Abstract
The orthopoxvirus serpin SPI-1 is an intracellular serine protease inhibitor that is active against cathepsin G in vitro. Rabbitpox virus (RPV) mutants with deletions of the SPI-1 gene grow on monkey kidney cells (CV-1) but do not plaque on normally permissive human lung carcinoma cells (A549). This reduced-host-range (hr) phenotype suggests that SPI-1 may interact with cellular and/or other viral proteins. We devised a genetic screen for suppressors of SPI-1 hr mutations by first introducing a mutation into SPI-1 (T309R) at residue P14 of the serpin reactive center loop. The SPI-1 T309R serpin is inactive as a protease inhibitor in vitro. Introduction of the mutation into RPV leads to the same restricted hr phenotype as deletion of the SPI-1 gene. Second-site suppressors were selected by restoration of growth of the RPV SPI-1 T309R hr mutant on A549 cells. Both intragenic and extragenic suppressors of the T309R mutation were identified. One novel intragenic suppressor mutation, T309C, restored protease inhibition by SPI-1 in vitro. Extragenic suppressor mutations were mapped by a new procedure utilizing overlapping PCR products encompassing the entire genome in conjunction with marker rescue. One suppressor mutation, which also rendered the virus temperature sensitive for growth, mapped to the DNA polymerase gene (E9L). Several other suppressors mapped to gene D5R, an NTPase required for DNA replication. These results unexpectedly suggest that the host range function of SPI-1 may be associated with viral DNA replication by an as yet unknown mechanism.
Members of the serine proteinase inhibitor (serpin) superfamily are widely distributed in nature and are diverse in function (44). Among viruses, only poxviruses have been found to encode inhibitory serpins. Some cellular serpins have gained ancillary functions by interactions with regions of the serpin not related to protease inhibition (4, 5, 11, 14). One orthopoxvirus serpin, cowpox SPI-3, has been shown to be bifunctional, because it acts together with the virus-encoded hemagglutinin to inhibit infected cell-cell fusion independent of protease inhibition activity (51). Rabbitpox virus (RPV) SPI-1 is an intracellular serpin, which functions in vitro as an inhibitor of human neutrophil cathepsin G (39). During RPV infection in cell culture, SPI-1 appears to inhibit a caspase-independent form of apoptosis in certain restrictive cell types, and deletion of the gene leads to a reduced-host-range (hr) phenotype. Infection of nonpermissive cells by RPV lacking SPI-1 (RPVΔSPI-1) is characterized by a relatively normal pattern of gene expression but formation of relatively few virions and early signs of apoptosis (1, 7, 39). Deletion of SPI-1 from vaccinia virus, which also leads to a loss of host range, produces a somewhat different phenotype characterized by a reduced synthesis of intermediate and late transcripts (43). The association of SPI-1 with host range suggests that this serpin may interact with either cellular and/or viral proteins in order to achieve this function. However, any potential serpin interaction with an unknown second protein cannot be evaluated strictly by assaying the behavior of the homogeneous, purified serpin.
Serpins that have retained inhibitory activity typically contain a small uncharged amino acid at the P14 residue of the reactive center loop (23). This ensures proper mobility of the loop and allows the conformational changes required for inhibition of protease activity (25, 34, 42, 45). Naturally noninhibitory serpins, such as ovalbumin and angiotensinogen, have a charged amino acid at the P14 position of the reactive center loop, which compromises this mobility (46). Thus, to explore the interactions of SPI-1 with other viral or cellular proteins, we introduced a T309R mutation at the P14 residue of SPI-1, which, as expected, rendered it inactive as a protease inhibitor in vitro. When the mutation was then introduced into RPV, the resultant virus (RPV SPI-1 T309R) had a reduced host range identical to that of the virus with the SPI-1 deletion, including loss of the ability to form plaques on A549 cells (39). This suggests that protease inhibition by SPI-1 is essential for a productive infection by RPV in these nonpermissive cells, which is most likely mediated by interaction with an intracellular protease.
We then selected for second-site suppressor mutations which restored the plaque-forming ability of RPV SPI-1 T309R on otherwise restrictive human A549 cells. This approach has allowed us to isolate viruses containing suppressor mutations which map both inside and outside of the SPI-1 gene. The existence of extragenic suppressors suggests that the interaction of SPI-1 with other viral proteins may be critical for full and robust SPI-1 protein activity. Using a novel mapping procedure, we have shown that many of these extragenic suppressors reside in genes E9L and D5R, which encode proteins essential for DNA replication and recombination (19-21).
MATERIALS AND METHODS
Cells and viruses.
CV-1 African green monkey kidney cells and human A549 cells were obtained from the American Type Culture Collection. HeLa-S1a cells were obtained from Toni Antalis (Jerome H. Holland Laboratory for the Biomedical Sciences, American Red Cross) (17). All viruses used are derivatives of RPV (rabbitpox virus, Utrecht strain). RPV strains with SPI-1 gene mutations (ΔSPI-1, T309R, and F322A), which cannot form plaques on A549 cells, have been described previously (39) and were provided by K. Moon. To determine virus titers, confluent monolayers of cells in six-well plates were infected with serial dilutions of virus and incubated at 37°C, and the cells were stained with crystal violet solution (2% crystal violet, 2% ethanol, 0.08% ammonium oxalate).
Selection for viral suppressor mutants by restored host range.
Spontaneous phenotypic revertants of RPV SPI-1 T309R were isolated by plaque purification three times on A549 cells and then amplified on CV-1 cells in six-well plates. Restoration of host range for each revertant virus clone was verified by plaque assay on A549 and CV-1 cells. DNA was isolated from each revertant with the DNeasy tissue kit (QIAGEN) according to the manufacturer's protocol, and the SPI-1 gene was amplified by PCR and sequenced. Revertant viruses that retained the parental SPI-1 T309R allele were presumed to have an extragenic suppressor mutation.
Gel mobility shift assay for protease-serpin interaction.
Wild-type (wt) or mutant SPI-1 DNA (T309R or T309C) was cloned into the pTM1 expression vector (18), which contains a T7 promoter and an internal ribosome entry site, and verified by DNA sequencing. pTM1 SPI-1 DNA from each clone provided the template for both synthesis and labeling of SPI-1 protein in the transcription and translation (TNT) system (TNT-T7 Quick coupled in vitro transcription and translation system, Promega). Each SPI-1 TNT product (2 μl) was incubated at 37°C for 90 min with purified human cathepsin G (Athens Biotech) in the range of 0 to 3 × 10−4 U per sample in a buffer of 100 mM Tris, pH 8, 10 mM CaCl2, analyzed by electrophoretic mobility shift in 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels (49), and detected by autoradiography.
Marker rescue of a temperature-sensitive mutant (sup-2).
Fragments of wild-type virus DNA (1 μg) were mixed with Lipofectamine PLUS lipid reagent (Invitrogen) according to the manufacturer's protocol. Confluent monolayers of CV-1 cells in six-well plates were then transfected with the lipid-treated DNA and infected at 37°C with the RPV sup-2 host range mutant at a multiplicity of 0.003 PFU per cell. After 3 h of incubation, the medium was replaced with 0.5% agarose in MEM with 5% fetal bovine serum. The cells were incubated at 41°C for 4 days and then stained with crystal violet.
Site-directed mutagenesis of RPV DNA polymerase (E9L).
A 1-kb fragment of wild-type RPV E9L DNA flanking the His142 codon was synthesized by PCR using the oligonucleotide primers 5′-GGG AAT TCC ACT ACG TGA TGG ATG TTC GGT GCA TT-3′ and 5′-GGG AAT TCC ATA TGT TAT TAA CGT GAA ACG-3′ to generate restriction endonuclease sites for cloning into the pAlterEx1 vector (pAltE9), which is required for mutagenesis. E ORF E (RPV homolog of VACVgp085, GenBank accession no. NP_063713) is embedded within E9L in the opposite orientation. The mutagenic oligonucleotide for disruption of E ORF E was FS359 (5′phosphorylated-GGA CGA GCA ATA TTT AAC GAA GAT TAA CAA TGG A-3′), which leads to a Cys-to-Arg mutation at codon 9 followed by a stop codon. A unique MboII restriction site is also produced. Mutagenesis of pAltE9 DNA with the FS359 oligonucleotide to effectively eliminate E ORF E was performed using the Altered Sites system (Promega), and the E9LΔE ORF E allele was cloned into RPV by ts marker rescue of the RPV sup-2 mutant (SPI-1 T309R, E9ΔH142) on CV-1 cells at 41°C. This yielded RPV SPI-1 T309R/ΔE ORF E, which was then plaque purified and verified by DNA sequencing.
DNA synthesis assay.
CV-1 cells in 60-mm dishes were infected with each virus at a multiplicity of 10 PFU per cell. The cells were incubated at either 31°C or 41°C, and samples were then harvested by scraping at time intervals spanning 24 h of infection. Cells and virus were collected by centrifugation, resuspended in 0.6 ml of 10× SSC (1.5 M NaCl, 0.15 M sodium citrate)-1 M NH4OAc per 60-mm dish, and stored at −80°C. After three freeze/thaw cycles, an equal volume of 10× SSC-1 M NH4OAc was added, and cellular debris was removed from the samples by brief centrifugation. Each sample (20 μl) was mixed with an equal volume of 0.8 M NaOH-20 mM EDTA, denatured at 99°C for 10 min, cooled to 4°C for 1 min, and diluted with 100 μl of 0.4 M NaOH-10 mM EDTA. Each sample (105 cell equivalents) was transferred to a positively charged nylon membrane (HyBond) with a vacuum manifold (Dot Blot apparatus, Bio-Rad), and the DNA was immobilized by UV cross-linking (Stratalinker, Stratagene). To detect the viral DNA, RPV genomic DNA was nonradioactively labeled with digoxigenin-dUTP by random priming using a DIG DNA labeling and detection kit (Roche Molecular Biochemicals).
Reconstruction of the RPV sup-2 mutant by marker rescue.
Confluent monolayers of A549 cells were infected with RPV SPI-1 T309R at a multiplicity of infection (MOI) of 5 PFU per cell and transfected with PCR product (1 μg) containing the sup-2 mutation (E9ΔH142). The cells were incubated at 37°C for 24 h, then harvested by scraping, and lysed by three cycles of freeze/thaw, and titers of RPV were determined on A549 cell monolayers. Revertant plaques were picked and screened for temperature sensitivity (ts). Revertants that were ts were then serially plaque purified three times on A549 cells and amplified on CV-1 cells for DNA preparation, from which E9L PCR products were generated for sequencing.
Development of 5-kb PCR libraries for RPV and VV-WR.
Virtually the entire genome of both RPV and vaccinia virus WR strain (VV-WR) was represented in 42 and 40 overlapping 5-kb PCR products, respectively. Only noncoding DNA within the inverted terminal repeats was omitted, using the first open reading frame (ORF) (RPXV-UTR 001 and 184) as boundaries for the library. The sequences of the entire set of PCR primers for RPV and VV-WR are available upon request. Primer sequences were based on both VV-WR (provided by B. Moss) and RPV (Utrecht strain, provided by M. Buller and C. Upton) genomic sequences obtained prior to publication. PCR primers were designed in pairs using Vector NTI (InforMax, Inc.) with the following optimal parameters: 26-mer length, melting temperature (Tm) of each primer was 58°C, difference in Tm between primers was less than 2°C, and the difference in the percent G+C was less than 2%. PCR products were 4.5 to 5.5 kb in length, with 5 kb preferred. Primers entirely within a single ORF were given priority, such that PCR products overlap by ∼0.5 kb (0.35 kb minimum) within the same ORF. PCRs included Vent DNA polymerase at 0.032 U per μl in 1× ThermoPol Buffer (New England Biolabs), which contains 2 mM MgSO4, with 0.3 μM concentrations of each primer, 0.5 mM deoxynucleoside triphosphates, and 2 ng genomic wild-type RPV or VV DNA per μl. For extragenic suppressor mutants, template DNA was prepared from infected CV-1 cells using the DNeasy tissue kit (QIAGEN) and diluted to ∼120 cell equivalents per μl. The following PCR conditions were used: 94°C for 3 min (denaturing), then 10 cycles of 94°C for 15 s, 53°C for 30 s (annealing), and 68°C for 5 min (extension), followed by 30 cycles with no change in denaturation or annealing conditions but with 5 s added to the extension time per cycle. PCRs concluded with an incubation at 68°C for 7 min. To remove excess primers for DNA quantitation, up to 100 μl of each PCR product was purified with a 30-kDa-cutoff filter (Ultrafree-MC, Millipore) by two cycles of dilution with 350 μl H2O and centrifugation at 5,000 × g for 5 min. DNA retained by each filter was diluted in 100 μl H2O and stored at −20°C. The quantity of DNA in each PCR product was measured by fluorometry (TD-700 fluorometer, Turner Designs) according to the manufacturer's suggested protocol.
Host range marker rescue.
Confluent monolayers of A549 cells were infected at 37°C with 0.01 PFU per cell of either RPV SPI-1 F322A or RPVΔSPI-1 or 0.0003 PFU of RPV SPI-1 T309R per cell. Lower levels of RPV SPI-1 T309R were used because the phenotype was slightly leaky. RPV SPI-1 F322A contains a mutation from phenylalanine to alanine in the reactive center loop which destroys protease inhibitory activity. The cells were cotransfected with 2 μg PCR product using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol, and incubated at 37°C. Additional minimal essential medium plus 10% fetal bovine serum was added 5 h postinfection, and incubation continued for 120 h at 37°C. The cells were harvested by scraping, disrupted by three freeze/thaw cycles, and stored at −80°C. Titers of recombinant viruses were determined on A549 cells by plaque assay.
Western blot analysis.
Proteins from 6 × 105 cell equivalents were resolved on 8% or 10% SDS-polyacrylamide gels (32), transferred to nitrocellulose membranes, and immunoblotted with a polyclonal antibody against either the vaccinia virus D5R or E9L protein (gifts of Paula Traktman). All blots used the ECL-PLUS system (Amersham) for chemiluminescent detection of secondary antibody and were exposed to either X-OMAT Blue (Kodak) or Hyperfilm-ECL (Amersham).
RESULTS
Isolation of suppressors which restore host range to RPV SPI-1 T309R mutants.
RPV containing the SPI-1 T309R mutation fails to form plaques on A549 cells, which illustrates RPV dependence on the serpin activity of SPI-1 in certain restrictive cell types. If interactions of RPV SPI-1 with other viral proteins are also required for growth in restrictive cells, these interactions could be identified based on restoration of wild-type host range from second-site suppressor mutations. Thus, second-site suppressors found in viral genes other than SPI-1 would identify these candidate viral proteins.
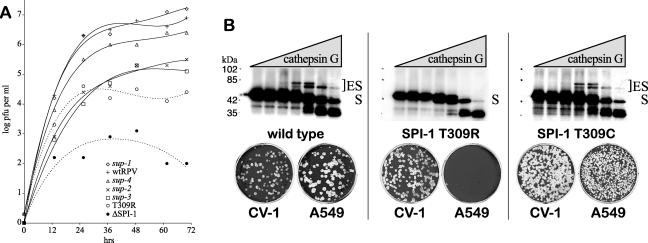
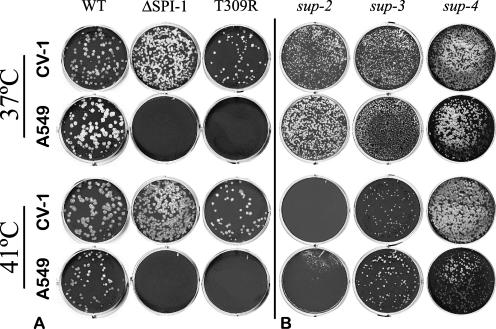
We were able to isolate spontaneous phenotypic revertants of RPV SPI-1 T309R at a frequency of ∼2 × 10−4, based on the ability to form plaques on A549 cells. Individual plaques were then plaque purified and amplified. We initially identified four phenotypic revertant viruses named the RPV sup-1, sup-2, sup-3, and sup-4 mutants for further characterization and genetic mapping of the suppressor loci. In each mutant, normal expression of the T309R SPI-1 protein during infection was verified by Western blot analysis (data not shown). The ability of each suppressor mutant to replicate and spread in A549 cells was determined at 12-h intervals by determining virus titers on CV-1 cells. Compared to wild-type virus, the parental RPV SPI-1 T309R mutant is restricted but viable, while the null deletion virus RPVΔSPI-1 is reduced by 10,000-fold (Fig. 1A). The growth of each of the revertants was variably enhanced over the parental T309R mutant; however, only the sup-1 mutant showed an increase that was comparable to wild-type RPV.
FIG. 1.
RPV host range can be restored by suppression of the SPI-1 T309R mutation. (A) Growth and spread of RPV mutants on A549 cells. Monolayers of A549 cells in six-well dishes were infected with ∼30 PFU of each RPV mutant under liquid medium at 37°C. Samples were harvested at 12-h intervals over 72 h, and the titers of the virus were determined on CV-1 cells. The titer of each virus with respect to time is shown. The initial concentration of virus was approximately 10 PFU per ml. RPVΔSPI-1 has a deletion of the SPI-1 ORF. T309R is the parental RPV SPI-1 P14 hr mutant. The RPV sup-1, sup-2, sup-3, and sup-4 mutants each contain suppressors of the SPI-1 T309R mutation. All suppressor isolates were selected on A549 cells by restored host range. Trend lines were drawn for each series as a fourth-order polynomial (Excel, Microsoft). Dotted trend lines indicate viruses which do not form plaques on A549 cells. While there is clearly growth of the T309R mutant in A549 cells relative to RPVΔSPI-1, the virus does not plaque on these cells. (B) Characterization of the SPI-1 T309C intragenic suppressor. Electrophoretic mobility shift assays with labeled SPI-1 protein (top panel) and the corresponding RPV host range phenotype in plaque assays (bottom panel) are shown. Wild-type SPI-1, SPI-1 T309R, or SPI-1 T309C DNA was cloned into the pTM1 vector for in vitro transcription and translation (TNT) to generate 35S-labeled SPI-1 protein. Increasing amounts of purified human cathepsin G (serine protease) were incubated at 37°C for 90 min with a constant amount of the labeled SPI-1 TNT product. The mixture was then resolved by 10% SDS-PAGE. Wild-type SPI-1 protein (∼45 kDa) generates two isoforms of a stable 1:1 complex with cathepsin G (∼25 kDa), where ES is enzyme-substrate. Formation of this complex is consistent with protease inhibitory function by a competitive and partially irreversible mechanism common to all inhibitory serpins. RPV containing the wild-type SPI-1 gene forms plaques on both CV-1 and A549 cells. The SPI-1 T309R mutation (middle) affects protease inhibition in vitro and leads to a reduced host range of the virus. Spontaneous reversion of RPV SPI-1 T309R to T309C (right) was isolated by genetic selection on A549 cells to restore plaque-forming ability. SPI-1 T309C protein shows a restored ability to form inhibitory complexes with cathepsin G in vitro that is comparable with wild-type SPI-1 protein.
Identification of a SPI-1 T309C pseudorevertant.
To identify any potential second-site mutations within the SPI-1 gene (intragenic suppression), DNA was isolated from CV-1 cells infected with each purified revertant virus, and the SPI-1 gene was amplified by PCR and sequenced. We identified a novel T309C mutation within the SPI-1 gene of the RPV sup-1 mutant. The host range phenotype for T309C (sup-1) is demonstrated in Fig. 1B. Wild-type RPV plaques on both CV-1 and A549 cells, while the SPI-1 mutant T309R plaques on CV-1 cells but is restricted on A549 cells. The T309C reversion restores the ability of the mutant virus to grow on A549 cells. We cloned the SPI-1 gene from wild-type RPV, RPV SPI-1 T309R, and RPV SPI-1 T309C (revertant) into an expression vector and synthesized radiolabeled proteins in a coupled transcription/translation system. The proteins were incubated with increasing amounts of the target protease, cathepsin G, and analyzed by SDS-polyacrylamide gel electrophoresis. Wild-type SPI-1 forms inhibitory enzyme complexes with cathepsin G which are abolished by the T309R mutation (Fig. 1B) as observed previously (39). The T309C mutation restores not only host range but also protease-serpin complex formation and thus presumably the inhibitory activity of SPI-1 protein in vitro (Fig. 1B). None of the other revertants had any additional mutations in the SPI-1 gene other than the parental T309R mutation. Hence, these remaining viruses must have second-site mutations that lie outside of the SPI-1 gene which restore host range on A549 cells.
The RPV sup-2 mutant contains a ts mutation for growth which maps to the viral DNA polymerase.
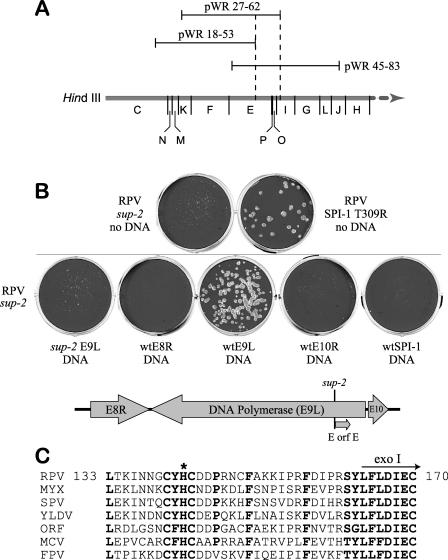
We showed that one of the suppressor mutants, the RPV sup-2 mutant, is temperature sensitive (ts) for growth and fails to plaque on either CV-1 or A549 cells at 41°C (Fig. 2B). By initially assuming that temperature sensitivity and restored host range were related properties, the temperature sensitivity of the sup-2 mutation was used to map the presumably linked suppressor mutation through the use of classical marker rescue techniques in conjunction with a cosmid library of wild-type vaccinia virus DNA (13). It is important to note that, despite its name, rabbitpox virus is now known to be a strain of vaccinia virus based on genomic sequence data (M. Buller and Chris Upton, personal communication). Preliminary mapping of the ts mutation was achieved by marker rescue as described in Materials and Methods. Briefly, CV-1 cells were infected with the RPV sup-2 mutant and transfected with each of the 11 individual cosmids from a vaccinia virus genomic library at 41°C. Cosmids pWR27-62 and pWR45-83 both rescued the sup-2 mutant but pWR18-53 did not, which mapped the ts mutation within a 10-kb region containing the HindIII E, P, O, and I fragments (Fig. 3A). Additional experiments indicated that rescue could be achieved by a cloned VV HindIII E fragment (not shown). Final mapping was achieved by additional marker rescue experiments with PCR fragments representing each of the complete HindIII E8R, E9L, and E10R contiguous ORFs (Fig. 3B). As expected, cells infected with the sup-2 mutant gave no growth at 41°C, whereas the RPV SPI-1 T309R parental mutant grew well on CV-1 cells at 41°C. Rescue was readily achieved with wt RPV ORF E9L DNA, placing the ts mutation of the RPV sup-2 mutant within the E9L ORF, which encodes the viral DNA polymerase (27). Marker rescue did not occur with E9L DNA from the RPV sup-2 mutant or wt RPV ORF E8R, ORF E10R, or wild-type SPI-1 DNA. Sequencing of the RPV sup-2 ORF E9L DNA revealed the deletion of a highly conserved histidine codon at position 142 (E9ΔH142) (Fig. 3C).
FIG. 2.
Temperature sensitivity of RPV extragenic suppressor mutants. Equivalent dilutions of each RPV isolate were plaqued on monolayers of CV-1 and A549 cells under agarose media in six-well plates at either 37°C or 41°C to assay for temperature sensitivity (ts). (A) Deletion of the SPI-1 gene from RPVΔSPI-1 or inactivation of the SPI-1 gene by the T309R mutation reduces RPV host range on A549 cells but does not affect its ability to form plaques at 41°C on permissive cells (CV-1). (B) Extragenic suppressor mutations sup-2, sup-3, and sup-4 all restore plaque formation to the RPV SPI-1 T309R mutant on A549 cells. The RPV sup-2 mutant is temperature sensitive (ts) for growth. The RPV sup-3 mutant has a slightly reduced ability to form plaques at 41°C. The RPV sup-4 mutant exhibits no temperature sensitivity.
FIG. 3.
Mapping of the sup-2 mutation by marker rescue. The ts property of the RPV sup-2 mutant was used to map the sup-2 mutation by marker rescue. (A and B) Monolayers of CV-1 cells in six-well plates were infected with the RPV sup-2 mutant at an MOI of 0.003 PFU per cell (10,000 PFU per well). Cells were then transfected with individual wild-type virus DNA fragments from either the vaccinia virus (A) or RPV (B) genome, and recombinant viruses were identified by restored plaque formation (rescue) at 41°C. (A) Using a cosmid library of the wild-type vaccinia virus (WR) genome, only fragments pWR27-62 and pWR45-83 rescued RPV sup-2 mutant infections at 41°C (data not shown), which mapped the ts mutation to within 10 kb (indicated by dotted lines). (B) RPV sup-2 mutant- and RPV SPI-1 T309R-infected CV-1 cells incubated at 41°C in the absence of transfected DNA are shown for comparison (top). To refine the marker rescue analysis, DNA from the SPI-1 gene and the complete open reading frames E8R, E9L, and E10R (contiguous ORFs shown below) was amplified by PCR from wild-type RPV genomic DNA. Transfection of RPV sup-2 mutant-infected CV-1 cells with wild-type E9L DNA (center) rescues plaque formation at 41°C, which was not seen in any other transfection. As a negative control, mutant E9L PCR product amplified from RPV sup-2 mutant genomic DNA (sup-2 E9L, middle left) does not rescue the infection. Failure to rescue the RPV sup-2 mutant at 41°C upon transfection with wt SPI-1 DNA (middle right) also indicates that the ts phenotype conferred by sup-2 is independent of the parental SPI-1 mutation. A mutation within the E9L gene of the RPV sup-2 mutant, E9ΔH142, was later identified by DNA sequencing. This mutation (sup-2, bottom right) is at the junction of E9L and a hypothetical ORF named E ORF E. (C) Amino acid alignment of vertebrate poxvirus DNA polymerases from various genera illustrates that codon His142 (*) of RPV E9L is within a highly conserved C(Y/F)HC motif (shown in bold). RPV, rabbitpox (orthopox); MYX, myxoma (leporipox); SPV, swinepox (suipox); YLDV, Yaba-like disease virus (yatapox); ORF, orf virus (parapox); MCV, molluscum contagiosum virus (molluscipox); FPV, fowlpox (avipox). The start of a proposed 3′-5′ exonuclease domain (exo I) within the DNA polymerase protein is also indicated for reference (30, 47).
Temperature sensitivity of the RPV sup-2 mutant DNA polymerase and suppression of restricted host range are linked.
The temperature sensitivity of the RPV sup-2 mutant could represent a second independent mutation unrelated and unlinked to suppression of restricted host range. To demonstrate linkage, we first analyzed the ability of the RPV sup-2 mutant to plaque on A549 cells after rescue of the ts phenotype with wt RPV ORF E9L DNA as described above. This virus, designated the sup-2 mutant(wtE9), was selected for the acquired ability to plaque on CV-1 cells at 41°C but was found unable to plaque on A549 cells at either temperature, like the parental RPV T309R mutant (Fig. 4A). This clearly indicates that both acquisition of temperature sensitivity and restoration of host range phenotypes are likely due to the same mutation within the E9L ORF.
FIG. 4.
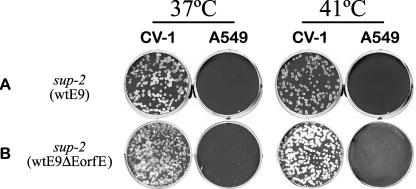
RPV host range mutant suppression by sup-2 does not depend on the hypothetical E ORF E gene. (A) Rescue of the RPV sup-2 mutant at 41°C in CV-1 cells by transfection with wt E9L PCR product replaces the E9ΔH142 allele in the RPV sup-2 mutant with wild-type E9L sequence (sup-2 wtE9). This recombinant virus does not grow in A549 cells, because it is a reconstruction of the parental RPV SPI-1 T309R mutant. Similarly, the RPV sup-2 mutant was later reconstructed from the RPV SPI-1 T309R mutant by replacing its wt E9L allele with mutant E9ΔH142 DNA using a variation on the marker rescue technique based on host range phenotype (hr) in A549 cells (data not shown). (B) DNA sequence analysis revealed that the sup-2 mutation also deletes the initiating Met1 codon from a hypothetical ORF designated E ORF E, which is embedded in the opposite orientation within the E9L ORF. To confirm that removal of the E ORF E gene by the sup-2 mutation has no effect on RPV host range, the E9ΔH142 allele of the RPV sup-2 mutant was replaced with wtE9ΔEORFE DNA by ts marker rescue. This removes E ORF E but restores the wt E9L gene. The phenotype of this recombinant (sup-2, wtE9ΔEORFE) is identical to the parental RPV SPI-1 T309R mutant.
Further confirmation of linkage of the two phenotypes was achieved by rescuing the restricted host range of the parental RPV T309R mutant in infected A549 cells by transfection with ORF E9L PCR product from RPV sup-2 mutant DNA to reconstruct the RPV sup-2 virus. Virus progeny was harvested 5 days postinfection, and recombinant viruses were selected by plaque assay on A549 cells. These recombinants were screened initially for temperature sensitivity at 41°C and then plaque purified on A549 cells at 37°C to maintain strictly the host range selection. DNA sequencing of ORF E9L PCR products from each recombinant isolate again revealed the same E9ΔH142 mutation seen in the original RPV sup-2 mutant (Fig. 3C). Therefore, based on these results and the high levels of recombinants observed with E9L DNA compared to mock-infected controls or controls containing other viral DNA fragments which produced no temperature-sensitive virus, we conclude that the sup-2 suppressor is temperature sensitive for growth. Furthermore, we conclude that temperature sensitivity is not the result of a second, independent, nonlinked mutation but is linked to restoration of host range.
During our phenotypic analysis of suppressor mutants we observed that the RPV sup-2 mutant, unlike most extragenic suppressor mutants selected on A549 cells, was also able to plaque on HeLa-S1a cells, which are normally nonpermissive for RPV SPI-1 T309R and RPVΔSPI-1 mutants (data not shown). Since our original selection for revertant viruses was done in A549 cells, it was likely that the restored host range of the RPV sup-2 mutant in HeLa-S1a cells was also due to the same ts mutation in DNA polymerase (E9ΔH142) identified by marker rescue. To confirm this, we compared the host range of the reconstructed RPV sup-2 mutant described above with both the parental RPV SPI-1 T309R mutant and wild-type RPV by plaque assays in HeLa-S1a cells. Again, the RPV sup-2 mutant showed a fully restored plaquing efficiency (data not shown). Thus, the restricted host range of RPV SPI-1 T309R in HeLa-S1a cells is also rescued by the E9ΔH142 mutation and may target a similar pathway found in A549 cells.
The E9L ORF contains an embedded ORF (E ORF E) initiated in the opposite direction by the same bases encoding histidine 142.
Inspection of the RPV and vaccinia virus E9L ORF (3 kb) reveals a hypothetical ORF (0.2 kb, 67 amino acids) encoded by the complementary strand in the opposite direction (Fig. 3B). To agree with an identical vaccinia virus (Copenhagen) genome entry (GenBank accession no. NP_063713), we also have designated this small ORF in RPV E ORF E. The theoretical start codon for E ORF E (Met1) uses the same three nucleotides that encode His142 of the E9L ORF. While no recognizable virus promoter or expressed protein has been identified for E ORF E, the restoration of host range could either be linked to the loss of this theoretical protein or be from mutation of the E9L ORF itself. To address this issue, site-directed mutagenesis was used to create a cysteine-to-glutamine mutation at codon 9 of the theoretical E ORF E protein followed by a stop codon, which are silent mutations in the E9L ORF. This mutant allele of the E9L ORF was designated wtE9ΔEORFE. Transfection of the wtE9ΔEORFE gene into RPV sup-2 mutant-infected cells gave recombinants which were no longer temperature sensitive and failed to plaque on A549 cells (Fig. 4B). These data show that both the original restored host range and ts phenotypes (Fig. 2B) were the direct result of the His142 deletion in the viral DNA polymerase and not the loss of the hypothetical E ORF E-encoded protein.
The RPV sup-2 mutant is temperature sensitive for DNA synthesis.
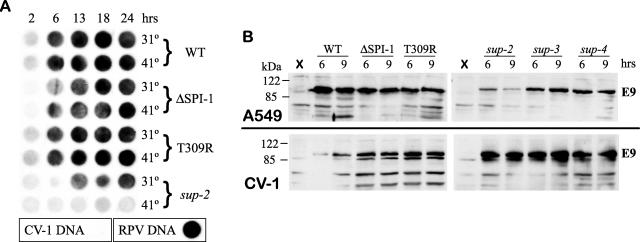
The RPV sup-2 mutant contains a deletion of His142 in the viral DNA polymerase which restores host range but creates temperature sensitivity for growth at 41°C. To examine the effect of the mutation on DNA synthesis, RPV sup-2 mutant-infected permissive CV-1 cells were assayed by dot blot analysis for DNA synthesis at both 31°C and 41°C. Viral DNA synthesis was not temperature sensitive in wt RPV-, ΔSPI-1-, or SPI-1 T309R mutant-infected cells as expected; however, DNA synthesis in RPV sup-2 mutant-infected cells was defective at 41°C (Fig. 5A). Thus, the deletion of His142 in the DNA polymerase gene is responsible for the temperature sensitivity of DNA synthesis.
FIG. 5.
Characterization of the RPV sup-2 mutant. (A) RPV DNA synthesis assay. CV-1 cells were infected with wild-type RPV or each designated mutant at an MOI of 10 PFU per cell at 31°C or 41°C. Samples were harvested at intervals up to 24 h postinfection, transferred to a nylon membrane by dot blot, and probed with digoxigenin-labeled RPV DNA to detect viral DNA synthesis by chemiluminescence. The CV-1 DNA and RPV DNA in the boxes contain control DNA samples for testing the hybridization specificity of the probe. (B) Western blot analysis of the E9L protein levels in infected cells. RPV-infected A549 (top) and CV-1 (bottom) cells were harvested at 6 and 9 h postinfection at 37°C. Steady-state levels of RPV DNA polymerase (∼116 kDa) were evaluated by Western blot analysis using a rabbit anti-VV-WR DNA polymerase antibody. X, uninfected cells.
Since SPI-1 is believed to be a protease inhibitor, we considered that increased protein turnover or degradation of virus proteins, such as the DNA polymerase, might occur in the absence of a functional SPI-1 protein in RPV SPI-1 T309R-infected cells. We examined the steady-state levels of RPV DNA polymerase expression at 37°C by Western blot analysis in infected A549 and CV-1 cells harvested at 6 and 9 h postinfection (Fig. 5B). Generally, steady-state levels of the DNA polymerase were similar in wt RPV-infected cells in the complete absence of SPI-1 protein (RPVΔSPI-1), in the presence of inactive SPI-1 protein (RPV SPI-1 T309R), or with two other extragenic suppressors of the SPI-1 T309R mutation (RPV sup-3 and sup-4). A significant decrease in DNA polymerase expression was observed in RPV sup-2 mutant-infected A549 cells, which is consistent with the fact that the sup-2 mutant is more temperature sensitive for plaque formation at 41°C in A549 cells than in CV-1 cells (data not shown).
Development of PCR-based genomic libraries for RPV and VV-WR.
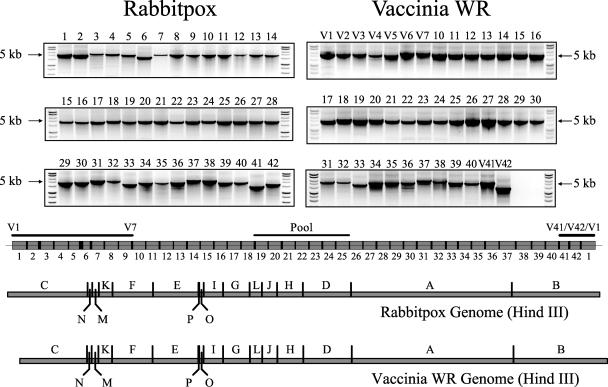
The mapping of the sup-2 mutation was greatly facilitated by the fact that the sup-2 mutant is also temperature sensitive for growth. The conventional mapping of ts mutants by marker rescue uses a single cosmid library that represents the entire wild-type vaccinia virus genome (13). Following marker rescue, the ts phenotype maps each ts mutation to the appropriate cosmids. In contrast, the mapping of hr mutation suppressors by standard marker rescue requires unique genomic libraries for each suppressor mutant and transfection with fragments from each library into nonpermissive cells (A549) infected with the parental mutant virus (RPV SPI-1 T309R) to restore host range. As an alternative to creating cosmid libraries for each viral mutant, we developed a simpler approach based on long PCR methods (10) to create “virtual” or “synthetic” genomic libraries for both RPV and VV. This method is useful not only for the mapping of our hr mutants but also for the mapping of virtually any mutant with a distinguishable phenotype. These “PCR libraries” comprise 40 to 42 overlapping 5-kb DNA fragments that span virtually the entire genome of each virus (Fig. 6). Most primers are derived from the more commonly studied vaccinia virus but work equally well with RPV because of the colinearity and overall sequence conservation between the two viruses. Nonetheless, some virus-specific primers were required at the relatively divergent termini of both genomes to complete each library. Terminal divergence is typical of all orthopoxviruses and in part serves to distinguish them. By choosing primers with similar characteristics throughout each entire genome, we successfully used the same conditions for all PCRs, which allowed complete libraries to be generated in one step. In preliminary mapping experiments, PCR products were also pooled to simulate larger DNA fragments during marker rescue (Fig. 6).
FIG. 6.
PCR-generated libraries of RPV and VV-WR. Virtually the entire genomes of rabbitpox (RPV) and vaccinia (VV-WR) viruses were divided into 42 and 40 overlapping PCR products, respectively, omitting only noncoding DNA at each terminus. Agarose gel electrophoresis of each PCR product is shown. The expected size and location of each RPV PCR product is shown in boxes (drawn to scale) that are aligned with the HindIII restriction maps for both RPV and VV-WR. Products designed specifically for VV-WR were given the prefix “V” (for distinction from RPV products) and are shown simply with a line. One example of a pool of PCR products used later for marker rescue is also shown. All primer sequences are available upon request.
Mapping of the extragenic suppressors.
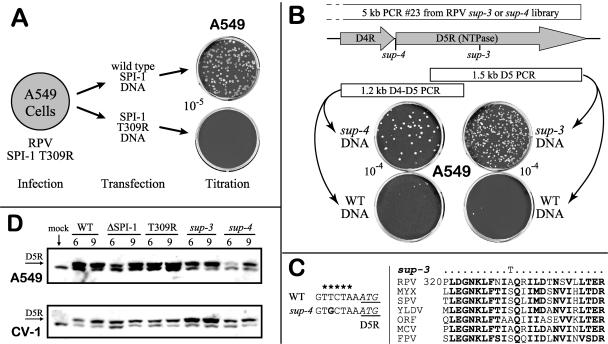
In order to map the remaining extragenic mutations responsible for restoring growth on A549 cells, we developed a two-step marker rescue protocol diagramed in Fig. 7A. We illustrate the technique based on the mapping of a known inactivating mutation in the SPI-1 gene, such as the T309R mutation, although any mutation with a phenotype could have been chosen. In the first step, A549 cells were infected with the RPV SPI-1 T309R hr mutant and cotransfected with a PCR product containing either the wt SPI-1 gene as a positive control to generate wild-type virus or the parental SPI-1 T309R mutant gene as a negative control, which should not generate recombinants. For mapping, cells infected with RPV SPI-1 T309R were transfected with either single PCR fragment products or a pool of fragments. The cells were then incubated at 37°C and harvested 5 days postinfection. In the second step, progeny recombinant viruses with a restored host range phenotype, over and above that observed for the parental mutant as a negative control, were detected on A549 cells by plaque assay (Fig. 7A). In this example, only PCR fragments encompassing the wt SPI-1 gene rescued the virus (data not shown). The strategy was used to map the other two extragenic suppressors, based again on rescue of the RPV SPI-1 T309R virus on A549 cells with PCR-generated genomic libraries from sup-3 and sup-4 viruses. Preliminary mapping was achieved from pools of six to seven PCR fragments (spanning 25 to 30 kb) to roughly localize the suppressor mutation within the 200-kb virus genome. For both sup-3 and sup-4, a pool of fragments 19 to 25 (Fig. 6) gave a significant rescue above background, and the use of individual fragments localized the mutations in both revertants to fragment 23 in the HindIII D fragment (data not shown). sup-3 was mapped to the D5R ORF using a 1.5-kb fragment of D5R (Fig. 7B). The D5R gene of the sup-3 mutant was sequenced, and a single mutation was found (A330T) within the open reading frame (Fig. 7C). The D5R protein is an 85-kDa NTPase that is essential for viral DNA replication and recombination and may participate in a multiprotein complex that includes the viral DNA polymerase (19). In contrast, mapping and sequencing of the sup-4 mutant D5R region showed that the suppressor mutation was not in the open reading frame, as might have been expected, but was rather in the upstream promoter region of the D5R gene (Fig. 7B and C).
FIG. 7.
Mapping and characterization of RPV sup-3 and sup-4. (A) Host range marker rescue. A549 cells were infected with RPV SPI-1 T309R and transfected with a 1-kb fragment of the SPI-1 gene amplified by PCR from either wild-type or SPI-1 T309R mutant RPV DNA. After 5 days of incubation under liquid growth medium at 37°C, viruses were harvested and their titers determined on A549. (B) Mapping of RPV sup-3 and sup-4. The host range marker rescue approach was applied using PCR-generated DNA libraries of either the RPV sup-3 or sup-4 mutant. In each case, only the 5-kb PCR product no. 23 was able to rescue host range (data not shown). ORFs D4R and D5R within the right terminus of this PCR product are shown (top). Only a 1.5-kb PCR product from the D5R gene of the RPV sup-3 mutant and a 1.2-kb PCR product from the region of overlap between the D4R and D5R genes of the RPV sup-4 mutant restored the host range of the RPV SPI-1 T309R mutant on A549 cells. As a control, homologous wild-type PCR products from either region did not restore host range under the same conditions. (C) DNA sequencing analysis. The RPV sup-3 mutant (right) contains an A330T mutation in the D5R gene. A partial alignment of orthologs of the RPV D5R gene from other poxviruses is shown flanking the RPV sup-3 mutation. MYX, myxoma; SPV, swinepox virus; YLDV, Yaba-like disease virus; ORF, orf virus; MCV, molluscum contagiosum virus; and FPV, fowlpox. The RPV sup-4 mutant (left) contains a mutation (T→G) just upstream of the ATG translation start site for D5R. The asterisk indicates transcription initiation sites mapped by S1 nuclease protection assays (35, 41). (D) D5R protein levels from RPV-infected A549 and CV-1 cells harvested at 6 and 9 h postinfection were detected by Western blot analysis using a rabbit anti-VV-WR D5R antibody. mock, uninfected cells; WT, wild-type RPV; ΔSPI-1, RPV with an SPI-1 gene deletion; T309R, RPV SPI-1 T309R.
D5R suppressor mutations and protein stability.
The D5R gene is highly conserved and considered to be an essential “core” gene found within all poxviruses (21). A ClustalW alignment of RPV D5R homologs from various vertebrate poxvirus genera in the region flanking the sup-3 mutation (A330T) demonstrates a high degree of conservation (Fig. 7C). The precise function of this region within the D5R protein is still unknown. However, because of the high conservation of the gene, one might expect that mutations in the D5R protein could potentially affect the function and perhaps the stability of this protein. In contrast, the sup-4 extragenic suppressor maps to the promoter region of D5R. Previous transcriptional analyses of the vaccinia D5R gene (RPV D5R homolog), using S1 nuclease protection assays, have shown that the transcription initiation site is in the upstream TTCTA sequence (35, 54) within which the sup-4 (T→G) mutation also maps, as indicated in Fig. 7C. The SPI-1 protein, a serpin, functions as a protease inhibitor. We considered that a nonfunctional SPI-1 gene, exemplified by the T309R mutant, could lead to enhanced cell-dependent proteolytic activity during an infection, which in turn could result in diminished steady-state levels of the viral proteins necessary for replication (including D5R) and cause a cell line-dependent failure of virus growth. Thus, both suppressors could lead to changes in the steady-state level of D5R expression responsible for suppressor activity.
To investigate these possibilities, we examined the steady-state levels of the D5R protein in wt and mutant infections present at 6 and 9 h (Fig. 7D), times at which normal expression of D5R occurs (20, 41). Generally, the levels of the D5R protein expressed in all cases were greater in A549 cells than in CV-1 cells. Neither the deletion of the SPI-1 gene nor the T309R inactivating mutation within the SPI-1 gene had any obvious effect on D5R levels in these cells. The expression of D5R from RPVΔSPI-1 did appear somewhat less in A549 cells at earlier times but not by 9 h postinfection. The D5R promoter mutation (sup-4) resulted in significantly lower levels of D5R protein in A549 cells at both 6 and 9 h postinfection. No evidence of altered proteolysis activity was seen in any of the samples. Thus, it would appear that the loss or inactivation of the SPI-1 protein has no significant effect on either the level or the stability of the D5R protein.
The sup-3 and sup-4 mutations do not show allelic specificity.
sup-3 and sup-4 were selected to suppress a single defective allele, SPI-1 T309R. If these suppressors specifically correct only this allele, then they should not restore host range to other SPI-1 mutants. This “allelic specificity” would provide genetic evidence consistent with direct interaction between SPI-1 and the D5R protein. To explore this possibility, we examined two additional types of SPI-1 mutants previously shown to have no activity against cathepsin G and to have a reduced host range (39): the P1 active-site mutant, RPV SPI-1 F322A, which inactivates protease activity, and RPVΔSPI-1, in which the SPI-1 gene has been deleted. Using the two-step marker rescue system, A549 cells were infected with either RPV SPI-1 F322A or RPVΔSPI-1 and transfected with D5R PCR products generated from wild-type, sup-3, or sup-4 viruses. Surprisingly, rescue was observed with both sup-3 and sup-4 D5R genes (Fig. 8) at approximately the same frequency originally observed for rescue of RPV SPI-1 T309R. Little if any rescue was observed in “mock”-transfected samples or samples transfected with viral DNA derived from other regions of the viral genome. To confirm that the rescued plaques were indeed D5R recombinants, three rescued plaques from each virus strain were purified on A549 cells and the D5R and SPI-1 genes sequenced. In all cases, the parental SPI-1 mutation was retained, and the expected D5R suppressor allele was present (data not shown). Thus, the suppression of SPI-1 mutations by sup-3 or sup-4 is not allele specific. These results tend to argue against any direct interaction between SPI-1 and D5R and suggest instead a more indirect role of D5R in host range mutant suppression.
FIG. 8.
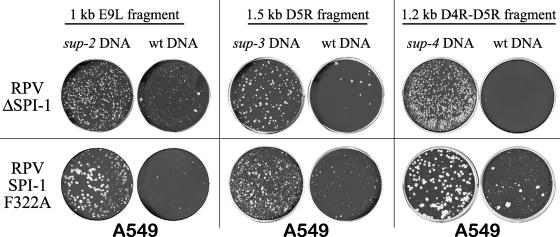
RPV sup mutations are not specific for the SPI-1 T309R allele. Host range of two additional RPV SPI-1 mutants (RPV SPI-1 F322A and RPVΔSPI-1) was partially to fully restored following transfection with PCR products from sup-2 (ORF E9L), sup-3 (ORF D5R), or sup-4 (ORFs D4R-D5R) compared with wild-type fragments of the same regions. Titers of virus progeny from transfected cells were determined on A549 cells by plaque assay.
Since sup-3 and sup-4 were capable of rescuing different defective alleles of SPI-1, we next asked, conversely, whether we could use these other SPI-1 mutants to select for more extragenic suppressors. We found that extragenic suppressors could be selected using either RPVΔSPI-1 or F322A virus. We have mapped a number of these suppressors, and many but not all map again to the D5R gene. The locations of the various suppressor mutations within the D5R gene are summarized in Fig. 9. It is particularly noteworthy that suppressors were isolated in the D5R gene from an RPV mutant in which the SPI-1 gene had been deleted. This argues further against suppression being caused by any direct interactions between SPI-1 and D5R.
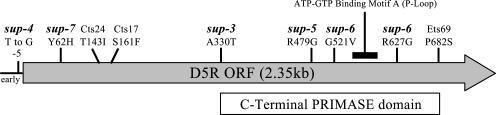
FIG. 9.
Positions of RPV sup mutations which map to the D5R gene. Map positions are shown for sup-3, sup-4, and three additional suppressor mutations in the D5R gene isolated in this study (sup-5, sup-6, and sup-7). Previously published studies of vaccinia virus have resulted in the isolation and mapping of ts mutants in the D5R gene which are also shown for reference (Cts17, Cts24, and Ets69) (12, 13, 21, 41). Mutations sup-5 (R479G) and sup-6 (G521V, R627G) were identified in two RPVΔSPI-1 revertant clones isolated in A549 cells and verified by marker rescue analysis (data not shown). The sup-7 (Y62H) mutation was identified by sequencing from a revertant of another SPI-1 host range mutant, RPV SPI-1 F322A. The RPV sup-6 mutant, like the sup-2 mutant, is a ts mutant and does not plaque at 41°C (data not shown). Motifs within the D5R gene include an ATP/GTP binding motif A (P-loop), as described by Evans et al. (19), and a C-terminal PRIMASE domain (P4 family) typically found in bacteria and phages, which encompasses many of the suppressor mutations shown (Hidden Markov Model accession no. TIGR01613 from The Institute for Genomic Research [www.tigr.org]).
DISCUSSION
Orthopoxviruses encode three serpins, SPI-1, SPI-2/crmA, and SPI-3. All three orthopoxvirus serpins are functional as protease inhibitors. SPI-2/crmA is active both against caspases and against granzyme B and is known to control inflammation and apoptosis (16, 28, 31, 36, 48, 52, 56, 57). SPI-3, active against plasmin, urokinase-like plasminogen activator, and tissue-type plasminogen activator, is required to prevent infected cell fusion (33, 50, 51, 55). SPI-1 has been shown to form inhibitory complexes with cathepsin G in vitro and is required for full host range (1, 7, 43). Some inhibitory serpins, such as antithrombin, are also known to be influenced by the presence of cofactors (15, 22, 38). Similarly, the cowpox virus SPI-3 protein has been shown to interact with the virus-encoded hemagglutinin (9).
The regulation of host range by SPI-1 presented a unique opportunity to apply a genetic approach to search for viral proteins which interact with or are otherwise involved in the biological activity of this serpin. Initially, we introduced a mutation at residue P14 (T309R) of the reactive center loop which compromises molecular flexibility but leaves other critical elements of the serpin backbone and the reactive center loop intact. Introduction of the SPI-1 T309R mutation into virus leads to loss of host range. Restoration of full host range provides a positive selection for potential extragenic mutations within the virus which can compensate for the SPI-1 T309R mutation. We report here the selection and mapping of such revertants and that they include both intragenic suppressors which map within the SPI-1 gene and extragenic suppressors which map to other virus genes. It would not be surprising if there were also cellular suppressor mutations which could compensate for SPI-1 mutant genes. We have not addressed that issue in this study.
We detected one intragenic suppressor by sequencing the revertant SPI-1 gene. In this intragenic mutant, the P14 residue we had previously mutated from T to R to inactivate the SPI-1 gene had mutated again to a cysteine (R→C). This mutant exhibited a restored host range and serpin inhibitory activity. A cysteine residue at the P14 residue of a serpin is extremely rare, and the only known naturally occurring serpin in which the P14 residue is cysteine is serpin 43Aa from Drosophila (24), although it is not known whether this serpin functions as an inhibitory serpin. However, a P14 mutant of the inhibitory human serpin antithrombin III (AT III), in which the naturally occurring P14 amino acid serine was changed to cysteine (P14 S→C), was recently constructed (26). Although some normal properties of AT III were altered, the mutant AT III was still active and able to form a 1:1 inhibitory complex with factor Xa, the natural target for AT III (26). To facilitate the mapping of extragenic suppressors, we developed a novel procedure based on the rescue of virus by transfection of infected cells with fragments from PCR-generated genomic libraries. The procedure has already proven to have profound advantages in the mapping of mutations and greatly simplifies the more conventional cosmid- or plasmid-based libraries in the mapping of other poxvirus mutations. Essentially, any selectable or identifiable genetic marker can be rapidly mapped with variations on the procedure in a fashion which negates the time-consuming necessity to prepare individual cosmid or plasmid libraries derived from each mutant of interest.
A second useful application of the PCR-based libraries should be noted. Sequencing of multiple orthopoxvirus genomes shows an internal, colinear, and overall well-conserved core of genes, whereas regions outside this core can be quite diverse (53). Preliminary data with several other orthopoxvirus templates show that while most of a given genome can be amplified with either the RPV or VV primer sets from conserved genes, there are some PCR primer pairs which are species specific. Comparing successes and failures, in combination with differences in product size, reveals a unique “fingerprint” for each species of orthopoxvirus which could form the basis of a rapid diagnostic strategy, readily adaptable to “field” conditions.
Many of the SPI-1 extragenic suppressors described here map to the essential viral D5R gene, which encodes a protein with primase motifs and NTPase function that is required for both DNA replication and recombination (19, 21). There were several other additional unanticipated findings which emerged during these studies. First was our finding that suppressors which correct a specific SPI-1 mutation (T309R) in the P14 residue have no allelic specificity and can also suppress virus with an SPI-1 deletion or virus with inactivating mutations in the P1 residue of the serpin. This lack of specificity argues against direct serpin-D5R protein interactions mediating host range function. The fact that host range suppressors can be isolated for virus with an SPI-1 deletion implies that inhibition of the protease activity which is linked to full host range can be bypassed by an as yet unknown mechanism.
Finally, we note that while many extragenic suppressors of the T309R mutation map to the D5R gene, another suppressor maps to the viral DNA polymerase (ORF E9L), and still other extragenic suppressors fail to map to either of these genes. The linkage of the replication apparatus to host range has been previously described. For instance, the host range of Bombyx mori and Autographa californica nuclear polyhedrosis viruses can be expanded by mutations in the DNA helicase (29, 37). Adenovirus host range mutants can expand host range with mutations in a DNA binding protein (2, 8, 40). Replication of the intertypic recombinant herpes simplex virus RB-1 is restricted in neurons due to mutations in the UL5 gene, which is a component of the helicase-primase complex implicated in DNA replication (3, 6). The exact role of the SPI-1 protein, not previously linked to DNA replication, as a comediator of host range in conjunction with the replication apparatus remains to be determined.
Acknowledgments
We are grateful to Bernie Moss, Chris Upton, and Mark Buller for making unpublished genomic sequences of vaccinia and rabbitpox viruses available to us for PCR primer development. We also thank Paula Traktman and Beth Unger for providing us with the E9L and D5R antibodies and recommended protocols for detection of this protein, R. Condit for discussions and providing us with the vaccinia virus cosmid library, and S. Moyer for constructive suggestions on the manuscript.
REFERENCES
- 1.Ali, A. N., P. C. Turner, M. A. Brooks, and R. W. Moyer. 1994. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology 202:306-314. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. W., M. M. Hardy, J. J. Dunn, and D. F. Klessig. 1983. Independent, spontaneous mutants of adenovirus type 2-simian virus 40 hybrid Ad2+ ND3 that grow efficiently in monkey cells possess indentical [sic] mutations in the adenovirus type 2 DNA-binding protein gene. J. Virol. 48:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera, I., D. Bloom, and M. Challberg. 1998. An intertypic herpes simplex virus helicase-primase complex associated with a defect in neurovirulence has reduced primase activity. J. Virol. 72:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becerra, S. P., A. Sagasti, P. Spinella, and V. Notario. 1995. Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J. Biol. Chem. 270:25992-25999. [DOI] [PubMed] [Google Scholar]
- 5.Bird, C. H., E. J. Blink, C. E. Hirst, M. S. Buzza, P. M. Steele, J. R. Sun, S. A. Jans, and P. I. Bird. 2001. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol. Cell. Biol. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom, D. C., and J. G. Stevens. 1994. Neuron-specific restriction of a herpes simplex virus recombinant maps to the UL5 gene. J. Virol. 68:3761-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, M. A., A. N. Ali, P. C. Turner, and R. W. Moyer. 1995. A rabbitpox virus serpin gene controls host range by inhibiting apoptosis in restrictive cells. J. Virol. 12:7688-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brough, D. E., S. A. Rice, S. Sell, and D. F. Klessig. 1985. Restricted changes in the adenovirus DNA-binding protein that lead to extended host range or temperature-sensitive phenotypes. J. Virol. 55:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brum, L. M., P. C. Turner, H. Devick, M. T. Baquero, and R. W. Moyer. 2003. Plasma membrane localization and fusion inhibitory activity of the cowpox virus serpin SPI-3 require a functional signal sequence and the virus encoded hemagglutinin. Virology 306:289-302. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, S., C. Fockler, W. M. Barnes, and R. Higuchi. 1994. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc. Natl. Acad. Sci. USA 91:5695-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang, Y. J., R. Swanson, S. M. Raja, and S. T. Olson. 2001. Heparin enhances the specificity of antithrombin for thrombin and factor Xa independent of the reactive center loop sequence. Evidence for an exosite determinant of factor Xa specificity in heparin-activated antithrombin. J. Biol. Chem. 276:14961-14971. [DOI] [PubMed] [Google Scholar]
- 12.Condit, R. C., and A. Motyczka. 1981. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology 113:224-241. [DOI] [PubMed] [Google Scholar]
- 13.Condit, R. C., A. Motyczka, and G. Spizz. 1983. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology 128:429-443. [DOI] [PubMed] [Google Scholar]
- 14.Darnell, G. A., T. M. Antalis, R. W. Johnstone, B. W. Stringer, S. M. Ogbourne, D. Harrich, and A. Suhrbier. 2003. Inhibition of retinoblastoma protein degradation by interaction with the serpin plasminogen activator inhibitor 2 via a novel consensus motif. Mol. Cell. Biol. 23:6520-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawes, J., K. James, and D. A. Lane. 1994. Conformational change in antithrombin induced by heparin probed with a monoclonal antibody against the 1C/4B region. Biochemistry 33:4375-4383. [DOI] [PubMed] [Google Scholar]
- 16.Dbaibo, G. S., and Y. A. Hannun. 1998. Cytokine response modifier A (CrmA): a strategically deployed viral weapon. Clin. Immunol. Immunopathol. 86:134-140. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson, J. L., E. J. Bates, A. Ferrante, and T. M. Antalis. 1995. Plasminogen activator inhibitor type 2 inhibits tumor necrosis factor alpha-induced apoptosis. Evidence for an alternate biological function. J. Biol. Chem. 270:27894-27904. [DOI] [PubMed] [Google Scholar]
- 18.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, E., N. Klemperer, R. Ghosh, and P. Traktman. 1995. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 69:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, E., and P. Traktman. 1987. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J. Virol. 61:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans, E., and P. Traktman. 1992. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma 102:S72-S82. [DOI] [PubMed] [Google Scholar]
- 22.Gettins, P., J. Choay, B. C. Crews, and G. Zettlmeiss. 1992. Role of tryptophan 49 in the heparin cofactor activity of human antithrombin III. J. Biol. Chem. 267:21946-21953. [PubMed] [Google Scholar]
- 23.Gettins, P. G. 2002. Serpin structure, mechanism, and function. Chem. Rev. 102:4751-4804. [DOI] [PubMed] [Google Scholar]
- 24.Green, C., E. Levashina, C. McKimmie, T. Dafforn, J. M. Reichhart, and D. Gubb. 2000. The necrotic gene in Drosophila corresponds to one of a cluster of three serpin transcripts mapping at 43A1.2. Genetics 156:1117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hood, D. B., J. A. Huntington, and P. G. Gettins. 1994. Alpha 1-proteinase inhibitor variant T345R. Influence of P14 residue on substrate and inhibitory pathways. Biochemistry 33:8538-8547. [DOI] [PubMed] [Google Scholar]
- 26.Huntington, J. A., and P. G. Gettins. 1998. Conformational conversion of antithrombin to a fully activated substrate of factor Xa without need for heparin. Biochemistry 37:3272-3277. [DOI] [PubMed] [Google Scholar]
- 27.Jones, E. V., and B. Moss. 1984. Mapping of the vaccinia virus DNA polymerase gene by marker rescue and cell-free translation of selected RNA. J. Virol. 49:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamada, S., Y. Funahashi, and Y. Tsujimoto. 1997. Caspase-4 and caspase-5, members of the ICE/CED-3 family of cysteine proteases, are CrmA-inhibitable proteases. Cell Death Differ. 4:473-478. [DOI] [PubMed] [Google Scholar]
- 29.Kamita, S. G., and S. Maeda. 1997. Sequencing of the putative DNA helicase-encoding gene of the Bombyx mori nuclear polyhedrosis virus and fine-mapping of a region involved in host range expansion. Gene 190:173-179. [DOI] [PubMed] [Google Scholar]
- 30.Knopf, C. W. 1998. Evolution of viral DNA-dependent DNA polymerases. Virus Genes 16:47-58. [DOI] [PubMed] [Google Scholar]
- 31.Komiyama, T., L. T. Quan, and G. S. Salvesen. 1996. Inhibition of cysteine and serine proteinases by the cowpox virus serpin CRMA. Adv. Exp. Med. Biol. 389:173-176. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Law, K. M., and G. L. Smith. 1992. A vaccinia serine protease inhibitor which prevents virus-induced cell fusion. J. Gen. Virol. 73:549-557. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, D. A., D. Ginsburg, D. E. Day, M. B. Berkenpas, I. M. Verhamme, J. Kvassman, and J. D. Shore. 1995. Serpin-protease complexes are trapped as stable acyl-enzyme intermediates. J. Biol. Chem. 270:25309-25312. [DOI] [PubMed] [Google Scholar]
- 35.Lee-Chen, G. J., N. Bourgeois, K. Davidson, R. C. Condit, and E. G. Niles. 1988. Structure of the transcription initiation and termination sequences of seven early genes in the vaccinia virus HindIII D fragment. Virology 163:64-79. [DOI] [PubMed] [Google Scholar]
- 36.Macen, J., A. Takahashi, K. B. Moon, R. Nathaniel, P. C. Turner, and R. W. Moyer. 1998. Activation of caspases in pig kidney cells infected with wild-type and CrmA/SPI-2 mutants of cowpox and rabbitpox viruses. J. Virol. 72:3524-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda, S., S. G. Kamita, and A. Kondo. 1993. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. J. Virol. 67:6234-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monkhouse, F. C., and S. Milojevic. 1968. Studies on the relation between plasma antithrombin and heparin-cofactor. Can. J. Physiol. Pharmacol. 46:347-350. [DOI] [PubMed] [Google Scholar]
- 39.Moon, K. B., P. C. Turner, and R. W. Moyer. 1999. SPI-1-dependent host range of rabbitpox virus and complex formation with cathepsin G is associated with serpin motifs. J. Virol. 73:8999-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, S. A., and D. F. Klessig. 1984. The function(s) provided by the adenovirus-specified, DNA-binding protein required for viral late gene expression is independent of the role of the protein in viral DNA replication. J. Virol. 49:35-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roseman, N. A., and D. E. Hruby. 1987. Nucleotide sequence and transcript organization of a region of the vaccinia virus genome which encodes a constitutively expressed gene required for DNA replication. J. Virol. 61:1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders, D. N., L. Jankova, S. J. Harrop, P. M. Curmi, A. R. Gould, M. Ranson, and M. S. Baker. 2001. Interaction between the P14 residue and strand 2 of beta-sheet B is critical for reactive center loop insertion in plasminogen activator inhibitor-2. J. Biol. Chem. 276:43383-43389. [DOI] [PubMed] [Google Scholar]
- 43.Shisler, J. L., S. N. Isaacs, and B. Moss. 1999. Vaccinia virus serpin-1 deletion mutant exhibits a host range defect characterized by low levels of intermediate and late mRNAs. Virology 262:298-311. [DOI] [PubMed] [Google Scholar]
- 44.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 45.Stein, P., and C. Chothia. 1991. Serpin tertiary structure transformation. J. Mol. Biol. 221:615-621. [DOI] [PubMed] [Google Scholar]
- 46.Stein, P. E., D. A. Tewkesbury, and R. W. Carrell. 1989. Ovalbumin and angiotensinogen lack serpin S-R conformational change. Biochem. J. 262:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taddie, J. A., and P. Traktman. 1993. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3′-5′ exonuclease domain. J. Virol. 67:4323-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tewari, M., W. G. Telford, R. A. Miller, and V. M. Dixit. 1995. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J. Biol. Chem. 270:22705-22708. [DOI] [PubMed] [Google Scholar]
- 49.Turner, P. C. 1996. Analysis of interactions between proteinases and serpins expressed in vitro using the TNT T7 Quick Syst. Promega Notes 60:11-13. [Google Scholar]
- 50.Turner, P. C., M. T. Baquero, S. Yuan, S. R. Thoennes, and R. W. Moyer. 2000. The cowpox virus serpin SPI-3 complexes with and inhibits urokinase-type and tissue-type plasminogen activators and plasmin. Virology 272:267-280. [DOI] [PubMed] [Google Scholar]
- 51.Turner, P. C., and R. W. Moyer. 1995. Orthopoxvirus fusion inhibitor glycoprotein SPI-3 (open reading frame K2L) contains motifs characteristic of serine proteinase inhibitors that are not required for control of cell fusion. J. Virol. 69:5978-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner, S., B. Kenshole, and J. Ruby. 1999. Viral modulation of the host response via crmA/SPI-2 expression. Immunol. Cell Biol. 77:236-241. [DOI] [PubMed] [Google Scholar]
- 53.Upton, C., S. Slack, A. L. Hunter, A. Ehlers, and R. L. Roper. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 77:7590-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villarreal, E. C., N. A. Roseman, and D. E. Hruby. 1984. Isolation of vaccinia virus mutants capable of replicating independently of the host cell nucleus. J. Virol. 51:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, J., X. Y. Sun, G. J. Fernando, and I. H. Frazer. 1992. The vaccinia virus K2L gene encodes a serine protease inhibitor which inhibits cell-cell fusion. Virology 189:678-686. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, Q., and G. S. Salvesen. 2000. Viral caspase inhibitors CrmA and p35. Methods Enzymol. 322:143-154. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, Q., S. Snipas, K. Orth, M. Muzio, V. M. Dixit, and G. S. Salvesen. 1997. Target protease specificity of the viral serpin CrmA—analysis of five caspases. J. Biol. Chem. 272:7797-7800. [DOI] [PubMed] [Google Scholar]