Abstract
UL12 is a 5′- to 3′-exonuclease encoded by herpes simplex virus type 1 (HSV-1) which degrades single- and double-stranded DNA. UL12 and the single-strand DNA binding protein ICP8 mediate a strand exchange reaction. We found that ICP8 inhibited UL12 digestion of single-stranded DNA but stimulated digestion of double-stranded DNA threefold. The stimulatory effect of ICP8 was independent of a strand exchange reaction; furthermore, the effect was specific to ICP8, as it could not be reproduced by Escherichia coli single-stranded DNA binding protein. The effect of ICP8 on the rate of UL12 double-stranded DNA digestion is attributable to an increase in processivity in the presence of ICP8.
The herpes simplex virus type 1 (HSV-1) contains a large double-stranded DNA (dsDNA) genome and appears to utilize a recombination-dependent pathway during DNA replication which is reminiscent of the large DNA bacteriophages (reviewed in reference 16). We have recently found that the HSV-1 single-stranded DNA (ssDNA) binding protein (SSB) ICP8 and the alkaline nuclease UL12 are capable of catalyzing an in vitro strand exchange reaction (10). In this reaction, the exonuclease resects DNA from an exposed double-stranded end, and the SSB anneals complementary single-stranded regions. Evidence from the analogous lambda Red and RecE/T systems indicates that the in vivo recombinase activity depends upon the specific interaction of the two components, as an exonuclease from one pair is unable to promote recombination with the synaptase of the other (7). This result suggests that the two proteins specifically influence the activity of one another. Here we investigate whether UL12 nuclease activity is affected by the presence of the synaptase ICP8 and whether any influence on activity coincides with the strand exchange reaction.
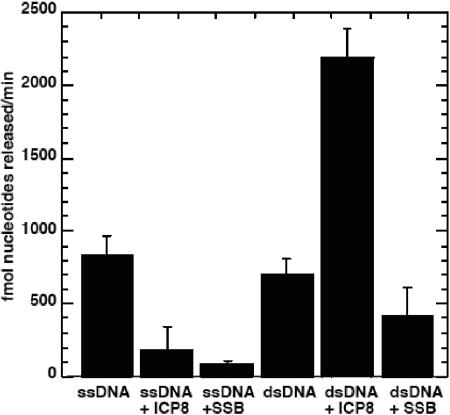
We assayed the in vitro nuclease and strand exchange activities of UL12 in the presence or absence of ICP8, using substrates and reaction conditions defined previously for these assays (10, 11). The substrate used was a PCR-generated 1.5-kb fragment, uniformly labeled by the inclusion of [32P]dCTP in the deoxynucleoside triphosphate mix. For measurements of digestion of single-stranded DNA, the substrate was prepared by boiling and quickly cooling the1.5-kb fragment. Figure 1 shows that the rates of digestion by UL12 were nearly the same for both dsDNA and ssDNA. In the presence of ICP8, however, the digestion of single-stranded DNA was greatly reduced. This is because ICP8 can coat the single-stranded DNA, making it inaccessible to UL12 digestion. In contrast, the rate of digestion of double-stranded DNA was tripled in the presence of ICP8. One explanation could be that ICP8 binds to the 3′-single-stranded tail that is produced by UL12 digestion and eliminates secondary structure that could interfere with further digestion by UL12. If this were the reason for the stimulation of nuclease activity, we would expect another SSB to have the same effect. As shown in Fig. 1, Escherichia coli SSB did not show any stimulatory effect on UL12 digestion of dsDNA. It did, however, inhibit digestion on ssDNA in a manner similar to ICP8. Thus, the stimulatory effect of ICP8 on UL12 was specific, since it could not be reproduced by another SSB.
FIG. 1.
UL12 nuclease activity in the presence of ICP8 or SSB. Reactions were performed in a buffer containing 20 mM Tris-Cl (pH 7.5), 1 mM MgCl2, 40 mM NaCl, 1 mM dithiothreitol, 20 ng 32P-labeled 1.5-kb dsDNA or ssDNA, 13.9 nM UL12, 1.75 μM ICP8, and 1.5 μM E. coli SSB, as indicated.
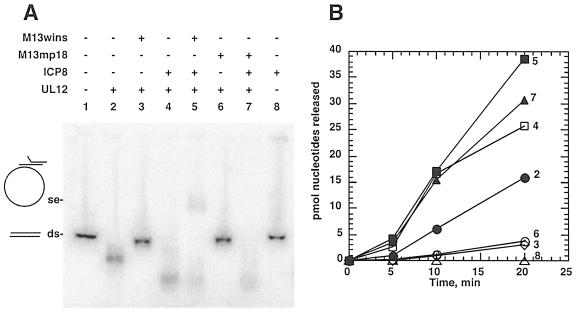
Since it has been proposed that synaptases modulate the activity of their cognate nucleases during strand exchange (15), we next examined the effect of ICP8 on UL12 during the strand exchange reaction. In this reaction, the 32P-labeled 1.5-kb fragment was paired with unlabeled single-stranded DNA that either possesses the 1.5-kb insert (M13wins) or does not share homology with the dsDNA substrate (M13pm18) (11). The strand exchange product was seen only when both UL12 and ICP8 were incubated together with the 1.5-kb fragment and the homologous ssDNA acceptor, M13wins (Fig. 2A, lane 5). Figure 2A also demonstrates the digestion of the dsDNA fragment by UL12. A modest shortening of the double-stranded fragment was observed upon incubation with UL12 alone (Fig. 2A, lane 2). When both the dsDNA and ssDNA substrates were present, however, dsDNA digestion was inhibited, probably due to the ability of the ssDNA to compete with the 1.5-kb fragment for UL12 binding and digestion (Fig. 2A, lanes 3 and 6). When ICP8 was added, in the presence or absence of ssDNA, the digestion of the 1.5-kb fragment was enhanced (Fig. 2A, lanes 4, 5, and 7). The digestion of the fragment appeared to be similar whether there was strand exchange (lane 5) or whether there was no strand exchange (lanes 4 and 7), indicating that the stimulatory effect of ICP8 is independent of the strand exchange reaction or the presence of ssDNA. ICP8 alone had no detectable nuclease activity (lane 8). Figure 2B confirms that ICP8 enhanced the nuclease activity of UL12 on dsDNA both in the presence and absence of the strand exchange process. Thus, UL12 apparently differs from other nuclease/synpatase members (15) in that the nuclease activity is apparently not modulated by homology during strand exchange.
FIG. 2.
ICP8 stimulates UL12 digestion of dsDNA in the presence or absence of strand exchange. (A) The strand exchange assay was performed, and products were separated by agarose gel electrophoresis. All reactions included the 32P-labeled 1.5-kb dsDNA substrate (20 ng). M13wins ssDNA (100 ng), M13mp18 ssDNA (100 ng), ICP8 (1.75 μM), and UL12 (13.9 nM) were added as indicated. Reaction conditions were as described for Fig. 1. Incubations were for 20 min at 37°C. The phosphorimage of the dried gel is shown. The positions of the double-stranded 1.5-kb substrate (ds) and the strand exchange products (se) are indicated. (B) Samples from the strand exchange reaction shown in panel A were removed at different time points for measurement of nuclease activity. All reactions included the 1.5-kb double-stranded 32P-labeled substrate. Numbers at the ends of the data curves refer to the lane numbers in panel A. Closed circles, UL12; open squares, UL12 + ICP8; filled squares, UL12 + ICP8 + ssM13wins; closed triangles, UL12 + ICP8 + ssM13mp18; open diamonds, UL12 + ssM13wins; open circles, UL12 + ssM13mp18; open triangles, ICP8.
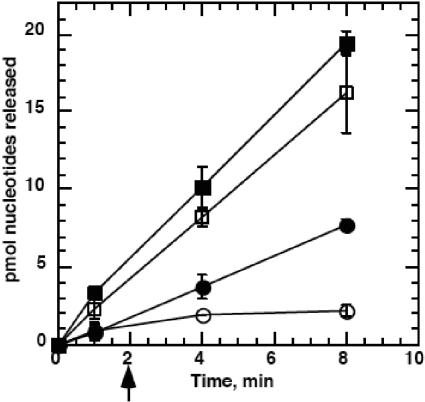
We next investigated the mechanism of the ICP8-mediated stimulation of UL12 nuclease activity. In their characterization of UL12, Hoffman and Cheng determined that UL12 is not a processive nuclease (3). If the initiation step of the UL12 nuclease reaction is slow, the effective rate of the reaction can be increased by enhancing the processivity of the enzyme. We therefore tested whether ICP8 increased the rate of UL12 digestion by increasing the processivity of the UL12 nuclease. We used heparin to trap the nuclease that had dissociated from the DNA. When heparin was added prior to UL12 or UL12/ICP8, no digestion of the substrates was observed, indicating that the amount of heparin used was sufficient to trap UL12 (data not shown). As shown in Fig. 3, the addition of heparin (marked with an arrow) completely arrested digestion by UL12 alone, consistent with its identification as a nonprocessive enzyme. In contrast, UL12 digestion in the presence of ICP8 was essentially unaffected by heparin, showing that UL12 did not dissociate from the DNA during the course of the assay. This demonstrates a significant increase in processivity in the presence of ICP8 and is likely to be the reason for the stimulation of UL12 nuclease activity. We cannot rule out that other factors such as direct stimulation of the nuclease or improved loading also contribute to the enhanced UL12 activity in the presence of ICP8, and further experiments will be required to analyze these possibilities.
FIG. 3.
ICP8 increases the processivity of UL12 on dsDNA. The nuclease assay was performed by first preincubating all reactants in the absence of Mg2+ for 2 min at 37°C, to allow for binding of the enzymes to the substrate DNA. The assay was initiated with the addition of MgCl2 to 1 mM. At 2 min (marked with arrow), 2 μl of 44 mg/ml heparin (4 mg/ml final) or water (controls) was added. Closed squares, UL12 + ICP8, no heparin; open squares, UL12 + ICP8 + heparin; closed circles, UL12 no heparin; open circles, UL12 + heparin. Protein concentrations were the same as in Fig. 1.
Many processivity-enhancing proteins rely upon topological tethering, often achieved by formation of rings which encircle the DNA. In contrast, HSV enzymes such as the polymerase processivity subunit, UL42, utilize an unusual mechanism to promote processive polymerase activity. UL42 binds DNA as a monomer (8), and it has been suggested that electrostatic attraction holds UL42 in close proximity to the DNA, providing processivity without formation of a ring structure around the DNA (17). In addition, in contrast to most replicative helicases which are ring-forming hexamers, the HSV replicative helicase is not (6). Another example is provided by the HSV-1 origin binding protein UL9, whose helicase activity is stimulated by ICP8 and is mediated via specific protein-protein interactions (1, 2). Thus, the HSV replicative proteins appear to have evolved mechanisms to ensure processivity without topological tethering via ring formation. The crystal structure of λ exonuclease indicates that it is a homotrimer with a channel large enough to accommodate DNA, providing a structural basis for its processivity (5). On the other hand, UL12 has been shown to be a dimer (4). Based on the previous reports by us and others that ICP8 and UL12 specifically interact with one another (9, 12-14) and the results presented in this paper, we propose that UL12 may achieve processivity via protein-protein interactions with ICP8, consistent with the virus' apparent avoidance of ring formation in establishing processivity.
We previously showed that potent ICP8 recombinase activity is revealed only upon resection of dsDNA substrate by a nuclease (11). These data, combined with the reported protein-protein interactions between ICP8 and UL12 and the specific stimulation of UL12 activity by ICP8 reported in this paper, support our hypothesis that UL12 and ICP8 act together to promote homologous recombination during HSV genome replication. The purification of ICP8 and UL12 mutant proteins that do not interact with one another is currently under way in order to further test this model.
Acknowledgments
We thank Nandakumar Balasubramarian for helpful comments on the manuscript.
This work was supported by grants to S.W. (A121747, A137549) and by a grant from the Damon Runyon Cancer Research Fund (DRG-1625) to N.B.R.
REFERENCES
- 1.Arana, M. E., B. Haq, N. Tanguy Le Gac, and P. E. Boehmer. 2001. Modulation of the herpes simplex virus type-1 UL9 DNA helicase by its cognate single-strand DNA-binding protein, ICP8. J. Biol. Chem. 276:6840-6845. [DOI] [PubMed] [Google Scholar]
- 2.Boehmer, P. E. 1998. The herpes simplex virus type-1 single-strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase. J. Biol. Chem. 273:2676-2683. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, P. J., and Y. C. Cheng. 1978. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J. Biol. Chem. 253:3557-3562. [PubMed] [Google Scholar]
- 4.Kehm, E., M. A. Goksu, and C. W. Knopf. 1998. Expression analysis of recombinant herpes simplex virus type 1 DNase. Virus Genes 17:129-138. [DOI] [PubMed] [Google Scholar]
- 5.Kovall, R., and B. W. Matthews. 1997. Toroidal structure of lambda-exonuclease. Science 277:1824-1827. [DOI] [PubMed] [Google Scholar]
- 6.Marintcheva, B., and S. K. Weller. 2001. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 70:77-118. [DOI] [PubMed] [Google Scholar]
- 7.Muyrers, J. P., Y. Zhang, F. Buchholz, and A. F. Stewart. 2000. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 14:1971-1982. [PMC free article] [PubMed] [Google Scholar]
- 8.Randell, J. C., and D. M. Coen. 2004. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol. 335:409-413. [DOI] [PubMed] [Google Scholar]
- 9.Reuven, N. B., S. Antoku, and S. K. Weller. 2004. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 78:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuven, N. B., S. Willcox, J. D. Griffith, and S. K. Weller. 2004. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 342:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas, M. S., M. Gao, D. M. Knipe, and K. L. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaughan, P. J., L. M. Banks, D. J. Purifoy, and K. L. Powell. 1984. Interactions between herpes simplex virus DNA-binding proteins. J. Gen. Virol. 65:2033-2041. [DOI] [PubMed] [Google Scholar]
- 15.Vellani, T. S., and R. S. Myers. 2003. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol. 185:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451-458. [DOI] [PubMed] [Google Scholar]
- 17.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]