ABSTRACT
Mycobacterium marinum is a well-known pathogenic mycobacterium for skin and soft tissue infections and is associated with fishes and water. Among nontuberculous mycobacteria (NTM), it is the leading cause of extrarespiratory human infections worldwide. In addition, there is a specific scientific interest in M. marinum because of its genetic relatedness to Mycobacterium tuberculosis and because experimental infection of M. marinum in fishes mimics tuberculosis pathogenesis. Microbiological characteristics include the fact that it grows in 7 to 14 days with photochromogenic colonies and is difficult to differentiate from Mycobacterium ulcerans and other mycolactone-producing NTM on a molecular basis. The diagnosis is highly suspected by the mode of infection, which is related to the hobby of fishkeeping, professional handling of marine shells, or swimming in nonchlorinated pools. Clinics distinguished skin and soft tissue lesions (typically sporotrichoid or subacute hand nodules) and lesions disseminated to joint and bone, often related with the local use of corticosteroids. In clinical microbiology, microscopy and culture are often negative because growth requires low temperature (30°C) and several weeks to succeed in primary cultivation. The treatment is not standardized, and no randomized control trials have been done. Therapy is a combination of surgery and antimicrobial agents such as cyclines and rifampin, with successful outcome in most of the skin diseases but less frequently in deep tissue infections. Prevention can be useful with hand protection recommendations for professionals and all persons manipulating fishes or fish tank water and use of alcohol disinfection after contact.
INTRODUCTION AND HISTORY
The first report of a mycobacterium isolated in fish, supposed to be Mycobacterium marinum, has been attributed to Bataillon et al. (1897), who isolated acid-fast bacilli named Mycobacterium piscium from a tuberculous lesion in a common carp (Cyprinus carpio) (1). M. marinum was then originally isolated and identified from marine fish at the Philadelphia Aquarium (2). M. marinum was initially thought to infect marine fishes only and was named accordingly, but it is now known to be a ubiquitous species. The above-mentioned original freshwater isolate of M. piscium could be a M. marinum variant. In the early literature, several other marine Mycobacterium species were described, such as M. platypoecilus, M. anabanti, and M. balnei. Comparative sugar fermentative reactions together with published morphological, cultural, and pathogenic data suggested that they were all synonymous with M. marinum (3) even if M. piscium has not been recognized as a species since its type culture is no longer available.
Human infection due to M. marinum was reported as a tuberculoid infection in people using public swimming pools in Sweden (1939) and in the United States (1951) (4). Linell and Norden identified the causative organism in 1954 after 80 persons had shown the same granulomatous skin lesions (4).These early findings led to the disease’s name of “swimming pool granuloma.” Today, however, due to sanitary chlorination practices, these kinds of outbreaks have disappeared. The names “fish tank granuloma” and “fish handler’s disease” are now used because of the association with home aquaria and water-related activities such as swimming in natural outdoor waters, fishing, and boating (5).
Scientific interest in M. marinum is mainly due to its genetic relatedness to M. tuberculosis and because experimental infection of M. marinum in the goldfish (Carassius auratus) mimics tuberculosis pathogenesis (6). More recently, M. marinum interest was linked with the emergence and burden of the M. ulcerans infection. Lastly, clinical interest in M. marinum infections may eventually emerge due to the expansion of aquarium-related activities.
FUNDAMENTAL BIOLOGY OF M. MARINUM
Taxonomy
M. marinum is one of the >150 species of the genus Mycobacterium (7–9), the only genus of the Mycobacteriaceae family. M. marinum is one of the nontuberculous mycobacteria (NTM), called also atypical mycobacteria or mycobacteria other than M. tuberculosis. According to Runyon’s classification, it belongs to group I, composed of photochromogenic species. Although it can grow in less than 7 days, its characteristics are far different from those of the so-called rapidly growing species of mycobacteria, such as M. chelonae or M. abscessus. Since it carries a single rRNA operon (10) and its 16S rRNA sequence contains the molecular signature of slow-growing mycobacteria (11), it definitely belongs to the slow-growing group of mycobacteria.
M. marinum is a pathogenic mycobacterium, which sets it, along with the related species M. ulcerans, apart from the other NTM that are opportunistic pathogens (12).
From phylogenetic analysis based on the 16S rRNA, rpoB, and hsp65, M. marinum appears close to the branch of the M. tuberculosis complex (13). DNA-DNA hybridization and mycolic acid studies confirm that M. marinum is one of the two species (along with M. ulcerans) most closely related to M. tuberculosis (14, 15).
Genetics
The M. marinum genome (strain M) was one of the genomes of major mycobacteria being sequenced. The length of the M. marinum genome is 6.5 Mb, which is larger than that of M. tuberculosis (4.4 Mb), that of M. leprae (3.3 Mb), and that of M. ulcerans (5.8 Mb) (for specific searches, see http://mycobrowser.epfl.ch/marinolist.html). The M. marinum genome is comparable to the M. ulcerans (16) and M. tuberculosis (17) genomes.
M. marinum shares more than 98% nucleotide sequence identity with the M. ulcerans genome. On the basis of sequences of the housekeeping and structural genes used for species identification, such as those of the ribosomal operon and those encoding RNA polymerase (rpoB), DNA gyrase (gyrA and gyrB), and the 65-kDa heat shock protein (hsp65), M. marinum cannot be differentiated from M. ulcerans, the nucleotide differences observed in some of these genes being related to strain variation (17–19). The major conclusion is that M. marinum seems to be an M. ulcerans ancestor (17, 20). Divergence would have occurred along with the gain by M. ulcerans of genes (grouped on the virulence pMUM megaplasmid) encoding the virulence factor mycolactone (21) and of copies of the insertion sequences IS2404 and IS2606 (20, 22). IS2404 expansion in the M. ulcerans genome has led to the inactivation of many genes through disruption of coding and promoter sequences and has mediated the deletion of approximately 1 Mbp of DNA from M. ulcerans compared to M. marinum (20).
Mycobacteria that contain genes for the production of mycolactone were designated “mycolactone-producing mycobacteria” (MPM). These strains are all closely related, and there is some justification for considering them to form the M. marinum complex or group (23). All these MPM are specialized variants of a common M. marinum progenitor and were adapted to live in restricted environments (24). M. pseudoshottsii, M. shottsii, M. shinshuense, and M. liflandii are MPM described for fishes only (25). Although M. marinum is devoid of the mycolactone-producing coding sequences as the one found in M. ulcerans, another mycolactone (mycolactone F) has been isolated from strains of M. marinum that have produced disease in fishes of the Red Sea (26, 27).
The molecular biology of M. marinum has been developed because it is less pathogenic and grows faster than M. tuberculosis, whereas it is a suitable model for tuberculosis pathogenesis (6, 28). Genetic manipulations such as transformation and transposition have been successful (29, 30). Virulence genes have been studied by gene expression in cultured macrophages or in in vitro models of granuloma (6, 29, 31). Mutations in the virulence genes were often complemented by the corresponding gene of M. tuberculosis that demonstrated the high relatedness of the genomes of the two species. Genome sequencing revealed that region of difference 1, which contains the ESAT-6- and CFP-10-encoding genes in M. tuberculosis, does exist also in M. marinum (32). The ESAT-6-like secretion system is probably a major secretion pathway for M. marinum, and this system is responsible for the secretion of recently evolved PE_PGRS and PPE proteins, which are especially abundant in M. marinum. The Esx secretion system is critical for virulence of both M. tuberculosis and M. marinum and is highly conserved between the two species (17).
Pathogenesis
The availability of fish models (goldfish and zebrafish) mimicking a natural mycobacterial infection (28, 33, 34) enables the study of the pathogen-host interaction. New models of infection have been described such as Drosophila melanogaster (in which M. marinum infection is lethal at a low dose [35]), intraperitoneal infection of adult leopard frogs (Rana pipiens), and infection of embryonic zebrafish (Danio rerio) (36). Infections in these animals are highly similar to M. tuberculosis infection in the human host. In particular, the formation of granuloma-like lesions in the host and the ability to cause both acute and chronic diseases are conserved (37, 38). However, these models were mostly used as surrogates for studying tuberculosis physiopathology or for screening new antituberculosis drugs (39) and, rarely, M. marinum infection itself (40).
M. marinum is an intracellular pathogen that proliferates within macrophages in a nonacid (pH 6.1 to 6.5) phagosome that does not fuse with the lysosome (41). Taking into account that the two species are also genetically related, it is probable that analogous molecular mechanisms are involved in the survival of these organisms in a hostile cell environment. M. marinum is therefore a very useful model system for studying intracellular survival of mycobacteria and possibly other host-pathogen interactions associated with tuberculosis (34, 42), including innate susceptibility (43). Ultrastructure studies have shown that M. marinum remains within activated macrophages in granulomas. Some of the genes that seem important in the capacity to replicate in macrophages and to explain the persistence in granulomas are homologous to the PE_PGRS family of genes discovered in the M. tuberculosis genome (31, 44).
The organism is able to survive, replicate in macrophages, and even escape from the phagosomes into the cytoplasm, where it can induce actin polymerization leading to direct cell spread (45, 46). It was shown recently that M. marinum bacteria are ejected from the cell through the ejectosome, an actin-based structure spreading mechanism that requires a cytoskeleton regulator from the host and an intact mycobacterial ESX-1 secretion system (46, 47). Recent findings using the zebrafish model infected by M. marinum but with various genetic and pharmacologically induced macrophage deficiencies try to explain the susceptibility of humans with mononuclear cytopenia to mycobacterial infections and highlight the therapeutic potential of myeloid growth factors in tuberculosis (48). Moreover, the virulence and immunomodulatory factors that were identified may constitute novel targets for antimycobacterial drugs (49). It has been also shown recently that EsxA contributes to mycobacterial virulence with its membrane-permeabilizing activity required for cytosolic translocation (50).
Experiments involving infection of zebrafish with different strains of M. marinum showed that strains can be divided into two types based on genetic diversity and virulence. Cluster I contains mainly the strains isolated from humans with fish tank granulomas, whereas the majority of cluster II strains were isolated from poikilothermic species. Acute disease progression was noted only with cluster I strains, whereas chronic-disease-causing isolates belonged to cluster II (51).
MICROBIOLOGICAL CHARACTERISTICS OF M. MARINUM
Microscopy and Cell Wall
Under the microscope, M. marinum cannot be distinguished from M. tuberculosis. It is a pleiomorphic rod (1.0 to 4.0 μm by 0.2 to 0.6 μm), not motile, true branching, and difficult to stain by usual methods but appears as an acid-fast bacillus after staining by the reference carbol fuchsin or Ziehl-Neelsen method (52).
The formation of microscopic cords has been described for M. marinum as classically described with M. tuberculosis, and a link between cording and virulence was observed in both species (53, 54).
The cell wall of M. marinum is mainly composed of keto-mycolates and methoxy-mycolates that differentiate it from those of M. tuberculosis and of other mycobacteria except M. ulcerans (15, 55).
Specific Characteristics of M. marinum
Culture
Like other mycobacteria, M. marinum is a strict aerobe. Its preferred carbon sources are glycerol, pyruvate, and glucose, but ethanol can be also used by M. marinum.
M. marinum has an optimal growth temperature of 30°C, whereas small colonies or no growth is observed at 37°C.
In primary culture, the growth rate might be low and positive culture may be obtained only after several weeks’ incubation. In subculture, the growth rate is between 1 and 2 weeks but can reach 4 to 5 days because of its rapid ability to adapt to laboratory conditions.
M. marinum is less exigent than M. tuberculosis for growth. It grows in all the media used for mycobacterial growth (egg based, broth, or agar based) without any additives or only 2 to 5% oleic acid-albumin-dextrose-catalase instead of 10% for M. tuberculosis, and it also grows on blood-containing agar. After subcultures, some of the strains may even grow on ordinary culture media.
Although its growth is dependent on oxygen like for other mycobacteria, 2% to 5% carbon dioxide in the gas phase above the medium improves the growth of M. marinum.
Phenotypic characteristics
Colonies of M. marinum are typically smooth or intermediate, white or beige when the media is kept in the dark, and yellow to orange after exposure to light (photochromogenic) (52) (Fig. 1). It is included in group I of Runyon’s classification along with M. kansasii. Differentiation between these two pathogenic NTM is done classically on the basis of biochemical characteristics. The absence of nitrate reductase production and growth on medium containing thiacetazone is in favor of M. marinum.
FIGURE 1.
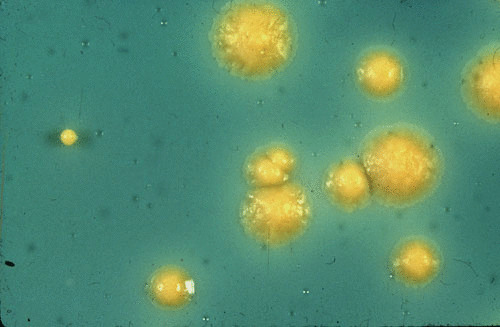
Typical photochromogenic colonies of Mycobacterium marinum grown on Lowenstein-Jensen solid medium.
Photochromogenicity is due to the active production of beta-carotene mediated by the gene crtB and can be inhibited by chloramphenicol (56).
Molecular identification
Molecular biology techniques have been successfully applied for the identification of mycobacteria. Molecular methods are an alternative for species identification offering the advantages of being rapid and accurate. PCR-based methods have been developed, and two of them are commercially available: INNOLiPA Mycobacteria v2 (Innogenetics), based on the amplification of the ribosomal gene spacer (16S-23S), and GenoType Mycobacteria CM/AS (Hain Lifescience), based on the amplification of the 23S rRNA gene (57, 58). Both use PCR coupled with reverse hybridization. So far, they cannot differentiate M. marinum from M. ulcerans since the rRNA sequences are similar (see “Genetics” above).
Specific PCR protocols have been described for detection in fishes (59).
Genotyping
Although infection due to M. marinum is not contagious between humans, strain genotyping has been undertaken for three reasons: first, to relate environmental strains to strains isolated in infected humans; second, in aquaculture, to differentiate strains isolated among fishes or water-living animals (26); and third, to demonstrate relapse or reinfection (60). The technique used was mainly pulsed-field gel electrophoresis since PCR-based methods showed low discriminative properties (61).
Mycobacterial interspersed repetitive unit–variable-number tandem-repeat typing, which is one of the reference typing methods for M. tuberculosis, has been applied to M. marinum and M. ulcerans. Unlike for M. ulcerans, the M. marinum genotypes were not clearly related to the geographic origins of the isolates, and genotyping does not discriminate between relapse and reinfection (62).
Multilocus sequence analysis applied to 22 M. marinum strains showed that the level of intraspecies nucleotide sequence divergence was higher than in M. ulcerans strains (20). M. marinum isolates from humans and fishes have also been compared by sequencing of the 16S rRNA and hsp65 genes, restriction mapping, and amplified fragment length polymorphism analysis (63). In that study, significant molecular differences separated clinical isolates from water-related isolates.
M. MARINUM INFECTION
Manifestations of M. marinum Disease
Fish disease
M. marinum disease in fishes is very common, especially in aquarium fishes. There is some evidence that the gastrointestinal tract could be the primary route of infection (64), and it has been demonstrated that poor diet and stress exacerbate mycobacterial infections in zebrafish (64–66). The severity of the infection ranges from chronic infection associated with a low mortality rate to a more acute form in which the entire population died. The acute fulminating disease is rare and is characterized by rapid morbidity and mortality with few clinical signs. M. marinum infection is more often a chronic progressive disease that may take years to develop into a noticeable illness. Affected fishes show comportmental changes, such as separating from other fishes and refusing food. They may have skin ulcerations or pigment alterations and develop spinal curvature. Unilateral or bilateral exophthalmia is also a typical feature. In fish, M. marinum infection is a systemic disease that can affect virtually any organ system but especially the spleen, kidneys, and liver (67, 68).
Human disease
Due to the optimal growth at 30°C and poor growth at 37°C, human infections with M. marinum are localized primarily to the skin. Infection in HIV-positive patients is usually not different from infection in HIV-negative ones. Scarce cases of M. marinum infection occurring in patients treated with tumor necrosis factor alpha (TNF-α) inhibitor therapy have been reported since 2002; cutaneous lesions displaying rapid sporotrichoid extension are more frequent than among other patients (69). Given the small number of cases reported (about 30), it is not possible yet to draw any conclusions regarding the frequency or the severity of M. marinum infection in this population (70–79). Nevertheless, these medicines should be stopped during the course of antibiotics. If not, the lesions may rapidly extend as already described (78). We also recommend applying preventive strategies (see “Prevention Strategies” below) especially for those patients.
M. marinum infection has different clinical presentations (Table 1). Most commonly, as described for about 60% of the cases, M. marinum is a cutaneous disease manifesting as a solitary papulonodular lesion on a finger or hand (80). In 25% of the cases, M. marinum disease takes on a sporotrichoid form (80, 81) (Fig. 2). This occurs when the infection spreads along the lymphatic vessels to the regional lymph nodes, producing multiple nodules resembling sporotrichosis. Occasionally, skin lesions appear as pustular, nodulo-ulcerative, granulomatous, or verrucous plaques.
TABLE 1.
Published studies of M. marinum infections that include more than 10 patientsa
| Reference | No. patients (no. with deep infection) | Fish exposure (%) | Cure (%) | Antibiotic treatmenta | Surgery (% cured) | |||
|---|---|---|---|---|---|---|---|---|
| Duration (mo) (mean) | Monotherapy | Combination therapy (no.) | ||||||
| No. of patients | Antibiotic and no. of patients (% cured) | |||||||
| Even-Paz, 1976 (106) | 10 (0) | 0 | 44 | NA | NA | NA | NA | |
| Chow et al., 1987 (122) | 24 (24) | 87.5b | 83 | 9 | 0 | 24 | 10 (70) | |
| Bonafe 1992 (145) | 27 (1) | 93 | >74 | 3.8 | NA | NA | NA | |
| Kozin and Bishop, 1994 (124) | 12 (6) | 100 | 100 | 6 | 7 | D = 5 (100), Co = 2 (100), R = 1 (100) | 5 | 12 (100) |
| Edelstein 1994 | 31 (0) | NA | 81 | 4 | 19 | M = 14 (71), D = 3 (67), T = 1 (100), Co = 1 (100) | 10 | NA |
| Ang et al., 2000 (5)c | 38 (NA) | 45 | 81 | 3.5 | 22 | Co = 19 (93), M = 3 (100) | 12 | 1 (100) |
| Casal et al., 2001 (139) | 39 | 90 | 99 | 2–4 | 20 | M = 12 (NA), R = 8 (NA) | 7 | NA |
| Aubry et al., 2002 (80) | 63 (18) | 84 | 87 | 3.5 | 23 | M = 19 (100), C = 4 (100) | 40 | 30 (16) |
| Ho 2006 (146) | 17 (NA) | 24 | 94 | 4.5 | 16 | D = 4 (NA), Co = 1 (NA), M = 11 (NA) | 1 | 0 |
| Johnson and Stout, 2015 (88) | 28 (19) | 20 | 95c | 5 | 2 | NA | 15 | 22 (79) |
| Sia et al., 2016 (94) | 29 (7) | 19 | 100 | NA | 3 | NA | 25 | 16 (100) |
| Blanc 2016 (147) | 23 (5) | NA | 91c | 3 | 11 | C = 9 (NA), Z = 1 (NA), D = 1 (NA) | 6 | 5 |
NA, not available; M, minocycline; C, clarithromycin; R, rifampin; D, doxycycline; T, tetracycline; Co, co-trimoxazole; Z, azithromycin.
100% swimming pool exposure.
Patients lost to follow-up were not assessed for the outcome.
FIGURE 2.
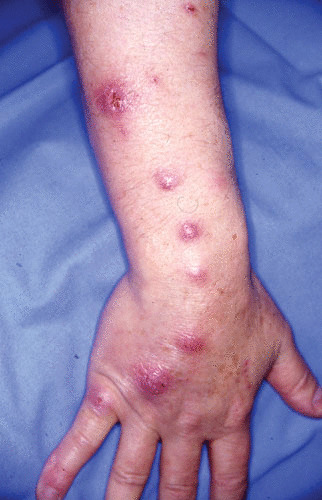
Sporotrichoid form of skin lesions typical of M. marinum infection. (Courtesy of Hervé Darie, Noisy le Grand, France.)
Deep infections, such as tenosynovitis (the most frequent), osteomyelitis, arthritis, and bursitis, occur in 20 to 40% of the cases (80–85). Unusual clinical presentations have also been reported, such as epididymo-orchitis (86) or destructive nasal lesions mimicking extranodal NK/T cell lymphoma (74) as well as severe osteomyelitis (87).
Deep infections result from the extension of a cutaneous infection or direct inoculation of the organism (88). Systemic dissemination is exceptional and has been reported to occur only in immunocompromised patients (e.g., solid-organ and hematopoietic stem cell transplant recipients or those on anti-TNF treatment [74, 89–91]), and general symptoms are lacking. Localized adenopathy is rare (15% of the cases in reference 80), and infection of deep organs, such as the lungs, is exceptional (92). In these cases, identification of M. marinum should be carefully checked.
There is often a several-month delay between the onset of the lesions and the patient’s seeking medical care (93, 94) because the lesions are subacute or chronic and usually painless, as for M. ulcerans. Moreover, the lesions can be self-limited and may heal spontaneously, though healing can take months to several years. Initial misdiagnosis of osteoarticular form of M. marinum infection can lead to intralesional injection of corticosteroid that favors local dissemination (83). These forms are often associated with a poor prognosis (8).
Extremities of the upper limbs, such as a finger or hand, are the most common site of infection in relation of fish or water animal exposure (94). Patients with skin lesions on the lower limbs are swimming pool cases or indirect cases. M. marinum is assumed to be introduced in the skin accidentally through preexisting wounds or abrasions. History of preceding minor trauma is common, and an occupation or hobby that resulted in a likely environmental water exposure is the rule. However, since the incubation period is very long, on average 3 weeks up to 9 months (80, 95), minor abrasions or wounds preceding the inoculation usually do not remain at the time of the diagnosis.
Immunity
Tuberculin skin testing is usually positive because of cross-reaction with M. tuberculosis (96). It may suggest a mycobacterial infection but does not distinguish M. marinum from tuberculosis or other mycobacterial infections and is therefore of little utility in the workup.
False positivity of the interferon gamma release assays performed today for the diagnosis of latent tuberculosis infection have been also reported with M. marinum infection (76). Indeed, as noted above, M. marinum shares with M. tuberculosis the specific antigens (ESAT-6 and CFP-10) used in these tests. However, interferon gamma release assays were not found to be useful as a diagnostic method for M. marinum infection (97, 98).
Diagnosis
Diagnosis can be difficult for the clinician (84) because the presentation is often insidious and nonspecific. If key historical information, such as fish exposure, is not obtained, the diagnosis is commonly delayed (95). Diagnosing an infection due to M. marinum requires a high index of suspicion, a properly obtained exposure history, and knowledge of the laboratory growth characteristics of the organism (94).
Differential diagnosis includes infection due to other NTM known to cause cutaneous infection (M. chelonae, M. ulcerans, M. ulcerans subsp. shinshuense [99], M. haemophilum, M. fortuitum, and M. tuberculosis), sporotrichosis, and noninfectious diseases such as sarcoidosis, skin tumors, and foreign-body reactions.
Though the diagnosis can be suspected clinically, especially when exposure is established, the diagnosis relies on isolation of a mycobacterium subsequently identified as M. marinum.
Clinical significance
Isolation of M. marinum has a clinical significance whatever the number of colonies or the smear positivity of the specimen, since it usually does not grow from the laboratory environment or from the uninfected human body. This important point differentiates M. marinum infection from infections due to other NTM such as M. abscessus or M. chelonae (100). Thus, correct identification is required (8, 52).
Bacteriological findings
Microscopic examination of the specimen after Ziehl-Neelsen staining is positive in only 30% of the cases. Even when positive, smear microscopy cannot distinguish M. marinum from other mycobacteria, including species of the M. tuberculosis complex.
A definite diagnosis is obtained by a positive culture. Cultures are reported as positive in 70 to 80% of the cases, but this number could be increased with attention paid to the specimen collection and proper temperature for incubation. The microbiologist should be aware of suspicion of M. marinum infection in order to perform cultures at 30°C.
Collection of specimens
Specimens containing M. marinum are taken from the skin, either as a skin biopsy sample or as aspirate pus. Swabs should be avoided for many reasons (81, 88). Other specimens are articular fluids or subcutaneous tissues and exudates often obtained via surgery (85, 87). Specimens must be collected before chemotherapy begins. The container should be sterile and should not contain any fixative or preservative. The collected specimen should be kept at 4°C if there are delays in delivery to the laboratory. Since M. marinum infection is not a multibacillary infection, it is necessary to collect the largest possible volume, especially in the case of skin biopsy or surgery.
Isolation procedures
Laboratory processing for M. marinum disease diagnosis is not an emergency. A level 2 laboratory might be sufficient for isolation and identification, although level 3 might be required for studies requiring large-volume subcultures (52).
Safety measures are required for handling specimens and cultures—wearing gloves, disinfection of the material and benches, and avoiding the use of needles—so that accidental inoculation does not occur.
Skin biopsy or wound fluids might be contaminated with skin flora, such as staphylococci, and consequently require a decontamination procedure (standard N-acetyl-L-cysteine 2% NaOH procedure or 4% HCl decontamination) prior to culture. Specimens from sterile deep structures (i.e., articular fluid) can be inoculated directly without decontamination (52, 101).
Since M. marinum grows poorly at 37°C, the aspirate or biopsy should be cultured at 30 to 32°C. Colonies grow within 5 to 14 days, but primo-culture requires a longer time and cultures should be kept for 6 to 12 weeks. Because other NTM can be responsible for the infection, specimens should also be incubated at 37°C.
After successful isolation of a photochromogenic mycobacterium, further differentiation between M. marinum and other organisms of Runyon group I is required (see “Microbiological Characteristics of M. marinum” above).
Methods based on molecular biology are limited by the high homology between M. marinum and M. ulcerans (see “Molecular identification” above). Matrix-assisted laser desorption ionization–time of flight mass spectrometry cannot distinguish these two species but can be performed as a rapid screening method before accurate identification (102). Neither the commercial INNO-LiPA v2 assay kits nor the GenoType Mycobacteria CM/AS allow differentiation between M. marinum, M. ulcerans, M. ulcerans subsp. shinshuense, M. shottsii, and M. pseudoshottsii (103).
Molecular identification using the analysis of conserved genes or DNA regions (hsp65, gyrA, rpoB, and 16S-23S internal transcribed spacer) associated with a 7-day culture of a photochromogenic colony is required to identify M. marinum. However, in the case of absence of photochromogen colonies or culture in broth, a gene sequence belonging to M. marinum or M. ulcerans (18, 19) coupled to the absence of IS2404 and IS2606 (22) may indicate M. marinum (104). We do recommend waiting for the colony morphology to confirm the identification. In the past few years, a mycolactone-producing subgroup of the M. marinum complex has been identified and analyzed. These IS2404-positive strains cause pathology in frogs and fish, but not in humans so far (25) (see “Fundamental Biology of M. marinum” above).
Histological findings
Tissue biopsy for histopathology is important but provides diagnosis of mycobacteriosis in only half of the cases, since histological findings depend on the age of the lesion.
Granulomas suggest diagnosis but are not pathognomonic and are also present in other mycobacterial infections. They are present in less than two-thirds of cases, and they may be confused with rheumatoid nodules (105). During the first months, there is a nonspecific inflammatory infiltrate. Later, granulomas with multinucleated giant cells are common findings (Fig. 3). Fibrinoid necrosis, but not true caseation, can be observed. Langhans giant cells are seen only occasionally. Hyperkeratosis with focal parakeratosis, hyperplasia, and liquefaction degeneration of the basal layer can be observed (88, 106, 107). Although acid-fast bacilli are seldom seen in the histological sections (67), staining should be attempted.
FIGURE 3.
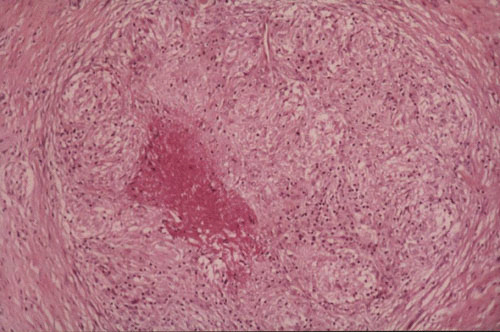
Active-disease histopathologic section of tissue from a patient with a M. marinum infection. The lesion shows granulomatous infiltrate with epithelioid and giant cells. (Courtesy of Bernard Cribier, Strasbourg, France.)
Importantly, in one case out of five, the infiltrate suggested no infectious origin, although deep skin biopsies and synovial biopsies provided more information. Therefore, for all forms of necrotic granuloma, whether or not accompanied by collections of neutrophils, a culture should be carried out in a specific medium, even in the absence of microscopic evidence of bacilli (105).
ANTIMICROBIAL SUSCEPTIBILITIES AND TREATMENT
Mode of Action and Resistance Mechanisms in M. marinum
The permeability of the M. marinum cell wall has not been investigated, but it has been shown to vary at least 10-fold between mycobacterial species, M. tuberculosis being 10-fold more permeable than M. chelonae (108). Considering the natural multidrug resistance of M. marinum (see below), the permeability of its cell wall could be close to that of M. chelonae. This low permeability probably allows survival in unfavorable environments.
In the genome, several genes encoding antibiotic resistance mechanisms have been reported. Genes encoding enzymes known to hydrolyze antibiotics were found for β-lactams (blaC), aminoglycosides (aac2′), and chloramphenicol (cph). Many potential efflux pumps and ABC transporters have been also described, including potential extruders for cyclines, macrolides, and aminoglycosides (17).
In Vitro Susceptibility to Antibiotics
From the studies dealing with a large number of strains and applying a standard method of testing, M. marinum has a natural multidrug resistance pattern (109, 110). Indeed, M. marinum has shown resistance to the antituberculosis drugs isoniazid, ethambutol, and pyrazinamide in most studies. Rifampin and rifabutin are the most active drugs in term of MICs. MICs of minocycline, doxycycline, clarithromycin, linezolid, sparfloxacin, moxifloxacin, imipenem, sulfamethoxazole, and amikacin are close to the susceptibility breakpoints, and thus, these drugs may have moderate activity. MICs of trimethoprim, azithromycin, telithromycin, quinupristin-dalfopristin, ciprofloxacin, gemifloxacin, ofloxacin, and levofloxacin are above the concentrations usually obtained in vivo, and consequently, M. marinum may be considered resistant to them (111–114).
All the strains have similar susceptibility patterns since for each drug the MIC50, geometric mean MIC, and modal MIC are very close (109, 111). This constitutes the natural or intrinsic susceptibility pattern of M. marinum (Table 2).
TABLE 2.
MICs of 17 antibiotics against 54 strains of Mycobacterium marinum determined by the agar dilution methoda
| MIC50 (μg/ml) | MIC90 (μg/ml) | Modal MIC (μg/ml) | Geometric mean ± SD (μg/ml) | Range (μg/ml) | |
|---|---|---|---|---|---|
| Rifampin | 0.25 | 0.5 | 0.25 | 0.24 ± 1.7 | 0.125–4 |
| Rifabutin | 0.06 | 0.06 | 0.06 | 0.06 ± 1.8 | 0.015–1 |
| Isoniazid | 4 | 8 | 4 | 5.6 ± 1.5 | 4–16 |
| Ethambutol | 2 | 4 | 2 | 1.7 ± 1.6 | 1–4 |
| Amikacin | 2 | 4 | 4 | 1 ± 1.7 | 1–8 |
| Doxycycline | 8 | 16 | 8 | 5.7 ± 2 | 0.5–16 |
| Minocycline | 2 | 4 | 2 | 2.9 ± 1.7 | 0.5–8 |
| Clarithromycin | 1 | 4 | 2 | 1.2 ± 2.3 | 0.5–4 |
| Azithromycin | 32 | 128 | 32 | NA | 8→128 |
| Ofloxacin | 4 | 16 | 4 | 6.1 ± 1.7 | 2–32 |
| Ciprofloxacin | 4 | 8 | 4 | 3.8 ± 1.8 | 1–16 |
| Levofloxacin | 4 | 8 | 4 | 4.5 ± 1.7 | 2–32 |
| Sparfloxacin | 1 | 2 | 1 | 1 ± 1.8 | 0.5–4 |
| Moxifloxacin | 0.5 | 1 | 0.5 | 0.6 ± 1.7 | 0.25–4 |
| Sulfamethoxazole | 8 | 128 | 8 | NA | 4→128 |
| Trimethoprim | 64 | 128 | 128 | 67.4 ± 2.3 | 16–512 |
| Imipenem | 2 | 8 | 2 | 2.6 ± 2.6 | 0.5–16 |
| Linezolid | 1 | 2 | NA | NA | 0.5–4 |
Acquired resistance has not been described in M. marinum so far for any of those antibiotics, even in relapsed cases. Slight differences from the above-described pattern of natural antibiotic susceptibility that might be observed are usually due to misidentification or differences in the method or the technique of antibiotic susceptibility testing (115).
Susceptibility Testing
Etest has been shown to be an accurate and precise method of MIC determination for bacteria other than mycobacteria, for rapidly growing mycobacterial species, and even for some of the slowly growing mycobacterial species (116–118). Different authors have questioned the reliability of the Etest for M. marinum and also for other mycobacteria, claiming that it may cause reports of false resistance (109, 111). Agreement between MICs, yielded by either the Etest method or the agar dilution method used as a reference, depends on the antibiotic and have been shown to be 83% for minocycline, 59% for rifampin, 43% for clarithromycin, and 24% for sparfloxacin (109, 119). Moreover, reproducibility with the Etest was low, in contrast to that with the agar dilution method. In conclusion, Etest is not recommended for M. marinum; antibiotic susceptibility testing and the agar dilution method remain the methods recommended.
Broth microdilution susceptibility testing is recommended by the CLSI and may use the commercially available Sensititre MIC plates (120). Although for M. marinum it may be recommended to use the MIC plate for slow-growing mycobacteria, we think that testing antibiotics contained in the MIC plate for rapidly growing mycobacteria may be more appropriate with regard to the antibiotics with low MICs (Table 2).
Since no primary (acquired) resistance has been described so far, routine susceptibility testing seems unnecessary except for relapsed cases as recommended for other atypical mycobacteria (80, 100).
Antimicrobial Therapy of M. marinum Infections
Patients infected with M. marinum are usually treated with antibiotics (Table 1). Different antibiotic regimens have been reported (83). The choice of the regimen appears to reflect more the personal experience and preference of individual authors than demonstrated efficacy. Antibiotic efficacy is unknown since (i) cases were reported separately in the literature, (ii) no therapeutic trial has been carried out, and (iii) M. marinum infection may be cured spontaneously (12, 106, 121).
A variety of antibiotics have been used, including cyclines, co-trimoxazole, rifampin plus ethambutol, and, more rarely, clarithromycin, levofloxacin, and amikacin (83, 121–123). Cure as well as failure has been described with all of these drugs (83, 121, 124). Overall, most of the patients are cured after therapy that includes cyclines or clarithromycin and rifampin, but failure cases have been also observed. Failure cases have rarely been observed with cyclines, but most of the patients treated with cyclines, and especially cyclines alone, have had mild infection limited to skin and soft tissues. In contrast, rifampin and rifabutin, which are the only antibiotics with low MICs close to those found for M. tuberculosis, were usually given in complicated cases with extension of the infection to the deeper structures, such as tenosynovitis and osteoarthritis, and failed to cure all cases. In our study, failure was related to deep infections (only 72% were cured) and to ulcer lesions (80).
Although MICs of the new fluoroquinolones, moxifloxacin and gatifloxacin, are lower than those of classical fluoroquinolones, and these compounds are very potent antituberculosis drugs, their in vivo activity against M. marinum has yet to be demonstrated. The efficacy of linezolid, also with low MICs, needs also to be tested in vivo.
We currently need results of in vivo experiments with an animal model or therapeutic trials with humans showing evidence of efficacy of some antibiotics. Until these are obtained, it is reasonable to recommend cyclines for M. marinum infection limited to the skin and the combination of rifampin and clarithromycin for infection extended to deeper structures.
In the literature, the duration of antimicrobial therapy in M. marinum infection varies from 2 weeks to 18 months, depending on several factors, such as the extension and severity of infection, the presence of underlying disorders, and the clinical response (83, 84). In many patients with mild disease, infection mostly resolves spontaneously, although complete resolution may take up to several years (81, 83). In our study (80), the duration of therapy ranged from 1 to 25 months and the median was 3.5 months. This duration was significantly longer for cases with infection spread to deeper structures except for the failure cases.
We may recommend to continue with antibiotherapy at least until the lesions heal and then for 2 additional months, especially in cases of infection extended to deeper structures.
Surgery
The place of surgery is controversial. For some authors, surgical debridement along with antimicrobial therapy is usually required for control (81). For others, surgical debridement should be limited to the cases with criteria known to be associated with a poor prognosis, including steroid injections into the lesion, a persistent drainage sinus tract after several months of antimicrobial therapy, and persistent pain (122). Most of the infections spread to deeper structures are treated with surgery, which seems reasonable (125). For infections limited to skin and soft tissue, there is no clear benefit of surgery and surgical side effects are unknown.
Other therapies, such as cryotherapy, X-ray therapy, and electrodessication, have been reported but have not been evaluated (83).
It should also be recommended to stop TNF-α inhibitor or other immunosuppressive therapy during the course of antibiotics when M. marinum infection occurs in patients treated with these medications. Indeed, despite the small number of cases described, it seems that the lesions may progress if these medications are not discontinued (78).
EPIDEMIOLOGY AND PREVENTION
Natural Habitats of M. marinum
M. marinum has been reported to affect a wide range of freshwater and marine fish species, suggesting an ubiquitous distribution. M. marinum is the main mycobacterium isolated from fish, although very little is known about its prevalence and impact on fisheries (126). Zanoni et al. reported that mycobacteria were found in 46.8% and 29.9% of the fishes imported into Italy and which died and that M. marinum represented 2.4 to 5.3% of the isolates (127), whereas Slany et al. reported the presence of M. marinum DNA in 41.7% of ornamental fishes of which 23.6% were culture positive in the Czech Republic (128).
M. marinum is transmitted in fishes through the consumption of contaminated feed, cannibalism of infected fish, aquatic detritus, or release of pathogens into the water due to gut or skin lesions or disintegration of infected fishes (67). In this respect, potential sources of infective material are numerous and include the soil and water, in which the bacterial cells remain viable for 2 years or more (67).
M. marinum infection in other aquatic vertebrates may be a source of infection to fishes. Frogs, snakes, and turtles may become involved in the transmission cycle. Snails are also thought to be a reservoir (67). Other invertebrate organisms, such as shellfishes or water fleas, have been shown to play a role in the transmission of this agent.
Findings of genetic studies comparing M. marinum isolates from humans and fish suggest that clinical cases may have derived from the ornamental fish trade associated with home aquaria and that only certain strains of M. marinum have zoonotic potential (63, 129).
The prevalence of NTM was evaluated in the environment of a swimming pool in Italy. M. marinum was isolated in only 4.5% of the water samples and pool edges, although 88.2% of pool water samples were positive for M. gordonae, M. chelonae, and M. fortuitum (130).
In addition to fish and fish tanks, reptiles and related poikilothermic animals and the vivaria in which such animals are maintained could be sources of M. marinum exposure. With millions of poikilothermic animals, such as tropical reptiles, being kept in close contact with people worldwide as new companion pets, one could predict that an increasing number of “vivarium granuloma” cases are likely to be diagnosed. Therefore, doctors should be aware that vivaria, in addition to fish tanks, are a source of M. marinum exposure for patients (131).
Epidemiology of M. marinum Infections
Like infections with other NTM, M. marinum infections are not contagious from human to human.
Before 1962, most cutaneous M. marinum infections reported in the literature involved swimming pool-associated injuries, including two large outbreaks involving almost 350 patients (4, 93). A possible explanation of the decline in pool-associated cases is the improvement in swimming pool water disinfection practices in recent decades. M. marinum survives only briefly after exposure to free chlorine concentrations of ≥0.6 μg/ml. There can be unexpected sources of exposure, such as coal mine water (132). Furthermore, outbreaks resulting from a common source of contamination have been described (94, 133).
M. marinum skin infection is now often acquired from aquarium maintenance and is called fish tank granuloma (134). Since M. marinum infection is an important zoonosis, there is a significant risk to all personnel working with fishes, water-living animals, or aquaria. M. marinum infection may be an occupational hazard for certain professionals (for example, for pet shop workers), but many infections occur in fish fanciers who keep an aquarium at home, hence the name “fish fanciers’ finger syndrome.” Although infection may be caused by direct injury from fish fins or bites, most are acquired during the handling of an aquarium, such as cleaning or changing the water (95, 96). Indirect infection due to a bath that was used to clean out fish tanks has also been described (80, 135, 136).
It is foreseen that the incidence of fish tank granuloma will increase (135) due to the extension of fishkeeping as a hobby and aquarium tourism. Fishkeeping is one of the most popular hobbies in many countries, such as Germany, the United States, and France, where about 10% of the population has an aquarium at home and business related to fishkeeping has increased every year.
The frequency of M. marinum isolation in laboratories is low, and M. marinum accounts for less than 1% of the mycobacterial clinical isolates (137). A recent survey in Europe showed that M. marinum represents about 1.3% of NTM isolated (138). In Spain, 39 cases were reported from 1991 to 1998 involving 21 laboratories (139). Less than half of the cases of M. marinum infection are bacteriologically confirmed. The incidence of M. marinum infection was estimated to be around 0.09 case per 100,000 inhabitants per year in France and between 0.05 and 0.27 in the United States.
Prevention Strategies
For the prevention of swimming pool granuloma, the Centers for Disease Control and Prevention recommend that concentrations of free chlorine be kept between 0.4 and 1 mg/liter in swimming pool water and between 2 to 5 mg/liter in spa and hot tub water (93).
Sanitation, disinfection, and destruction of carrier fishes are the primary methods of controlling M. marinum infection in fishes. This practice is mostly pursued for the food fish; however, with expensive fish species, this practice may be difficult to apply. Antimicrobial treatment is not able to eliminate M. marinum from affected fishes (140). Regarding importation of ornamental fish, there exists great variation in policy. For example, Europe decided on an EU Directive 2003/858/EC health certificate-derived template for all imported live fish (including tropical ornamentals) (141).
Individual prevention is the first line of defense for anyone involved with aquaria or anyone working or recreating in a marine environment. Preventive strategies should be developed for fish tank activity, such as wearing gloves when cleaning the tank (95). Common-sense measures include the following:
Bandage or dress any open wound or cut before exposure.
Clean hands thoroughly before and after exposure to aquarium water and components. Hydroalcoholic solutions may be used instead of hand washing.
Do not swallow aquarium water when checking for salinity or siphoning water.
Do not overcrowd aquaria, since this favors the multiplication of mycobacteria.
UV germicide lamps to treat aquarium water are efficient for mycobacteria as long as they are used in clean conditions at the correct flow rate (142).
Do not transfer tank filters or fishes in the bath that is used for humans, or carefully clean it with sodium hypochlorite (143).
The exposed population should be educated in order to recognize signs of M. marinum disease in fishes and in humans so they can inform medical staff, a point that will expedite the diagnosis.
Fish salespersons should be educated. Indeed, many tropical fish salespersons ignore warnings about fish tank granuloma. In France, although 20% of them are at risk of M. marinum infection, 95% of them immerse their hands without gloves in the fish tanks every day (144).
Some authors recommend against installing ornamental aquaria in hospital units, particularly units likely to receive immunocompromised patients.
REMAINING PROBLEMS AND CONCLUSION
Treatment evaluation requires large-scale trials, probably at an international level. Infections limited to the skin and soft tissue should be distinguished from infections extended to deeper structures. Antibiotics such as cyclines, rifampin, and clarithromycin need to be evaluated along with the new fluoroquinolones and linezolid. Surgery needs subsequent evaluation.
Surveillance of M. marinum infection, which is expected to increase due to fishkeeping as a hobby and aquarium tourism, should be undertaken at least in some highly exposed countries. A simple surveillance could be based on culture-confirmed cases. Bacteriology laboratories, dermatologists, and infectious disease physicians should play a crucial role in case finding.
Since there is no human-to-human transmission of M. marinum infection, the prevention of inoculation from the environment is the main strategy for eradicating the disease. Simple recommendations such as hand protection and hygiene measures and fish tank and aquarium maintenance seem worthy of being largely disseminated and evaluated.
Professionals should also take M. marinum risk into account and apply the recommendation for decreasing M. marinum infection among farm fishes and among professionals handling fishes.
Lastly, diagnosis should be expedited: think to (i) ask, “Do you possess a fish tank at home? Who cleans it, and how?”; (ii) sample the lesion for mycobacteriological analysis; (iii) inform the laboratory that there is a suspicion of M. marinum infection; and (iv) incubate at 30°C in addition to 37°C and wait for weeks until smooth photochromogenic colonies appear.
FIGURE 4.
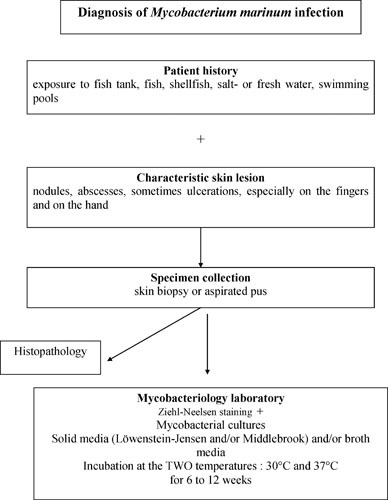
Microbiological diagnosis of human infection due to M. marinum.
Contributor Information
Alexandra Aubry, Centre National de Référence des mycobactéries et résistance des Mycobactéries aux antituberculeux; Sorbonne Université, Université Pierre et Marie Curie, AP-HP Hôpital Pitié-Salpêtrière; Centre d’Immunologie et des Maladies Infectieuses, Team 13, INSERM U1135, Paris, France.
Faiza Mougari, Centre National de Référence des mycobactéries et résistance des Mycobactéries aux antituberculeux; Laboratoire de Bactériologie, AP-HP Hôpital Lariboisière; Université Paris Diderot, IAME UMR 1137 Inserm, Paris, France.
Florence Reibel, Centre National de Référence des mycobactéries et résistance des Mycobactéries aux antituberculeux; Sorbonne Université, Université Pierre et Marie Curie, AP-HP Hôpital Pitié-Salpêtrière; Centre d’Immunologie et des Maladies Infectieuses, Team 13, INSERM U1135, Paris, France.
Emmanuelle Cambau, Centre National de Référence des mycobactéries et résistance des Mycobactéries aux antituberculeux; Laboratoire de Bactériologie, AP-HP Hôpital Lariboisière; Université Paris Diderot, IAME UMR 1137 Inserm, Paris, France.
David Schlossberg, Philadelphia Health Department, Philadelphia, PA.
REFERENCES
- 1.Bataillon E, Moeller A, Terre L. 1902. Über die identitat des Bacillus des Karpfens (Bataillon, Dubard et terre) und des bacillus der Blindsschleuche (Moeller). Zentralblatt für tuberculose 3:467–468. [PubMed] [Google Scholar]
- 2.Aronson J. 1926. Spontaneous tuberculosis in salt water fish. J Infect Dis 39:315–320. [PubMed] [Google Scholar]
- 3.Baker J, Hagan W. 1942. Tuberculosis of a Mexican platyfish (Platypoecilus maculatus). J Infect Dis 70:248–252. [Google Scholar]
- 4.Linell F, Norden A. 1954. Mycobacterium balnei, a new acid-fast bacillus occurring in swimming pools and capable of producing skin lesions in humans. Acta Tuberc Scand Suppl 33:1–84. [PubMed] [Google Scholar]
- 5.Ang P, Rattana-Apiromyakij N, Goh CL. 2000. Retrospective study of Mycobacterium marinum skin infections. Int J Dermatol 39:343–347. [DOI] [PubMed] [Google Scholar]
- 6.Ruley KM, Ansede JH, Pritchett CL, Talaat AM, Reimschuessel R, Trucksis M. 2004. Identification of Mycobacterium marinum virulence genes using signature-tagged mutagenesis and the goldfish model of mycobacterial pathogenesis. FEMS Microbiol Lett 232:75–81. [DOI] [PubMed] [Google Scholar]
- 7.Behr MA, Falkinham JO, III. 2009. Molecular epidemiology of nontuberculous mycobacteria. Future Microbiol 4:1009–1020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Wayne LG, Sramek HA. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev 5:1–25. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortoli E. 2014. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin Microbiol Rev 27:727–752. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helguera-Repetto C, Cox RA, Muñoz-Sànchez JL, Gonzalez-y-Merchand JA. 2004. The pathogen Mycobacterium marinum, a faster growing close relative of Mycobacterium tuberculosis, has a single rRNA operon per genome. FEMS Microbiol Lett 235:281–288. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Stahl DA, Urbance JW. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol 172:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolinsky E. 1992. Mycobacterial diseases other than tuberculosis. Clin Infect Dis 15:1–10. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Kurokawa S, Kabayama J, Hwang SD, Nho SW, Hikima J, Jung TS, Sakai M, Kondo H, Hirono I, Aoki T. 2013. Comparative genome analysis of fish and human isolates of Mycobacterium marinum. Mar Biotechnol (New York) 15:596–605. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Rogall T, Wolters J, Flohr T, Böttger EC. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol 40:323–330. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Tønjum T, Welty DB, Jantzen E, Small PL. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J Clin Microbiol 36:918–925. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PD, Davies JK, Jenkin GA, Small PL, Jones LM, Tekaia F, Laval F, Daffé M, Parkhill J, Cole ST. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res 17:192–200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Chae GT, Kim EC, Cha CY, Kook YH. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol 37:1714–1720. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauendorffer JN, Guillemin I, Aubry A, Truffot-Pernot C, Sougakoff W, Jarlier V, Cambau E. 2003. Identification of mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J Clin Microbiol 41:1311–1315. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinear TP, Jenkin GA, Johnson PD, Davies JK. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J Bacteriol 182:6322–6330. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A 101:1345–1349. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chemlal K, Huys G, Fonteyne PA, Vincent V, Lopez AG, Rigouts L, Swings J, Meyers WM, Portaels F. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J Clin Microbiol 39:3272–3278. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip MJ, Porter JL, Fyfe JA, Lavender CJ, Portaels F, Rhodes M, Kator H, Colorni A, Jenkin GA, Stinear T. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol 189:2021–2029. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doig KD, Holt KE, Fyfe JA, Lavender CJ, Eddyani M, Portaels F, Yeboah-Manu D, Pluschke G, Seemann T, Stinear TP. 2012. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics 13:258. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stragier P, Hermans K, Stinear T, Portaels F. 2008. First report of a mycolactone-producing Mycobacterium infection in fish agriculture in Belgium. FEMS Microbiol Lett 286:93–95. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Ucko M, Colorni A, Kvitt H, Diamant A, Zlotkin A, Knibb WR. 2002. Strain variation in Mycobacterium marinum fish isolates. Appl Environ Microbiol 68:5281–5287. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranger BS, Mahrous EA, Mosi L, Adusumilli S, Lee RE, Colorni A, Rhodes M, Small PL. 2006. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect Immun 74:6037–6045. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. 2003. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol Lett 225:177–182. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan L, Falkow S. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect Immun 62:3222–3229. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talaat AM, Trucksis M. 2000. Transformation and transposition of the genome of Mycobacterium marinum. Am J Vet Res 61:125–128. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Chan K, Knaak T, Satkamp L, Humbert O, Falkow S, Ramakrishnan L. 2002. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc Natl Acad Sci U S A 99:3920–3925. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Ingen J, de Zwaan R, Dekhuijzen R, Boeree M, van Soolingen D. 2009. Region of difference 1 in nontuberculous Mycobacterium species adds a phylogenetic and taxonomical character. J Bacteriol 191:5865–5867. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talaat AM, Reimschuessel R, Wasserman SS, Trucksis M. 1998. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect Immun 66:2938–2942. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trucksis M. 2000. Fishing for mycobacterial virulence genes: a promising animal model. ASM News 66:668–674. [Google Scholar]
- 35.Dionne MS, Ghori N, Schneider DS. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun 71:3540–3550. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cosma CL, Swaim LE, Volkman H, Ramakrishnan L, Davis JM. 2006. Zebrafish and frog models of Mycobacterium marinum infection. Curr Protoc Microbiol Unit 10B.2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. 2006. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun 74:6108–6117. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weerdenburg EM, Abdallah AM, Mitra S, de Punder K, van der Wel NN, Bird S, Appelmelk BJ, Bitter W, van der Sar AM. 2012. ESX-5-deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cell Microbiol 14:728–739. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Sridevi JP, Anantaraju HS, Kulkarni P, Yogeeswari P, Sriram D. 2014. Optimization and validation of Mycobacterium marinum-induced adult zebrafish model for evaluation of oral anti-tuberculosis drugs. Int J Mycobacteriol 3:259–267. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Cronan MR, Tobin DM. 2014. Fit for consumption: zebrafish as a model for tuberculosis. Dis Model Mech 7:777–784. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker LP, George KM, Falkow S, Small PL. 1997. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect Immun 65:1497–1504. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao LY, Groger R, Cox JS, Beverley SM, Lawson EH, Brown EJ. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect Immun 71:922–929. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobin DM, Vary JC, Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717–730. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh VK, Berry L, Bernut A, Singh S, Carrère-Kremer S, Viljoen A, Alibaud L, Majlessi L, Brosch R, Chaturvedi V, Geurtsen J, Drancourt M, Kremer L. 2016. A unique PE_PGRS protein inhibiting host cell cytosolic defenses and sustaining full virulence of Mycobacterium marinum in multiple hosts. Cell Microbiol 18:1489–1507. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Barker LP, Brooks DM, Small PL. 1998. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol Microbiol 29:1167–1177. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 198:1361–1368. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagedorn M, Rohde KH, Russell DG, Soldati T. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729–1733. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagán AJ, Yang CT, Cameron J, Swaim LE, Ellett F, Lieschke GJ, Ramakrishnan L. 2015. Myeloid growth factors promote resistance to mycobacterial infection by curtailing granuloma necrosis through macrophage replenishment. Cell Host Microbe 18:15–26. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohanty S, Jagannathan L, Ganguli G, Padhi A, Roy D, Alaridah N, Saha P, Nongthomba U, Godaly G, Gopal RK, Banerjee S, Sonawane A. 2015. A mycobacterial phosphoribosyltransferase promotes bacillary survival by inhibiting oxidative stress and autophagy pathways in macrophages and zebrafish. J Biol Chem 290:13321–13343. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, Wang D, Jiang G, Liu W, Deng Q, Li X, Qian W, Ouellet H, Sun J. 2016. EsxA membrane-permeabilizing activity plays a key role in mycobacterial cytosolic translocation and virulence: effects of single-residue mutations at glutamine 5. Sci Rep 6:32618. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Sar AM, Abdallah AM, Sparrius M, Reinders E, Vandenbroucke-Grauls CM, Bitter W. 2004. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect Immun 72:6306–6312. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cernoch P, Enns R, Saubolle M. 1994. Cumitechs 16A, Laboratory Diagnoses of the Mycobacterioses. Coordinating ed., Weissfeld AS. American Society for Microbiology, Washington, DC. [Google Scholar]
- 53.Glickman MS. 2008. Cording, cord factors, and trehalose dimycolate, p 63–73. In Daffe M, Reyrat J (ed), The Mycobacterial Cell Envelope. ASM Press, Washington, DC. [Google Scholar]
- 54.Julián E, Roldán M, Sánchez-Chardi A, Astola O, Agustí G, Luquin M. 2010. Microscopic cords, a virulence-related characteristic of Mycobacterium tuberculosis, are also present in nonpathogenic mycobacteria. J Bacteriol 192:1751–1760. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daffé M, Lanéelle MA, Lacave C. 1991. Structure and stereochemistry of mycolic acids of Mycobacterium marinum and Mycobacterium ulcerans. Res Microbiol 142:397–403. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Ramakrishnan L, Tran HT, Federspiel NA, Falkow S. 1997. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization, and gene disruption via homologous recombination. J Bacteriol 179:5862–5868. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russo C, Tortoli E, Menichella D. 2006. Evaluation of the new GenoType Mycobacterium assay for identification of mycobacterial species. J Clin Microbiol 44:334–339. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tortoli E, Nanetti A, Piersimoni C, Cichero P, Farina C, Mucignat G, Scarparo C, Bartolini L, Valentini R, Nista D, Gesu G, Tosi CP, Crovatto M, Brusarosco G. 2001. Performance assessment of new multiplex probe assay for identification of mycobacteria. J Clin Microbiol 39:1079–1084. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salati F, Meloni M, Fenza A, Angelucci G, Colorni A, Orrù G. 2010. A sensitive FRET probe assay for the selective detection of Mycobacterium marinum in fish. J Fish Dis 33:47–56. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Holmes GF, Harrington SM, Romagnoli MJ, Merz WG. 1999. Recurrent, disseminated Mycobacterium marinum infection caused by the same genotypically defined strain in an immunocompromised patient. J Clin Microbiol 37:3059–3061. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sechi LA, Colorni A, Duprè I, Molicotti P, Fadda G, Zanetti S. 2002. Strain variation in Mediterranean and Red Sea Mycobacterium marinum isolates. New Microbiol 25:351–356. [PubMed] [PubMed] [Google Scholar]
- 62.Stragier P, Ablordey A, Meyers WM, Portaels F. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J Bacteriol 187:1639–1647. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ucko M, Colorni A. 2005. Mycobacterium marinum infections in fish and humans in Israel. J Clin Microbiol 43:892–895. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harriff MJ, Bermudez LE, Kent ML. 2007. Experimental exposure of zebrafish, Danio rerio (Hamilton), to Mycobacterium marinum and Mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: a potential model for environmental mycobacterial infection. J Fish Dis 30:587–600. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Jacobs JM, Rhodes MR, Baya A, Reimschuessel R, Townsend H, Harrell RM. 2009. Influence of nutritional state on the progression and severity of mycobacteriosis in striped bass Morone saxatilis. Dis Aquat Organ 87:183–197. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Ramsay JM, Watral V, Schreck CB, Kent ML. 2009. Husbandry stress exacerbates mycobacterial infections in adult zebrafish, Danio rerio (Hamilton). J Fish Dis 32:931–941. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Decostere A, Hermans K, Haesebrouck F. 2004. Piscine mycobacteriosis: a literature review covering the agent and the disease it causes in fish and humans. Vet Microbiol 99:159–166. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Antuofermo E, Pais A, Polinas M, Cubeddu T, Righetti M, Sanna MA, Prearo M. 2 July 2016. Mycobacteriosis caused by Mycobacterium marinum in reared mullets: first evidence from Sardinia (Italy). J Fish Dis 10.1111/jfd.12515. [DOI] [PubMed] [Google Scholar]
- 69.Caron J, Michot C, Fabre S, Godreuil S, Guillot B, Dereure O. 2011. Aggressive cutaneous infection with Mycobacterium marinum in two patients receiving anti-tumor necrosis factor-alfa agents. J Am Acad Dermatol 65:1060–1062. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Danko JR, Gilliland WR, Miller RS, Decker CF. 2009. Disseminated Mycobacterium marinum infection in a patient with rheumatoid arthritis receiving infliximab therapy. Scand J Infect Dis 41:252–255. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Dare JA, Jahan S, Hiatt K, Torralba KD. 2009. Reintroduction of etanercept during treatment of cutaneous Mycobacterium marinum infection in a patient with ankylosing spondylitis. Arthritis Rheum 61:583–586. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Hess SD, Van Voorhees AS, Chang LM, Junkins-Hopkins JM, Kovarik CL. 2009. Subcutaneous Mycobacterium marinum infection in a patient with chronic rheumatoid arthritis receiving immunosuppressive therapy. Int J Dermatol 48:782–783. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Ramos JM, García-Sepulcre MF, Rodríguez JC, Padilla S, Gutiérrez F. 2010. Mycobacterium marinum infection complicated by anti-tumour necrosis factor therapy. J Med Microbiol 59:617–621. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Asakura T, Ishii M, Kikuchi T, Kameyama K, Namkoong H, Nakata N, Sugita K, Tasaka S, Shimizu T, Hoshino Y, Okamoto S, Betsuyaku T, Hasegawa N. 2016. Disseminated Mycobacterium marinum infection with a destructive nasal lesion mimicking extranodal NK/T cell lymphoma: a case report. Medicine (Baltimore) 95:e3131. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ribot E, Poisnel E, De Biasi C, Roudier J, Balandraud N. 2014. Atypical Mycobacterium marinum infection (aquarium granuloma) in a patient on TNFα antagonist therapy for psoriatic arthritis. Joint Bone Spine 81:272–273. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Kaneko S, Seishima M, Asano Y, Chinuki Y, Morita E. 2014. Mycobacterium marinum infection in a case of psoriasis treated with antitumor necrosis factor α antibody detected by QuantiFERON(®)-TB test. Int J Dermatol 53:e187–e189. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Kump PK, Högenauer C, Wenzl HH, Petritsch W. 2013. A case of opportunistic skin infection with Mycobacterium marinum during adalimumab treatment in a patient with Crohn’s disease. J Crohns Colitis 7:e15–e18. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Demitsu T, Yamada T, Umemoto N, Narita T, Kakurai M, Yoneda K. 2012. Cutaneous Mycobacterium marinum infection mimicking felon in a patient with psoriatic arthritis treated with infliximab. J Dermatol 39:970–971. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Alkhawaja S, Tammam N, Khalifa N. 2010. Mycobacterium marinum infection after infliximab therapy. Iran J Allergy Asthma Immunol 9:255–257. [PubMed] [PubMed] [Google Scholar]
- 80.Aubry A, Chosidow O, Caumes E, Robert J, Cambau E. 2002. Sixty-three cases of Mycobacterium marinum infection: clinical features, treatment, and antibiotic susceptibility of causative isolates. Arch Intern Med 162:1746–1752. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Gluckman SJ. 1995. Mycobacterium marinum. Clin Dermatol 13:273–276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Clark RB, Spector H, Friedman DM, Oldrati KJ, Young CL, Nelson SC. 1990. Osteomyelitis and synovitis produced by Mycobacterium marinum in a fisherman. J Clin Microbiol 28:2570–2572. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edelstein H. 1994. Mycobacterium marinum skin infections. Report of 31 cases and review of the literature. Arch Intern Med 154:1359–1364. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Zenone T, Boibieux A, Tigaud S, Fredenucci JF, Vincent V, Chidiac C, Peyramond D. 1999. Non-tuberculous mycobacterial tenosynovitis: a review. Scand J Infect Dis 31:221–228. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Riera J, Conesa X, Pisa J, Moreno J, Siles E, Novell J. 2016. Septic arthritis caused by Mycobacterium marinum. Arch Orthop Trauma Surg 136:131–134. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Macek P, Bodnarova M, Zavada J, Jezek P, Pavlik I, Slany M, Havelkova M, Stork J, Duskova J, Hanus T, Kocvara R. 2011. Mycobacterium marinum epididymoorchitis: case report and literature review. Urol Int 87:120–124. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Nguyen HH, Fadul N, Ashraf MS, Siraj DS. 2015. Osteomyelitis infection of Mycobacterium marinum: a case report and literature review. Case Rep Infect Dis 2015:905920. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson MG, Stout JE. 2015. Twenty-eight cases of Mycobacterium marinum infection: retrospective case series and literature review. Infection 43:655–662. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lacaille F, Blanche S, Bodemer C, Durand C, De Prost Y, Gaillard JL. 1990. Persistent Mycobacterium marinum infection in a child with probable visceral involvement. Pediatr Infect Dis J 9:58–60. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Streit M, Böhlen LM, Hunziker T, Zimmerli S, Tscharner GG, Nievergelt H, Bodmer T, Braathen LR. 2006. Disseminated Mycobacterium marinum infection with extensive cutaneous eruption and bacteremia in an immunocompromised patient. Eur J Dermatol 16:79–83. [PubMed] [PubMed] [Google Scholar]
- 91.Tchornobay AM, Claudy AL, Perrot JL, Lévigne V, Denis M. 1992. Fatal disseminated Mycobacterium marinum infection. Int J Dermatol 31:286–287. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Lai CC, Lee LN, Chang YL, Lee YC, Ding LW, Hsueh PR. 2005. Pulmonary infection due to Mycobacterium marinum in an immunocompetent patient. Clin Infect Dis 40:206–208. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Philpott JA, Jr, Woodburne AR, Philpott OS, Schaefer WB, Mollohan CS. 1963. Swimming pool granuloma: a study of 290 cases. Arch Dermatol 88:158–162. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Sia TY, Taimur S, Blau DM, Lambe J, Ackelsberg J, Yacisin K, Bhatnagar J, Ritter J, Shieh WJ, Muehlenbachs A, Shulman K, Fong D, Kung E, Zaki SR. 2016. Clinical and pathological evaluation of Mycobacterium marinum group skin infections associated with fish markets in New York City. Clin Infect Dis 62:590–595. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Jernigan JA, Farr BM. 2000. Incubation period and sources of exposure for cutaneous Mycobacterium marinum infection: case report and review of the literature. Clin Infect Dis 31:439–443. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Lewis FM, Marsh BJ, von Reyn CF. 2003. Fish tank exposure and cutaneous infections due to Mycobacterium marinum: tuberculin skin testing, treatment, and prevention. Clin Infect Dis 37:390–397. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Arend SM, van Meijgaarden KE, de Boer K, de Palou EC, van Soolingen D, Ottenhoff TH, van Dissel JT. 2002. Tuberculin skin testing and in vitro T cell responses to ESAT-6 and culture filtrate protein 10 after infection with Mycobacterium marinum or M. kansasii. J Infect Dis 186:1797–1807. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Kobashi Y, Mouri K, Yagi S, Obase Y, Miyashita N, Okimoto N, Matsushima T, Kageoka T, Oka M. 2009. Clinical evaluation of the QuantiFERON-TB Gold test in patients with non-tuberculous mycobacterial disease. Int J Tuberc Lung Dis 13:1422–1426. [PubMed] [PubMed] [Google Scholar]
- 99.Funakoshi T, Kazumi Y, Okada R, Nishimoto K, Saito M, Amagai M, Shimura H, Ohyama M. 2009. Intractable ulcer caused by Mycobacterium shinshuense: successful identification of mycobacterium strain by 16S ribosomal RNA 3′-end sequencing. Clin Exp Dermatol 34:e712–e715. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Wallace R, Glassroth J, Griffith DE, Olivier KN, Cook JL, Gordin F. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am J Respir Crit Care Med 156:S1–S25. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Pfyffer GE, Welscher HM, Kissling P, Cieslak C, Casal MJ, Gutierrez J, Rüsch-Gerdes S. 1997. Comparison of the Mycobacteria Growth Indicator Tube (MGIT) with radiometric and solid culture for recovery of acid-fast bacilli. J Clin Microbiol 35:364–368. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurokawa S, Kabayama J, Fukuyasu T, Hwang SD, Park CI, Park SB, del Castillo CS, Hikima J, Jung TS, Kondo H, Hirono I, Takeyama H, Aoki T. 2013. Bacterial classification of fish-pathogenic Mycobacterium species by multigene phylogenetic analyses and MALDI Biotyper identification system. Mar Biotechnol (New York) 15:340–348. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Pourahmad F, Thompson KD, Taggart JB, Adams A, Richards RH. 2008. Evaluation of the INNO-LiPA mycobacteria v2 assay for identification of aquatic mycobacteria. J Fish Dis 31:931–940. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Gauthier DT, Rhodes MW. 2009. Mycobacteriosis in fishes: a review. Vet J 180:33–47. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Cribier B, Aubry A, Caumes E, Cambau E, Jarlier V, Chosidow O. 2011. Histopathological study of Mycobacterium marinum infection. Ann Dermatol Venereol 138:17–22. (In French.) [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Even-Paz Z, Haas H, Sacks T, Rosenmann E. 1976. Mycobacterium marinum skin infections mimicking cutaneous leishmaniasis. Br J Dermatol 94:435–442. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.El-Khalawany MA. 2014. Atypical mycobacterial cutaneous infections in Egyptians: a clinicopathological study. J Dermatol 41:303–310. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aubry A, Jarlier V, Escolano S, Truffot-Pernot C, Cambau E. 2000. Antibiotic susceptibility pattern of Mycobacterium marinum. Antimicrob Agents Chemother 44:3133–3136. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Utrup LJ, Moore TD, Actor P, Poupard JA. 1995. Susceptibilities of nontuberculosis mycobacterial species to amoxicillin-clavulanic acid alone and in combination with antimycobacterial agents. Antimicrob Agents Chemother 39:1454–1457. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bråbäck M, Riesbeck K, Forsgren A. 2002. Susceptibilities of Mycobacterium marinum to gatifloxacin, gemifloxacin, levofloxacin, linezolid, moxifloxacin, telithromycin, and quinupristin-dalfopristin (Synercid) compared to its susceptibilities to reference macrolides and quinolones. Antimicrob Agents Chemother 46:1114–1116. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown-Elliott BA, Crist CJ, Mann LB, Wilson RW, Wallace RJ, Jr. 2003. In vitro activity of linezolid against slowly growing nontuberculous mycobacteria. Antimicrob Agents Chemother 47:1736–1738. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rhomberg PR, Jones RN. 2002. In vitro activity of 11 antimicrobial agents, including gatifloxacin and GAR936, tested against clinical isolates of Mycobacterium marinum. Diagn Microbiol Infect Dis 42:145–147. [DOI] [PubMed] [Google Scholar]
- 114.Vera-Cabrera L, Brown-Elliott BA, Wallace RJ, Jr, Ocampo-Candiani J, Welsh O, Choi SH, Molina-Torres CA. 2006. In vitro activities of the novel oxazolidinones DA-7867 and DA-7157 against rapidly and slowly growing mycobacteria. Antimicrob Agents Chemother 50:4027–4029. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Parrish N, Luethke R, Dionne K, Carroll K, Riedel S. 2011. Case of Mycobacterium marinum infection with unusual patterns of susceptibility to commonly used antibiotics. J Clin Microbiol 49:2056–2058. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wallace RJ, Jr, Wiss K. 1981. Susceptibility of Mycobacterium marinum to tetracyclines and aminoglycosides. Antimicrob Agents Chemother 20:610–612. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lebrun L, Onody C, Vincent V, Nordmann P. 1996. Evaluation of the Etest for rapid susceptibility testing of Mycobacterium avium to clarithromycin. J Antimicrob Chemother 37:999–1003. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Flynn CM, Kelley CM, Barrett MS, Jones RN. 1997. Application of the Etest to the antimicrobial susceptibility testing of Mycobacterium marinum clinical isolates. J Clin Microbiol 35:2083–2086. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Werngren J, Olsson-Liljequist B, Gezelius L, Hoffner SE. 2001. Antimicrobial susceptibility of Mycobacterium marinum determined by E-test and agar dilution. Scand J Infect Dis 33:585–588. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Chazel M, Marchandin H, Keck N, Terru D, Carrière C, Ponsoda M, Jacomo V, Panteix G, Bouzinbi N, Bañuls AL, Choisy M, Solassol J, Aubry A, Godreuil S. 2016. Evaluation of the SLOMYCO Sensititre(®) panel for testing the antimicrobial susceptibility of Mycobacterium marinum isolates. Ann Clin Microbiol Antimicrob 15:30. [PubMed] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huminer D, Pitlik SD, Block C, Kaufman L, Amit S, Rosenfeld JB. 1986. Aquarium-borne Mycobacterium marinum skin infection. Report of a case and review of the literature. Arch Dermatol 122:698–703. [PubMed] [PubMed] [PubMed] [Google Scholar]
- 122.Chow SP, Ip FK, Lau JH, Collins RJ, Luk KD, So YC, Pun WK. 1987. Mycobacterium marinum infection of the hand and wrist. Results of conservative treatment in twenty-four cases. J Bone Joint Surg Am 69:1161–1168. [PubMed] [PubMed] [PubMed] [Google Scholar]
- 123.Iijima S, Saito J, Otsuka F. 1997. Mycobacterium marinum skin infection successfully treated with levofloxacin. Arch Dermatol 133:947–949. [DOI] [PubMed] [Google Scholar]
- 124.Kozin SH, Bishop AT. 1994. Atypical Mycobacterium infections of the upper extremity. J Hand Surg Am 19:480–487. [DOI] [PubMed] [Google Scholar]
- 125.Bhatty MA, Turner DP, Chamberlain ST. 2000. Mycobacterium marinum hand infection: case reports and review of literature. Br J Plast Surg 53:161–165. [DOI] [PubMed] [Google Scholar]
- 126.Heckert RA, Elankumaran S, Milani A, Baya A. 2001. Detection of a new Mycobacterium species in wild striped bass in the Chesapeake Bay. J Clin Microbiol 39:710–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zanoni RG, Florio D, Fioravanti ML, Rossi M, Prearo M. 2008. Occurrence of Mycobacterium spp. in ornamental fish in Italy. J Fish Dis 31:433–441. [DOI] [PubMed] [Google Scholar]
- 128.Slany M, Makovcova J, Jezek P, Bodnarova M, Pavlik I. 2014. Relative prevalence of Mycobacterium marinum in fish collected from aquaria and natural freshwaters in central Europe. J Fish Dis 37:527–533. [DOI] [PubMed] [Google Scholar]
- 129.Broutin V, Bañuls AL, Aubry A, Keck N, Choisy M, Bernardet JF, Michel C, Raymond JC, Libert C, Barnaud A, Stragier P, Portaels F, Terru D, Belon C, Dereure O, Gutierrez C, Boschiroli ML, Van De Perre P, Cambau E, Godreuil S. 2012. Genetic diversity and population structure of Mycobacterium marinum: new insights into host and environmental specificities. J Clin Microbiol 50:3627–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leoni E, Legnani P, Mucci MT, Pirani R. 1999. Prevalence of mycobacteria in a swimming pool environment. J Appl Microbiol 87:683–688. [DOI] [PubMed] [Google Scholar]
- 131.Bouricha M, Castan B, Duchene-Parisi E, Drancourt M. 2014. Mycobacterium marinum infection following contact with reptiles: vivarium granuloma. Int J Infect Dis 21:17–18. [DOI] [PubMed] [Google Scholar]
- 132.Huaman MA, Ribes JA, Lohr KM, Evans ME. 2015. Mycobacterium marinum infection after exposure to coal mine water. Open Forum Infect Dis 3:ofv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Feng Y, Xu H, Wang H, Zhang C, Zong W, Wu Q. 2011. Outbreak of a cutaneous Mycobacterium marinum infection in Jiangsu Haian, China. Diagn Microbiol Infect Dis 71:267–272. [DOI] [PubMed] [Google Scholar]
- 134.Swift S, Cohen H. 1962. Granulomas of the skin due to Mycobacterium balnei after abrasions from a fish tank. N Engl J Med 267:1244–1246. [DOI] [PubMed] [Google Scholar]
- 135.Dobos KM, Quinn FD, Ashford DA, Horsburgh CR, King CH. 1999. Emergence of a unique group of necrotizing mycobacterial diseases. Emerg Infect Dis 5:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.King AJ, Fairley JA, Rasmussen JE. 1983. Disseminated cutaneous Mycobacterium marinum infection. Arch Dermatol 119:268–270. [PubMed] [Google Scholar]
- 137.Good RC. 1980. From the Center for Disease Control. Isolation of nontuberculous mycobacteria in the United States, 1979. J Infect Dis 142:779–783. [DOI] [PubMed] [Google Scholar]
- 138.van der Werf MJ, Ködmön C, Katalinić-Janković V, Kummik T, Soini H, Richter E, Papaventsis D, Tortoli E, Perrin M, van Soolingen D, Zolnir-Dovč M, Ostergaard Thomsen V. 2014. Inventory study of non-tuberculous mycobacteria in the European Union. BMC Infect Dis 14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Casal M, Casal MM, Spanish Group of Mycobacteriology. 2001. Multicenter study of incidence of Mycobacterium marinum in humans in Spain. Int J Tuberc Lung Dis 5:197–199. [PubMed] [Google Scholar]
- 140.Astrofsky KM, Schrenzel MD, Bullis RA, Smolowitz RM, Fox JG. 2000. Diagnosis and management of atypical Mycobacterium spp. infections in established laboratory zebrafish (Brachydanio rerio) facilities. Comp Med 50:666–672. [PubMed] [Google Scholar]
- 141.Passantino A, Macrì D, Coluccio P, Foti F, Marino F. 2008. Importation of mycobacteriosis with ornamental fish: medico-legal implications. Travel Med Infect Dis 6:240–244. [DOI] [PubMed] [Google Scholar]
- 142.Dailloux M, Laurain C, Weber M, Hartemann P. 1999. Water and nontuberculous mycobacteria. Water Res 33:2219–2228. [Google Scholar]
- 143.Parent LJ, Salam MM, Appelbaum PC, Dossett JH. 1995. Disseminated Mycobacterium marinum infection and bacteremia in a child with severe combined immunodeficiency. Clin Infect Dis 21:1325–1327. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Schmoor P, Descamps V, Bouscarat F, Grossin M, Belaïch S, Crickx B. 2003. Tropical fish salesmen’s knowledge and behaviour concerning “fish tank granuloma.” Ann Dermatol Venereol 130:425–427. (In French.) [PubMed] [PubMed] [Google Scholar]
- 145.Bonafe JL, Grigorieff-Larrue N, Bauriaud R. 1992. Atypical cutaneous mycobacterium diseases. Results of a national survey. Ann Dermatol Venereol 119:463–470. [PubMed] [PubMed] [Google Scholar]
- 146.Ho MH, Ho CK, Chong LY. 2006. Atypical mycobacterial cutaneous infections in Hong Kong: 10-year retrospective study. Hong Kong Med J 12:21–26. [PubMed] [PubMed] [Google Scholar]
- 147.Blanc P, Dutronc H, Peuchant O, Dauchy FA, Cazanave C, Neau D, Wirth G, Pellegrin JL, Morlat P, Mercie P, Tunon-de-Lara JM, Doutre MS, Pelissier P, Dupon M. 2016. Nontuberculous mycobacterial infections in a French hospital: a 12-year retrospective study. PLoS One 11:e0168290. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


