Abstract
The hypothesis that the intrapatient emergence of cytotoxic T-lymphocyte escape variants contributes to the evolution of human immunodeficiency virus type 1 at the population (interpatient) level was tested using the HLA-A*0201-restricted gag p17 epitope SLYNTVATL. Using a simple experimental design, we investigated the evolutionary processes operating within this epitope among patients while compensating for the confounding influence of intrapatient natural selection. Using this approach, we revealed a pattern of A*0201-driven escape within patients, followed by the sustained transmission of these escape variants among patients irrespective of their HLA type.
During infection with human immunodeficiency virus type 1 (HIV-1), the cytotoxic T-lymphocyte (CTL) response plays a central role in controlling viral replication (3, 16, 17, 27) and has been shown to drive the emergence of escape variants capable of evading CTL recognition (4, 13, 22). If these escape variants are continually transmitted, it is possible that the CTL response will also influence HIV-1 genetic variation at the population (interpatient) level. The accumulation of escape variants across the infected population is supported by the observation that rare HLA supertypes are associated with improved control of viral replication and slower disease progression (26). However, although the horizontal transmission of escape variants has been inferred from the detection of CTL escape mutants early in the acute phase of infection (1, 7, 11, 18), whether this process has a major effect on HIV-1 evolution at the population level is unclear. If escape variants carry a fitness cost in patients unable to mount a response against the epitope in question (i.e., who lack the appropriate HLA type), then the persistent transmission of these variants is unlikely. Indeed, the reversion of CTL escape mutations in patients with an incompatible genetic background has been demonstrated (18), and the need for compensatory mutations suggests that some CTL escape mutations incur a cost (15). In the analogous situation of drug-resistant variants, compensatory mutations may enhance the fitness of resistant strains (9) but fail to prevent reversion to susceptible genotypes upon transmission to drug-naive recipients (8).
How intrapatient CTL escape contributes to evolution at the population level is crucial in understanding long-term patterns of change in HIV. To address this question, we examined the HLA-A*0201-restricted gag p17 epitope SLYNTVATL (SL9) in subtype B patients. CTL escape has been intensively studied in this epitope (5, 10, 12, 14), and the A*0201 HLA type is by far the most common within Caucasians (44% phenotype frequency) (19), making it a strong candidate to influence evolution at the population level.
By combining multiple patients within a single analysis, it is possible to analyze evolutionary dynamics at the population level. However, it is first necessary to exclude the confounding effect of selection operating within each of the individual patients. This was achieved by only analyzing those patients unable to mount a CTL response against SL9, which enabled us to determine whether A*0201+-associated escape mutants were transmitted through the population because of their enhanced fitness or emerged within each patient individually.
In our initial analysis, we confirmed that intrapatient positive selection within SL9 is limited to A*0201+ patients. Sequences were obtained from two previously published studies (5, 6) (accession numbers AF017813 to AF017980, AF028563 to AF028587, AF060031 to AF060073, and AF073382 to AF073441), totaling 18 HLA-typed patients, with the number of clones from each ranging from 7 to 43 (average, 15.6) (Table 1). First, sequences were aligned manually using the Se-Al program (23). Maximum-likelihood phylogenetic trees were then estimated using the PAUP* package (25) assuming the general time-reversible (GTR + I + Γ) model of nucleotide substitution, with all parameters estimated from the data (available on request). For each patient phylogeny, an ancestral reconstruction using maximum likelihood was performed with the DATAMONKEY program (21). The number of synonymous (dS) and nonsynonymous (dN) substitutions per site for each codon was then determined. Under positive selection, we expect dN > dS. A per-site comparison of the difference between dN and dS in the A*0201+ and A*0201− patient groups allowed us to test whether the strength of positive selection was the same in each group. For amino acids 71 to 91 in gag p17, we found a significant difference only at T84 (A*0201− mean dN − dS = −0.22; A*0201+ mean dN − dS = 0.60; P = 0.010 using a one-tailed t test) (Table 1). Although not an explicit test of intrapatient positive selection, this demonstrates that nonsynonymous changes at T84 are limited largely to A*0201+ patients. Changes at this site have been shown to facilitate escape from A*0201 (10).
TABLE 1.
Intrapatient HIV-1 data sets
| Subtype B consensus | na | lb | G71 | S72 | E73 | E74 | L75 | R76 | S77 | L78 | Y79 | N80 | T81 | V82 | A83 | T84 | L85 | Y86 | C87 | V88 | H89 | Q90 | R91 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A2− patients | |||||||||||||||||||||||
| 35i | 10 | 273 | —c | — | — | — | — | — | — | — | F | — | — | — | — | — | — | — | — | — | — | — | Q |
| 053i | 10 | 570 | — | — | — | — | — | K | — | — | F | — | — | — | — | V | — | — | — | — | — | — | — |
| 11497 | 10 | 234 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | F | S | — | R | — | — |
| 11841 | 7 | 126 | — | — | — | — | — | — | — | — | F | — | — | — | — | — | — | — | — | — | — | — | — |
| 11850 | 16 | 273 | — | — | — | — | — | Q | — | — | — | — | — | I | — | V | — | — | — | — | — | — | K |
| 13632 | 13 | 180 | — | — | — | — | — | K | — | — | F | — | — | I | — | — | — | — | — | — | — | — | — |
| 13997 | 8 | 264 | — | — | — | — | — | K | — | — | — | — | — | — | — | — | — | — | — | — | — | — | K |
| 14532 | 14 | 495 | — | — | — | — | — | K | — | — | — | — | — | I | — | V | — | — | — | — | — | — | — |
| 15160 | 13 | 357 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| A2+ patients | |||||||||||||||||||||||
| 115i | 14 | 447 | — | — | — | — | — | — | — | — | F | — | — | — | — | — | — | — | — | — | — | — | K |
| 221L | 24 | 330 | — | — | — | — | — | K | — | — | — | — | — | — | — | V | — | — | — | — | — | — | — |
| 11504 | 11 | 411 | — | — | — | — | — | K | — | — | F | — | — | — | — | — | — | — | — | — | — | — | — |
| 11914 | 7 | 249 | — | — | — | — | — | K | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 13010 | 17 | 315 | — | — | — | — | — | — | — | — | — | — | — | — | — | V | — | — | — | — | — | — | K |
| 13070 | 12 | 222 | — | — | — | — | I | K | — | — | — | — | — | I | — | V | — | — | — | — | — | — | — |
| 15626 | 12 | 195 | — | — | — | — | — | — | — | — | — | — | — | I | — | — | — | — | — | — | — | — | — |
| 18030 | 39 | 420 | — | — | — | — | — | K | — | — | — | — | — | I | — | — | — | — | — | — | — | — | G |
| VI06 | 43 | 498 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | K |
n, number of sequences.
l, sequence length in base pairs.
—, no change.
We next examined whether transmission of T84 variants influences evolution at the population level. We combined sequences from multiple A*0201− patients within a single analysis (accession numbers AY656030 to AY656080) (18). By excluding A*0201+ patients, this ensures that any selection observed is unlikely to be the consequence of multiple independent selection events operating within the patient group. Full-length gag sequences were collected from a total of 36 subtype B-infected patients visiting clinics in and around the London area (18). Of these, 17 were A*0201− and should therefore be incapable of mounting an SL9-restricted response. We tested for positive selection among the A*0201− patients by estimating dN/dS for each codon using the CODEML program (28). A comparison of two models of codon evolution allows investigation of selection pressures based on their fit to the data. In model 8 (M8), codons are distributed into 11 categories of dN/dS, 10 of which have a value of ≤1, with an extra category for which dN/dS is estimated freely and can therefore be >1 (implying positive selection). This was compared to the neutral M8N model in which the 11th dN/dS category is fixed at 1 (implying neutrality), using a likelihood ratio test with one degree of freedom. Evidence for positive selection is provided if M8 is a significantly better description of the data than M8N and dN/dS for the 11th category is >1. The posterior probability that the assignment of sites to this category is correct is estimated using Bayesian inference. To ensure that our analysis is not compromised by frequent recombination (2), we tested for genetic exchange using the GENECONV program prior to selection analysis (24). No evidence for recombination was found in the A*0201− data set (results available on request). We found evidence for positive selection at site T84 (dN/dS = 1.8; posterior probabilities, >0.95; Table 2; the result from the analysis of all 36 patients is also shown). Selection was also observed at Y79, a site which is similarly associated with escape from A*0201 (10). This supports the notion that mutations at these sites are advantageous in A*0201+ patients and sustained in the wider population through interpatient transmission.
TABLE 2.
Interpatient HIV-1 selection analysis
| Subtype B consensus | na | lb | G71 | S72 | E73 | E74 | L75 | R76 | S77 | L78 | Y79 | N80 | T81 | V82 | A83 | T84 | I85 | Y86 | C87 | V88 | H89 | Q90 | R91 | dN/dS | LRTcP value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A*0201+/− | 36 | 1,140 | —d | — | — | — | — | K | — | — | F | — | — | — | — | — | — | — | — | — | — | — | — | 2.26 | <0.05 |
| A*0201− | 17 | 1,140 | — | — | — | — | — | K | — | — | F | — | — | — | — | — | — | — | — | — | — | — | — | 1.81 | <0.05 |
| A*01−; A*0201−; A*1101−; A*3002−; B*0801− | 21 | 1,140 | — | — | — | — | — | K | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 2.43 | <0.05 |
n, number of sequences.
l, sequence length in base pairs.
dN/dS values are only shown in cases where the likelihood ratio test (LRT) of positive-selection model M8, compared to neutral model M8N, yielded a significant result. Positively selected sites, with a >95% posterior probability of being in the selected class of sites, are in boldface type.
—, no change.
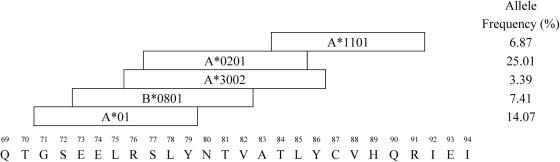
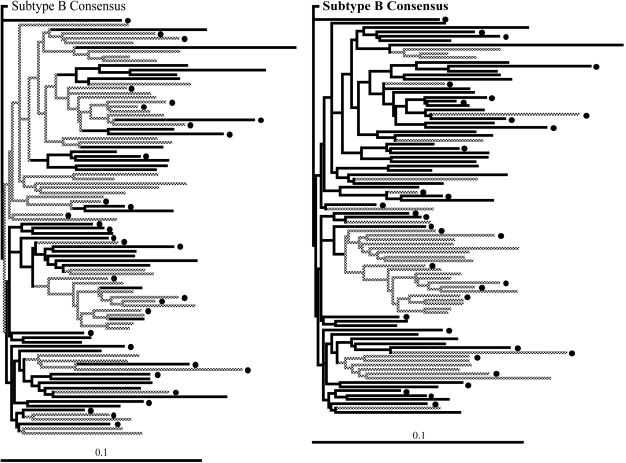
SL9 is also overlapped by epitopes restricted by A*01, A*1101, A*3002, and B*0801 (Fig. 1). To exclude their possible influence, we repeated the analyses using patients negative for all these alleles, as well as A*0201. To obtain the necessary numbers, we combined sequences from multiple cohorts in the United Kingdom, Boston, and Barbados, using 21 patients out of a total of 92 (18). This analysis again revealed evidence for selection at sites Y79 and T84 (dN/dS = 2.4; posterior probabilities, >0.95; Table 2), confirming that the selection observed is operating at a level above the individuals from whom the sequences were derived. To further validate our hypothesis of population level evolution, we tested the assumption of sustained transmission by mapping the selected sites onto a phylogenetic tree. Using the principle of parsimony, we were able to show that changes at both the Y79 and T84 sites were located on branches that precede the divergence between patients, consistent with our assumption that these variants are transmitted (Fig. 2).
FIG. 1.
HLA alleles restricting epitopes within the region of SLYNTVATL. Allele frequencies were obtained from reference 19.
FIG. 2.
Phylogenetic trees showing the evolutionary relationships among 92 patients within the United Kingdom cohort. A dot indicates A*0201+. The trees were rooted using the subtype B consensus sequence. Horizontal branch lengths are drawn to a scale of substitutions per site. The locations of changes within the SLYNTVATL epitope are shown as grey branches. Panels: left, Y79; right, T84.
Although we were able to detect positive selection operating within SL9 at the population level, that A*0201 is the allele responsible for driving this diversification is only clear for T84, since nonsynonymous changes at this site are limited to A*0201+ patients. Nevertheless, the high frequency of A*0201 compared to other alleles that restrict overlapping epitopes (Fig. 1) (19), and the well-documented role of A*0201 in selecting for intrapatient escape through variation in Y79 and T84 (10, 14, 20), is supportive of our hypothesis of positive selection at both sites.
In summary, our analyses demonstrate that escape from the A*0201-restricted CTL response is also having a major effect on evolution within the SL9 epitope at the wider population level. Our study therefore illustrates the potential impact of intrapatient immune selection on the long-term evolution of HIV-1.
Acknowledgments
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimova, M., R. Nielsen, and Z. Yang. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Brander, C., K. E. Hartman, A. K. Trocha, N. G. Jones, R. P. Johnson, B. Korber, P. Wentworth, S. P. Buchbinder, S. Wolinsky, B. D. Walker, and S. A. Kalams. 1998. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Investig. 101:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brander, C., O. O. Yang, N. G. Jones, Y. Lee, P. Goulder, R. P. Johnson, A. Trocha, D. Colbert, C. Hay, S. Buchbinder, C. C. Bergmann, H. J. Zweerink, S. Wolinsky, W. A. Blattner, S. A. Kalams, and B. D. Walker. 1999. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 73:10191-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furutsuki, T., N. Hosoya, A. Kawana-Tachikawa, M. Tomizawa, T. Odawara, M. Goto, Y. Kitamura, T. Nakamura, A. D. Kelleher, D. A. Cooper, and A. Iwamoto. 2004. Frequent transmission of cytotoxic-T-lymphocyte escape mutants of human immunodeficiency virus type 1 in the highly HLA-A24-positive Japanese population. J. Virol. 78:8437-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandhi, R. T., A. Wurcel, E. S. Rosenberg, M. N. Johnston, N. Hellmann, M. Bates, M. S. Hirsch, and B. D. Walker. 2003. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin. Infect. Dis. 37:1693-1698. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Lerma, J. G., S. Nidtha, K. Blumoff, H. Weinstock, and W. Heneine. 2001. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc. Natl. Acad. Sci. USA 98:13907-13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., C. Pasquier, E. C. Holmes, B. Liang, Y. Tang, J. Izopet, K. Saune, E. S. Rosenberg, S. K. Burchett, K. McIntosh, M. Barnardo, M. Bunce, B. D. Walker, C. Brander, and R. E. Phillips. 2001. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol. Lett. 79:109-116. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P. J., A. K. Sewell, D. G. Lalloo, D. A. Price, J. A. Whelan, J. Evans, G. P. Taylor, G. Luzzi, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1997. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J. Exp. Med. 185:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 19.Marsh, S. G. E., P. Parnham, and L. D. Barber. 2000. The HLA facts book. Academic Press, Inc., New York, N.Y.
- 20.Oxenius, A., D. A. Price, A. Trkola, C. Edwards, E. Gostick, H. T. Zhang, P. J. Easterbrook, T. Tun, A. Johnson, A. Waters, E. C. Holmes, and R. E. Phillips. 2004. Loss of viral control in early HIV-1 infection is temporally associated with sequential escape from CD8+ T cell responses and decrease in HIV-1-specific CD4+ 61 and CD8+ T cell frequencies. J. Infect. Dis. 190:713-721. [DOI] [PubMed] [Google Scholar]
- 21.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 22.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut, A. 2002. Se-Al. Sequence alignment editor v2.0a11. [Online.] http://evolve.zoo.ox.ac.uk/.
- 24.Sawyer, S. A. 1999. GENECONV: a computer package for the statistical detection of gene conversion. [Online.] http://www.math.wustl.edu/∼sawyer.
- 25.Swofford, R. 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4 ed. Sinauer Associates, Sunderland, Mass.
- 26.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 27.Walker, B. D., S. Chakrabarti, B. Moss, T. J. Paradis, T. Flynn, A. G. Durno, R. S. Blumberg, J. C. Kaplan, M. S. Hirsch, and R. T. Schooley. 1987. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328:345-348. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biol. Sci. 13:555-556. [DOI] [PubMed] [Google Scholar]