Abstract
Respiratory syncytial virus (RSV) subverts the antiviral interferon (IFN) response, but the mechanism for this evasion was unclear. Here we show that RSV preferentially inhibits IFN-α/β signaling by expression of viral NS1 and NS2. Thus, RSV infection or expression of recombinant NS1 and NS2 in epithelial host cells causes a marked decrease in Stat2 levels and the consequent downstream IFN-α/β response. Similarly, NS1/NS2-deficient RSV no longer decreases Stat2 levels or IFN responsiveness. RSV infection decreased human but not mouse Stat2 levels, so this mechanism of IFN antagonism may contribute to viral host range, as well as immune subversion.
Paramyxoviruses appear to effectively antagonize the interferon (IFN) system through either inhibition of IFN generation or blockade of IFN signal transduction components such as Stat1 and Stat2 (8). For Sendai and measles viruses, the viral V and C proteins inhibit Stat1 expression and activation (7, 11, 14, 22), whereas the V protein of human parainfluenza virus 2 affects Stat2 responses (1, 15). However, these proteins are not present in respiratory syncytial virus (RSV), the most common cause of serious respiratory infection in childhood (12). Nonetheless, RSV is also relatively resistant to the IFN-induced antiviral state (2). Recombinant deletion mutants of bovine RSV demonstrated that the nonstructural proteins NS1 and NS2 are cooperatively necessary for this resistance (21). These proteins also appear to inhibit induction of IFN, since deletion mutants of NS1 and NS2 for both human and bovine RSV induce more IFN-α/β production than does wild-type RSV (23, 24). However, these previous approaches have not addressed a possible action of RSV proteins on IFN signaling that might also inhibit the initial induction of an IFN-dependent antiviral state.
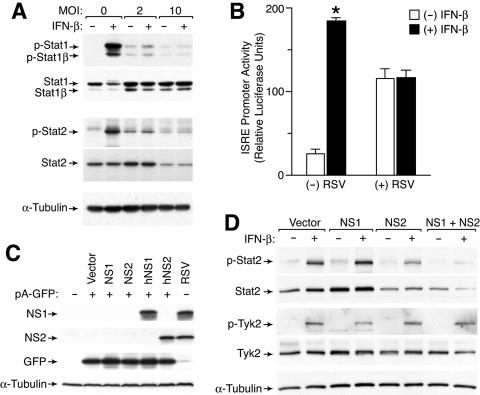
In the present study, we found that infection of primary-culture human tracheobronchial epithelial cells (hTECs) (13) or a human alveolar epithelial cell line (A549 cells) with a low inoculum (multiplicity of infection [MOI] of 2) of RSV slightly increased baseline Stat1 and Stat2 expression and tyrosine phosphorylation but markedly suppressed any subsequent response to IFN-β (Fig. 1A and data not shown). A higher inoculum of RSV (MOI of 10) decreased the level of Stat2 expression and fully inhibited IFN-β responsiveness, as described by others (19). Neither level of RSV inoculation caused any significant decrease in Stat1 expression or IFN-γ responsiveness (Fig. 1A and data not shown). In concert with these effects, RSV infection also increased baseline ISRE promoter activity without affecting control Renilla luciferase activity and suppressed IFN-β responsiveness of the ISRE promoter (Fig. 1B). Thus, at a low inoculum, RSV-driven increases in baseline Stat1 and Stat2 expression and phosphorylation are likely due to endogenous IFN-β production acting in a paracrine manner on uninfected cells. A high inoculum of RSV causes selective down-regulation of Stat2 with consequent inhibition of IFN-α/β signaling that becomes apparent when Stat2 expression is inhibited in nearly all cells.
FIG. 1.
Effect of RSV infection on IFN-β responsiveness and Stat2 levels. (A) Western blot analysis for tyrosine-phosphorylated and total Stat1 and Stat2 using lysates from A549 cells that were infected with RSV Long for 24 h and then incubated with IFN-β (1,000 U/ml) for 30 min. (B) Promoter transactivation analysis from A549 cells that were transfected with an ISRE-driven luciferase reporter plasmid and a control Renilla luciferase reporter and then infected with RSV (MOI of 2) for 24 h, incubated with IFN-β (1,000 U/ml) for 12 h, and subjected to luciferase assay. Values represent luciferase light units normalized to a Renilla luciferase control (which was no different between infected and uninfected conditions), and a significant difference from the untreated condition is indicated by an asterisk. (C) Western blot analysis for NS1 and NS2 using 293T cells that were transiently transfected with 0.5 μg of pA-GFP and 1.5 μg of pcDNA5-empty, pcDNA5-NS1, pcDNA5-NS2, pcDNA5-hNS1, or pcDNA5-hNS2. Equivalent transfection efficiency was verified by flow cytometric analysis for GFP expression. RSV-infected A549 cells were included as a control. (D) Western blot analysis for phosphorylated and total Stat2 and Tyk2 using A549 cells stably expressing vector alone or hNS1, hNS2, or hNS1 plus hNS2 that were incubated with IFN-β for 30 min. Stable cell lines were generated by successive transduction first with MSCV-IRES-GFP containing no hNS, hNS1, or hNS2 insert and then with MSCV-IRES-Thy1.1 containing the appropriate hNS insert, followed by sorting for GFP and Thy1.1 expression.
To define the roles of NS1 and NS2 in antagonizing IFN-α/β action, we aimed to express these proteins in host cells in the absence of viral infection and determine the effect on IFN-β responsiveness. Initial attempts using a variety of expression vectors were unsuccessful. Because RSV transcription and replication take place entirely within the cytoplasm, we reasoned that transcription of the cloned viral sequences from plasmids within the nucleus might subject them to RNA processing events not normally encountered when viral mRNAs are produced in the cytoplasm. The NS1 and NS2 open reading frames (ORFs), like much of the RSV genome, are unusually AT rich. We therefore constructed “humanized” sequences with redundant codons more frequently used in mammalian expression (and therefore more GC rich) by annealing synthetic oligonucleotides covering the NS1 and NS2 ORFs, followed by PCR amplification. The amino acid sequences of the new ORFs, designated hNS1 and hNS2, were unaffected. Polyclonal antibodies against NS1 and NS2 were derived by immunizing chickens or rabbits, respectively, with purified, histidine-tagged recombinant protein expressed in Escherichia coli (East Coast Biologics). Transfection of hNS1 and hNS2 using the pcDNA backbone (Invitrogen), in contrast to the original sequences, resulted in significant expression detectable by Western blot analysis (Fig. 1C). This humanization approach will likely also improve expression of cDNAs from genes from other normally cytoplasmic viruses using standard expression vectors.
We next established stable NS1 and NS2 expression in A549 cells using murine stem cell virus (MSCV) retroviral vectors carrying green fluorescent protein (GFP) and Thy1.1 expression markers (20). Flow cytometric sorting for GFP and Thy1.1 generated four cell lines, each expressing these markers and either hNS1, hNS2, hNS1 plus hNS2, or a control without hNS1 or hNS2. Western blot analysis of these cell lines with and without IFN-β treatment demonstrated that hNS2 markedly suppressed Stat2 expression, and while hNS1 had little effect by itself, the combination of hNS1 and hNS2 further suppressed Stat2 expression even below baseline levels (Fig. 1D). Neither hNS1 nor hNS2 expression directly interfered with IFN-β activation of Stat2 (since the ratio of phosphorylated Stat2 to total Stat2 was constant across conditions). Similarly, hNS1 and hNS2 expression did not alter upstream IFNAR-associated Tyk2 phosphorylation (Fig. 1D). Nonetheless, the NS1/NS2-driven decrease in Stat2 levels caused a consequent decrease in the net intensity of the Stat2 activation signal.
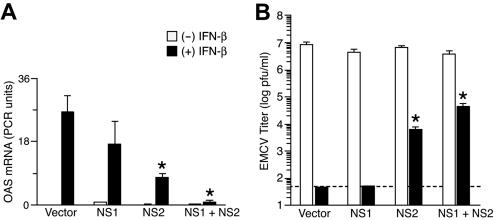
In accord with the pattern of Stat2 activation, expression of hNS1 and hNS2 also inhibited induction of IFN-β target genes as assessed by quantitative real-time PCR for 2′-5′ oligoadenylate synthetase mRNA levels (Fig. 2A). In this case as well, hNS2 was more effective than hNS1 and the combination was most effective in suppressing the actions of IFN-β. To characterize the functional consequence of hNS1 and hNS2 expression on viral replication, we monitored IFN sensitivity of encephalomyocarditis virus (EMCV). Pretreatment with IFN-β was completely effective in inhibiting EMCV replication in the absence of hNS1 and hNS2 expression or in the presence of hNS1 expression, but this antiviral effect of IFN-β was significantly decreased in cells expressing hNS2 and was downregulated even further in cells expressing hNS1 and hNS2 (Fig. 2B).
FIG. 2.
Effect of hNS1/hNS2 expression on IFN-β-dependent gene expression and viral clearance. (A) Real-time PCR analysis of 2′-5′ oligoadenylate synthetase (OAS) mRNA levels in NS-expressing A549 cells that were incubated with IFN-β (1,000 U/ml) for 6 h. Cells were transduced as described in the legend to Fig. 1. Values represent the mean ± the standard error of the mean normalized to a glyceraldehyde-3-phosphate dehydrogenase control. (B) Titers of EMCV in NS-expressing A549 cells that were treated with IFN-β (1,000 U/ml) for 12 h and then infected with EMCV (MOI of 0.1) for 24 h. Cell supernatants were used to determine EMCV titers by plaque assay. Values represent the mean ± the standard error of the mean, and dotted line indicates the lower limit of detection for the assay. Significant difference from the vector-alone condition is indicated by an asterisk.
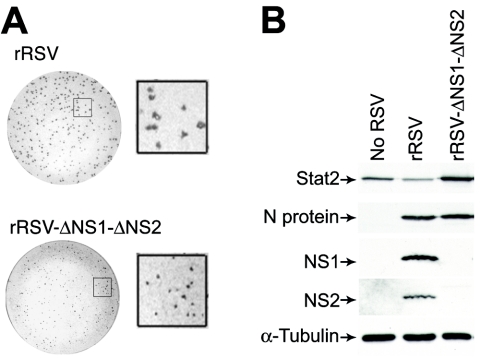
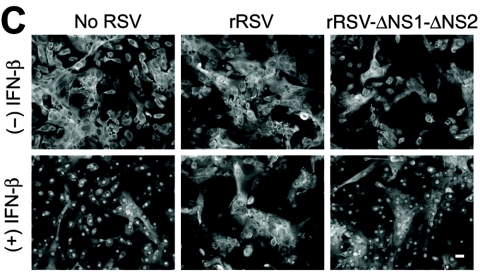
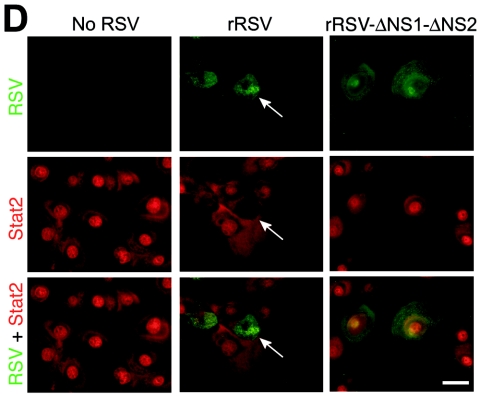
To define whether loss of native NS1 and NS2 might impact the capacity of RSV for antagonism of IFN-β signaling, we compared the behaviors of wild-type and NS-deficient RSVs. Reverse genetics have been used to study the RSV A2 strain (4, 10), but to maintain consistency with our experimental model, we developed a comparable system for RSV Long. The genomic sequence of wild-type RSV Long was used to generate plasmids encoding antigenomic cDNAs for wild-type and NS1/NS2 mutant RSV Long. Antigenomic and N, P, L, and M2 plasmids were cotransfected into BHK-SR19 cells expressing T7 polymerase from a Sindbis virus replicon, and rescued viruses were propagated in Vero cells. In accordance with previous findings (9), the ΔNS1-ΔNS2 mutant virus was attenuated in plaque size and replication rate even in IFN-deficient Vero cells (Fig. 3A). Nonetheless, using a higher inoculum (MOI of 10) and earlier timing (24 h postinoculation), we achieved levels of viral infection and protein expression that were comparable for wild-type recombinant RSV (rRSV) and rRSV-ΔNS1-ΔNS2 (Fig. 3B and data not shown). To better approximate natural RSV infection, we compared rRSV to rRSV-ΔNS1-ΔNS2 infection using primary-culture airway epithelial cells (hTECs). In this system, wild-type rRSV infection also decreased the Stat2 expression level while rRSV-ΔNS1-ΔNS2 infection resulted in an increase in Stat2 levels (Fig. 3B). These findings are consistent with NS1/NS2-dependent inhibition of Stat2 expression during infection with wild-type virus, but when this inhibition is removed, the ΔNS1-ΔNS2 mutant virus also causes an increase in IFN-β levels (23) and thereby also increases IFN-dependent Stat2 expression. In concert with these effects on Stat2 expression, we also found that infection with wild-type rRSV inhibited IFN-β-dependent translocation of Stat2 to the nucleus, whereas rRSV-ΔNS1-ΔNS2 infection no longer allowed this anti-IFN effect (Fig. 3C and D). Moreover, two-color immunofluorescence to colocalize RSV and Stat2 showed that Stat2 expression and nuclear translocation were decreased in cells infected with wild-type rRSV whereas both of these endpoints were intact in cells infected with rRSV-ΔNS1-ΔNS2 (Fig. 3D).
FIG. 3.
Requirement of NS1/NS2 for an RSV-dependent decrease in Stat2 expression and IFN-β responsiveness. (A) Plaque number and size for rRSV with or without NS1/NS2 (upper and lower parts, respectively) inoculated onto Vero cells for 7 days, followed by peroxidase immunostaining for RSV. Insets indicate approximately fourfold enlargement. (B) Western blot analysis for indicated proteins using hTECs that were left uninfected or infected with rRSV with or without NS1/NS2 (MOI of 10). (C) Immunofluorescence microscopy for Stat2 using hTECs that were infected with rRSV with or without NS1/NS2 (MOI of 5) for 24 h and then incubated with IFN-β (1,000 U/ml). (D) Same conditions as panel C but immunostaining for Stat2 (red, CY3 fluorescence) and RSV G protein (green, fluorescein isothiocyanate fluorescence) after incubation with IFN-β. Arrow indicates RSV-infected cell with decreased nuclear translocation of Stat2; neighboring uninfected cells and rRSV-ΔNS1-ΔNS2-infected cells exhibit normal IFN-β-driven translocation of Stat2. Bars = 20 μm.
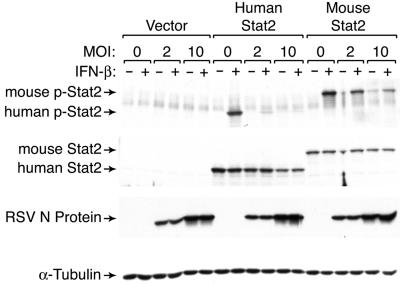
Unlike other components of IFN signal transduction pathways, Stat2 is not well conserved between species and the C-terminal portion of Stat2 contains microsatellite repeats that vary between species and mouse strains (5, 6, 17, 18). This variability has suggested that Stat2 antagonism may contribute to the species specificity of other paramyxoviruses (16), so we reasoned that Stat2 could also confer specificity of RSV infection. We therefore complemented Stat2-deficient U6A cells with MSCV encoding IRES-GFP, human Stat2-IRES-GFP, or mouse Stat2-IRES-GFP and were able to demonstrate that U6A cells expressing human Stat2 were more susceptible than cells expressing mouse Stat2 to downregulation of phosphorylated and total Stat2 levels during RSV infection (Fig. 4). This finding suggests that targeting Stat2 may be a determinant of RSV tropism for humans and provides an explanation for previous evidence of human NS1 and NS2 conferring species specificity in studies of recombinant bovine RSV (3). Under the present conditions, the retroviral long terminal repeat rather than the endogenous promoter controls Stat2 expression, so RSV inhibition of Stat2 expression is likely due at least in part to a posttranscriptional mechanism. RSV infection in mice can be inhibited by delivery of small interfering RNA nanoparticles targeting the viral NS1 gene (25). Thus, further development of antiviral therapeutic targets will also benefit from a better understanding of RSV NS1 and NS2 function.
FIG. 4.
Effect of RSV infection on human versus mouse Stat2 levels. Western blot analysis for phosphorylated and total Stat2 using U6A cells stably expressing no Stat2 (vector) or human or mouse Stat2 and incubated without or with IFN-β (1,000 U/ml). Stable cell lines were generated by transduction with MSCV-IRES-GFP containing no Stat2 or human or mouse Stat2 insert, followed by sorting for GFP. Expression of control RSV N protein was not significantly different between cell lines.
Nucleotide sequence accession number.
The genomic sequence of wild-type RSV Long was deposited in GenBank under accession number AY911262. The amino acid sequences of the ORFs hNS1 and hNS2 were deposited in GenBank under accession numbers AY904040 and AY904041, respectively.
Acknowledgments
We thank D. Farrar for human and murine Stat2 constructs, K. Murphy for MSCV retroviral vectors, G. Stark for U6A cells, and Apath, LLC, for BHK-SR19 cells. We also gratefully acknowledge L. Shornick, E. Agapov, and A. Pekosz for helpful discussions. R.M.B. thanks M. Garcia-Blanco, J. Heitman, and G. Prince for generous support.
This research was supported by grants from the National Institutes of Health, the Martin Schaefer Fund, and the Alan A. and Edith L. Wolff Charitable Trust.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atreya, P. L., and S. Kulkarni. 1999. Respiratory syncytial virus strain A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology 261:227-241. [DOI] [PubMed] [Google Scholar]
- 3.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrar, J. D., J. D. Smith, T. L. Murphy, S. Leung, G. R. Stark, and K. M. Murphy. 2000. Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nat. Immunol. 1:65-69. [DOI] [PubMed] [Google Scholar]
- 6.Farrar, J. D., J. D. Smith, T. L. Murphy, and K. M. Murphy. 2000. Recruitment of Stat4 to the human interferon-alpha/beta receptor requires activated Stat2. J. Biol. Chem. 275:2693-2697. [DOI] [PubMed] [Google Scholar]
- 7.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2001. Paramyxovirus accessory proteins as interferon antagonists. Microbiol. Immunol. 45:787-800. [DOI] [PubMed] [Google Scholar]
- 9.Jin, H., X. Cheng, V. L. Traina-Dorge, H. J. Park, H. Zhou, K. Soike, and G. Kemble. 2003. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 21:3647-3652. [DOI] [PubMed] [Google Scholar]
- 10.Jin, H., D. Clarke, H. Z. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leader, S., and K. Kohlhase. 2003. Recent trends in severe respiratory syncytial virus (RSV) among US infants, 1997 to 2000. J. Pediatr. 143:S127-S132. [DOI] [PubMed] [Google Scholar]
- 13.Look, D. C., W. T. Roswit, A. G. Frick, Y. Gris-Alevy, D. M. Dickhaus, M. J. Walter, and M. J. Holtzman. 1998. Direct suppression of Stat1 function during adenoviral infection. Immunity 9:871-880. [DOI] [PubMed] [Google Scholar]
- 14.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 16.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, C., M. J. Lecomte, and C. Schindler. 1999. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 27:4191-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulson, M., S. Pisharody, L. Pan, S. Guadagno, A. L. Mui, and D. E. Levy. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343-25349. [DOI] [PubMed] [Google Scholar]
- 19.Ramaswamy, M., L. Shi, M. M. Monick, G. W. Hunninghake, and D. C. Look. 2004. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 30:893-900. [DOI] [PubMed] [Google Scholar]
- 20.Ranganath, S., W. Ouyang, D. Bhattarcharya, W. C. Sha, A. Grupe, G. Peltz, and K. M. Murphy. 1998. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161:3822-3826. [PubMed] [Google Scholar]
- 21.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaffer, J. A., W. J. Bellini, and P. A. Rota. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389-397. [DOI] [PubMed] [Google Scholar]
- 23.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and gamma interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, W., H. Yang, X. Kong, S. Mohapatra, H. S. Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal small interfering RNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56-62. [DOI] [PubMed] [Google Scholar]