Abstract
Coxsackievirus B3 (CVB3) is a common human pathogen that is endemic throughout the world. There is currently no vaccine available, although the virus is known to be highly lethal to newborns and has been associated with heart disease and pancreatitis in older children and adults. Previously, we showed that the virulence of CVB3 is reduced by a lysine-to-arginine substitution in the capsid protein VP2 (K2168R) or a glutamic acid-to-glycine substitution in VP3 (E3060G). In this report, we show that the double mutant virus CVB3(KR/EG) displays additional attenuation, particularly for the pancreas, in A/J mice. In addition, two other attenuating mutations have been identified in the capsid protein VP1. When either the aspartic acid residue D1155 was replaced with glutamic acid or the proline residue P1126 was replaced with methionine, the resulting mutant also possessed an attenuated phenotype. Moreover, when either of these mutations was incorporated into CVB3(KR/EG), the resulting triple mutant viruses, CVB3(KR/EG/DE) and CVB3(KR/EG/PM), were completely noncardiovirulent and caused only small foci of damage to the pancreas, even at a high dose. Both triple mutants were found to be immunogenic, and a single injection of young A/J mice with either was found to protect them from a subsequent lethal challenge with wild-type CVB3. These findings indicate that the triple mutants could be exploited for the development of a live attenuated vaccine against CVB3.
The group B coxsackieviruses (CVBs) belong to the family Picornaviridae and the genus Enterovirus, a group of human viruses that includes the coxsackieviruses, polioviruses, and echoviruses. The CVBs are associated with a wide range of clinical manifestations that range from mild febrile illness to severe life-threatening diseases, including aseptic meningitis, myocarditis, and pancreatitis. The association of CVBs with human heart disease is particularly noteworthy, as these viruses are the most common cause of acute viral myocarditis and are involved in as many as 25% of cases in some studies (2, 13). Children are particularly susceptible to CVB infection, with aseptic meningitis and myocarditis being the major causes of morbidity and mortality. In fact, the mortality rate for infants with myocarditis alone is in the range of 30 to 50% and is even higher when other organs are involved (20). While acute CVB myocarditis is rarely lethal to adults, it can lead to persistent myocardial inflammation, which in turn may progress to dilated cardiomyopathy (5, 12, 18, 23). The incidence of dilated cardiomyopathy is about 6 cases per 100,000 people in North America, with approximately 100,000 new cases occurring each year, accounting for 50% of all cardiac transplant patients (7). However, despite the wealth of evidence of the role of CVBs in cardiomyopathy as well as potentially fatal disease in infants, there is currently no vaccine available. This is due in part to the fact that there are six serotypes of CVB, suggesting that a hexavalent vaccine would be necessary. However, the more cardiovirulent serotypes are those that bind to the secondary receptor DAF (decay accelerating factor), namely, CVB1, CVB3, and CVB5, indicating that a vaccine directed at these three strains would have the potential to greatly reduce CVB-associated cardiomyopathy. As a first step in the process of developing such a trivalent vaccine, we have constructed two triple mutants of CVB3 and show here that they are capable of protecting young mice against a subsequent lethal challenge with the wild-type (wt) strain.
The CVBs are small, nonenveloped viruses with a positive-strand RNA genome of about 7.4 kilobases. This consists of a single long open reading frame flanked by two nontranslated regions (NTRs), which contain signals that control both the translation and replication of the viral genome. In infected cells, a polyprotein is produced from the single open reading frame and posttranslationally cleaved to produce 11 proteins, including the four capsid proteins, VP1 to VP4. One copy of each capsid protein forms a protomer, and 60 protomers form the dodecahedral protein shell that encapsidates the viral genome. The main immunogenic areas of the CVBs lie in the surface-exposed “loops” of VPs 1, 2, and 3, and amino acid substitutions within these areas are largely responsible for defining the different serotypes of the virus. Specifically, the DE and EF loops are the major surface loops of VP1 and VP2, respectively, while the knob region is the main surface protrusion of VP3 (21). These three sites have been found to contain the major neutralizing sites for the coxsackieviruses as well as for other picornaviruses (1).
At the present time, there are two regions of the CVB3 genome in which attenuating mutations have been identified. The first is the 5′ nontranslated region (5′ NTR), which several reports have demonstrated contains determinants of cardiovirulence. Initially, it was shown that exchanging the 5′ NTR of a cardiovirulent strain with that from a nonmyocarditic strain resulted in a derivative that did not cause heart damage in mice (9, 15). A single mutation at nucleotide 234 in the 5′ NTR was subsequently demonstrated to confer myocardicity in a laboratory strain (29), but this mutation was not found to play a role in natural variants of the virus (11, 12). Recently, by sequencing the 5′ NTR of several CVB3 isolates, Dunn et al. have shown that mutations clustered within stem loop II of the 5′ NTR affect myocardicity, further attesting to the importance of this region in determining cardiovirulence (8). The other major location that has been shown to contain cardiovirulence determinants is the P1 region that encodes the capsid proteins. In 1996, a single amino acid substitution in the puff region of VP2 (asparagine to aspartate at amino acid 165) was found to reduce the myocardicity of a CVB3 mutant, H310A1 (14). Recently, we identified two mutations in the capsid proteins that affected cardiovirulence in studies of a panel of “escape” mutants (EMs 1 to 10) derived by treating a parental myocarditic strain, CVB3(RK), with a highly neutralizing monoclonal antibody (26). Two mutations were identified that were associated with both a lack of neutralization by the monoclonal antibody and reduced cardiovirulence. One of these was a lysine-to-arginine substitution at amino acid 158, also located in the puff region of VP2 in EM1. The second was a mutation of a glutamic acid to glycine at amino acid 60 in the knob region of VP3. A subsequent analysis of the positions of these two mutations in a stereographical image of the capsid structure showed that they were located in close proximity to each other and within the footprint identified for DAF binding for the related picornavirus echovirus 7 (10).
For this report, we exploited this previous information for the development of two triple mutants of CVB3 that could form the basis for a highly attenuated vaccine. The triple mutants contain the previously characterized VP2 (KR) and VP3 (EG) mutations and a third mutation in VP1 (one of several mutations in VP1 that we have found to be attenuating). By combining these mutations, we constructed triple mutants that are stable upon passaging and are capable of protecting susceptible A/J mice from a challenge with high titers of the virulent CVB3(T7) strain. Moreover, the triple mutants are not only noncardiovirulent but also attenuated for pancreatic tissue, which is highly susceptible to damage by CVB3 viruses. In view of the high morbidity of newborns infected with coxsackieviruses as well as the fact that these viruses may be involved in 25% of cases of viral myocarditis in patients of all ages, an attenuated vaccine would be a significant health benefit. Our identification of four mutations that greatly reduce the virulence of CVB3 could form the basis of such a vaccine. Moreover, by extrapolation of these findings to other group B coxsackieviruses, a similar strategy involving site-directed mutagenesis and reverse genetics could be used to construct a multivalent vaccine against the most virulent serotypes.
MATERIALS AND METHODS
Cells, viruses, and mice.
Vero cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium-F12 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin-neomycin solution. The CVB3(T7) strain was derived from an infectious clone, pCVB3(T7), of the Nancy strain (a generous gift from Reinhardt Kandolf, Germany). Virus stocks were prepared in Vero cells at a multiplicity of infection of 0.1 PFU per cell. After freeze-thawing of the stocks to release intracellular viruses, the supernatants were stored at −70°C. Virus titration was performed on Vero cells by standard techniques. Three-week-old A/J mice were obtained from Jackson Laboratory (Bar Harbor, Maine) and maintained in a level 2 containment facility at the University of British Columbia for about 1 week prior to viral inoculation at 4 to 5 weeks of age. For experiments to assess the vaccine potential of the triple mutants, the mice were immunized at 3 weeks of age and challenged 22 days later with a cardiovirulent virus.
Construction of infectious clones and recovery of infectious viruses.
The construction of the infectious clones pCVB3(KR) and pCVB3(EG) was previously described (26). The plasmid pCVB3(KR) has a single mutation of A to G at nucleotide 1421, resulting in a lysine (K)-to-arginine (R) substitution at amino acid 158 of the capsid protein VP2 (K2158R), while pCVB3(EG) contains a single mutation of A to G at nucleotide 1916, resulting in a glutamic acid (E)-to-glycine (G) substitution at amino acid 60 of VP3 (E3060G). In order to construct the double mutant pCVB3 (KR/EG), we digested both clones with BssHII and BsiWI to give a 7.5-kb fragment as well as the 2.7-kb fragment that harbored the A-to-G mutation from pCVB3(EG). By ligating the 2.7-kb fragment from pCVB3(EG) to the 7.5-kb fragment containing the A-to-G mutation from pCVB3(KR), we obtained the construct pCVB3 (KR/EG).
For the construction of pCVB3(DE), a single mutation (U to A) at nucleotide 2916 that caused an asparatate (D)-to-glutamine (E) substitution at amino acid 1155 of VP1 (D1155E) was incorporated into a PCR primer (5′-GTA CCA CCA GGT GGA CCT GTA CCA GAT AAA GTT GAA TCA TAC GT-3′) (the mutation is shown in bold). This was used to generate a 1,568-bp PCR fragment with the primer 5′-CGA CTT GCC GGC ACC AGG GCT CCC-3′, using pCVB3(T7) as a template. The plasmid pCVB3(T7) was digested with the restriction enzymes PflMI and BssHII to generate the linearized vector pCVB3(T7)/PflMI-BssHII and a fragment comprising nucleotides 2146 to 2884 (1,568 bp) with a PflMI site at both ends. The 1,568-bp PCR fragment was digested with PflMI and BssHII to generate a 1,354-bp fragment corresponding to a section of the viral cDNA from nucleotides 2884 to 4238. This was cloned into the linearized vector pCVB3(T7)/PflMI-BssHII to obtain the subclone pCVB3(DE)(2884-2146). This 2146-2884 fragment was then cloned into the linearized subclone pCVB3(DE)(2884-2146)/PflMI to generate pCVB3(DE). For the construction of pCVB3(KR/EG/DE), the same strategy was used except that pCVB3(T7) was replaced with pCVB3 (KR/EG). The aspartate-to-glycine mutation (shown in bold in the primer sequence) in pCVB3(DG) was created in the same way, using the primers 5′-GTA CCA CCA GGT GGA CCT GTA CCA GAT AAA GTT GGT TCA TAC GT-3′ and 5′-CGA CTT GCC GGC ACC AGG GCT CCC-3′ to generate a 1,568-bp PCR fragment for incorporation into CVB3(T7).
Plasmids pCVB3(PM) and pCVB3(KR/EG/PM) were generated by site-directed mutagenesis. The mutagenic primers PMF (5′-CAA GTA CTC AAC AGA TGT CAA CCA CAC AGA ACC-3′) and PMB (5′-GGT TCT GTG TGG TTG ACA TCT GTT GAG TAC TTG-3′) were used for pCVB3 (PM) and pCVB3 (KR/EG/PM), using either pCVB3 (T7) or pCVB3 (KR/EG), respectively, as the template. The nucleotides in bold in the primer sequences represent the inserted mutations, and their presence in each plasmid construct was confirmed by DNA sequencing. To produce the corresponding mutant virus, we linearized each plasmid construct containing a mutation(s) with SalI. Viral RNAs were produced by in vitro transcription with T7 and were transfected into Vero cells. After 48 or 72 hours of incubation at 37°C, the virus was harvested and stored at −70°C.
Determination of viral phenotype in A/J mice.
Male A/J mice (4 to 5 weeks old) were injected intraperitoneally with phosphate-buffered saline (PBS) (controls) or 105 PFU of either CVB3(T7) or a mutant virus. Three mice from each group were anesthetized and sacrificed on days 3, 7, and 10 postinfection. The heart and pancreas were removed from each mouse and divided into two pieces. One piece was snap-frozen for viral titration and the other was fixed in 4% paraformaldehyde for histological analysis. These tissue samples were embedded in paraffin, and 3-μm sections were mounted on slides. The tissue sections were either stained with Masson's trichrome stain or used to detect the viral genome by in situ hybridization, which was performed as described previously (26).
Determination of the stability of mutant viruses.
The stability of the mutants was tested by reverse transcription-PCR (RT-PCR) sequencing using primers that flanked each of the inserted mutations. Initially, stability was confirmed following passaging in vitro prior to the preparation of stocks of each mutant. In addition, the stability was determined in vivo, 3 and 7 days after the inoculation of A/J mice with each mutant. Total cellular RNAs were isolated from the hearts of three mice, and a cDNA fragment containing the mutation(s) was amplified from the viral RNAs by RT-PCR as described above. The presence of each mutation was then confirmed by DNA sequencing. For further evaluation of the stability of the triple mutants CVB3(KR/EG/DE) and CVB3(KR/EG/PM), viruses isolated on day 3 postinoculation (p.i.) from the pancreases of mice injected with each mutant were inoculated into naïve mice (n = 3) at a multiplicity of infection of 105 PFU. The original stock virus of each triple mutant was also injected into three mice for comparison. As before, the heart and pancreas were removed on day 3 p.i., and a portion of each was used for viral titration, while RNAs were extracted from the remainder for RT-PCR sequencing.
Evaluation of the ability of CVB3(KR/EG/DE) and CVB3(KR/EG/PM) to protect mice from lethal challenge.
Three-week-old male A/J mice in groups of 16 were inoculated by the intraperitoneal route with 105 PFU of either the CVB3(KR/EG/DE) or CVB3(KR/EG/PM) virus in 0.2 ml of PBS. The mice were observed for 21 days for morbidity and mortality. On day 21 p.i., the mice were bled from the tail vein in order to measure CVB3-specific neutralizing antibodies by an enzyme-linked immunosorbent assay. The mice were challenged by intraperitoneal inoculation on day 22 after injection with 106 PFU of CVB3(T7) virus, one of the triple mutants, or PBS as a control. On day 1 postchallenge, the mice were bled again to determine the level of circulating virus in the serum. On days 3 and 10 postchallenge, three mice from each group were sacrificed in order to assess the degree of viral replication as well as the amount of tissue damage in both the pancreas and the heart. The remaining mice were monitored up to day 28 postchallenge and were then sacrificed for pathological examinations. The levels of CVB-specific neutralizing antibodies in the serum were also measured on days 3, 7, 10, 14, 21, and 28 postchallenge.
Quantitation of CVB3-specific antibodies.
The titers of virus-specific antibodies in the sera of CVB3-infected mice were determined by an enzyme-linked immunosorbent assay. Briefly, 96-well plates were coated overnight at 4°C with CVB3(T7). The plates were washed with PBS-0.05% Tween 20, blocked with bovine serum albumin, and then incubated with dilutions of sera from the inoculated animals for 2 h at room temperature. Biotin-conjugated rabbit anti-mouse immunoglobulin G (Chemicon International Inc., Calif.) was used as the secondary antibody, and bound secondary antibodies were subsequently visualized by the use of streptavidin-horseradish peroxidase and the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate system (Sigma). The absorbance was read at 450 nm. The mean absorbance was plotted versus the dilution of antiserum, and the antibody titer was defined as the highest dilution in which the minimal response was 0.05 optical density units above the negative control, which comprised pooled sera from uninfected, age-matched A/J mice.
RESULTS
A double mutant of CVB3 shows highly attenuated cardiovirulence but causes some damage to the pancreas.
Previously, we have shown that mutants with VP2 (K2158R) and VP3 (E3060G) mutations in CVB3 are less cardiovirulent than the parental CVB3(T7) strain (26). As a first step in constructing a highly attenuated strain that could be exploited as a vaccine, we made a double mutant containing both of these mutations. Specifically, a DNA restriction fragment containing the codon for the E3060G mutation from pCVB3(EG) was used to replace the corresponding fragment of pCVB3(KR) to generate pCVB3(KR/EG). The virus derived from this infectious clone replicated in Vero cells and gave similar plaques to those of the wild-type virus CVB3(T7) in this cell type.
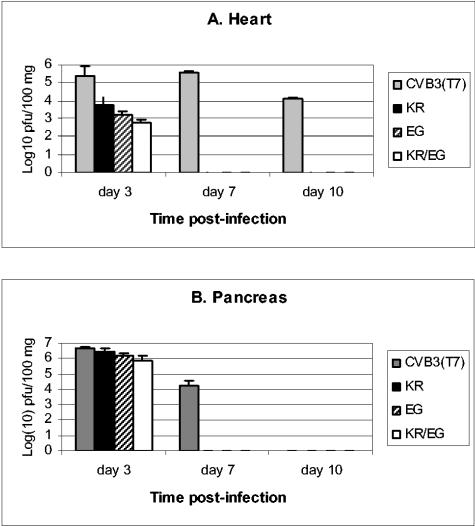
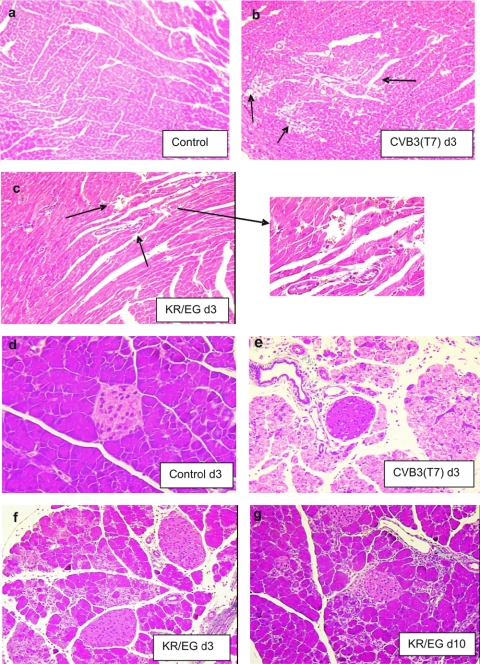
To determine whether the double mutant virus CVB3(KR/EG) was more attenuated than either of the single mutants assessed previously (26), we infected susceptible A/J mice with 105 PFU of CVB3(KR/EG) and compared the outcome of infection with that obtained in mice infected with an equivalent amount of the parental strain CVB3(T7), each single mutant, or PBS as a control. On day 3, 7, and 10 postinfection, hearts and pancreases were harvested for viral titration and histological analysis. The titers of the double mutant CVB3(KR/EG) relative to those of the wt strain CVB3(T7) and the single mutants in the heart and pancreas are shown in Fig. 1. In the heart, the wt virus CVB3(T7) replicated to a titer of over 105 PFU/100 mg of tissue by day 3 postinfection. On day 7, this high viral titer was maintained, and even on day 10, the virus was still present at about 104 PFU/100 mg heart tissue. In contrast, the double mutant virus CVB3(KR/EG) was detected at a titer of slightly less than 103 PFU/100 mg of tissue in the heart on day 3, which was 100-fold less than that of the wt strain. Furthermore, CVB3(KR/EG) was rapidly eliminated, with no virus detectable on days 7 and 10 (Fig. 1A). In a comparison with the single mutants, the titers of CVB3(KR/EG) were approximately five- to eightfold lower in the heart than those obtained with CVB3(KR) but only two- to threefold lower than those obtained with CVB3(EG). This indicates that no substantial increase in attenuation of the virus in heart tissue was obtained by combining the two mutations, which might be expected if both mutations reduce cardiovirulence by a similar mechanism, for example, by affecting the affinity for the coreceptor DAF. In pancreatic tissues, the titers of CVB3(KR/EG) were reduced three- to fivefold relative to that of the wild-type virus on day 3 but were not significantly different from the CVB3(EG) titer. In addition, the double mutant and both single mutants were rapidly eliminated, with no virus detected on either day 7 or day 10 p.i. In contrast, the CVB3(T7) virus was still detected, at about 104 PFU/100 mg, on day 7. A histological analysis of Masson's trichrome-stained tissues from mice infected with CVB3(T7) showed large areas of myocyte necrosis and subsequent tissue fibrosis in heart tissue, comprising between 20 to 30% of the sections examined (Fig. 2b). In contrast, only tiny foci of damaged myocytes were seen in some heart sections from the mice infected with the double mutant CVB3(KR/EG) (Fig. 2c). The wt strain also caused massive necrosis to pancreatic exocrine tissue, although the islets and ductal tissue were noticeably resistant to infection (Fig. 2e). CVB3(KR/EG) caused significant damage, with widespread foci of infection seen on day 3 (Fig. 2f). However, by day 10, the pancreas had largely recovered in mice that received the double mutant (Fig. 2g), while widespread necrosis was still present in mice infected with CVB3(T7) (not shown). These findings were supported by the level of viral genomes found in each case by in situ hybridization, as shown in Fig. 3. In mice infected with CVB3(T7), large foci of cells containing viral genomes accumulated in the heart, while much smaller amounts of CVB3(KR/EG) were seen in tiny pockets of infected cells (Fig. 3g and h). In the pancreas, massive amounts of viral genome were seen with the wt strain (b), while the CVB3(KR/EG) mutant was detected at much lower levels in discrete foci of infection (c). These data show that the replication of the CVB3(KR/EG) double mutant was notably attenuated in A/J mice and that this virus was also cleared more rapidly from infected tissues.
FIG. 1.
Viral titers of CVB3(T7) in the heart and pancreas compared with those of the single mutants KR and EG and the combined mutant KR/EG. On days 3, 7, and 10 postinfection, hearts and pancreases were harvested from male A/J mice that had been inoculated with either the wt or a mutant virus. The titers of virus in the heart were about 20-fold lower for the KR mutant and 100-fold lower for both the EG and KR/EG mutants than for the wt strain on day 3. None of the mutant viruses were detected on days 7 and 10, while the mean titer of the wt CVB3(T7) strain was highest on day 7 and residual virus was still detected on day 10. In the pancreas, the titers of all of the mutants were not significantly different from that of the wt on day 3 p.i., but the mutant viruses were more rapidly eliminated than the wt strain.
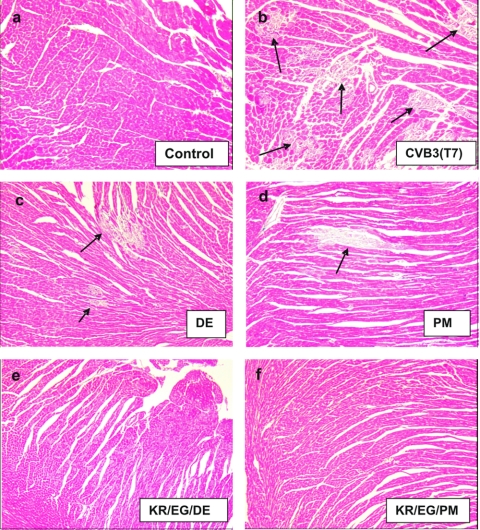
FIG. 2.
Histopathology of a heart and pancreas infected with CVB3(T7) and KR/EG on day 3 p.i. (Masson's trichrome stain). By day 3, large foci of tissue damage were visible in the heart tissue of mice infected with the wt strain (b), and these were further increased in size up to day 10 p.i. (see Fig. 5b). The KR/EG mutant was highly attenuated for heart tissue, with only rare foci of myocyte damage encompassing small numbers of cells (c). In the pancreas, the CVB3(T7) strain caused almost complete necrosis of the acinar tissue, while islets and the endothelial cells surrounding blood vessels were preserved (e). The KR/EG mutant caused noticeably less damage to the exocrine pancreas, although the infection was widespread on day 3. However, the acinar cells had largely regenerated by day 10 (g), while no such recovery was seen after CVB3(T7) infection.
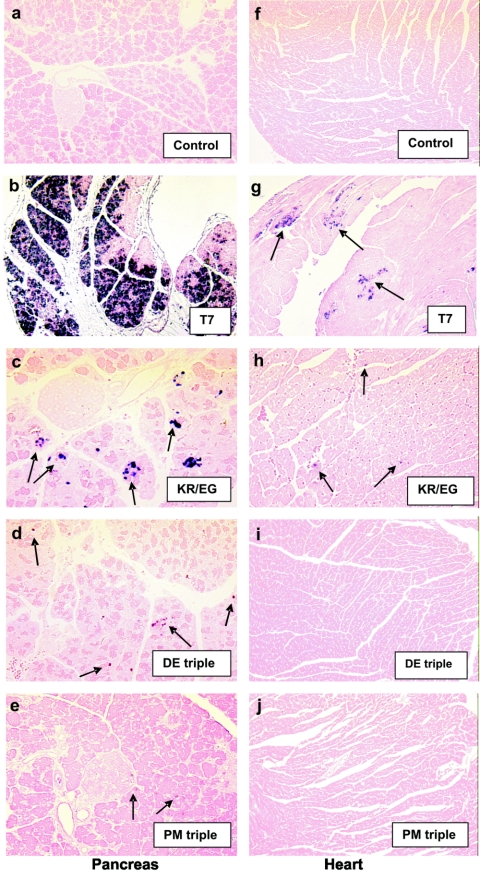
FIG. 3.
Detection of viral genomes in the heart and pancreas on day 3 postinfection by in situ hybridization of CVB3(T7) and mutant viruses. The left panels show pancreatic sections, while the right panels depict heart sections. In panel b, the large amount of viral genome in a pancreas infected with the wt (T7) strain is shown. In contrast, only focal regions containing genome of CVB3(KR/EG) can be seen scattered throughout the section (c). These were further reduced in number and size in the case of the two triple mutants, CVB3(KR/EG/DE) (d) and CVB3(KR/EG/PM) (e). On the right side, foci containing viral genomes were seen throughout the T7-infected heart sections. These expanded up to day 7 p.i., and on day 10, damaged areas with low levels of viral genome around the periphery were seen (not shown). In contrast, only tiny foci encompassing a few cells containing genomes were detected in hearts from mice infected with the double mutant, KR/EG. No viral genome was seen in the case of either triple mutant.
Identification of additional mutations in VP1 that attenuate CVB3.
Since the CVB3(KR/EG) double mutant caused significant damage to the exocrine pancreas on day 3 p.i., the introduction of a third mutation was performed. Several mutations in VP1 were tested for the ability to further attenuate the virus. A previous report had identified a D1155G substitution in the EF loop as a candidate for the attenuation of a natural isolate of CVB3 (4), so this was the first mutation tested. In addition, a mutation of the proline at amino acid 126 in the DE loop of VP1 was selected, extrapolating from data on the virulence determinants of a CVB4 strain (17). To determine whether D1155 and P1126 of VP1 are attenuating sites in CVB3, we mutated aspartic acid 155 in the EF loop of VP1 to glycine (G) and to a more conservative substitution, glutamic acid (E), by site-directed mutagenesis of pCVB3(T7). This enabled the generation of the mutant viruses CVB3(DE) and CVB3(DG) by the in vitro transcription and transfection of Vero cells. In addition, proline 126 in the DE loop of VP1 was mutated to methionine (M) to generate the mutant virus CVB3(PM).
The phenotypic characteristics of the different mutants are summarized in Table 1. The CVB3(DG) mutant virus formed very tiny plaques on Vero cell monolayers. Although the mutation in the virus was originally identified in a natural isolate (4), it was found to be unstable. The sequences of the viruses isolated from heart tissue on day 3 after the infection of A/J mice with CVB3(DG) showed that 30% of the mice contained a virus with a reversion from glycine to aspartic acid on day 3 p.i., while on days 7 and 10, 100% of the mice contained the revertant virus. Although the triple mutant CVB3(KR/EG/DG) showed reduced virulence, no further experimentation has been done with this mutant in view of the potential for reversion. The second mutant tested, CVB3(DE), was found to give smaller plaques but only slightly reduced yields of virus relative to CVB3(T7) on Vero cells. In contrast to CVB3(DG), the CVB3(DE) mutant was found to be stable after in vitro passaging and inoculation into mice. Specifically, viruses isolated from both the pancreas and the heart on day 3 p.i. were shown to have retained the DE mutation. In addition, this mutant was found to be less cardiovirulent than CVB3(T7) in A/J mice, replicating in the heart to a titer that was approximately 10-fold lower than that of the wild-type virus on day 3, while on days 7 and 10, no virus was detected (Fig. 4A). In agreement with the plaque assay data, significantly less heart damage was caused by CVB3(DE) than by the wild-type virus (Fig. 5c), with only one or two foci of tissue damage seen on each section by day 7, representing <5% of the tissue [as opposed to >30% of the tissue with CVB3(T7)]. In the pancreas, CVB3(DE) caused a widespread necrosis of acinar tissue on day 3 which was associated with only three- to fivefold less infectious virus than the wild-type infection. However, the mutant virus was cleared more quickly, with no detectable viral titer on day 7. Pockets of surviving acinar tissue were observed in sections stained with Masson's trichrome, and by day 10, a significant recovery of the tissue was seen (Fig. 6c and d). The CVB3(PM) mutant was also found to be stable following passaging in vitro and in vivo, as described above. Similar to the CVB3(DE) mutant, CVB3(PM) displayed attenuated replication in the heart, with viral titers on day 3 that were >50-fold lower than those of the wild-type virus. There was also less heart damage observed, although occasional foci of fibrosis were observed on day 10 (Fig. 5d). In the pancreas, early damage was seen on day 3 with CVB3(PM), similar to that seen with CVB3(DE), as shown in Fig. 6e, but by day 7, no remaining virus was detected by a plaque assay and large areas of surviving or regenerated exocrine tissue were seen (Fig. 6f). These two VP1 mutations and the EG mutation in VP3 are the only ones that we have identified that are associated with significantly less damage to the exocrine pancreas, a tissue that is extraordinarily permissive to CVB3 infection.
TABLE 1.
Characterisation of mutants
| Virus | Mutation(s) in CVB3(T7) parent strain | In vitro properties | Pathogenic properties in 5-week-old A/J mice |
|---|---|---|---|
| CVB3(T7) | None | Large plaques | Highly virulent. Causes widespread damage to heart and complete necrosis of pancreas. Lethal in 3-week-old A/J mice |
| CVB3(DE) | D1155E in EF loop of VP1 | Large plaques | Attenuated. Only foci of damage to heart but widespread necrosis of pancreas |
| CVB3(KR/EG/DE) | K2158R in puff of VP2 E3060G in knob of VP3 D1155E in EF loop of VP1 | Small plaques | Highly attenuated. No detectable damage to heart and only small foci in pancreas on day 3 p.i. |
| CVB3(DG) | D1155G in EF loop of VP1 | Pinpoint plaques | Virus rapidly reverts in vivo; 30% of infected mice contain reverted virus on day 3 p.i., 100% on days 7 and 10 |
| CVB3(KR/EG/DG) | K2158R in puff of VP2 E3060G in knob of VP3 D1155G in EF loop of VP1 | Pinpoint plaques | No virus detectable in either heart or pancreas on day 3 postinfection |
| CVB3(PM) | P1126M in DE loop of VP1 | Small plaques | Attenuated for heart but some pancreatic damage on day 3 |
| CVB3(KR/EG/PM) | K2158R in puff of VP2 E3060G in knob of VP3 P1126M in DE loop of VP1 | Small plaques | Highly attenuated. No damage to heart and only tiny foci in pancreas on day 3 p.i. |
FIG. 4.
Viral titers in the heart and pancreas following infection with the triple mutants, the single mutants CVB3(DE) and CVB3(PM), and the parental CVB3(T7) strain. In heart tissue, titers of wt CVB3(T7) were over 105 PFU/100 mg tissue on days 3 and 7 p.i. The single mutants CVB3(DE) and CVB3(PM) showed around a 10-fold reduction in viral titer on day 3 and were eliminated by day 7 p.i. Both triple mutants were highly attenuated in the heart, with only 102 to 103 PFU/100 mg found on day 3 for the CVB3(KR/EG/DE) strain, while no virus was detected in the case of CVB3(KR/EG/PM) (i.e., <10 PFU/100 mg tissue) on any of the days tested.
FIG. 5.
Histopathology of the heart on day 10 p.i. with the VP1 single mutants DE and PM, the triple mutants KR/EG/DE and KR/EG/PM, and the CVB3(T7) strain. Large areas of tissue fibrosis were visible in the hearts of mice infected with CVB3(T7), encompassing up to 20 to 30% of each section by day 10 p.i. (b). In contrast, only one or two foci of damaged tissue were present in heart sections from mice infected with either single mutant (c and d), and no heart damage at all was seen with either triple mutant (e and f) at any time.
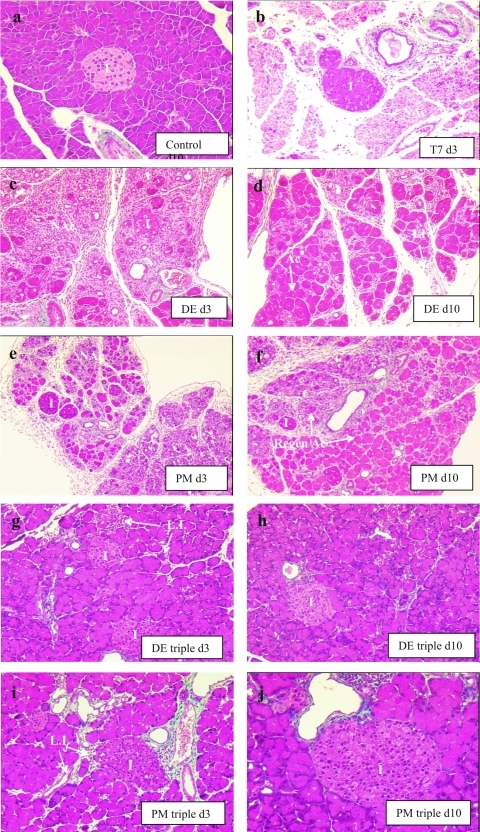
FIG. 6.
Histopathology of DE and PM single mutants and their corresponding triple mutants in the pancreas (Masson's trichrome stain). As described before, CVB3(T7) caused a complete necrosis of acinar tissue, and no regeneration was seen up to day 10 p.i. Both the DE (c) and PM (e) single mutants caused widespread damage to the exocrine pancreas, but islands of intact acini were present, and by day 10 these had enlarged to encompass >80% of the tissue (d and f, respectively). The triple mutants caused only minor damage to the pancreas, which was largely limited to tiny clusters of infected acinar cells associated with some lymphocytic infiltration on day 3 and essentially complete recovery of the tissue by day 10.
The data above indicate that certain mutations at either D1155 or P1126 of VP1 attenuate the virulence of CVB3(T7). While all of the substitutions tested gave viable viruses when grown in vitro, certain mutations, such as the D1155G mutation, were not tolerated in vivo and reverted rapidly to the original sequence.
The triple mutants CVB3(KR/EG/DE) and CVB3(KR/EG/PM) are highly attenuated in both the pancreas and the heart.
After the single mutations D1155E and P1126M of VP1 had been demonstrated to be attenuating and stable, we tested whether incorporating either of these mutations into the double mutant CVB3(KR/EG) further attenuated virulence, particularly that for the pancreas, which had shown significant levels of exocrine damage during the acute stage of infection with CVB3(KR/EG). Infectious clones containing the DE and PM mutations in VP1 were constructed with the template pCVB3(KR/EG), and the corresponding triple mutant viruses CVB3(KR/EG/DE) and CVB3(KR/EG/PM) were derived from these.
Low titers of the CVB3(KR/EG/DE) strain in heart tissues of A/J mice were detected on day 3 postinfection (between 102 and 103 PFU/100 mg of tissue) (Fig. 4A), but this was not associated with any observed cardiomyopathy (Fig. 5e). The titers were >500-fold lower than those obtained with the wild-type virus and >10-fold lower than those obtained with the single DE mutant (Fig. 4A). In conjunction with the low viral titers, no viral genomes were detected in heart tissues on either day 3 (Fig. 3i) or day 7 (not shown). In comparison, the second triple mutant, CVB3(KR/EG/PM), appeared to be completely noncardiotropic, with <10 PFU of virus/100 mg tissue detected by a plaque assay on day 3 or day 7 postinfection (Fig. 4) and no genomes observed by in situ hybridization (Fig. 3). In the pancreas, small amounts of damage in the acinar tissue were observed with both triple mutants on day 3, and this damage was associated with lymphocytic infiltration (Fig. 6). In addition, extremely low levels of viral genomes were detectable in tiny foci of infection by in situ hybridization (Fig. 3). On days 7 and 10, when the pancreases of mice infected with the wt virus were almost completely necrotic, in the case of the two triple mutants, there had been some regeneration of the damaged exocrine tissue and only small pockets of inflammation remained. Moreover, the damage caused by either triple mutant was notably less than that seen in mice infected with CVB3(KR/EG) (compare Fig. 6g to j with Fig. 2f and g). These findings demonstrate that both VP1 mutations, D1155E and P1126M, further attenuate the virulence of the double mutant CVB3(KR/EG) for both the heart and pancreatic tissue. While the pancreas still supported low levels of viral replication after a high-dose inoculum, the infections were less pathogenic, and recovery of the tissue was almost complete by day 10 p.i.
Stability of triple mutants following in vivo passaging.
To evaluate whether any reversion occurred at the mutated sites following passaging in vivo, we isolated viruses on day 3 p.i. from the pancreases of mice injected with CVB3(KR/EG/DE) or CVB3(KR/EG/PM) and used them to inoculate naïve 4- to 5-week-old A/J mice (n = 3) at a multiplicity of infection of 105 PFU. The original stock virus of each triple mutant was also injected into mice for comparison. On day 3 p.i., the titers of virus in the pancreas were found to be similar for the original stock and the passaged virus. For the CVB3(DE) triple mutant, the relative mean titers were 9.3 × 104 PFU (original) and 8.2 × 104 PFU (passaged), while for the CVB3(PM) triple mutant, they were 1.7 × 103 PFU (original) and 2.4 × 103 (passaged). In addition, RNAs were extracted from part of each pancreas and subjected to RT-PCR sequencing. In each case, the three mutations were found to be retained in all three inoculated mice.
The triple mutants can protect mice against a lethal dose of CVB3(T7).
To determine whether immunization with either of the triple mutants, CVB3(KR/EG/DE) or CVB3(KR/EG/PM), stimulates a protective immune response, we challenged immunized mice with a lethal dose of CVB3(T7). Specifically, A/J mice (at 3 weeks of age, when they are highly susceptible to the wt virus) were infected intraperitoneally with 105 PFU of CVB3(KR/EG/DE) or CVB3(KR/EG/PM) or injected with PBS as a control. The mice were bled on day 21 to determine the antibody titers in sera prior to the challenge and were then injected with 106 PFU of CVB3(T7) the following day. The viral titer in the serum was measured on day 1 postchallenge in order to assess the degree of viral dissemination. On days 3 and 10 postchallenge, three mice from each group were sacrificed, and their hearts and pancreases were removed for histological examinations and viral titration. Another group of animals were observed until day 28 postchallenge and then bled from the tail vein to monitor antibody levels in the serum.
After immunization with either triple mutant, CVB3(KR/EG/DE) or CVB3(KR/EG/PM), none of the 3-week-old A/J mice showed symptoms of CVB3 infection, despite their young age at the time of injection, when the wt virus is lethal, even at an inoculum of 102 PFU (J. Chantler, unpublished observation). Both triple mutant viruses did, however, induce immune responses, with antibody titers of 1:8,000 for CVB3(KR/EG/DE) and 1:6,000 for CVB3(KR/EG/PM) measured on day 21 postimmunization (Table 2). Upon challenge with a lethal dose of virulent wt CVB3(T7), all of the immunized mice survived without any symptoms. No virus was detected in the serum on day 1 postchallenge or in the target tissues, the heart and pancreas, on day 3 postchallenge. However, the antibody levels in serum were boosted to levels of 1:96,000 for CVB3(KR/EG/DE) and 1:64,000 for CVB3(KR/EG/PM) by day 7 and to 1:128,000 by day 10 postchallenge. These high antibody titers were maintained through day 28 (data not shown). In addition, histological examinations of both the hearts and the pancreases of the mice immunized with either triple mutant sacrificed on day 3, day 10, or day 28 postchallenge demonstrated the complete protection of these tissues from CVB3(T7)-induced damage (Fig. 7). In contrast, for the group inoculated with PBS, viral titers of about 106 PFU per ml of serum were detected on day 1 postchallenge (Table 2), and all of the mice showed signs of acute infection, including ruffled fur, light sensitivity, huddling, and a loss of appetite. By day 3, the titers of CVB3(T7) were >107 PFU/100 mg tissue in both the heart and the pancreas. Although these mice mounted an antibody response, the level of tissue damage seen by day 7 was substantial (Fig. 7b and c), and all of the animals were dead by day 10 postinjection.
TABLE 2.
Efficacy of preimmunization with triple mutants on subsequent challenge with a lethal dose of CVB3(T7)
| Immunizing virus | Dose inoculated (PFU) | Morbidity after i.p. inoculation | Neutralizing antibody titer in pooled sera on day 21 | Response to the challenge of 1 × 106 PFU of CVB3(T7)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Viral titers at indicated day (d)
|
Antibody titer in sera at day
|
Morbidity or mortality | |||||||
| Serum d 1 (PFU/ml) | Heart d 3 (PFU/g) | Pancreas d 3 (PFU/g) | 7 | 10 | |||||
| PBS | PBS | 0/16 | NDa | 1.1 × 106 | 1.69 × 108 | 5.87 × 108 | 1: 8,000 | NAb | 16/16 |
| CVB3(KR/EG/DE) | 1 × 105 | 0/16 | 1: 8,000 | ND | ND | ND | 1: 96,000 | 1: 128,000 | 0/16 |
| CVB3(KR/EG/PM) | 1 × 105 | 0/16 | 1: 6,000 | ND | ND | ND | 1: 64,000 | >1: 128,000 | 0/16 |
ND, not detectable.
NA, not available.
FIG. 7.
Assessment of vaccine potential of triple mutants by immunization of mice with either KR/EG/DE or KR/EG/PM and subsequent challenge with CVB3(T7). The histology of the heart and the pancreas on days 3, 10, and 28 postchallenge is shown. Mice that were immunized with either triple mutant showed no tissue damage following challenge with 106 PFU of CVB3(T7) on day 21 postimmunization, either on day 3 when pancreatic damage is maximal or on day 10 when fibrosis of the heart is normally extensive. In contrast, widespread damage was seen in mice that had been inoculated with PBS and then challenged with equivalent amounts of wt virus (b, c, k, and l). (a) Control pancreas. (b) CVB3(T7) infection on day 3 p.i. Massive acinar necrosis (N.Ac) and a condensed islet (I) are present. (c) CVB3(T7) infection on day 10 p.i. Acinar necrosis and substantial lymphocytic infiltration are present. (d to f) CVB3(DE) triple mutant infection on days 3, 10, and 28 p.i. The pancreas is intact and no necrosis is visible. (g to i) CVB3(PM) triple mutant infection on days 3, 10, and 28 p.i. No damage to acinar tissue or islets is visible. (j) Control heart. (k) CVB3(T7) infection on day 3 p.i. Foci of acute viral damage and cell necrosis are indicated with arrows. (l) CVB3(T7) infection on day 10 p.i. Large areas of fibrosis are present (arrows). (m to o) CVB3 (DE) triple mutant infection on day 3, day 10, and day 28 p.i. No heart damage is present. (p to r) CVB3(PM) triple mutant infection on day 3, day 10, and day 28 p.i. No heart damage is present.
DISCUSSION
Previously, we characterized two antibody escape mutants of CVB3, EM1 and EM10 (26), that were found to be attenuated for heart tissue while still causing complete necrosis of the pancreas. With the aim of constructing a more attenuated strain that could form the basis for a CVB3 vaccine, we incorporated the mutations characterized in EM1 and EM10 into a double mutant virus, CVB3(KR/EG). This strain was found to be further attenuated for the heart, although significant damage to pancreatic tissue was still seen. To address this, we examined the ability of mutations at other sites in the viral genome to reduce virulence. Several mutations in the surface loops of VP1 were tested, and two of these, D1155E and P1126M, were shown to be attenuating when incorporated into the wt background. We next incorporated each of these mutants into the CVB3(KR/EG) mutant to give two triple mutants, CVB3(KR/EG/DE) and CVB3(KR/EG/PM). These two mutants were found to be genetically stable, lacked cardiovirulence, and showed a greatly reduced tropism for the pancreas. In addition, they maintained the ability to produce protective neutralizing antibodies and totally protected susceptible mice against a subsequent challenge with a lethal dose of wt virus.
The underlying mechanism for the attenuation of CVB3(KR/EG/DE) and CVB3(KR/EG/PM) is not yet entirely established. Since the K2158R, E3060G, D1155E, and P1126M mutations are all located on the surface of the virus capsid, it is possible that they affect the interaction of the virus particle with one of the two cellular receptors, CAR (coxsackievirus-adenovirus receptor) and DAF (decay-accelerating factor). Alternatively, the mutations may alter the stability of the virus particles, as previously shown for the K2158R and E3060G single mutants (26). When each of the four mutations was incorporated individually into the parental CVB3(T7) background, the resulting single mutants displayed various degrees of attenuation. Our characterizations of the K2158R and E3060G mutations have been reported previously and will only be summarized briefly here (26). K2158 is located in the middle of the “puff” region (or EF loop), comprising amino acids 129 to 180, which is the largest, most variable surface loop of VP2. In addition to our finding that replacing lysine (K) with arginine (R) at amino acid 158 reduced the cardiovirulence of CVB3(T7), a second mutation in this region, namely, a replacement of asparagine with aspartate at amino acid 165, has also been found to be attenuating (14), corroborating the importance of the “puff” region in the infectious process. The second attenuating site that we identified (E3060G) is in the “knob” region, the major surface protrusion of VP3. In the case of other picornaviruses such as rhinoviruses and polioviruses, the “knob” region has been identified as the location of a major neutralization domain (19, 22, 24, 25). However, our findings are the first to associate a mutation here with effects on virulence (26). In recent studies of the related echoviruses, the footprint for DAF binding has been shown to encompass amino acid residues in both the VP2 puff and the VP3 knob loops (10), suggesting that both the KR and EG mutations may affect binding to the coreceptor DAF and thus influence viral entry. This possibility of differential binding of the mutants to DAF is currently under investigation. In order to identify other mutations that might further reduce CVB3's virulence, we examined several mutations in VP1 that we predicted might influence binding to the primary receptor CAR. The D1155E mutation is within the EF loop (1152-1158) of VP1, which is located at the top of the binding cleft for CAR (18). Cameron-Wilson et al. (4) had previously identified a glycine at position 1155 as a potentially attenuating mutation in the EF loop by an analysis of a strain whose reversion to cardiovirulence was associated with a substitution of glycine for aspartic acid (D) at this site. However, the introduction of this mutation alone into a wt background was not performed in their study. We now show that position 1155 is indeed an attenuating site, but the replacement of D with G leads to a highly unstable virus that rapidly reverts in mice. Subsequent analyses of other substitutions at this site showed that replacing the aspartic acid with serine in VP1 of the double mutant CVB3(KE/EG) resulted in a nonviable virus, while a more conservative replacement of aspartic acid (D) with glutamic acid (E) produced a stable mutation that further attenuated the KR/EG double mutant. The other attenuating site within VP1 of CVB3 that we identified was P1126, which is located in the DE loop (residues 1125 to 1137). This is the most prominent exterior loop near the fivefold axes of all picornaviruses, and residues within the DE loop are thought to comprise ion binding sites along the fivefold axis (21) that may help to stabilize the virus pentamers. In addition to the putative stabilizing role of the DE loop, Caggana et al. (3) have identified a single amino acid substitution (Thr1129) within the VP1 DE loop of CVB4 that affects virulence. Our observation that a proline-to-methionine substitution at amino acid 126 in VP1 of CVB3 affects viral replication in the heart and pancreatic tissue suggests that the DE loop also plays a role in the pathogenicity of CVB3. Both triple mutants, CVB3(KR/EG/DE) and CVB3(KR/EG/PM), caused no observable damage to heart tissue; in fact, no virus was detected in the heart by a plaque assay with the latter mutant. However, each of the triple mutants maintained the ability to mount a protective immune response by day 21 postinfection. Moreover, when immunized mice were challenged with a lethal dose of wild-type virulent CVB3(T7), no virus was detected in the serum on day 1, suggesting that the circulating antibodies induced by the triple mutants were capable of protecting the mice from reinfection. In addition, the immunized mice showed a secondary antibody response to challenge, with neutralizing antibodies boosted to very high titers by day 7 and maintained through day 28. As shown in Table 2 and Fig. 7, the immunized mice were completely protected from challenge with the wild-type CVB3(T7) virus, while all of the mice injected with PBS died. This suggests that both CVB3(KR/EG/DE) and CVB3(KR/EG/PM) have the potential to be developed into live attenuated CVB3 vaccine candidates. Such live attenuated vaccines have a major advantage over subunit vaccines in that they activate all components of the immune system, notably both antibody responses and cytotoxic T cells, as well as stimulating a broad protective response to all antigenic epitopes of the virus. This helps to circumvent disease potentiation that may arise following immunization with subunit vaccines, DNAs encoding selective protective antigens of viruses (11), or chimeric viruses engineered to express specific viral epitopes (6).
Since the risks of reversion and second-site compensatory mutations are a concern in the development of live virus vaccines (16, 17, 27, 28), we also monitored the incidence of reversion in mice immunized with our mutants. When CVB3(DG) was injected into mice, the reversion from G to D at amino acid position 155 of VP1 was detected in 33% of the infected mice on day 3 postinfection and in 100% of infected mice on days 7 and 10 p.i. This mutation was therefore discarded as highly unstable. In contrast, we found that the KR/EG double mutant is very stable, possibly because the two mutations are both located in the putative footprint for coreceptor DAF binding. Therefore, both sites would need to be altered simultaneously to give the mutant a significant growth advantage. This concept of incorporating two mutations, both of which limit viral replication by a similar mechanism, could be extended to our VP1 mutations by combining the DE and PM substitutions into a quadruple mutant. This has not yet been tested; however, both of the VP1 mutants were found to be stable in our mouse model when incorporated into the KR/EG double mutant. Since the likelihood of reversion is greatly increased when the virus undergoes high levels of replication, the limited growth of the triple mutants in vivo reduces the opportunity for reversion to occur. Thus, with each additional mutation, reversion at a single site will not give a sufficient growth advantage for the reverted strain to establish itself. While this suggests that the incorporation of even more mutations would be desirable, the vaccine candidate must be allowed to retain the capacity to replicate to a sufficient degree to allow an immune response to be elicited.
In conclusion, we have constructed two triple mutants, both of which are highly attenuated in 3-week-old mice that succumb to infection with even low titers of the parental CVB3(T7) strain. All of the young mice survived following immunization with either triple mutant and showed no overt symptoms. The immunized mice were shown to have serum antibodies to CVB3, and upon a subsequent challenge with high titers of virulent virus, these immunized mice were totally protected, while age-matched mice that had not received one of the triple mutants were uniformly killed. Our conclusion is that the two triple mutants, CVB3(KR/EG/DE) and CVB3(KR/EG/PM), could be used to develop a highly attenuated CVB3 vaccine. In addition, the similarity between serotypes that we exploited in choosing surface regions of the structural proteins to mutate could be utilized to produce attenuated strains of other virulent group B coxsackieviruses, using a similar strategy of site-directed mutagenesis and reverse genetics.
Acknowledgments
We thank Daniel Harvey and other members of the staff of the Animal Unit at the British Columbia Research Institute for Children's and Women's Heath for their assistance with the animal experiments. We also thank Julie Chow and Vlady Pavlova in the Histology Unit of the Department of Pathology at UBC for tissue processing.
This research was supported in part by a grant from the Heart and Stroke Foundation of Canada.
REFERENCES
- 1.Appleyard, G., S. M. Russell, B. E. Clarke, S. A. Speller, M. Trowbridge, and J. Vadolas. 1990. Neutralization epitopes of human rhinovirus type 2. J. Gen. Virol. 71:1275-1282. [DOI] [PubMed] [Google Scholar]
- 2.Baboonian, C., M. J. Davies, J. C. Booth, and W. J. McKenna. 1997. Coxsackie B viruses and human heart disease. Curr. Top. Microbiol. Immunol. 223:31-52. [DOI] [PubMed] [Google Scholar]
- 3.Caggana, M., P. Chan, and A. Ramsingh. 1993. Identification of a single amino acid residue in the capsid protein VP1 of coxsackievirus B4 that determines the virulent phenotype. J. Virol. 67:4797-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron-Wilson, C. L., Y. A. Pandolfino, H. Y. Zhang, B. Pozzeto, and L. C. Archard. 1998. Nucleotide sequence of an attenuated mutant of coxsackievirus B3 compared with the cardiovirulent wild type: assessment of candidate mutations by analysis of a revertant to cardiovirulence. Clin. Diagn. Virol. 9:99-105. [DOI] [PubMed] [Google Scholar]
- 5.Cetta, F., and V. V. Michels. 1995. The autoimmune basis of dilated cardiomyopathy. Ann. Med. 27:169-173. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, N. M., K. S. Kim, S. Tracy, J. Jackson, K. Hofling, J. S. Leser, J. Malone, and P. Kolbeck. 2000. Coxsackievirus expression of the murine secretory protein interleukin-4 induces increased synthesis of immunoglobulin G1 in mice. J. Virol. 74:7952-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codd, M. B., D. D. Sugrue, B. J. Gersh, and L. J. Melton III. 1989. Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975-1984. Circulation 80:564-572. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, J. J., S. S. Bradrick, N. M. Chapman, S. M. Tracy, and J. R. Romero. 2003. The stem loop II within the 5′ nontranslated region of clinical coxsackievirus B3 genomes determines cardiovirulence phenotype in a murine model. J. Infect. Dis. 187:1552-1561. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, J. J., N. M. Chapman, S. Tracy, and J. R. Romero. 2000. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: localization to the 5′ nontranslated region. J. Virol. 74:4787-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, Y., F. Lin, P. R. Chipman, C. M. Bator, T. S. Baker, M. Shoham, R. J. Kuhn, M. E. Medof, and M. G. Rossmann. 2002. Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex. Proc. Natl. Acad. Sci. USA 99:10325-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henke, A., E. Wagner, J. L. Whitton, R. Zell, and A. Stelzner. 1998. Protection of mice against lethal coxsackievirus B3 infection by using DNA immunization. J. Virol. 72:8327-8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandolf, R. 1993. Molecular biology of viral heart disease. Herz 18:238-244. [PubMed] [Google Scholar]
- 13.Kim, K. S., G. Hufnagel, N. M. Chapman, and S. Tracy. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11:355-368. [DOI] [PubMed] [Google Scholar]
- 14.Knowlton, K. U., E. S. Jeon, N. Berkley, R. Wessely, and S. Huber. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70:7811-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C., E. Maull, N. Chapman, S. Tracy, and C. Gauntt. 1997. Genomic regions of coxsackievirus B3 associated with cardiovirulence. J. Med. Virol. 52:341-347. [PubMed] [Google Scholar]
- 16.Macadam, A. J., G. Ferguson, D. M. Stone, J. Meredith, J. W. Almond, and P. D. Minor. 2001. Live-attenuated strains of improved genetic stability. Dev. Biol. (Basel) 105:179-187. [PubMed] [Google Scholar]
- 17.Macadam, A. J., S. R. Pollard, G. Ferguson, R. Skuce, D. Wood, J. W. Almond, and P. D. Minor. 1993. Genetic basis of attenuation of the Sabin type 2 vaccine strain of poliovirus in primates. Virology 192:18-26. [DOI] [PubMed] [Google Scholar]
- 18.Martino, T. A., P. Liu, and M. J. Sole. 1994. Viral infection and the pathogenesis of dilated cardiomyopathy. Circ. Res. 74:182-188. [DOI] [PubMed] [Google Scholar]
- 19.Minor, P. D., A. J. Macadam, D. M. Stone, and J. W. Almond. 1993. Genetic basis of attenuation of the Sabin oral poliovirus vaccines. Biologicals 21:357-363. [DOI] [PubMed] [Google Scholar]
- 20.Modlin, J. F., and H. A. Rotbart. 1997. Group B coxsackie disease in children. Curr. Top. Microbiol. Immunol. 223:53-80. [DOI] [PubMed] [Google Scholar]
- 21.Muckelbauer, J. K., and M. G. Rossmann. 1997. The structure of coxsackievirus B3. Curr. Top. Microbiol. Immunol. 223:191-208. [DOI] [PubMed] [Google Scholar]
- 22.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose, N. R., A. Herskowitz, and D. A. Neumann. 1993. Autoimmunity in myocarditis: models and mechanisms. Clin. Immunol. Immunopathol. 68:95-99. [DOI] [PubMed] [Google Scholar]
- 24.Sherry, B., A. G. Mosser, R. J. Colonno, and R. R. Rueckert. 1986. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J. Virol. 57:246-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry, B., and R. Rueckert. 1985. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J. Virol. 53:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadnick, E., M. Dan, A. Sadeghi, and J. K. Chantler. 2004. Attenuating mutations in coxsackievirus B3 map to a conformational epitope that comprises the “puff” region of VP2 and the “knob” of VP3, part of a putative binding site for coreceptor. J. Virol. 78:13987-14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardy-Panit, M., B. Blondel, A. Martin, F. Tekaia, F. Horaud, and F. Delpeyroux. 1993. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 67:4630-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatem, J. M., C. Weeks-Levy, A. Georgiu, S. J. DiMichele, E. J. Gorgacz, V. R. Racaniello, F. R. Cano, and S. J. Mento. 1992. A mutation present in the amino terminus of Sabin 3 poliovirus VP1 protein is attenuating. J. Virol. 66:3194-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu, Z., N. M. Chapman, G. Hufnagel, S. Tracy, J. R. Romero, W. H. Barry, L. Zhao, K. Currey, and B. Shapiro. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J. Virol. 69:4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]