Abstract
The ERV3 locus at chromosome 7q11 is a much studied human endogenous retroviral (HERV) sequence, owing to an env open reading frame (ORF) and placental RNA and protein expression. An analysis of the human genome demonstrated that ERV3 is one of a group of 41 highly related elements (ERV3-like HERVs) which use proline, isoleucine, or arginine tRNA in their primer binding sites. In addition to elements closely related to ERV3, the group included the previously known retinoic acid-inducible element, RRHERVI, also referred to as HERV15, but was separate from the related HERV-E elements. The ERV3-like elements are defective. The only element with an ORF among gag, pro, pol, and env genes was the env ORF of the original ERV3 locus. A search in dbEST revealed ERV3 RNA expression in placenta, skin, carcinoid tumor, and adrenal glands. Expression was also studied with newly developed real-time quantitative PCRs (QPCR) of ERV3 and HERV-E(4-1) env sequences. Results from a novel histone 3.3 RNA QPCR result served as the expression control. QPCR results for ERV3 were compatible with previously published results, with a stronger expression in adrenal gland and placenta than in 15 other human tissues. The expression of the envelope (env) of ERV3 at chromosome 7q11 was also studied by using stringent in situ hybridization. Expression was found in corpus luteum, testis, adrenal gland, Hassal's bodies in thymus, brown fat, pituitary gland, and epithelium of the lung. We conclude that ERV3 env is most strongly expressed in adrenal and sebaceous glands as well as in placenta.
A substantial part of the human genome consists of retroelements in which the human endogenous retroviruses (HERVs) are included. Depending on the definition of a retrovirus, the HERVs occupy between 2 and 8% of the human genetic makeup (57, 70). Three decades of HERV research has generated a number of outstanding issues regarding their role in genomic evolution, physiology, and pathology (10, 44). Nearly all HERVs known at present integrated up to 100 million years ago. They retain the proviral structure of exogenous retroviruses. Inherited from generation to generation, a few of the HERVs have open reading frames (ORFs) that encode known or putative proteins, while others are highly mutated. Activation of these elements through their long terminal repeats (LTRs) has been observed at the mRNA level in selected tissues and different human cell lines (reviewed in references 10 and 44). The main focus has been on HERV expression in relation to disease, for example, HERV-W and HERV-H in multiple sclerosis and schizophrenia and HERV-K in testis tumors (13, 17, 25, 29, 56). Moreover, downregulation of ERV3 is reported in choriocarcinoma (25, 29). The presence of conserved ORFs is circumstantial evidence for a functional role for HERV-encoded proteins, exemplified by Rec/cORF, syncytin, and ERV3 Env (11, 45, 49). Furthermore, it is established that HERV LTRs also control transcription of cellular genes such as those for phospholipase A2/otoconin (22, 35, 79) and pleiotrophin (64). In general, HERVs are expressed as RNA at a low level in most studied tissues (43, 47, 48) but at higher levels in others (67). Although protein expression is less studied, a few HERV proteins are more or less well known (49, 78). In a recent study, Schön et al. reported that the polymerase gene of HERVs is active in an organ-specific way due to the tissue specificity of the LTRs (63), analogous to other promoters. The envelope gene of ERV3 is highly expressed in certain tissues, like sebaceous and adrenal glands (3, 31).
ERV3 is one of the most studied HERVs (3-5, 11, 15, 18, 19, 26, 29-31, 41, 51, 59, 60, 78, 86). It is sometimes referred to as HERV-R. We dislike this term for reasons discussed below. ERV3 is a class I HERV which clusters with viruses belonging to the genus Gammaretrovirus (10, 77). It integrated 30 to 40 million years ago and is present in humans and other higher primates, with the exception of gorillas (26). It was originally thought to be a solitary provirus (47). A few later observations indicated the existence of a group of ERV3-like sequences (8, 53). Despite this, a detailed description of this group, its expression, and the extent of ERV3-like sequences is lacking. This study describes the structure of the original ERV3 locus at chromosome band 7q11, identifies several novel ERV3-like sequences in the human genome, and defines an ERV3-like HERV group. Transcription factor binding sites unique to the 5′ LTR which possibly are involved in ERV3 7q11 expression were identified. Expressed sequence tag (EST) sequences probably transcribed from the ERV3 7q11 locus were searched for in different cell types and used to corroborate previously reported cap, splice, and polyadenylation sites (30). We developed real-time quantitative PCRs (QPCRs) that quantitatively detect transcripts from the SU of env from HERV-E(4-1) and ERV3 7q11 in comparison with nonretroviral reference genes. The QPCR was used to study the tissue-specific pattern of RNA expression of the env genes of ERV3 7q11 and a related group, HERV-E. We also checked ERV3 env mRNA expression at the tissue and cellular level by an in situ hybridization (ISH) technique.
MATERIALS AND METHODS
Search for ERV3-like sequences.
ERV3-like sequences were gathered by two different methods. First, the SU part of Env was used to search for ERV3 7q11- and HERV-E(4-1)-like sequences in the htgs (high-throughput genomic sequences) and nr (nonredundant) databases at the National Institute of Biotechnology Information website, using the TBLASTN program (1). The 50 sequences which had scores of at least 200 were included for further work. Proviral sequences were extracted from the corresponding genomic clone sequences by use of the program RetroTector (G. O. Sperber and J. Blomberg, unpublished data). The 11 most highly scoring and nonredundant ERV3 7q11 SU-like sequences and 6 HERV-E SU-like sequences were aligned at the protein and nucleotide levels by use of the program CLUSTAL W (75) (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html). Later, an additional ERV3 7q11 SU-like sequence (in BAC AL356804) was found (Table 1). Data from the alignment were used for constructing an unrooted tree using the programs DNAdist, BioNJ, and Drawtree as implemented at http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html (21). This tree included human sequences available in March 2003.
TABLE 1.
Selected ERV3 env-like loci from the human genome version hg15
| Clone | PBS | Chromosomal band (position in chromosome) | Position in clone | RT scorea | Gene |
|---|---|---|---|---|---|
| AC073210b | R | 7q11 (63858202) | 63888-54276 | 1887 | LTR gag pro pol env LTR |
| AC013429c | P | 7q32 (137651301) | 90435-99714 | 1373 | LTR gag pro pol env LTR |
| AB019437_IgHCVrd | ? | 14q32 (105225894) | 35323-45272 | 1229 | LTR gag pro pol env LTR |
| AC005023 | P | Xq25 (117198092) | 141024-132534 | 1173 | LTR gag pro pol env LTR |
| AF290422 (HERV15Yq1)e | I | Yq11 (13743185) | 15-9731 | 1047 | LTR gag pro pol env LTR |
| AF290423 (HERV15Yq2)e | I | Yq11 (14533115) | 15-9953 | 1183 | LTR gag pro pol env LTR |
| M64936 (HUMRIRT, RRHERVI)e | I | Multiple locations | 1-3357 | 233 | LTR gag pro pol env LTR |
| AL356804f | 14q24 (68899917) | 183361-180474 | 485 | pol env | |
| AL133512 | Xq21 (86869080) | 119293-110957 | 711 | pro pol env | |
| AC006032-AC007379-AC022486g | Yp11 (3010257) | 169763-162558 | 712 | gag pro pol env | |
| AC008175g | Yq11 (23865959) | 123584-114042 | 752 | gag pro pol env LTR | |
| AF072711_lipaseh | 9q34.3 (129305366) | 24330-17768 | 480 | pro pol env LTR | |
| SY-3 ERV3-like HERVi | 13q321 (98255835) | NAj | 170 | pol env |
Score for the respective element identified by RetroTector (see Materials and Methods).
AC073210 (NT_030723) close to human plk, a Krüppel-like zinc finger gene. Mosaic ERV3-human plk transcripts are produced through alternative splicing. This is the ERV3 locus, which gave rise to the ERV3 pol-env clone of Cohen et al. (13).
AC013429 (NT_007680) close to the TIF1 gene (the gene for the transcriptional intermediary factor 1, a multi-zinc finger transcription factor, locus identifier 8805, positions 524084 to 397969; the ERV-like sequence is at positions 417766 to 427065, between exons 9 and 10 of TIF1, in the opposite direction from TIF1). The protein encoded by this gene mediates transcriptional control by interaction with the activation function 2 (AF2) region of several nuclear receptors, including the estrogen, retinoic acid, and vitamin D3 receptors. It contains three zinc-binding domains. ERV18Y colocalizes with this element.
This element resides in the immunoglobulin heavy-chain variable region on chromosome 14.
The two HERVI65 integrations on chromosome Yq11 occur close to each other and cause loss of a gene through homologous recombination. There is a striking concentration of RRHERVI integrations at Yq11. There are also five ERV3-like integrations in the same portion of chromosome Y. HUMRIRT (RRHERVI) integrations are split up. There is no exact match to HUMRIRT, which is RNA based. There are multiple hits on chromosomes 16p12, Xq24, Yq11(=HERVI5y), and others.
This element was detected after the search on which Fig. 2A was based.
Not only are AC008175 and AC006032 very similar, but there are other sequences in this portion of chromosome Y which have closely matching sequences on chromosome Y.
The ERV3-like element occurs in the 5′ UTR of a lipase gene on chromosome 9.
The SY-3 locus is between exons 5 and 6 of a hypothetical gene (locus 121919).
NA, not applicable. Clone information for Sy-3 is not available.
When version hg15 (April 2003), the first assembly of the human genome, became available at the UCSC website (http://genome.ucsc.edu), a genomewide search for retroviral sequences was performed. A second selection method, based on Pol similarity, was used to single out ERV3-related sequences among these sequences. Altogether, 76 such pol-containing sequences were selected from the total collection of 3,661 pol-containing RetroTector-defined retroviral sequences. RetroTector is described further below. Their relation to each other and to other gammaretroviral sequences was investigated using Clustal W and a crosswise similarity table. The latter was produced using a program written by J. Blomberg (see below).
EST sequences were searched in dbEST (National Center for Biotechnology Information) during the spring of 2004 by using BLAST (1). EST sequences were selected according to percent identity to the ERV3 7q11 search sequence, as indicated in Results.
Characterization of ERV3-like sequences.
The programs RetroTector and RetroTector Shell (G. O. Sperber and J. Blomberg, unpublished data) were used with the April 2003 version (hg15) of the human genome to identify likely retroviral sequences. Most of the ALU sequences were removed prior to the analysis. The programs identified 3,661 elements with a pol gene. These were characterized and classified by the programs. Briefly, the program function RetrovID searches for LTR-like sequences and conserved retroviral motifs from gag, pro, pol, and env and parses them together into chains by the use of distance tables and other heuristics. The function ORFID constructs likely protein sequences (puteins) from the machine-identified gag, pro, pol, and env gene candidates. It uses codon statistics, frequency of stop codons, and alignment to proteins of known retroviruses belonging to the same genus to approximate the original ORF. The Xonid function searches for possible ORFs in stretches not used by ORFID. The RetroTector interpretations were automatically matched with sequences in an annotated list of retroviral sequences from GenBank, using similarity to Pol proteins, and in a list of repetitive elements from Repbase (27), using similarity to the entire nucleotide sequence of the elements. Both similarity searches were performed with word-based BLAST-like algorithms (1) written by J. Blomberg. This initial screening was followed by alignments and clustering using pol and env nucleotides and Pol and Env proteins with the Clustal W 1.83 program. Similarity matrices were generated from all alignments. The Kimura and PAM250 score matrices were used for nucleotides and proteins, respectively. Both alignments and similarity matrices were based on the pairwise deletion technique. This minimizes the influence of insertions and deletions in the retroviral elements. Thus, similarities were not much affected by gaps. The cladogram shown below (see Fig. 2B) was based on the guide neighbor-joining tree produced by Clustal. It was processed with TreeView (courtesy of R. Page, Taxonomy and Systematics, Division of Environmental and Evolutionary Institute of Biomedical and Life Sciences, University of Glasgow; available at http://taxonomy.zoology.gla.ac.uk/rod/rod.html) and tree-modifying programs written by J. Blomberg. The final classification of retroviral elements was consequently based on several independent methods. RetroTector also has an XonID function, which localizes ORFs whose products are longer than 50 amino acids (aa) which do not overlap the four major ones (gag, pro, pol, and env). XonID prioritizes ORFs which begin just after a predicted splice acceptor and end at a predicted splice donor site. It also uses the same codon statistical functions as ORFID. It was applied to the ERV3 locus at 7q11.
FIG. 2.
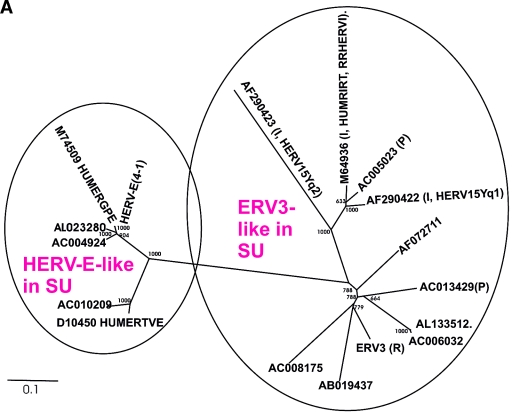
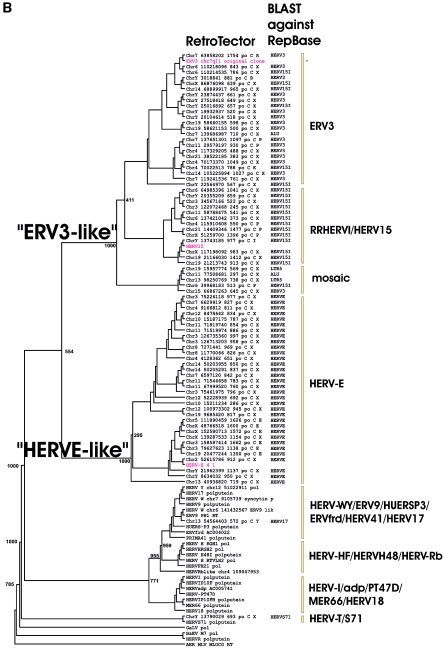
(A) Dendrogram of ERV3- and HERV-E-like sequences based on the nucleotide alignment of the SU part of env, partly shown in Fig. 3. PBS were analyzed by using the RetroTector program (see Materials and Methods) (G. O. Sperber and J. Blomberg, unpublished data). Sequences with an identifiable PBS are shown in parentheses, giving the amino acid (using the one-letter code) corresponding to parts of the tRNA sequence. (B) Pol amino acid-based cladogram, derived from the alignment shown in Fig. S3a (see the supplemental material). The neighbor-joining method was used. For each element, the following RetroTector-derived data are shown: chromosomal position, chain score, PBS usage, sum of stops and frame shifts, and the most similar RepBase element. The bootstrap value (1,000 replicates) for the branch which divides the ERV3-like and the HERVH HERV-E-like elements is also shown.
Transcription factor binding sites (TFBS) were studied in the 5′ and 3′ LTRs, primarily using ConSite (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite/). ConSite predictions were based on longer weight matrices than those of other available TFBS prediction methods. This gives a higher statistical significance, and ConSite predictions were therefore used. The vertebrate TFBS subset of the JASPAR database used by ConSite was selected.
RNA samples from diverse tissues and cDNA synthesis.
A commercial RNA panel, Human Total RNA Master Panel II (Clontech Laboratories, Palo Alto, CA), was used for cDNA synthesis with subsequent QPCR. Human Total RNA Master Panel II contains RNA from 20 different tissues, most of which consist of pooled RNA from two or more persons. RNA from tissues from the brain cerebellum, whole brain, heart, liver, and lung originated from a single person. Samples derived from a single person of Asian origin, except for the whole brain, which was of Caucasian origin. The sources of pooled RNA varied between 2 and 84 persons, all of Caucasian origin. During the work, it was discovered that three of the RNA samples, from heart, lung, and liver, probably were degraded. They yielded almost no signal with both HERV and housekeeping gene QPCRs. They were therefore excluded from further analysis. The RNA described above was used as a template for cDNA synthesis with 2 μg RNA in each reaction mixture. Synthesis of cDNA was made in a 50-μl reaction mixture containing Stratascript reverse transcription (RT; 1 U/μl) (Stratagene, Amsterdam, The Netherlands), 1× Stratascript buffer (Stratagene, Amsterdam, The Netherlands), 0.01 M dithiothreitol (Promega, Madison, WI), 0.8 mM each deoxynucleoside triphosphate (Applied Biosystems, PE Europe, The Netherlands), random hexamers (10.6 ng/μl; Amersham Pharmacia Biotech, Uppsala, Sweden), and RNasin (1.6 U/μl; Promega, Madison, WI). The cDNA reaction mixture was incubated at 25°C for 10 min, 37°C for 90 min, and 70°C for 15 min and then stored at −20°C. According to the manufacturer, the RNA contained virtually no genomic DNA. However, a control for DNA contamination, a reaction mixture without RT, was made for every RNA sample. The signal was usually negative. In cases where the negative reaction became positive, it was at least 10 times weaker than the RT-positive signal. It was then subtracted from the RT-positive signal. Two microliters of each cDNA was used in each PCR. We prepared five samples in addition to the 16 tissues included in the RNA panel. The samples included three from different placentas (three individuals) and two samples from skeletal muscle, one from a single person and one pooled from five different persons. Total RNA was isolated from 30 mg of tissue. The QIAGEN RNA isolation kit was used according to the manufacturer′s recommendations (Merck Eurolab, Stockholm, Sweden). Total RNA was DNase treated using a DNA-free kit from Ambion (Austin, TX), following the protocol from the manufacturer. After DNase treatment, RNA was used for cDNA synthesis, which was performed as described above in a 50-μl reaction mixture. Using this cDNA, 4 μl was used for each PCR.
Primer design and QPCR.
Primers and reporter-quencher (R-Q) probes were designed with the help of the Primer3 program (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3). The midpoint temperature (Tm) was calculated by using the Cybergene program (http://www.cybergene.se/). The primers were synthesized by Interactiva (Thermo Hybaid GmbH, Ulm, Germany), and the three R-Q probes were synthesized by Scandinavian Gene Synthesis (SGS, Köping, Sweden). Primer pairs and probes were all high-performance liquid chromatography purified by the manufacturer. All R-Q probes were labeled with 6-carboxyfluorescein as a reporter at their 5′ ends and an internal Dark Quencher attached to a thymidine in a middle position of the probe sequence (12 to 16 bp from the 5′ end). We primarily chose histone 3.3 (M11353) for use as a reference gene since it is evenly expressed in many cell types, regardless of cell cycle stage (80, 82, 83). Histone 3.3 mRNA has previously been used as a reference in other RNA expression studies (6, 47, 48). Histone 3.3 mRNA is polyadenylated, unlike other histone mRNAs (80). There is room for further studies of its expression in different tissues. The following histone 3.3 primers were based on those originally designed by Mats Lindeskog (Lund University) (48) but were modified by us to fit new PCR conditions: forward primer 5′ CCTCTACTGGAGGGGTGAAGAA 3′ (Tm = 59.4°C) and reverse primer 5′ TGCCTCCTGCAAAGCACCGATA 3′ (Tm = 59.4°C). The probe, 5′ CTCTGGAAGCGCAGATCTGTTTTAAAGTCCT 3′, is situated 6 nucleotides (nt) from the reverse primer, with a Tm that is 10°C higher than that of the primer pair (Tm = 69.7°C). By the use of dilutions of a clone of an amplimer from a human DNA sample (L. Hu and D. Uzhameckis, unpublished data), the sensitivity for the histone PCR was found to be 1 to 10 target gene equivalents per PCR. The integrity of the plasmid was ascertained by sequencing. Optimization was first made with an iCycler (Bio-Rad Laboratories AB, Sundbyberg, Sweden). Normalization of mRNA expression against cell number, DNA content, total protein, and mRNAs from housekeeping genes, like those for actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and histone 3.3, has been used by us or others in numerous studies (50, 62, 76). These genes were selected because of their previously observed wide and approximately even expression levels in many tissues. Besides histone 3.3 RNA, RNAs from three widely used housekeeping genes, i.e., the GAPDH, hypoxanthine phosphoribosyltransferase 1, and ubiquitin C (UBC) genes (GenBank accession number M26880) (76), were also determined (data not shown). The mean of these three last-mentioned reference RNAs with the Clontech tissue RNA panel was 260 eq per ng of total RNA, with a standard error of the mean of 40.9%, while a histone 3.3 average was 19.5 eq per ng of total RNA, with a standard error of the mean of 15.2%. Thus, histone 3.3 RNA was more evenly expressed. HERV QPCR data were therefore normalized to histone 3.3 RNA expression only. In the following, the word “equivalents” is used instead of “copies” for description of the amount of RNA or DNA detected in QPCR because of probable slight variations in amplification efficiency in samples.
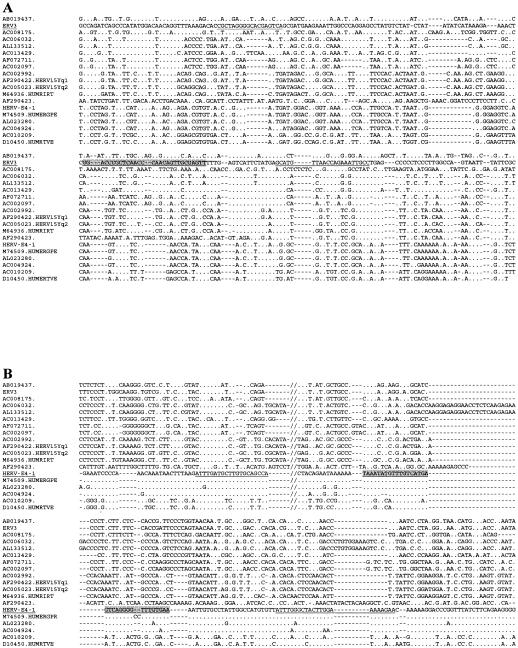
Primer pairs and probes for HERV-E(4-1) and ERV3 7q11 were designed in the SU region where the related HERVs are most divergent and thus should generate specific probes. The chosen primers and probes for HERV-E (HUMER41, accession no. M10976) and ERV3 (HUMERVA34A, accession no. M12140.1) are marked in the original sequence as shown below (see Fig. 3A and B, respectively). The Tm for the ERV3 probe (68.5°C), 5′ AACTCGCAACTGTTGGGTTGAGCGGGTCC 3′, was 15°C higher than the Tms of its primer pair, namely, forward 5′ CGCTAGGGGCACGAGTCA 3′ (Tm = 53.4°C) and reverse 5′ AGGCAATTTCTGGTTAACATGCT 3′ (Tm = 59.2°C).
FIG. 3.
Parts of the SU region of the ERV3- and HERV-E-related sequences aligned with the annotated repetitive sequences for HUMRIRT, HUMERGPE, HERV15Yq1, and HERV15Yq2. QPCR primers are indicated for ERV3 (A) and HERV-E (B) in the alignment by underlining and probe sequences are indicated by underlining and shading. Double slashes in the alignment in panel B show where 60 nt were omitted in the figure due to space limitations. Periods denote identity to the master sequence, which is underlined.
The Tm difference between primers and probe for HERV-E was 9°C. The sequences were as follows: probe, 5′ TCACAAACCCCTGACTCATGACAAACATATTTA 3′ (Tm = 68.4°C); forward primer, 5′ ATTTGATGCTTGTGCAGCCATTA 3′ (Tm = 59.2°C); and reverse primer, 5′ TTCTTTTTCCAAGTAGCCCAAAT 3′ (Tm = 58.8°C). Thus, the annealing and extension temperatures were relatively stringent for both ERV3 and HERV-E SU PCRs. Initially, ERV3 and HERV-E QPCRs were optimized by using human DNA, the above-mentioned plasmids, and an iCycler iQ multicolor real-time PCR detection system (Bio-Rad Laboratories AB, Sundbyberg, Sweden). The ERV3 clone, php 1.7, is a 1.7-kb HindIII-PstI subclone and was a kind gift from Maurice Cohen. The HERV-E plasmid was a kind gift from Arnold Rabson and Malcolm Martin and contained a 1.1-kb HindIII-BamHI subclone from clone 4-1. All subsequent PCRs were run on a Rotor-Gene real-time PCR cycler (Corbett Research, Corbett Robotics, Eight Mile Plains, QLD, Australia) with the following temperature profile: 120 s at 50°C, 600 s at 95°C, and cycling for 15 s at 95°C and 60 s at 54°C. Primer pairs, R-Q probes, and distilled water were added to the twice-concentrated TaqMan Universal PCR Master Mix containing AmpliTaq Gold DNA polymerase, AmpErase uracil N-glycosylase (UNG), deoxynucleoside triphosphates with UTP, Passive reference 1, and optimized buffer components such as MgCl2 (Applied Biosystems, PE Europe, The Netherlands). The threshold for background noise was calculated at cycle 11. We used both plasmid and genomic DNA, isolated from normal human blood, to create standard curves, typically giving a correlation coefficient ranging from 0.970 to 1.000. The standard curves for the histone 3.3, ERV3 7q11 env, and HERV-E env QPCRs were essentially straight lines over the range of 106 to 101 target gene equivalents per PCR. Although we describe ERV3-like sequences (Table 1; see Fig. 2), it is likely that under the relatively stringent PCR conditions presented here, most of the amplimers will be derived from the ERV3 7q11 sequence itself. Thus, one haploid genome corresponds to one target gene in the ERV3 QPCR. The number of target genes for the HERV-E QPCR is harder to estimate, due to an unknown degree of sequence variation in the amplifiable target genes (env). Judging from a genomewide RetroTector HERV survey (unpublished data), there are approximately 10 copies of SU-containing highly HERV-E(4-1)-related proviruses per haploid genome.
Tissues used for ISH with ERV3 env probe.
Normal human tissues were obtained from the Department of Clinical Pathology, Uppsala University Hospital. They were treated according to routine clinical procedures, following surgery, as briefly described below. All tissues were investigated by the same pathologist and judged normal and without remarks. With permission, pieces of the following organs were taken from a 50-year-old woman who died from a ruptured aortic aneurysm: aorta, breast, brain, liver, lung, esophagus, spleen, skeletal muscle, kidney, thyroid, bladder, and stomach. The other tissues (mentioned below) were taken from anonymous routine cases, which were judged as histologically normal. Tissues were fixed in 4% phosphate-buffered formaldehyde and processed for routine histopathology and archived as paraffin blocks. The following tissues were derived from one (not necessarily the same) individual: aorta, bladder, bone marrow, breast, brown fat (from a child), brain (cerebellum), colon, kidney, liver (from a newborn), esophagus, parathyroid gland, pituitary gland (adeno part), skeletal muscle (striated), and seminal vesicle. The following tissues were derived from two or more individuals: adrenal gland, brain, gastric mucosa, heart, lung and bronchial epithelia, lymph node, ovary (corpus luteum), pancreas, parotid gland, placenta (from >10 individuals), prostate, skin (from >10 individuals), testis (slightly atrophic), thymus, thyroid gland, and uterus (endometrium).
ISH.
ISH concerning ERV3 7q11 was performed as described previously (3, 5). Briefly, paraffin-embedded tissues were sectioned (4-μm thick) and mounted on 3-aminopropyltrietoxylsilane-coated slides (Sigma, St. Louis, MO). Sections were pretreated with 0.2 M HCl for 10 min and permeabilized with 2 μg/ml Proteinase K (Merck, Darmstadt, Germany) at 37°C for 15 min prior to hybridization. Hybridization conditions were stringent. Tissue sections were hybridized with 35S-labeled riboprobes produced as described before (3) according to a Promega standard protocol for in vitro transcription. Hybridization was then continued overnight at 56°C, and samples were washed in 2× standard saline citrate and 50% formamide prior to treatment with RNase A (Boehringer Mannheim, Germany), at 100 μg/ml, at 37°C for 30 min. Application of NTB2 photographic emulsion (KODAK AB, Järfälla, Sweden) diluted 1:1 in distilled water was followed by exposure at 4°C for 2 to 4 weeks. Slides were developed and counterstained with Mayer′s hematoxylin and mounted with Pertex (Histolab Products AB, Göteborg, Sweden). Photos were taken in a bright field with a 40× oil immersion SPLAN objective through a Vanox Zeiss microscope.
Grading of ISH.
The grading system used has been previously described (4, 5). The selected tissues were screened by use of a microscope for elevated levels of silver grain density, representing ERV3 env mRNA. A semiquantitative grading was done using scores ranging from − to +++, and sample slides were compared to their respective positive control slides with sections of placenta and skin. Most tissues found positive were tested at least twice and from more than one individual (see Table 3).
TABLE 3.
Tissues used for ISH with ERV3 env probe
| Tissue | Gradea | Cell type |
|---|---|---|
| From one individual or hybridized once | ||
| Aorta | − | |
| Bladder | − | |
| Bone marrow | +/− | Few cells, not identified |
| Breast | − | |
| Brown fatc | ++ | |
| Brain (cerebellum) | − | |
| Gastric mucosa | − | |
| Heart | − | |
| Kidney (newborn) | − | |
| Liver | +/− | Hepatocytes |
| Esophagus | − | |
| Parathyroid gland | − | |
| Pituitary (adenohypophysis part) | ++ | All cells |
| Seminal vesicle | +/− | Secretory epithelium |
| From two or more individuals | ||
| Adrenal gland | ++ | All layers |
| Brain | +/− | Few cells |
| Lung and bronchial epithelium | + | Bronchial epithelial cells |
| Lymph node | − | |
| Ovary (corpus luteum) | +++ | Progesterone-producing cells |
| Pancreas | − | |
| Parotid gland | +/− | |
| Placentab | +++ | Syncytiotrophoblasts |
| Prostate | + | Glandular epithelium |
| Skinb | +++ | Sebaceous glands (+differentiating keratinocytes) |
| Testis (slightly atrophic) | ++ | Not identified (mature sperm cells reduced) |
| Thymus | ++ | Cells in the Hassal's bodies |
| Thyroid gland | + | Follicle epithelium |
| Uterus (endometrium) | − | |
| Stomach (ventricle) | − |
Semiquantitative grading of in situ hybridization results. The positive control was placenta.
Tissues tested from more than 10 individuals
This tissue was hybridized several times.
RESULTS
The aim of this study was to bioinformatically define ERV3 7q11 and any ERV3-like sequences and to analyze the expression of ERV3 7q11 in normal human tissues using ESTs, real-time PCR, and ISH. The last two techniques were focused on the env gene. As a reference, the expression of the related HERV-E(4-1) env was also studied. ERV3 has earlier been considered a solitary integration. This is unusual for HERVs, which tend to occur in multiple copies. The recent expansion of sequence information made it possible to ascertain whether ERV3 is a single-copy HERV or if there is a group of ERV3-like HERVs. This question is answered below (see “ERV3-like sequences based on SU protein sequence”).
Structure of the ERV3-locus on chromosome 7, band q11.
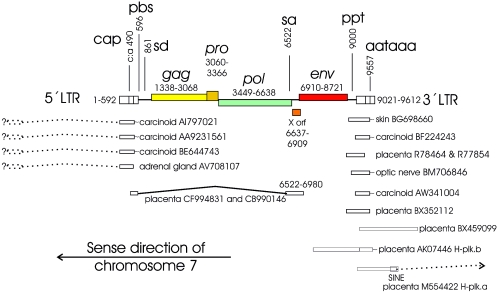
We found that the clone AC073210.8 (Table 1; see Fig. 3A) in GenBank contains not only the annotated ERV3 pol env 3′ LTR sequence (accession no. M12140.1), submitted by Cohen et al. (15) but also a 5′ LTR. Its absolute position on chromosome 7 is antisense to the assigned chromosomal direction, with the 5′ LTR starting at position 63858202 (human genome version hg15, April 2003) (Fig. 1). The LTRs are 91% identical. The provirus has a primer binding site (PBS) complementary to arginine tRNA, and gag, pro, pol, and env sequences similar to those of other members of genus Gammaretrovirus. The only longer ORFs are in pro and env. A detailed interpretation done with RetroTector is available as supplemental material (see Fig. S1). The distance between the PBS and the probable start of the gag sequences is unusually long (900 bp) but has no likely ORF, as studied with XonID (Fig. 1). The predicted Gag polyprotein contains a matrix protein (MA) with an MGQ myristylation signal and a PPPY motif necessary for “late” functions, i.e., budding (54, 55, 81). A capsid protein (CA) with a major homology region (MHR) related to that of MLV, HERV-H, and HERV-E and a nucleocapsid protein (NC) with one zinc finger are also present. At the predicted end of Pol is a GPY/F motif, called IN7 in RetroTector. This motif occurs in several gammaretroviruses as well as in gypsy (46) and chromovirus (23) elements, both widespread retrotransposons. In the latter two, the GPY/F motif may border a chromodomain. Both of these C-terminal integrase portions may influence integration specificity by interaction with DNA-binding proteins (69).
FIG. 1.
Overview of the ERV3 locus at chromosome 7q11 as interpreted by the RetroTector computer program (G. O. Sperber and J. Blomberg, unpublished data) from the human genome version hg15 (April 2003). Positions of selected ESTs and cDNAs and observed or probable splices (indicated by questions marks) are also shown. Positions are given relative to the first nucleotide of the 5′ LTR. Note that the element is antisense to the ascribed start of chromosome 7. SINE, short interspersed element. Sequences are identified by GenBank accession numbers.
Between the predicted 3′ end of pol (chromosome position 63851564) and the 5′ end of env (chromosome position 63851292) is a 255-bp (Fig. 1) ORF (corresponding to 85 amino acids) of unknown significance, here referred to as the ERV3 XORF. It was suggested by the RetroTector XonID function. Theoretically, it could start at the methionine at nt 6623 (chromosome position 63851579), in frame 1, requiring a shift to frame 3 at approximately nt 6738 (chromosome position 63851464). Alternatively, the putative XORF protein could have an internal ribosomal entry start after this frameshift, or it could represent an unusually long DNA-binding domain of the ERV3 integrase. The putative ERV3 XORF protein is histidine rich and has a moderate similarity to a toxin receptor. It is further analyzed in the supplementary material (see Fig. S1 in the supplemental material).
According to the RetroTector interpretation, Env starts with MLGMNMLLITLFLLLPLSMLK, 40 amino acids upstream of MTKTLLYHTYYECAGTCLGTC, the Env start suggested by Cohen et al. (15). We favor the first sequence. It includes a hydrophobic von Heijne signal sequence motif typical of the amino terminus of retroviral membrane proteins. Other groups (19, 26) made interpretations similar to ours.
ERV3 7q11 ESTs, splicing, and the LTR structure of the ERV3 7q11.
A BLAST search with three nucleotide sequences from three portions of the ERV3 7q11 provirus in the human EST database yielded 25 to 86 hits which were >95% identical to the ERV3 7q11 sequence (Table 2). The hit frequency for a tissue depends on the number of hits per tissue cDNA library. Some tissues have many libraries and thus give many hits. Additionally, the cloning strategy, using 5′, 3′, or random priming, determines the likelihood that a certain query sequence will find a hit. Thus, this statistic is at best semiquantitative. However, it demonstrates in which tissues ERV3 is expressed. Hits for placenta, testis, skin, and brain were especially frequent. Notable were also six ESTs from carcinoid, a multiple endocrine tumor with a strong genetic dependence. Illustrative ESTs are depicted together with the ERV3 7q11 provirus (Fig. 1). Four of the carcinoid ESTs were almost identical (98%) to the 5′ LTR of ERV3 7q11 and clearly different from the 3′ LTR. The simplest explanation for this is that these ESTs are polyadenylated at the 5′ LTR of ERV3 at 7q11, which is unusual. It is unlikely that these transcripts were polyadenylated at single ERV3 LTRs. We did not find any single ERV3 LTRs which were as similar to the transcripts as the ERV3 7q11 5′ LTR is. The start site of these transcripts is not known. According to this interpretation it should be upstream of ERV3 7q11.
TABLE 2.
ESTs detected by a BLAST search with four different nucleotide sequencesa
| Tissue | No. of hits with nt sequence for:
|
|||
|---|---|---|---|---|
| R-U5 (120 bp)b | U3 (471 bp)b | TM+PPT (715 bp)c | Total | |
| Placenta | 10 | 14 | 8 | 32 |
| Skin | 2 | 3 | 5 | |
| Adrenal gland | 1 | 2 | 4 | 7 |
| Testis | 3 | 3 | ||
| Ovary | 2 | 2 | ||
| Pancreatic islet | 3 | 3 | 6 | |
| Kidney | 3 | 9 | 12 | |
| Brain | 7 | 8 | 1 | 16 |
| Optic nerve | 2 | 2 | ||
| Prostate | 3 | 1 | 4 | |
| Cartilage | 4 | 2 | 6 | |
| Lung | 1 | 1 | 2 | |
| Fetal lung | 1 | 1 | 2 | |
| Fetal liver/spleen | 3 | 6 | 1 | 10 |
| Fetal heart | 1 | 2 | ||
| Fetal retina | 1 | 1 | 2 | |
| Colon | 1 | 1 | ||
| Bone marrow | 1 | 1 | 2 | |
| B cells from germinal center | 1 | 2 | ||
| Alveolar macrophage | 1 | 1 | ||
| Total fetus | 4 | 3 | 7 | |
| Carcinoid | 6 | 6 | ||
| Colon tumor | 5 | 4 | 1 | 10 |
| Colon adenocarcinoma | 1 | 1 | ||
| Gastric carcinoma | 2 | 3 | 5 | |
| Endometrial adenocarcinoma | 1 | 1 | ||
| Uterus tumor | 1 | 1 | 2 | |
| Cervical tumor | 1 | 2 | 3 | |
| Kidney tumor | 2 | 2 | ||
| Oligodendroglioma | 5 | 1 | 1 | 7 |
| Neuroblastoma | 1 | 4 | 5 | |
| Leiomyosarcoma | 1 | 1 | ||
| Total | 56 | 86 | 25 | 167 |
Hits with a score of more than 46 and a percent identity to ERV3 7q11 of more than 95% are shown. Searches with gag and pol sequences did not yield any ERV3 7q11 specific hits (not shown).
An LTR consists of two unique sequences (U3 and U5) and one repeat sequence (R).
TM, transmembrane protein nucleotide sequence; PPT, polypurine tract.
Many of the ESTs were polyadenylated. Their sequence revealed the 3′ border of the R region of the ERV3 3′ LTR. It was situated 15 nt from the end of the polyadenylation signal (AAATAAAA; which started at nt 537 (chromosome position 63857665) in the 5′ LTR and at nt 9021 (chromosome position 63849181) in the 3′ LTR). This is a typical distance for gammaretroviruses (14). It conforms exactly with the experimentally derived polyadenylation site (29). The 5′ border of R was harder to verify. Two ESTs (CF994831 and CB990146) which began close to this site were detected by a BLAST search with the predicted RU5 of the 5′ LTR. They started with 20 dissimilar non-ERV3 nucleotides. Their ERV3 similarity commenced at nt 494 (chromosome position 63857708) of the 5′ LTR, 13 nt downstream of the end of a typical TATAAAA box. The earlier reported experimentally obtained cap site at nt 498 (30, 51) is 4 nt beyond the probable EST-derived cap site. The somewhat corrupted start of the ESTs makes it impossible to determine an exact cap site based on them. The ESTs were spliced at a predicted canonical splice donor at nt 861 (chromosome position 63857341) to an acceptor at nt 6522 (chromosome position 63851680). These splice sites were exactly the same as those found earlier (30).
Using EST data, and RetroTector and TFBS predictions, a map of the ERV3 7q11 5′ and 3′ LTRs was made (see Fig. S5 and S6 in the supplemental material). The 5′-3′ LTR nonidentity of 9% indicates that many of the preintegrational TFBS should have been damaged by random mutation. If indeed there was selection for expression of any ERV3 transcripts, it is likely that TFBS essential for their expression were spared or created postintegration. Despite the 9% divergence between the LTRs, the ERV3 7q11 env ORF, covering 1,812 nt, has been maintained. Observations of an ERV3 protein by Western blotting and immunohistochemistry with anti-ERV3 sera (78) also support the existence of an ERV3 Env protein. A possible selection for production of the ERV3 Env protein in certain tissues should be mirrored by selection at the 5′ LTR, and less so at the 3′ LTR, if the 5′ LTR has the dominating promoter. Comparing the 5′ and 3′ LTRs, a region upstream of the TATAA box was almost spared of mutation and may be especially important for ERV3 promoter activity. Its downstream adjacent region was more extensively mutated (see Fig. S5 and S6b in the supplemental material). To better understand any selective forces on the 5′ LTR, we mapped TFBS in the 5′ and 3′ LTRs. Using ConSite at moderate stringency (10 bits), we determined that TFBS with more than one predicted occurrence in the 5′ LTR included 12 SOX-5, 5 HFH-2, 4 RORα1, 3 Sox17, and 3 Thing1/E47 sites. Of these, three SOX-5, two RORα1, and three SOX17 sites were unique to the 5′ LTR relative to the 3′ LTR. For comparison, five random control sequences of equal lengths (492 nt) and with approximately the same ATGC content as that of the 5′ LTR of ERV3 (27% A, 29% T, 20% G, and 24% C) were also analyzed. SOX-5 sites occurred 2, 3, 3, 4, and 5 times (average, 3.4), while SOX17 occurred 1, 3, 3, 3, and 4 times (average, 2.8), and RORα1 occurred 0, 0, 1, 2, and 2 times (average, 1.0) in each of these five control sequences, respectively. The SOX5 frequency thus deviated most from random prediction (P = 0.01, Mann-Whitney test) and was the most frequent among the TFBS unique to the 5′ LTR. The predicted SOX-5 sites (12 in the 5′ LTR and 10 in the 3′ LTR) clustered in the middle of U3. Although several ERV3-like ESTs mapping to sequences downstream of the canonical splice donor and upstream of the canonical splice acceptor were found, none of them probably came from ERV3 7q11, judging from the degree of nucleotide identity. A scarcity of full-length mRNAs from the ERV3 7q11 provirus was also noted earlier (30).
Among the more completely recorded mRNAs, only one encompassing most of ERV3 7q11 env, the placental H-plk.a, was found. Most other env-containing ESTs encompassed only the TM domain. A chimeric cDNA clone starting close to the 3′ LTR of ERV3 7q11 (the placental H-plk.b) runs through the ERV3 7q11 3′ LTR into an adjacent long interspersed element and is then spliced from an ensuing short interspersed element into a Krüppel zinc finger, as described earlier (30).
ERV3-like sequences based on SU protein sequence.
ERV3 has been described as a single-copy HERV (47, 78). However, even if the SU part is one of the most variable in a retrovirus, many ERV3-like sequences were found when searches for matches were done at the protein level, using the SU part of Env. These sequences were subsequently aligned at the nucleotide level. A neighbor-joining unrooted tree was then built (Fig. 2A). This SU-based tree shows the relationship with four other class 1 HERVs, HUMRIRT (M64936), HERV15yq1 (AF290422), HERV15yq2 (AF290423), and HUMERGPE (M74509). HERV-E(4-1) formed a cluster together with other known HERV-E sequences (e.g., HUMERGPE). Close to ERV3 were sequences from clone AC013429, which have a proline tRNA as the PBS, and from clones AB019437 and AC008175, which did not have an identifiable PBS. More distant from ERV3, among the cluster of ERV3 SU-like proviral sequences, was HUMRIRT, also known as RRHERVI (28). The branching pattern was similar to that of the Pol amino-acid based tree (Fig. 2B; discussed below).
Overall, ERV3 SU-like proviruses with an identifiable PBS had a PBS complementary to proline, arginine, or isoleucine tRNA (Table 1). ERV3 itself uses arginine tRNA. The use of an arginine PBS is not confined to ERV3-like elements (see below). We therefore could not call the group “HERV-R.”
Delineation of ERV3-like sequences based on similarity in Pol.
When the RetroTector version 010 and the RetroTector Shell version 01 programs became available, we selected all ERV3-like sequences in the human genome version 15 using an ERV3 Pol protein-based search. Out of 76 retrieved ERV3-related sequences, 41 ERV3-like sequences based on Pol similarity formed a separate cluster at 80% similarity or higher, using the PAM250 score matrix (Fig. 2B; see Fig. S3b and Table S4a in the supplemental material). Although the 11 ERV3 SU-like elements of Fig. 2A and the 12 elements in Table 1 had a pol gene, several were defective in pol and therefore hard to classify by Pol. However, an alignment showed that all could be accommodated in the larger ERV3-like group, which was Pol based (data not shown). Thus, env and pol genes evolved together in this gammaretroviral group. In the following, the 41 Pol-based ERV3-like elements are simply called “ERV3-like.” However, the 80% limit did not completely separate ERV3-like elements from HERV-E-like ones, neither in Pol nor in Env (see Tables S4a and 4b, respectively, in the supplemental material). The neighbor-joining tree derived from the Clustal alignment of the 76 elements (Fig. 2B) also demonstrated the ERV3- and HERV-E-like clusters, with bootstrap support. The similarity contingency table (see Table S4b in the supplemental material) showed a somewhat incomplete separation of ERV3-like and HERV-E-like elements, possibly indicating recombination and/or a common evolutionary origin of the groups. As shown in Fig. S3b (see the supplemental material), the ERV3-like elements consisted of 23 elements highly similar to ERV3 7q11 in Pol, hereafter called “ERV3 elements,” and 18 elements highly similar to RRHERVI, hereafter called “RRHERVI elements.” In the first group, 13 elements had a recognized env gene, while in the second group, 11 elements had such a gene. Out of the 41 ERV3-like sequences, only one element, the ERV3 locus on 7q11, had an env ORF. Among the 76 elements, HERV-E elements had the most complete pol gene, based on the presence of conserved motifs. The LTR divergence of the 41 ERV3-like elements was on average 11.4%, with a range of 5.1 to 24.2%. The corresponding figures for the HERV-E elements shown in Fig. 3B were 15.1% with a range of 7.0 to 30.5%. Some of the more complete ERV3 elements were analyzed in greater depth (Table 1 and Fig. 3B).
PBS tRNA usage: HERV-R in Repbase (27).
Of the 41 ERV3-like elements, 2 used arginine, 2 used proline, and 1 used isoleucine tRNA as a PBS. A weak lysine tRNA-like PBS was also predicted. The PBS of the remaining elements was not identified by RetroTector, for unknown reasons. A RetroTector Shell search indicated that the human genome contains three ERV3-related elements with an arginine PBS. Two of them (ERV3 7q11 itself and chrY.3018841) belonged to the 41 ERV3-like elements, while the third (chr19.21399629) was classified as “HUERSP3-like,” a gammaretroviral sequence which is more distant from ERV3 than HERV-E (data not shown).
Altogether, RetroTector detected 72 retroviral elements with an arginine PBS in the human genome. The great majority of them, 69, thus fell outside of the ERV3-like group. Of the 69, 3 were HERVRB-like, 5 were HUERSP3-like, 2 were MER41-like, 2 were HERV19-like, and 2 were HERVFc-like, based on RepBase nomenclature. The rest have not yet been classified, but some of them contained stretches similar to the HERV9 and the little-studied MER41, MER51, and MER66 sequences. This is an example of “PBS promiscuity” (see Discussion). RepBase (27) contains a sequence called HERV-R. This HERV-R was 100% identical to the baboon endogenous retrovirus (74) and therefore is not a human ERV. This is a further reason to avoid using the term HERV-R.
Structure and localization of ERV3-like elements.
ERV3-like elements were common on chromosome Y. Six elements were located there, while two occurred on each of chromosomes 2, 6, and 19. Chromosomes 14 and X contained one ERV3-like element each. A complete list of the ERV3-like elements is available in the supplemental material (Table S2). Table 1 contains a partial list of the ERV3-like elements.
The ERV3-like sequences harbored not only env genes but also other proviral structures (Table 1). The LTRs of HERV-E contain a promoter known to affect transcription of several cellular genes. Moreover, HERV-E transcripts have been seen in both normal and diseased tissues (52, 65). To retrieve HERV-E(4-1) Env-encoding sequences, the same type of TBLASTN search that was initially made with ERV3 Env was also made with HERV-E Env. The results are shown with the ERV3-like SU sequences in Fig. 3. The nucleotide alignments presented in Fig. 3 illustrate how the subsequently designed real-time primers and probe fit with the ERV3-like sequences. As anticipated, most similarities were observed in the conserved transmembrane region (TM). Therefore, we chose the SU region of env for designing specific primers. As shown in the alignment, point mutations, insertions, and deletions favor a PCR specific for the ERV3 7q11 locus. A similar narrow specificity was also expected of the HERV-E(4-1) SU-derived primers and probe. The searches for ERV3-like sequences revealed interesting details regarding the genetic environment at their integration sites, mentioned in footnotes to Table 1. It is beyond the scope of this paper to discuss the possible effects of ERV3-like elements on the surrounding genes.
QPCR results.
In order to study the HERV expression quantitatively, specific real-time PCRs were developed for ERV3 and HERV-E. The ERV3 QPCR method had a sensitivity of around 10 haploid human genomes and around 10 equivalents of ERV3 plasmid DNA per PCR. The HERV-E QPCR method could detect 5 to 10 haploid genomes and around 10 equivalents of HERV-E(4-1) plasmid DNA. The specificity for both methods measured as the absence of cross-amplification from the HERV-E(4-1) and ERV3 plasmids was 100%.
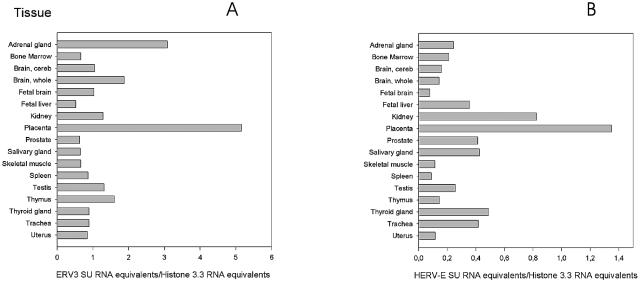
The ERV3 and HERV-E SU QPCRs confirmed previous data showing high expression in adrenal gland and placenta (30, 33).
QPCR with ERV3 and HERV-E.
The two graphs showing the results of QPCR with ERV3 and HERV-E (Fig. 4) reveal great differences in mRNA expression between organs, but also between the SU of ERV3 (Fig. 4A) and HERV-E (Fig. 4B). The high expression of ERV3 in the adrenal gland confirmed previous ISH results. Other organs with high expression levels were whole brain (ERV3), placenta (ERV3 and HERV-E), kidney (ERV3 and HERV-E), thymus (ERV3), thyroid gland (HERV-E), prostate (HERV-E), and trachea (HERV-E). However, all organs had some expression of ERV3 and HERV-E SU.
FIG. 4.
ERV3 (A) and HERV-E (B) env mRNA expression detected by QPCR specific for the SU region of HERV-E and ERV3, respectively. Each value represents a mean of the results for at least two experiments. cDNA from an RNA panel (Clontech) from several human tissues was analyzed. The values for placenta and skeletal muscle are averages based on samples from a commercial tissue RNA panel and RNA extracted from local patient samples, as described in Materials and Methods. All values are expressed relative to the histone 3.3 signal of the same sample. Note that the scale for HERV-E is different from that of ERV3. cereb, cerebellum.
ISH results with an ERV3 probe.
Based on our previous Northern blot analysis of ERV3 and RT-PCRs of ERV3 7q11 and HERV-E(4-1) of others, we knew that there was a difference in expression both between organs and between individuals (4, 66). Since a tissue is composed of several different cell types, existing in different ratios, we wanted to know which of the cell types expressed high levels of the retroviral transcript. This question was addressed by ISH using a 1.7-kb ERV3 7q11 env probe under high-stringency conditions. The results are presented in Table 3 and Fig. 5.
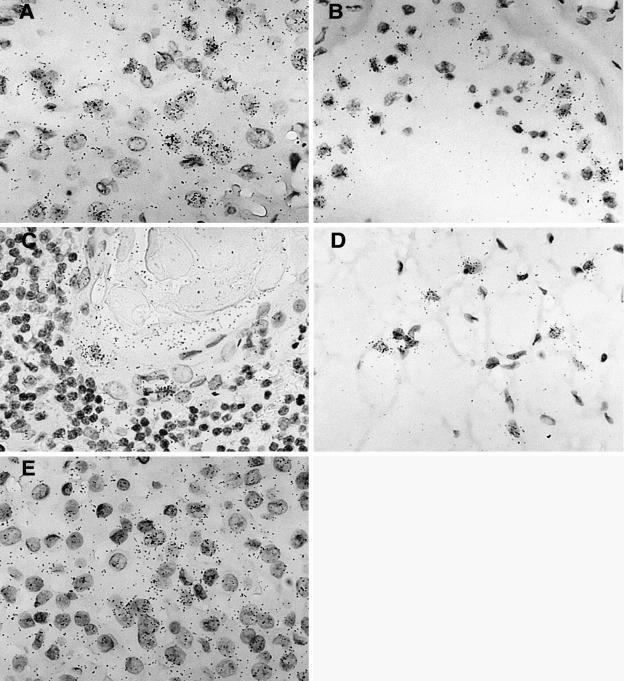
FIG. 5.
Hybridization signal obtained with the ERV3 antisense SU probe in five different tissues. (A) Tissue section from a corpus luteum demonstrating a positive and elevated ERV3 signal in progesterone-producing luteinized follicular cells. (B) Positive signal in yet unidentified cells obtained from a tissue section from a human, partly atrophic, testis with reduced spermatogenesis. (C) Tissue section from thymus with an elevated signal in cells in the outer part of a Hassal's body. (D) Brown fat section showing a high ERV3 message in typical multivacuolated brown fat cells. (E) Tissue section from a human pituitary gland (adenohypophysis portion), where an elevated ERV3 message is visible in all cell types.
Table 3 shows that several tissues have elevated levels of ERV3 7q11 env mRNA. In summary, all layers of the adrenal gland, cells in brown fat, bronchial epithelial cells of lung, progesterone-producing cells of corpus luteum, all cells in the adeno part of the pituitary gland, glandular epithelium of the prostate, sebaceous gland of the skin, cells in testis (not Leydig cells), Hassal's bodies of the thymus, and follicle cells of the thyroid gland contained ERV3 SU mRNA. Adrenal gland, skin, and placenta (a positive control) have previously been reported to harbor high concentrations of ERV3 transcripts (3, 31). A selection of tissues containing cells with the highest levels of detected mRNA are illustrated in Fig. 5, i.e., corpus luteum, testis, thymus, brown fat, and pituitary gland.
DISCUSSION
This study is a systematic analysis of a class I HERV genome, ERV3 7q11, sometimes in contrast to the related HERV-E. Both have been frequently studied before. However, a systematic analysis using the full information from the human genome databases and quantitative RNA detection methods has not been done.
ERV3-like sequences and the ERV3 locus.
ERV3 is a rather typical gammaretrovirus. Most of the structures of MLV are discernible in it. However, unlike MLV, there is no indication of a pp12 Gag protein. In addition, a space separates the predicted pol from the predicted env. It contains a putative ORF, here referred to as ERV3 XORF. Theoretically, it could encode a protein of its own, an extension of the Pol protein, or an unusual leader sequence for the Env protein. The position between predicted pol and env is typical of sequences for regulatory proteins of complex retroviruses. This putative protein or protein subdomain should be searched for in tissues with high ERV3 expression using antisera and proteomic techniques. If it is a C-terminal extension of the integrase new to gammaretroviruses, a participation in chromatin binding (69) should be investigated.
Mutilated and recombinant HERVs create taxonomic problems. The somewhat incomplete separation in the similarity matrices (see Fig. S3a and S3b in the supplemental material) may be symptoms of recombination. Gene conversion is an often invoked but rarely proven recombinatory mechanism for highly related HERVs (71). Nevertheless, the Pol-based classification used in this paper was to a large extent consistent with the most similar RepBase (27) sequence, found by a BLAST algorithm with the nucleotide sequence of the whole element. The older PBS-based classification (37) was sometimes congruent. For example, the HERV-E group was well delineated from other HERV groups. A glutamine tRNA PBS is almost exclusively associated with the HERV-E group (work with RetroTector; data not shown). Meanwhile, neutral group names referring to established loci are preferable to minimize confusion. We therefore chose the name “ERV3-like” for the group of elements comprising ERV3 and RRHERVI elements. They share a Pol protein similarity of 80% or higher, as estimated with the PAM250 matrix. Based on a comparison of RepeatMasker (A. Smit, unpublished data) output and its corollary, the HERVd database (53), both of which are based on RepBase, and RetroTector output, from the human genome version 15 (April 2003), the 41 ERV3-like elements defined here were classified either as HERV3 or HERV15 by RepeatMasker (J. Blomberg and G. O. Sperber, unpublished data). The two independent classifications thus were approximately concordant.
ERV3 7q11 has been described as a single-copy gene, related to the higher-copy group HERV-E (51). We here show that it is part of a group of 41 elements. A few of the ERV3-like sequences were previously described, some of which occur on the Y chromosome (34). The env gene of orthologs of the human ERV3 7q11 locus had an ORF that was maintained in several Old World monkeys. However, the locus was absent in gorillas; hence, it may not perform an exclusive essential function in primates (26). Single-nucleotide polymorphisms have been reported in the original ERV3 provirus, where not only env but also LTR polymorphic sequences were discovered by Rasmussen et al. (59, 60). Additionally, de Parseval et al., at the same time, discovered a single-nucleotide polymorphism in the carboxy terminus of ERV3 SU, introducing a stop codon in 1% of Caucasians, which would create a truncated Env protein (18, 19). It is, however, not known if the premature stop in the carboxy-terminal surface region would preclude a function or not. Regardless, women with the mutation had normal pregnancies (19). In previous studies of ERV3, Lin et al. showed that when ERV3 Env was expressed in the trophoblastic cell line BeWo, the trophoblasts differentiated, fused, and produced beta-human chorionic gonadotropin (41). A similar effect was attributed to the HERV-W Env protein syncytin (49), which can fuse cells in placenta, possibly forming the syncytiotrophoblastic layer.
Interestingly, some of the ERV3-like sequences were found to be integrated in the vicinity of genes encoding regulatory proteins, including an estrogen and retinoic acid receptor regulatory protein TIF1 (Table 2), with unknown effects. Although TFBS predictions must be judged with caution, the frequent occurrence (12 hits) of predicted SOX-5 sites in the 5′ LTR of ERV3 requires a comment. Sox (Sry-type high-mobility-group box) proteins are a subfamily of DNA-binding proteins with a high-mobility-group domain. Sry is the testis-determining factor (68). Sox proteins have functions in sex determination and neurogenesis (38). SOX-5 is highly expressed in testis, especially in spermatids (9, 12, 73, 84). Expression in the germ line is relevant for an endogenous retrovirus. The other frequently predicted sites were HFH2 and Thing1. HFH2 (FoxD3; 5 hits in the 5′LTR) drives the development of the neural crest and production of parathyroid hormone (58), and Thing1 (Hand1; 3 hits) is important for trophoblast transition to syncytiotrophoblast and neural crest development (16). Carcinoid tumors, here reported to express ERV3 RNA, have been considered to be a neural crest derivative (36). RORα1 (3 hits) is expressed in sebaceous glands (61) and belongs to the retinoic acid receptor family. Retinoids influence lipid synthesis in sebaceous glands (72) and activate RRHERV-I, which belongs to the ERV3-like HERVs (Table 1) (28). As shown here, ERV3 RNA is abundant in placenta, testis, and sebaceous glands. It is tempting to see a pattern of ERV3 expression corresponding to the known activities of these transcription factors. However, random sequences of similar nucleotide compositions were also predicted to bind some of the same factors. Their possible involvement in promotion of transcription from ERV3 at 7q11 should be experimentally verified in transient-transfection experiments.
Tissue-specific expression of ERV3 and related sequences.
ESTs which likely were encoded at ERV3 7q11 were found in cDNA libraries from normal tissues like placenta, testis, and adrenal gland. A reason for the relative scarcity of unspliced transcripts described previously (30), and in this work, could be nonsense-mediated decay of the full-length mRNAs, which are relatively more defective than the env mRNAs (39). Some malignancies, primarily carcinoid and colon tumors, also contained ERV3 cDNA. As mentioned above, carcinoid is a neuroendocrine cancer with an inheritable predisposition. The observation of multiple ERV3 ESTs in two different cDNA libraries from the neuroendocrine tumor carcinoid should be followed up.
QPCR showed high levels of ERV3 7q11 env RNA expression in adrenal gland, skeletal muscle, brain, placenta, thymus, and testis. HERV-E(4-1) env RNA was present primarily in placenta, skeletal muscle, kidney, thyroid gland, prostate, and trachea. Thus, tissues with a function in reproduction as well as endocrine functions tended to have a higher expression of ERV3 7q11 env. The expression was higher than that for env of the related HERV-E(4-1) and had a different tissue profile.
Our ISH studies of ERV3 confirmed previous Northern blot analyses showing that ERV3 is active in some specific cell types, such as syncytiotrophoblasts in placenta and cells in sebaceous glands (3, 30). In addition, high activities were found in brown fat, corpus luteum, testis, and Hassal's bodies in thymus. Several of these tissues are under strong hormonal influence. The EST and QPCR data also showed high expression levels in adrenal gland. The possible functions of a retroviral protein in these tissues are unknown. In animals, a high expression of ERVs is common in reproductive tissues like seminiferous tubules, vesicula seminales, testis, and placenta (20, 24, 25, 32, 40). The results of EST searches, QPCR, and ISH from this investigation and previous Northern blot analyses were similar. They cover different aspects of RNA expression. dbEST covers a broad range of normal and pathological tissues, but quantification is imperfect. QPCR gives an approximate number of HERV RNA molecules per nanogram of RNA or per household gene transcript but is sensitive to mutations in primer and probe sequences. ISH identifies which cell types express the RNA but is not as quantitative as QPCR. Northern blot analysis describes the molecular weights of the RNAs but is less quantitative than QPCR. Since ISH was performed with fixed tissues and PCR was done with fresh ones, the procedures are not completely comparable. Besides, it is known that external factors such as ischemia and anoxia can increase the activities of selected retroelements such as VL-30 (2). Further, polymorphism and epigenetic events such as imprinting could lead to both inter- and intraindividual variation in HERV expression. An interindividual variation in RNA expression has been shown for several HERVs (7, 42, 48, 85).
The results presented here demonstrate that both ERV3 7q11 and HERV-E(4-1) are expressed in most organs, as reported by Sibata et al. (66). Although an exact comparison cannot be made, ERV3 expression was higher than HERV-E expression in several tissues. Similar to the results presented here, expression of ERV3 env has been previously reported to be high in adrenal and sebaceous glands (3, 4) and in placenta (11, 41), whereas HERV-E env expression was found to be high in placenta (66).
In conclusion, this investigation provides a description of ERV3 structure, ERV3-like elements in the human genome, and the degree of ERV3 7q11 and HERV-E(4-1) env RNA expression in normal tissues. Measurements of organ-, cell type-, and transcript-specific expression by QPCR, ISH, and previous Northern blot analysis gave concordant results. The techniques described here will allow further studies of the pattern of HERV expression in selected model systems and in diseased persons.
Supplementary Material
Acknowledgments
This work was supported by grants (projects 2037-B99-16BB and 4239-B00-02XBB) from the Swedish Cancer Society to Erik Larsson and Jonas Blomberg, respectively, and by a grant from the Swedish Scientific Council to Jonas Blomberg (521-2001-6520).
We thank Anna Forsman, Lijuan Hu, Patric Jern, and Dmitrijs Uzhameckis for valuable assistance.
Footnotes
Supplementary material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, G. R., D. L. Stoler, and L. A. Scarcello. 1989. Retrotransposon-like VL30 elements are efficiently induced in anoxic rat fibroblasts. J. Mol. Biol. 205:765-769. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, A.-C., M. Merza, P. Venables, F. Ponten, J. Sundström, M. Cohen, and E. Larsson. 1996. Elevated levels of the endogenous retrovirus ERV3 in human sebaceous glands. J. Investig. Dermatol. 106:125-128. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, A.-C., A.-C. Svensson, C. Rolny, G. Andersson, and E. Larsson. 1998. Expression of human endogenous retrovirus ERV3 (HERV-R) mRNA in normal and neoplastic tissues. Int J. Oncol. 12:309-313. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, A.-C., P. J. W. Venables, R. R. Tönjes, J. Scherer, L. Eriksson, and E. Larsson. 2002. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology 297:220-225. [DOI] [PubMed] [Google Scholar]
- 6.Andersson, M.-L., M. Lindeskog, P. Medstrand, B. Westley, F. May, and J. Blomberg. 1999. Diversity of human endogenous retrovirus class II-like sequences. J. Gen. Virol. 80:255-260. [DOI] [PubMed] [Google Scholar]
- 7.Andersson, M. L., P. Medstrand, H. Yin, and J. Blomberg. 1996. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res. Hum. Retroviruses. 12:833-840. [DOI] [PubMed] [Google Scholar]
- 8.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaise, R., J. Grober, P. Rouet, G. Tavernier, D. Daegelen, and D. Langin. 1999. Testis expression of hormone-sensitive lipase is conferred by a specific promoter that contains four regions binding testicular nuclear proteins. J. Biol. Chem. 274:9327-9334. [DOI] [PubMed] [Google Scholar]
- 10.Blomberg, J., D. Uzhameckis, and P. Jern. 2005. Evolutionary aspects of human endogenous retroviral sequences (HERVs) and disease, p. 204-238. In E. D. Sverdlov (ed.), Retroviruses and primate genome evolution. Landes Bioscience, Georgetown, Tex.
- 11.Boyd, M. T., C. M. Bax, B. E. Bax, D. L. Bloxam, and R. A. Weiss. 1993. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology 196:905-909. [DOI] [PubMed] [Google Scholar]
- 12.Budde, L. M., C. Wu, C. Tilman, I. Douglas, and S. Ghosh. 2002. Regulation of IkappaBbeta expression in testis. Mol. Biol. Cell. 13:4179-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, T., P. Dissing Sørensen, H. Riemann, H. J. Hansen, M. Munch, S. Haahr, and A. Møller-Larsen. 2000. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol. Scand. 101:229-238. [DOI] [PubMed] [Google Scholar]
- 14.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [PubMed]
- 15.Cohen, M., M. Powers, C. O'Connell, and N. Kato. 1985. The nucleotide sequence of the env gene from the human provirus ERV3 and isolation and characterization of an ERV3-specific cDNA. Virology 147:449-458. [DOI] [PubMed] [Google Scholar]
- 16.Cserjesi, P., D. Brown, G. E. Lyons, and E. N. Olson. 1995. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 170:664-678. [DOI] [PubMed] [Google Scholar]
- 17.Deb-Rinker, P., T. A. Klempan, R. L. O'Reilly, E. F. Torrey, and S. M. Singh. 1999. Molecular characterization of a MSRV-like sequence identified by RDA from monozygotic twin pairs discordant for schizophrenia. Genomics 61:133-144. [DOI] [PubMed] [Google Scholar]
- 18.de Parseval, N., G. Forrest, P. J. Venables, and T. Heidmann. 1999. ERV-3 envelope expression and congenital heart block: what does a physiological knockout teach us. Autoimmunity 30:81-83. [DOI] [PubMed] [Google Scholar]
- 19.de Parseval, N., and T. Heidmann. 1998. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J. Virol. 72:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupressoir, A., and T. Heidmann. 1996. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol. 16:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5. University of Washington, Seattle.
- 22.Feuchter-Murthy, A. E., J. D. Freeman, and D. L. Mager. 1993. Splicing of a human endogenous retrovirus to a novel phospholipase A2 related gene. Nucleic Acids Res. 21:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorinsek, B., F. Gubensek, and D. Kordis. 2004. Evolutionary genomics of chromoviruses in eukaryotes. Mol. Biol. Evol. 21:781-798. [DOI] [PubMed] [Google Scholar]
- 24.Harris, J. R. 1998. Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. Bioessays 20:307-316. [DOI] [PubMed] [Google Scholar]
- 25.Herbst, H., M. Sauter, and N. Müeller-Lantzsch. 1996. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am. J. Pathol. 149:1727-1735. [PMC free article] [PubMed] [Google Scholar]
- 26.Herve, C. A., G. Forrest, R. Lower, D. J. Griffiths, and P. J. Venables. 2004. Conservation and loss of the ERV3 open reading frame in primates. Genomics 83:940-943. [DOI] [PubMed] [Google Scholar]
- 27.Jurka, J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- 28.Kannan, P., R. Buettner, D. R. Pratt, and M. A. Tainsky. 1991. Identification of a retinoic acid-inducible endogenous retroviral transcript in the human teratocarcinoma-derived cell line PA-1. J. Virol. 65:6343-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, N., E. Larsson, and M. Cohen. 1988. Absence of expression of a human endogenous retrovirus is correlated with choriocarcinoma. Int. J. Cancer 41:380-385. [DOI] [PubMed] [Google Scholar]
- 30.Kato, N., S. Pfeifer-Ohlsson, M. Kato, E. Larsson, J. Rydnert, R. Ohlsson, and M. Cohen. 1987. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J. Virol. 61:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsumata, K., H. Ikeda, M. Sato, H. Harada, A. Wakisaka, M. Shibata, and T. Yoshiki. 1998. Tissue-specific high-level expression of human endogenous retrovirus-R in the human adrenal cortex. Pathobiology 66:209-215. [DOI] [PubMed] [Google Scholar]
- 32.Kiessling, A. A., R. Crowell, and C. Fox. 1989. Epididymis is a principal site of retrovirus expression in the mouse. Proc. Natl. Acad. Sci. USA 86:5109-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamura, M., N. Maruyama, T. Shirasawa, R. Nagasawa, K. Watanabe, M. Tateno, and T. Yoshiki. 1994. Expression of an endogenous retroviral gene product in human placenta. Int. J. Cancer. 58:836-840. [DOI] [PubMed] [Google Scholar]
- 34.Kjellman, C., H. O. Sjögren, and B. Widegren. 1995. The Y chromosome: a graveyard for endogenous retroviruses. Gene 161:163-170. [DOI] [PubMed] [Google Scholar]
- 35.Kowalski, P. E., J. D. Freeman, and D. L. Mager. 1999. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics 57:371-379. [DOI] [PubMed] [Google Scholar]
- 36.Kvols, L. K. 2003. Neoplasms of the diffuse endocrine system: endocrine neoplasms of the gastroenteropancreatic (GEP) system. In D. Kufe, R. Pollock, R. Weichselbaum, R. Bast, Jr., T. Gansler, J. Holland, and E. Frei III (ed.), Holland-Frei cancer medicine, 6th ed. BC Decker, Hamilton, Ontario, Canada.
- 37.Larsson, E., N. Kato, and M. Cohen. 1989. Human endogenous proviruses. Curr. Top. Microbiol. Immunol. 148:115-132. [DOI] [PubMed] [Google Scholar]
- 38.Laudet, V., D. Stehelin, and H. Clevers. 1993. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 21:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeBlanc, J. J., and K. L. Beemon. 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 78:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lecher, P., A. Bucheton, and A. Pelisson. 1997. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J. Gen. Virol. 78:2379-2388. [DOI] [PubMed] [Google Scholar]
- 41.Lin, L., B. Xu, and N. S. Rote. 1999. Expression of endogenous retrovirus ERV-3 induces differentiation in BeWo, a choriocarcinoma model of human placental trophoblast. Placenta 20:109-118. [DOI] [PubMed] [Google Scholar]
- 42.Lindeskog, M., and J. Blomberg. 1997. Spliced human endogenous retroviral HERV-H env transcripts in T-cell leukaemia cell lines and normal leukocytes: alternative splicing pattern of HERV-H transcripts. J. Gen. Virol. 78:2575-2585. (Erratum, 79:212, 1998.) [DOI] [PubMed] [Google Scholar]
- 43.Lindeskog, M., P. Medstrand, and J. Blomberg. 1993. Sequence variation of human endogenous retrovirus ERV9-related elements in an env region corresponding to an immunosuppressive peptide: transcription in normal and neoplastic cells. J. Virol. 67:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Löwer, R., J. Löwer, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magin, C., R. Löwer, and J. Löwer. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medstrand, P., and J. Blomberg. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J. Virol. 67:6778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medstrand, P., M. Lindeskog, and J. Blomberg. 1992. Expression of human endogenous retroviral sequences in peripheral blood mononuclear cells of healthy individuals. J. Gen. Virol. 73:2463-2466. [DOI] [PubMed] [Google Scholar]
- 49.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 50.Nystrom, K., M. Biller, A. Grahn, M. Lindh, G. Larson, and S. Olofsson. 2004. Real time PCR for monitoring regulation of host gene expression in herpes simplex virus type 1-infected human diploid cells. J. Virol. Methods 118:83-94. [DOI] [PubMed] [Google Scholar]
- 51.O'Connell, C., S. O'Brian, W. Nash, and M. Cohen. 1984. ERV3, a full-length human endogenous provirus: chromosomal localization and evolutionary relationships. Virology 138:225-235. [DOI] [PubMed] [Google Scholar]
- 52.Ogasawara, H., T. Hishikawa, I. Sekigawa, H. Hashimoto, N. Yamamoto, and N. Maruyama. 2000. Sequence analysis of human endogenous retrovirus clone 4-1 in systemic lupus erythematosus. Autoimmunity 33:15-21. [DOI] [PubMed] [Google Scholar]
- 53.Paces, J., A. Pavlicek, R. Zika, V. V. Kapitonov, J. Jurka, and V. Paces. 2004. HERVd: the human endogenous retroviruses database: update. Nucleic Acids Res. 32(Database issue):D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patnaik, A., and J. W. Wills. 2002. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76:2789-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perron, H., J. A. Garson, F. Bedin, F. Beseme, G. Paranhos-Baccala, F. Komurian-Pradel, F. Mallet, P. W. Tuke, C. Voisset, J. L. Blond, B. Lalande, J. M. Seigneurin, B. Mandrand, et al. 1997. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc. Natl. Acad. Sci. USA 94:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prak, E. T., and H. H. Kazazian, Jr. 2000. Mobile elements and the human genome. Nat. Rev. Genet. 1:134-144. [DOI] [PubMed] [Google Scholar]
- 58.Qin, L., P. Qiu, L. Wang, X. Li, J. T. Swarthout, P. Soteropoulos, P. Tolias, and N. C. Partridge. 2003. Gene expression profiles and transcription factors involved in parathyroid hormone signaling in osteoblasts revealed by microarray and bioinformatics. J. Biol. Chem. 278:19723-19731. [DOI] [PubMed] [Google Scholar]
- 59.Rasmussen, H. B., and J. Clausen. 1998. Large number of polymorphic nucleotides and a termination codon in the env gene of the endogenous human retrovirus ERV3. Dis. Markers. 14:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rasmussen, H. B., A. Heltberg, G. Lisby, and J. Clausen. 1996. Three allelic forms of the human endogenous retrovirus, ERV3, and their frequencies in multiple sclerosis patients and healthy individuals. Autoimmunity 23:111-117. [DOI] [PubMed] [Google Scholar]
- 61.Reichrath, J., M. Mittmann, J. Kamradt, and S. M. Muller. 1997. Expression of retinoid-X receptors (-alpha,-beta,-gamma) and retinoic acid receptors (-alpha,-beta,-gamma) in normal human skin: an immunohistological evaluation. Histochem J. 29:127-133. [DOI] [PubMed] [Google Scholar]
- 62.Ropenga, A., A. Chapel, M. Vandamme, and N. M. Griffiths. 2004. Use of reference gene expression in rat distal colon after radiation exposure: a caveat. Radiat. Res. 161:597-602. [DOI] [PubMed] [Google Scholar]
- 63.Schön, U., W. Seifarth, C. Baust, C. Hohenadl, V. Erfle, and C. Leib-Mösch. 2001. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology 279:280-291. [DOI] [PubMed] [Google Scholar]
- 64.Schulte, A. M., S. Lai, A. Kurtz, F. Czubayko, A. T. Riegel, and A. Wellstein. 1996. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 93:14759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiroma, T., J. Sugimoto, T. Oda, Y. Jinno, and F. Kanaya. 2001. Search for active endogenous retroviruses: identification and characterization of a HERV-E gene that is expressed in the pancreas and thyroid. J. Hum. Genet. 46:619-625. [DOI] [PubMed] [Google Scholar]
- 66.Sibata, M., H. Ikeda, K. Katumata, K. Takeuchi, A. Wakisaka, and T. Yoshoki. 1997. Human endogenous retroviruses: expression in various organs in vivo and its regulation in vitro. Leukemia 11(Suppl.):145-146. [PubMed] [Google Scholar]
- 67.Simpson, G. R., C. Patience, R. Löwer, R. R. Tönjes, H. D. Moore, R. A. Weiss, and M. T. Boyd. 1996. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology 222:451-456. [DOI] [PubMed] [Google Scholar]
- 68.Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths, M. J. Smith, J. W. Foster, A. M. Frischauf, R. Lovell-Badge, and P. N. Goodfellow. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346:240-244. [DOI] [PubMed] [Google Scholar]
- 69.Singleton, T. L., and H. L. Levin. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot. Cell 1:44-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 71.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 72.Strauss, J. S., M. E. Stewart, and D. T. Downing. 1987. The effect of 13-cis-retinoic acid on sebaceous glands. Arch. Dermatol. 123:1538a-1541. [PubMed] [Google Scholar]
- 73.Takamatsu, N., H. Kanda, I. Tsuchiya, S. Yamada, M. Ito, S. Kabeno, T. Shiba, and S. Yamashita. 1995. A gene that is related to SRY and is expressed in the testes encodes a leucine zipper-containing protein. Mol. Cell. Biol. 15:3759-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamura, T., M. Noda, and T. Takano. 1981. Structure of the baboon endogenous virus genome: nucleotide sequences of the long terminal repeat. Nucleic Acids Res. 9:6615-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 77.van Regenmortel, M., C. Fauquet, D. Bishop, E. Carsten, M. Estes, S. Lemon, J. Maniloff, M. Mayo, D. McGeoch, C. Pringle, and R. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 78.Venables, P. J., S. M. Brookes, D. Griffiths, R. A. Weiss, and M. T. Boyd. 1995. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology 211:589-592. [DOI] [PubMed] [Google Scholar]
- 79.Wang, Y., P. E. Kowalski, I. Thalmann, D. M. Ornitz, D. L. Mager, and R. Thalmann. 1998. Otoconin-90, the mammalian otoconial matrix protein, contains two domains of homology to secretory phospholipase A2. Proc. Natl. Acad. Sci. USA 95:15345-15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wells, D., and L. Kedes. 1985. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc. Natl. Acad. Sci. USA 82:2834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu, R. S., and W. M. Bonner. 1985. Mechanism for differential sensitivity of the chromosome and growth cycles of mammalian cells to the rate of protein synthesis. Mol. Cell. Biol. 5:2959-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu, R. S., and W. M. Bonner. 1981. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell 27:321-330. [DOI] [PubMed] [Google Scholar]
- 84.Wunderle, V. M., R. Critcher, A. Ashworth, and P. N. Goodfellow. 1996. Cloning and characterization of SOX5, a new member of the human SOX gene family. Genomics 36:354-358. [DOI] [PubMed] [Google Scholar]
- 85.Yin, H., P. Medstrand, M. L. Andersson, A. Borg, H. Olsson, and J. Blomberg. 1997. Transcription of human endogenous retroviral sequences related to mouse mammary tumor virus in human breast and placenta: similar pattern in most malignant and nonmalignant breast tissues. AIDS Res. Hum. Retrovir. 13:507-516. [DOI] [PubMed] [Google Scholar]
- 86.Yuan, C. C., W. Miley, and D. Waters. 2001. A quantification of human cells using an ERV-3 real time PCR assay. J. Virol. Methods 91:109-117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.