Abstract
Mannose binding lectin (MBL) is a central component of the innate immune response and thus may be important for determining hepatitis B virus (HBV) persistence. Since single-nucleotide polymorphisms (SNPs) in the gene encoding MBL (mbl2) alter the level of functional MBL, we hypothesized that mbl2 genotypes are a determinant of HBV persistence or recovery from viral infection. We tested this hypothesis by using a nested case control design with 189 persons with HBV persistence matched to 338 individuals who had naturally recovered from HBV infection. We determined genotypes of two promoter and three exon 1 SNPs in mbl2 and grouped these genotypes according to the amount of functional MBL production. We found that the promoter SNP −221C, which leads to deficient MBL production, was more common in those subjects with viral persistence (odds ratio [OR], 1.38; 95% confidence interval [CI], 1.01 to 1.89; P = 0.04). Those subjects homozygous for the combination of promoter and exon 1 genotypes associated with the highest amount of functional MBL had significantly increased odds of recovery from infection (OR, 0.55; 95% CI, 0.37 to 0.84; P = 0.005). Conversely, those homozygous for the combination of promoter and exon 1 genotypes which produce the lowest amount of functional MBL were more likely to have viral persistence (OR, 1.76; 95% CI, 1.02 to 3.01; P = 0.04). These data are consistent with the hypothesis that functional MBL plays a central role in the pathogenesis of acute hepatitis B.
Chronic hepatitis B infection affects 400 million people and is the most common cause of cirrhosis and hepatocellular carcinoma worldwide. Although most adults recover from acute hepatitis B virus (HBV) infection, it is not fully understood why some develop chronic hepatitis B. The strength and breadth of the host immune response are important (3, 30), and recent data from chimpanzees and transgenic mice suggest that differences in innate immunity affect recovery from HBV infection (11, 16).
Mannose binding lectin (MBL) plays a central role in the innate immune response (32). mbl2, which encodes MBL, is located on chromosome 10 and consists of four exons (Fig. 1). Several single-nucleotide polymorphisms (SNPs) in the gene's promoter and in exon 1 that ultimately reduce the level of functional MBL have been described (8). Reduced MBL concentrations have been linked with diminished responses to several infectious diseases, including human immunodeficiency virus (HIV) infection and recurrent infections, and higher levels have been linked with inflammatory outcomes such as vascular complications of diabetes mellitus (5, 12, 24, 29). Although one exon 1 SNP has been associated with HBV persistence (31), this gene and its functionally different haplotypes have not been comprehensively examined in persons infected with HBV.
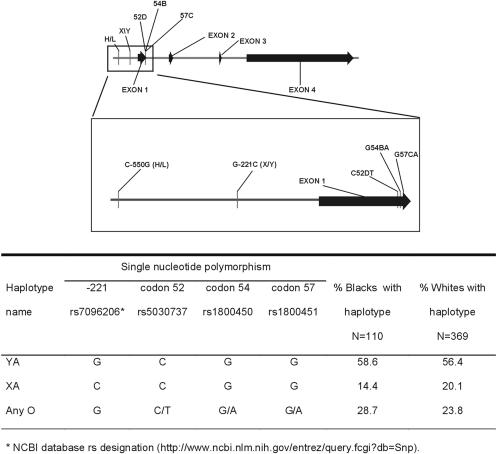
FIG. 1.
Schematic diagram of mbl2 including the gene's promoter region, which spans 7,100 bases and consists of four exons. An enlarged panel illustrates the relative positions of the promoter and exon 1 SNPs genotyped in this study. The SNPs in the promoter are designated by a minus sign and the exon 1 SNPs are designated by their codon numbers, which is the standard nomenclature for this gene. The wild-type and mutant alleles are designated before and after the SNP numbers, respectively. Below the diagram is a table delineating haplotype names, allele composition of each haplotype, and frequencies in blacks and whites. The rs numbers for each SNP are indicated in the table and are taken from the Entrez SNP website (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Snp). The rs number for the −550 SNP is 11003125.
We hypothesized that the mbl2 SNPs that reduce functional MBL levels would be found more often in persons with HBV persistence and vice versa. To test this hypothesis, we determined genotypes of the promoter SNPs at −550 and −221 and the three exon 1 SNPs at codons 52, 54, and 57 by using a well-characterized cohort of individuals with either HBV persistence or recovery.
MATERIALS AND METHODS
Study participants.
Subjects in this study were participants in one of two other studies: (i) the AIDS Link to Intravenous Experience (ALIVE) study, which is an ongoing study of 2,921 injection drug users enrolled in Baltimore, Md., from February 1988 to March 1989, as previously described (33), or (ii) the Multicenter AIDS Cohort Study (MACS), which is an ongoing study of 5,622 homosexual men enrolled in one of four U.S. cities between 1984 and 1985 and between 1987 and 1991 (4, 18).
To investigate the hypothesis that mbl2 SNPs may be associated with recovery from acute hepatitis B, a nested case control design was used. When possible, participants from the cohorts with persistent HBV infections were matched to two persons from the same cohort who had recovered from HBV infection but who were otherwise similar with regard to nongenetic factors. If two matched controls were not available, then one control was matched. Matching criteria included geographic location and factors that have been associated with recovery from hepatitis B, including age within 10 years, gender, ethnicity, and human immunodeficiency virus type 1 (HIV-1) infection status (9, 15). Subjects were considered persistently infected with HBV if their sera or plasma tested positive for HBV surface antigen (HBsAg) at two visits separated by a minimum of 6 months. Testing for antibodies against HBV core antigen (anti-HBc) and HBsAg (anti-HBs) was performed as needed to exclude primary HBV infection. Individuals recovered from hepatitis B were positive for anti-HBc and anti-HBs without the presence of HBsAg at two time points separated by a minimum of 6 months. HBV infection statuses of HIV-positive subjects were determined before antiretroviral therapy was available.
Informed consent was obtained from all participants, and the study was approved by the institutional review boards at all participating institutions.
Serologic testing.
All serum specimens were stored at −70°C until testing. HIV-1 antibody testing was done by enzyme immunoassay, with reactive results confirmed as positive by Western blotting as previously reported (4, 18, 33). HBsAg, anti-HBs, and anti-HBc testing was done using commercially available kits according to the specifications of the manufacturer (AUSZYME, AUSAB, and CORZYME, respectively; Abbott Laboratories, Abbott Park, IL).
DNA extraction and mbl2 genotyping.
For each individual, cell lines transformed with Epstein-Barr virus were established, and genomic DNA was extracted from these cell lines by using phenol-chloroform.
The mbl2 SNPs at −550, −221, codon 52, and codon 57 were genotyped using the AcycloPrime-FP SNP detection assay (Perkin Elmer, Boston, MA), a single-base extension method performed according to the manufacturer's specifications (14). In this method, a 200-bp fragment containing the SNP of interest is amplified (primers are listed in Table 1) with cycling conditions of 95°C for 10 min; 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 60 s; and then a final extension step of 72°C for 10 min. After amplification, the excess primer and deoxynucleoside triphosphates are degraded with shrimp alkaline phosphatase and exonuclease I according to the manufacturer's specifications. In the final step, one of two fluorescent terminators representing the alleles present at the SNP of interest is added to a primer ending immediately upstream of the SNP site (extension primer). The cycling protocol for the single-base extension step is 95°C for 2 min followed by 15 cycles of 95°C for 15 s and 55°C for 30 s. The results are identified by the amount of fluorescence polarization (FP) of each allele as determined by the Victor2V instrument (Perkin Elmer, Boston, MA). We verified the FP results with direct sequencing for 10 samples from each SNP and found no errors.
TABLE 1.
Primers used for FP and sequence-specific primers (SSP)
| Primer namea | Sequence (5′ → 3′) |
|---|---|
| FP primers | |
| Exon 1 forward | TGGTGGCAGCGTCTTACTCA |
| Exon 1 reverse | CCCAGGCAGTTTCCTCTGGA |
| 52D extension (C/T) | CAACGGCTTCCCAGGCAAAGATGGG |
| 57C extension (C/T) | CAACACGTACCTGGTTCCCCCTTTTCT |
| −550 forward | ACTCTGCCAGGGCCAACGTA |
| −550 reverse | CAGCTGATTCCCCTCCAGGAC |
| −550 extension (G/C) | GAAAATGCTTACCCAGGCAAGCCTGT |
| −221 forward | GGGATTCAGGTGGCAGATGG |
| −221 reverse | TGATGAGCAGTGGGGATCCTA |
| −221 extension (G/C) | CGGTCCCATTTGTTCTCACTGCCAC |
| SSP | |
| 54B mutant forward | CCCCCTTTTCTCCCTTGGTGT |
| 54B wild type forward | CCCCCTTTTCTCCCTTGGTGC |
| 54B reverse | CTGCACCCAGATTGTAGGACAGAG |
Letters in parentheses indicate bases at polymorphic alleles.
The SNP at codon 54 was genotyped by PCR sequence-specific primers with which two complementary reactions were run for each SNP. The two reactions shared a common primer, but the second primers differed at their 3′-terminal ends by one base, which represents the bases at the polymorphic alleles. The PCR products were run on a 1% agarose gel, and an allele was assigned as present if a band was detected. The procedure was verified by sequencing the SNPs from 10 individuals with no errors. Ambiguous genotypes from either method were assigned based on direct sequencing.
Statistical analysis.
The first analysis consisted of examining the individual SNPs. Allele frequencies for each SNP were calculated from the genotypes and compared with respect to recovery from infection and viral persistence by using conditional logistic regression (SAS version 10; SAS, Cary, NC). An odds ratio (OR) of >1 was associated with viral persistence, and an OR of <1 was associated with recovery. Hardy-Weinberg equilibrium was assessed for each SNP by using the chi-square test with 1 degree of freedom.
Since examining individual SNPs does not fully account for differences in the amounts of functional MBL produced, we analyzed subjects grouped by genotypes that have been found to correlate with the amount of functional MBL (8). The group designation system follows previously established nomenclature for the gene. Groups were established by determining haplotypes consisting of exon 1 and −221 genotypes. Haplotypes were constructed using software designed for population-based studies, PHASE version 2.0 (http://www.stat.washington.edu/stephens/software.html) (28). The exon 1 genotype with the C allele at codon 52, the G allele at codon 54, and the G allele at codon 57 was designated A since this produces normal levels of functional MBL (Fig. 1 table). If the genotype included one or more of the alternate alleles, T allele at codon 52, A allele at codon 54, or A allele at codon 57, it was designated O since any one of these significantly reduces the amount of functional MBL. The analysis was then expanded to include the −221 SNP. The genotype with the G allele at −221, which produces normal levels of functional MBL, was designated Y, whereas that with the C allele, which profoundly down-regulates the amount of functional MBL, was designated X. If the exon 1 genotype was O, the −221 SNP was not included since the amount of functional MBL was already so low that it was not significantly altered by the −221 genotype. Thus, the possible haplotypes were YA, XA, and O. The frequencies of these haplotypes were similar in whites and blacks (Fig. 1 table), so the analyses were done with the entire cohort and the results were then stratified by ethnicity if significant. Since each person has two haplotypes, ultimately, six mbl2 genotypes were possible for each subject: YA/YA, YA/XA, XA/XA, YA/O, XA/O, and O/O. For the analysis, we divided patients into high-MBL (YA/YA) and low-MBL or MBL-deficient (XA/O and O/O) groups based on published data regarding MBL concentrations measured in persons with these genotypes (8). All other genotypes were considered intermediate, and these groups were compared with respect to recovery from infection and viral persistence by using conditional logistic regression. The haplotype analyses were also extended to include the −550 SNP to determine the effect of the −550 promoter SNP.
A P of <0.05 was considered significant in all analyses, and results for any genotype that met this criterion were stratified by the individuals' HIV infection statuses to exclude HIV association. Genotypes were also stratified by subjects' ethnicity (i.e., black versus white) to evaluate ethnic differences. Since those subjects in the other-ethnicity category were heterogeneous and small in number, they were excluded from the ethnicity analysis.
RESULTS
Study subjects.
The study group was composed of 189 persons with chronic hepatitis B and 338 persons who had recovered from HBV infection (40 chronically infected persons had only one match), for a total of 378 and 676 alleles in each group, respectively. No significant differences were detected between those recovered from hepatitis B and those with HBV persistence with respect to the matching criteria: 64% were HIV positive, the mean age was 34 years, and 98% were male. The majority of the study group was white (76%), with 22% black and 2% of other ethnicity.
Individual SNPs and haplotypes.
In univariate analysis of the −550, −221, codon 52, codon 54, and codon 57 SNPs, only −221C (X allele) was associated with viral persistence (OR, 1.38; 95% confidence interval [CI], 1.01 to 1.89; P = 0.04) (Table 2). Homozygosity for the X allele was infrequent (3.7%) and did not strengthen this relationship. Homozygosity for the Y allele was associated with viral clearance, but the association was not stronger than that for Y alone (OR, 0.65; P = 0.03). None of the other SNPs in either the homozygous or heterozygous state were associated with outcome. The association with the X allele was the same when results were stratified by black and white ethnicities and when they were stratified by HIV infection statuses (Table 4).
TABLE 2.
Frequencies of mbl2 SNPs and haplotypes in persons recovered from hepatitis B and those with viral persistence
| Genotype | % Of chromosomesa with the indicated genotype in:
|
OR | 95% CI | P | |
|---|---|---|---|---|---|
| Recovered subjects (n = 648-676) | Subjects with viral persistence (n = 372-378) | ||||
| SNPs | |||||
| −550G | 33.0 | 29.3 | 0.82 | 0.61-1.09 | 0.17 |
| −221C (X allele) | 17.7 | 23.4 | 1.38 | 1.01-1.89 | 0.04 |
| Codon 52T | 6.7 | 6.6 | 0.97 | 0.58-1.61 | 0.90 |
| Codon 54A | 10.0 | 13.4 | 1.35 | 0.90-2.03 | 0.15 |
| Codon 57A | 6.8 | 5.9 | 0.95 | 0.53-1.68 | 0.85 |
| Haplotypes | |||||
| YA | 59.8 | 51.9 | 0.73 | 0.56-0.94 | 0.02 |
| XA | 17.5 | 23.0 | 1.40 | 1.03-1.92 | 0.03 |
| O | 40.9 | 44.1 | 1.14 | 0.78-1.66 | 0.50 |
n is the total number of chromosomes for which genotypes were determined and varies slightly due to the failure of some specimens to amplify for a particular SNP.
TABLE 4.
ORs for significant SNPs and haplotypes stratified by ethnicity and HIV infection status
| Genotype(s)a | OR (95% CI) for subjects with the following characteristicb:
|
|||
|---|---|---|---|---|
| black (n = 110) | white (n = 369) | HIV positive (n = 330) | HIV negative (n = 189) | |
| −221C | 1.53 (0.71-3.30) | 1.41 (0.99-2.00) | 1.34 (0.90-2.01) | 1.55 (0.89-2.69) |
| YA/YA | 0.61 (0.27-1.37) | 0.50 (0.31-0.82) | 0.55 (0.32-0.94) | 0.49 (0.24-1.00) |
| XA/O or O/O | 1.36 (0.42-4.41) | 1.97 (1.06-3.64) | 1.85 (0.86-4.00) | 2.24 (0.93-5.40) |
Genotypes are defined in Materials and Methods.
n, number of study subjects with the indicated characteristic.
Haplotype analysis demonstrated that those with the YA haplotype were more likely to recover from an HBV infection (OR, 0.73; 95% CI, 0.56 to 0.94; P = 0.02). Since the X allele at −221 occurred only with the A haplotype, the XA haplotype did not differ from X alone. The O haplotype was not associated with either recovery or viral persistence.
Genotypic groups based on expected functional MBL levels.
Several studies have demonstrated a consistent relationship between MBL levels and mbl2 genotypes (8, 27). Individuals homozygous for YA (YA/YA) have the highest levels, whereas persons with an X allele at −221 on one chromosome and an O on the other chromosome (XA/O) or who are homozygous for O (O/O) have the lowest levels (8). Using these previously established data, we divided our genotypes into one of three groups: (i) YA/YA, (ii) XA/O or O/O, and (iii) all other combinations, which included those with MBL levels intermediate between those of the two former groups. Those subjects with the YA/YA genotype had significantly increased odds of recovery from infection (OR, 0.55; 95% CI, 0.37 to 0.84; P = 0.005), whereas those with XA/O or O/O were more likely to have viral persistence (OR, 1.76; 95% CI, 1.02 to 3.01; P = 0.04) (Table 3). A clear trend between mbl2 genotype and HBV infection outcome was noted in these three groups (P of 0.002 for Maentel-Haenszel test for trend). These relationships were not altered with stratification by HIV infection status or ethnicity (Table 4).
TABLE 3.
Proportion of individuals with genotypes yielding high, intermediate, and low MBL levels among those recovered from hepatitis B and those with viral persistence
| MBL level (haplotypes)a | % Of subjectsb with the indicated MBL level among:
|
OR | 95% CI | P | |
|---|---|---|---|---|---|
| Recovered subjects (n = 301) | Subjects with viral persistence (n = 179) | ||||
| High (YA/YA) | 37.9 | 24.6 | 0.55 | 0.37-0.84 | 0.005 |
| Intermediate (YA/XA, YA/O, or XA/XA) | 49.2 | 56.4 | 1.29 | 0.88-1.87 | 0.19 |
| Low or none (XA/O or O/O) | 13.0 | 19.0 | 1.76 | 1.02-3.01 | 0.04 |
The MBL haplotypes are as defined in Materials and Methods.
n, number of study subjects.
Since a small effect of the −550 promoter on MBL levels has been demonstrated (8), we divided those with the YA genotype into two groups based on the −550 SNP. There was no difference in the frequencies of recovery from infection or viral persistence between those with either C or G at −550 (OR, 0.81 for both groups). Homozygosity for YA with a C at −550 and for YA with a G at −550 also led to similar outcomes (OR, 0.66 and 0.60, respectively).
DISCUSSION
In this study, we find that mbl2 genotypes correlating with high MBL levels are associated with recovery from an HBV infection whereas those correlating with lower levels are associated with viral persistence. The major role of MBL is activation of complement; however, complement does not play a known role in HBV pathogenesis. Thus, these data suggest either a novel mechanism of immune response to HBV involving complement or an alternative role for MBL in HBV pathogenesis.
Although direct binding of MBL to HBV has not been demonstrated, it could occur since the HBV envelope contains the required N-acetylglucosamine and mannose recognition moieties (23). Once bound, the precise role of MBL in clearance of HBV is not known; however, a variety of potential mechanisms are plausible. A major function of MBL is activation of the classical complement cascade through the cleavage of C4 and C2 by the MBL-associated serine protease (7). Thus, whether complement plays a role in HBV pathogenesis deserves investigation. MBL has been associated with antiviral responses including interference in attachment of influenza A virus to host cells, viral spread, and viral release (17). In HIV infection, MBL has been shown to bind to gp120, which prevents binding of the virus to its CD4 receptor (6). It is possible that such mechanisms may occur with HBV. Another plausible biological role for MBL is via up- or down-regulation in cytokine production, which occurs in vitro with fungi, parasites, and bacteria (2, 22, 25). The precise nature of the cytokine response is dependent upon the organism, but alteration of levels of tumor necrosis factor alpha, which is a cytokine important for noncytolytic clearance of HBV (10), has been described. Further work is needed to determine the precise contribution of MBL to the control of HBV infection.
It is important that the results of genotype and haplotype analyses were consistent with the relationships found when individuals were grouped by presumed functional MBL levels. The −221C (X) allele and the XA haplotype were associated with viral persistence, and individuals with these genotypes were found only in groups of subjects producing intermediate or low levels of MBL. On the other hand, the YA haplotype was more common in those who recovered from hepatitis B, which is consistent with the fact that this haplotype was restricted to the groups with either high or intermediate MBL levels.
It is also notable that the ORs for the significant SNPs and SNP combinations were similar in both blacks and whites, the two major ethnic groups in this population. This consistent trend suggests that MBL is involved directly in HBV clearance rather than indirectly through linkage disequilibrium with neighboring loci.
A few other studies have examined mbl2 with respect to recovery from HBV infection, but the data are inconsistent (1, 13, 26, 31). In the largest of these studies, investigators genotyped the codon 52, 54, and 57 SNPs in 180 Gambians with persistent HBV infections and 157 who had recovered from HBV infections (1). The frequency of the codon 57 homozygous mutant was higher in those with viral persistence (8.9% versus 5.7%) but did not reach statistical significance. The other SNPs were not present in the homozygous mutant state. Since the −221 promoter SNP was not examined in this study, the investigators could not analyze the YA/YA or XA/O genotype. Differences between studies may also be related to the age of subjects at HBV acquisition. In The Gambia, transmission of HBV generally occurs in early childhood (34), whereas in the United States, acquisition is usually in adulthood. Since children are more likely to have viral persistence after HBV infection (20), different aspects of the immune system may be relatively more important depending on whether HBV was acquired before or during adulthood. Of the other studies, one found an association between the codon 52 mutation and HBV persistence (31).
It would have been helpful to be able to correlate the precise levels of MBL in these subjects with the MBL genotypes and HBV infection outcomes. However, since MBL is an acute-phase reactant, the levels in sera when subjects were enrolled in this study would not be expected to be representative of levels at the time when HBV persistence or recovery from infection was determined. In addition, the fact that persons with chronic hepatitis B have an ongoing infection would serve as a strong confounder in a posthoc evaluation of MBL levels. A meaningful measurement of MBL levels in our cohort would require a sample taken prior to HBV infection, which is not available. Fortunately, there is a well-established relationship between genotypes and MBL levels, as has been documented for over 1,000 individuals (8).
Approximately half of our subjects were HIV infected, but this could not have affected the results of our study for several reasons. HBV infection occurs prior to HIV infection in most cases, so the outcome of the viral hepatitis infection is determined prior to acquisition of HIV (19, 21). Even more convincingly, those recovered from infection and those with viral persistence were matched according to HIV infection status, and stratification of results of the analysis with respect to HIV infection status did not significantly alter the results.
In summary, this is the first study to clearly demonstrate that genetically determined differences in mbl2 are important determinants of recovery from HBV infection. These data support the importance of the innate immune response in this process, but further study is needed to explain the precise role of MBL in HBV pathogenesis.
Acknowledgments
This work was funded by National Institutes of Health grants DA00441, DA12568, DA04334, and DK56415. C.L.T. was additionally supported, in part, by the Investigators in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund. The Multicenter AIDS Cohort Study (MACS) is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute: grants UO1-AI-35042, 5-MO1-RR-00722 (GCRC), UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
We thank the participants in the study for making this work possible and Abbott Laboratories for donating HBsAg and anti-HBs kits.
REFERENCES
- 1.Bellamy, R., C. Ruwende, K. P. McAdam, M. Thursz, M. Sumiya, J. Summerfield, S. C. Gilbert, T. Corrah, D. Kwiatkowski, H. C. Whittle, and A. V. Hill. 1998. Mannose binding protein deficiency is not associated with malaria, hepatitis B carriage nor tuberculosis in Africans. Q. J. Med. 91:13-18. [DOI] [PubMed] [Google Scholar]
- 2.Chaka, W., A. F. Verheul, V. V. Vaishnav, R. Cherniak, J. Scharringa, J. Verhoef, H. Snippe, and A. I. Hoepelman. 1997. Induction of TNF-alpha in human peripheral blood mononuclear cells by the mannoprotein of Cryptococcus neoformans involves human mannose binding protein. J. Immunol. 159:2979-2985. [PubMed] [Google Scholar]
- 3.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 4.Chmiel, J. S., R. Detels, R. A. Kaslow, M. Van Raden, L. A. Kingsley, and R. Brookmeyer. 1987. Factors associated with prevalent human immunodeficiency virus (HIV) infection in the Multicenter AIDS Cohort Study. Am. J. Epidemiol. 126:568-577. [DOI] [PubMed] [Google Scholar]
- 5.Eisen, D. P., and R. M. Minchinton. 2003. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin. Infect. Dis. 37:1496-1505. [DOI] [PubMed] [Google Scholar]
- 6.Ezekowitz, R. A., M. Kuhlman, J. E. Groopman, and R. A. Byrn. 1989. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J. Exp. Med. 169:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadjeva, M., K. Takahashi, and S. Thiel. 2004. Mannan-binding lectin—a soluble pattern recognition molecule. Mol. Immunol. 41:113-121. [DOI] [PubMed] [Google Scholar]
- 8.Garred, P., F. Larsen, H. O. Madsen, and C. Koch. 2003. Mannose-binding lectin deficiency—revisited. Mol. Immunol. 40:73-84. [DOI] [PubMed] [Google Scholar]
- 9.Gilson, R. J., A. E. Hawkins, M. R. Beecham, E. Ross, J. Waite, M. Briggs, T. McNally, G. E. Kelly, R. S. Tedder, and I. V. Weller. 1997. Interactions between HIV and hepatitis B virus in homosexual men: effects on the natural history of infection. AIDS 11:597-606. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti, L. G., K. Ando, M. V. Hobbs, T. Ishikawa, L. Runkel, R. D. Schreiber, and F. V. Chisari. 1994. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. USA 91:3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, T. K., L. Tarnow, S. Thiel, R. Steffensen, C. D. Stehouwer, C. G. Schalkwijk, H. H. Parving, and A. Flyvbjerg. 2004. Association between mannose-binding lectin and vascular complications in type 1 diabetes. Diabetes 53:1570-1576. [DOI] [PubMed] [Google Scholar]
- 13.Hohler, T., M. Wunschel, G. Gerken, P. M. Schneider, K. H. Meyer zum Buschenfelde, and C. Rittner. 1998. No association between mannose-binding lectin alleles and susceptibility to chronic hepatitis B virus infection in German patients. Exp. Clin. Immunogenet. 15:130-133. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, T. M., X. Chen, S. Duan, R. D. Miller, and P. Y. Kwok. 2001. Universal SNP genotyping assay with fluorescence polarization detection. BioTechniques 31:560, 562, 564-560, 568, passim. [DOI] [PubMed] [Google Scholar]
- 15.Hyams, K. 1995. Risks of chronicity following acute hepatitis B virus infection: a review. Clin. Infect. Dis. 20:992-1000. [DOI] [PubMed] [Google Scholar]
- 16.Kakimi, K., L. G. Guidotti, Y. Koezuka, and F. V. Chisari. 2000. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 192:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kase, T., Y. Suzuki, T. Kawai, T. Sakamoto, K. Ohtani, S. Eda, A. Maeda, Y. Okuno, T. Kurimura, and N. Wakamiya. 1999. Human mannan-binding lectin inhibits the infection of influenza A virus without complement. Immunology 97:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaslow, R. A., D. G. Ostrow, R. Detels, J. P. Phair, B. F. Polk, and C. R. Rinaldo. 1987. The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am. J. Epidemiol. 126:310-318. [DOI] [PubMed] [Google Scholar]
- 19.Kingsley, L. A., C. R. Rinaldo, Jr., D. W. Lyter, R. O. Valdiserri, S. H. Belle, and M. Ho. 1990. Sexual transmission efficiency of hepatitis B virus and human immunodeficiency virus among homosexual men. JAMA 264:230-234. [PubMed] [Google Scholar]
- 20.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 21.Levine, O. S., D. Vlahov, R. Brookmeyer, S. Cohn, and K. E. Nelson. 1996. Differences in the incidence of hepatitis B and human immunodeficiency virus infections among injecting drug users. J. Infect. Dis. 173:579-583. [DOI] [PubMed] [Google Scholar]
- 22.Santos, I. K., C. H. Costa, H. Krieger, M. F. Feitosa, D. Zurakowski, B. Fardin, R. B. Gomes, D. L. Weiner, D. A. Harn, R. A. Ezekowitz, and J. E. Epstein. 2001. Mannan-binding lectin enhances susceptibility to visceral leishmaniasis. Infect. Immun. 69:5212-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiraishi, H., R. Shirachi, K. Akiyama, and N. Ishida. 1980. Glycopeptide composition of hepatitis B surface antigen. J. Gen. Virol. 48:31-38. [DOI] [PubMed] [Google Scholar]
- 24.Soborg, C., H. O. Madsen, A. B. Andersen, T. Lillebaek, A. Kok-Jensen, and P. Garred. 2003. Mannose-binding lectin polymorphisms in clinical tuberculosis. J. Infect. Dis. 188:777-782. [DOI] [PubMed] [Google Scholar]
- 25.Soell, M., E. Lett, F. Holveck, M. Scholler, D. Wachsmann, and J. P. Klein. 1995. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-alpha release. J. Immunol. 154:851-860. [PubMed] [Google Scholar]
- 26.Song, L. H., V. Q. Binh, D. N. Duy, S. Juliger, T. C. Bock, A. J. Luty, P. G. Kremsner, and J. F. Kun. 2003. Mannose-binding lectin gene polymorphisms and hepatitis B virus infection in Vietnamese patients. Mutat. Res. 522:119-125. [DOI] [PubMed] [Google Scholar]
- 27.Steffensen, R., S. Thiel, K. Varming, C. Jersild, and J. C. Jensenius. 2000. Detection of structural gene mutations and promoter polymorphisms in the mannan-binding lectin (MBL) gene by polymerase chain reaction with sequence-specific primers. J. Immunol. Methods 241:33-42. [DOI] [PubMed] [Google Scholar]
- 28.Stephens, M., N. J. Smith, and P. Donnelly. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summerfield, J. A., S. Ryder, M. Sumiya, M. Thursz, A. Gorchein, M. A. Monteil, and M. W. Turner. 1995. Mannose binding protein gene mutations associated with unusual and severe infections in adults. Lancet 345:886-889. [DOI] [PubMed] [Google Scholar]
- 30.Thimme, R., S. Wieland, C. Steiger, J. Ghrayeb, K. A. Reimann, R. H. Purcell, and F. V. Chisari. 2003. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas, H. C., G. R. Foster, M. Sumiya, D. McIntosh, D. L. Jack, M. W. Turner, and J. A. Summerfield. 1996. Mutation of gene of mannose-binding protein associated with chronic hepatitis B viral infection. Lancet 348:1417-1419. [DOI] [PubMed] [Google Scholar]
- 32.Turner, M. W. 2003. The role of mannose-binding lectin in health and disease. Mol. Immunol. 40:423-429. [DOI] [PubMed] [Google Scholar]
- 33.Vlahov, D., J. C. Anthony, A. Muñoz, J. Margolik, D. D. Celentano, L. Solomon, and B. F. Polk. 1991. The ALIVE Study: a longitudinal study of HIV-1 infection in intravenous drug users: description of methods. J. Drug Issues 21:759-776. [PubMed] [Google Scholar]
- 34.Whittle, H., H. Inskip, A. K. Bradley, K. McLaughlan, F. Shenton, W. Lamb, J. Eccles, B. A. Baker, and A. J. Hall. 1990. The pattern of childhood hepatitis B infection in two Gambian villages. J. Infect. Dis. 161:1112-1115. [DOI] [PubMed] [Google Scholar]