soc B 371:20150465. 10.1098/rstb.2015.0465. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langwig KE, Voyles J, Wilber MQ, Frick WF, Murray KA, Bolker BM, Collins JP, Cheng TL, Fisher MC, Hoyt JR, Lindner DL, McCallum HI, Puschendorf R, Rosenblum EB, Toothman M, Willis CKR, Briggs CJ, Kilpatrick AM. 2015. Context-dependent conservation responses to emerging wildlife diseases. Front Ecol Environ 13:195–202. 10.1890/140241 [DOI] [Google Scholar]
- 50.Bosch J, Sanchez-Tomé E, Fernández-Loras A, Oliver JA, Fisher MC, Garner TWJ. 2015. Successful elimination of a lethal wildlife infectious disease in nature. Biol Lett 11:20150874. 10.1098/rsbl.2015.0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Slocombe R, Ragan MA, Hyatt AD, McDonald KR, Hines HB, Lips KR, Marantelli G, Parkes H. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA 95:9031–9036. 10.1073/pnas.95.15.9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, Ballmann AE, Coleman JTH, Redell DN, Reeder DM, Blehert DS. 2011. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480:376–378. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- 53.Langwig KE, Frick WF, Hoyt JR, Pariseb KL, Drees KP, Kunz TH, Foster JT, Kilpatrick AM. 2016. Drivers of variation in species impacts for a multi-host fungal disease of bats. Philos Trans R Soc B 371:20150456. 10.1098/rstb.2015.0456. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, Butchkoski CM, Kunz TH. 2010. An emerging disease causes regional population collapse of a common North American bat species. Science 329:679–682. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- 55.Rajkumar SS, Li X, Rudd RJ, Okoniewski JC, Xu J, Chaturvedi S, Chaturvedi V. 2011. Clonal genotype of Geomyces destructans among bats with white nose syndrome, New York, USA. Emerg Infect Dis 17:1273–1276. 10.3201/eid1707.102056 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.May RC, Stone NRH, Wiesner DL, Bicanic T, Nielsen K. 2016. Cryptococcus: from environmental saprophyte to global pathogen. Nat Rev Microbiol 14:106–117. 10.1038/nrmicro.2015.6 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 58.Pyrgos V, Seitz AE, Steiner CA, Prevots DR, Williamson PR. 2013. Epidemiology of cryptococcal meningitis in the US: 1997–2009. PLoS One 8:e56269. 10.1371/journal.pone.0056269 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78:16–48. 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 60.Byrnes EJ III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6:e1000850. 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meis J, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc B 371:20150460. 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC. 2015. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. MBio 6:e00536. 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Linden JW, Snelders E, Kampinga GA, Rijnders BJ, Mattsson E, Debets-Ossenkopp YJ, Kuijper EJ, Van Tiel FH, Melchers WJ, Verweij PE. 2011. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg Infect Dis 17:1846–1854. 10.3201/eid1710.110226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Netea MG, Latz E, Mills KH, O’Neill LA. 2015. Innate immune memory: a paradigm shift in understanding host defense. Nat Immunol 16:675–679. 10.1038/ni.3178 [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Misra BB, Chaturvedi R. 2015. When plants brace for the emerging pathogens. Physiol Mol Plant Pathol 92:181–185. 10.1016/j.pmpp.2015.03.004 [DOI] [Google Scholar]
- 66.Piotrowska MJ, Ennos RA, Fountaine JM, Burnett FJ, Kaczmarek M, Hoebe PN. 2016. Development and use of microsatellite markers to study diversity, reproduction and population genetic structure of the cereal pathogen Ramularia collo-cygni. Fungal Genet Biol 87:64–71. 10.1016/j.fgb.2016.01.007 [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Ahmed S, de Labrouhe DT, Delmotte F. 2012. Emerging virulence arising from hybridisation facilitated by multiple introductions of the sunflower downy mildew pathogen Plasmopara halstedii. Fungal Genet Biol 49:847–855. 10.1016/j.fgb.2012.06.012 [DOI] [PubMed] [Google Scholar]
- 68.Stukenbrock EH. 2013. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol 199:895–907. 10.1111/nph.12374 [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Linde CC, Zhan J, McDonald BA. 2002. Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology 92:946–955. 10.1094/PHYTO.2002.92.9.946 [DOI] [PubMed] [Google Scholar]
- 70.Fones H, Gurr S. 2015. The impact of Septoria tritici Blotch disease on wheat: an EU perspective. Fungal Genet Biol 79:3–7. 10.1016/j.fgb.2015.04.004 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suffert F, Sache I, Lannou C. 2011. Early stages of Septoria tritici blotch epidemics of winter wheat: build-up, overseasoning, and release of primary inoculum. Plant Pathol 60:166–177. 10.1111/j.1365-3059.2010.02369.x [DOI] [Google Scholar]
- 72.Schotanus K, Soyer JL, Connolly LR, Grandaubert J, Happel P, Smith KM, Freitag M, Stukenbrock EH. 2015. Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin 8:41. 10.1186/s13072-015-0033-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Wang J, Schierup MH. 2011. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res 21:2157–2166. 10.1101/gr.118851.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Croll D, McDonald BA. 2012. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog 8:e1002608. 10.1371/journal.ppat.1002608 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhillon B, Gill N, Hamelin RC, Goodwin SB. 2014. The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola. BMC Genomics 15:1132. 10.1186/1471-2164-15-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, Taga M, White GJ, Zhou S, Schwartz DC, Freitag M, Ma LJ, Danchin EG, Henrissat B, Coutinho PM, Nelson DR, Straney D, Napoli CA, Barker BM, Gribskov M, Rep M, Kroken S, Molnár I, Rensing C, Kennell JC, Zamora J, Farman ML, Selker EU, Salamov A, Shapiro H, Pangilinan J, Lindquist E, Lamers C, Grigoriev IV, Geiser DM, Covert SF, Temporini E, Vanetten HD. 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5:e1000618. 10.1371/journal.pgen.1000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stukenbrock EH. 2013. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol 199:895–907. 10.1111/nph.12374 [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Stukenbrock EH, McDonald BA. 2008. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 46:75–100. 10.1146/annurev.phyto.010708.154114 [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10:417–430. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Sperschneider J, Gardiner DM, Thatcher LF, Lyons R, Singh KB, Manners JM, Taylor JM. 2015. Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol Evol 7:1613–1627. 10.1093/gbe/evv092 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong S, Raffaele S, Kamoun S. 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev 35:57–65. 10.1016/j.gde.2015.09.001 [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Ma LJ, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373. 10.1038/nature08850 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF Jr. 2007. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol 16:1701–1711. 10.1111/j.1365-294X.2007.03225.x [DOI] [PubMed] [Google Scholar]
- 85.Nunes MJL. 2011. Magnaporthe oryzae, the blast pathogen: current status and options for its control. Plant Sci Rev 264:233–240. [Google Scholar]
- 86.Malaker PK, Barma NC, Tiwari TP, Collis WJ, Duveiller E, Singh PK, Joshi AK, Singh RP, Braun HJ, Peterson GL, Pedley KF. 2016. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis 100:2330. 10.1094/PDIS-05-16-0666-PDN [DOI] [Google Scholar]
- 87.Brown JK, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537–541. 10.1126/science.1072678 [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Nagarajan S, Singh H, Joshi LM, Saari EE. 1976. Meteorological conditions associated with long distance dissemination and deposition of Puccinia graminis tritici uredospores in India. Phytopathology 66:198–203. 10.1094/Phyto-66-198 [DOI] [Google Scholar]
- 89.Heuch J. 2014. What lessons need to be learnt from the outbreak of Ash Dieback Disease, Chalara fraxinea in the United Kingdom? Arboricult J 36:32–44. 10.1080/03071375.2014.913361 [DOI] [Google Scholar]
- 90.Liebhold AM, Brockerhoff EG, Garrett LJ, Parke JL, Britton KO. 2012. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front Ecol Environ 10:135–143. 10.1890/110198 [DOI] [Google Scholar]
- 91.Roy BA, Alexander HM, Davidson J, Campbell FT, Burdon JJ, Sniezko R, Brasier C. 2014. Increasing forest loss worldwide from invasive pests requires new trade regulations. Front Ecol Environ 12:457–465. 10.1890/130240 [DOI] [Google Scholar]
- 92.U.S. Fish and Wildlife Service. Listing salamanders as injurious due to risk of salamander chytrid fungus. https://www.fws.gov/injuriouswildlife/salamanders.html.
- 93.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. 10.1038/nature10947 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, Chandelier A, Cech T, Chira D, Diamandis S, Gaitniekis T, Hantula J, Holdenrieder O, Jankovsky L, Jung T, Jurc D, Kirisits T, Kunca A, Lygis V, Malecka M, Marcais B, Schmitz S, Schumacher J, Solheim H, Solla A, Szabò I, Tsopelas P, Vannini A, Vettraino AM, Webber J, Woodward S, Stenlid J. 2013. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol 197:238–250. 10.1111/j.1469-8137.2012.04364.x [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Stukenbrock EH, Bataillon T. 2012. A population genomics perspective on the emergence and adaptation of new plant pathogens in agro-ecosystems. PLoS Pathog 8:e1002893. 10.1371/journal.ppat.1002893 [PubMed][CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langwig KE, Feng J, Parise KL, Kath J, Kirk D, Frick WF, Foster JT, Kilpatrick AM. 2015. Invasion dynamics of white-nose syndrome fungus, midwestern United States, 2012–2014. Emerg Infect Dis 21:1023–1026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20:238–244. 10.1016/j.tree.2005.02.009 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bartoli C, Lamichhane JR, Berge O, Guilbaud C, Varvaro L, Balestra GM, Vinatzer BA, Morris CE. 2015. A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: the example of kiwifruit canker. Mol Plant Pathol 16:137–149. 10.1111/mpp.12167 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bebber DP, Ramotowski MA, Gurr SJ. 2013. Crop pests and pathogens move polewards in a warming world. Nat Clim Chang 3:985–988. 10.1038/nclimate1990 [DOI] [Google Scholar]
- 100.Bebber DP, Holmes T, Gurr SJ. 2014. The global spread of crop pests and pathogens. Glob Ecol Biogeogr 23:1398–1407. 10.1111/geb.12214 [DOI] [Google Scholar]
- 101.Clare FC, Halder JB, Daniel O, Bielby J, Semenov MA, Jombart T, Loyau A, Schmeller DS, Cunningham AA, Rowcliffe M, Garner TWJ, Bosch J, Fisher MC. 2016. Climate forcing of an emerging pathogenic fungus across a montane multihost community. Philos Trans R Soc B 371:20150454. 10.1098/rstb.2015.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ploetz RC. 2000. Panama disease: a classic and destructive disease of banana. Plant Health Prog 10:1–7. [Google Scholar]
- 103.Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, Thomma BP, Ploetz RC, Kema GH. 2015. Worse comes to worst: bananas and Panama disease – when plant and pathogen clones meet. PLoS Pathog 11:e1005197. 10.1371/journal.ppat.1005197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.García-Bastidas F, Ordóñez N, Konkol J, Al-Qasim M, Naser Z, Abdelwali M, Salem N, Waalwijk C, Ploetz RC, Kema GH. 2016. First report of Fusarium oxysporum f. sp. cubense tropical race 4 associated with Panama disease of banana outside Southeast Asia. Plant Dis 98:694–694. [DOI] [PubMed] [Google Scholar]
- 105.Chavez VA, Parnell S, Bosch FVD. 2015. Designing strategies for epidemic control in a tree nursery: the case of ash dieback in the UK. Forests 6:4135–4145. 10.3390/f6114135 [DOI] [Google Scholar]
- 106.Pautasso M, Aas G, Queloz V, Holdenrieder O. 2013. European ash (Fraxinus excelsior) dieback: a conservation biology challenge. Biol Conserv 158:37–49. 10.1016/j.biocon.2012.08.026 [DOI] [Google Scholar]
- 107.Mitchell RJ, Pakeman RJ, Broome A, Beaton JK, Bellamy PE, Brooker RW, Ellis CJ, Hester AJ, Hodgetts NG, Iason GR, Littlewood NA, Pozsgai G, Ramsay S, Riach D, Stockan JA, Taylor AFS, Woodward S. 2016. How to replicate the functions and biodiversity of a threatened tree species? The case of Fraxinus excelsior in Britain. Ecosys 19:573. 10.1007/s10021-015-9953-y [DOI] [Google Scholar]
- 108.Gross A, Hosoya T, Queloz V. 2014. Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol Ecol 23:2943–2960. 10.1111/mec.12792 [DOI] [PubMed] [Google Scholar]
- 109.Kraj W, Zarek M, Kowalski T. 2012. Genetic variability of Chalara fraxinea, dieback cause of European ash (Fraxinus excelsior L.). Mycol Prog 11:37–45. 10.1007/s11557-010-0724-z [DOI] [Google Scholar]
- 110.Burokiene D, Prospero S, Jung E, Marciulyniene D, Moosbrugger K, Norkute G, Rigling D, Lygis V, Schoebel CN. 2015. Genetic population structure of the invasive ash dieback pathogen Hymenoscyphus fraxineus in its expanding range. Biol Invas 17:2743–2756. 10.1007/s10530-015-0911-6 [DOI] [Google Scholar]
- 111.Gross A, Sieber TN. 2016. Virulence of Hymenoscyphus albidus and native and introduced Hymenoscyphus fraxineus on Fraxinus excelsior and Fraxinus pennsylvanica. Plant Pathol 65:655–663. 10.1111/ppa.12450 [DOI] [Google Scholar]
- 112.Gross A, Han JG. 2015. Hymenoscyphus fraxineus and two new Hymenoscyphus species identified in Korea. Mycol Prog 14:19. 10.1007/s11557-015-1035-1 [DOI] [Google Scholar]
- 113.Gross A, Hosoya T, Zhao YJ, Baral HO. 2015. Hymenoscyphus linearis sp. nov: another close relative of the ash dieback pathogen H. fraxineus. Mycol Prog 14:20. 10.1007/s11557-015-1041-3 [DOI] [Google Scholar]
- 114.Carrari E, Capretti P, Santini A, Luchi N. 2015. Hymenoscyphus fraxineus mycelial growth on media containing leaf extracts of different Oleaceae. For Pathol 45:540–543. 10.1111/efp.12238 [DOI] [Google Scholar]
- 115.Friday JB, Keith L, Hughes F. 2015. Rapid ‘Ōhi’a death (ceratocystis wilt of ‘Ōhi’a). 1University of Hawaii College of Tropical Agriculture. http://www.ctahr.hawaii.edu/oc/freepubs/pdf/PD-107.pdf [Google Scholar]
- 116.Keith LM, Hughes RF, Sugiyama LS, Heller WP, Bushe BC, Friday JB. 2015. First report of ceratocystis wilt on ‘Ōhi’a (Metrosideros polymorpha). Plant Dis 99:1276. 10.1094/PDIS-12-14-1293-PDN [DOI] [Google Scholar]
- 117.Mueller-Dombois D. 1988. Community organization and ecosystem theory. Can J Bot 66:2620–2625. 10.1139/b88-357 [DOI] [Google Scholar]
- 118.West Hawaii Today. 2015. Lava-loving ohia lehua: a pioneer plant in peril. http://westhawaiitoday.com/news/volcano-update/lava-loving-ohia-lehua-pioneer-plant-peril.
- 119.Jacobs DF, Selig MF, Severeid LR. 2009. Aboveground carbon biomass of plantation-grown American chestnut (Castanea dentata) in absence of blight. For Ecol Manag 258:288–294. [Google Scholar]
- 120.Smith KG, Lips KR, Chase J. 2009. Selecting for extinction: nonrandom disease-associated extinction homogenizes amphibian biotas. Ecol Lett 12:1069–1078. 10.1111/j.1461-0248.2009.01363.x [DOI] [PubMed] [Google Scholar]
- 121.Boyles JG, Cryan PM, McCracken GF, Kunz TH. 2011. Economic importance of bats in agriculture. Science 332:41–42. 10.1126/science.1201366 [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Brown JK, Kema GH, Forrer HR, Verstappen EC, Arraiano LS, Brading PA, Foster EM, Fried PM, Jenny E. 2001. Resistance of wheat cultivars and breeding lines to Septoria tritici blotch caused by isolates of Mycosphaerella graminicola in field trials. Plant Pathol 50:325–338. 10.1046/j.1365-3059.2001.00565.x [DOI] [Google Scholar]
- 123.Torriani SF, Melichar JP, Mills C, Pain N, Sierotzki H, Courbot M. 2015. Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet Biol 79:8–12. 10.1016/j.fgb.2015.04.010 [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Snelders E, Huis In ’t Veld RA, Rijs AJ, Kema GH, Melchers WJ, Verweij PE. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl Environ Microbiol 75:4053–4057. 10.1128/AEM.00231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andrade AC, Del Sorbo G, Van Nistelrooy JG, De Waard MA. 2000. The ABC transporter AtrB from Aspergillus nidulans mediates resistance to all major classes of fungicides and some natural toxic compounds. Microbiology 146:1987–1997. 10.1099/00221287-146-8-1987 [DOI] [PubMed] [Google Scholar]
- 126.Gauthier GM, Keller NP. 2013. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol 61:146–157. 10.1016/j.fgb.2013.08.016 [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Kilaru S, Schuster M, Latz M, Das Gupta S, Steinberg N, Fones H, Gurr SJ, Talbot NJ, Steinberg G. 2015. A gene locus for targeted ectopic gene integration in Zymoseptoria tritici. Fungal Genet Biol 79:118–124. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fones HN, Mardon C, Gurr SJ. A role for the asexual spores in infection of Fraxinus excelsior by the ash-dieback fungus Hymenoscyphus fraxineus. Sci Rep 6:34638. 10.1038/srep34638 [DOI] [PMC free article] [PubMed] [Google Scholar]


