Abstract
Human immunodeficiency virus type 1 (HIV-1) efficiently enters cells of Old World monkeys but encounters a block before reverse transcription. This restriction is mediated by a dominant repressive factor. Recently, a member of the tripartite motif (TRIM) family proteins, TRIM5α, was identified as a blocking factor in a rhesus macaque cDNA library. Among Old World monkey cell lines, the African green monkey kidney cell line CV1 is highly resistant to not only HIV-1 but also simian immunodeficiency virus SIVmac infection. We analyzed TRIM5α of CV1 cells and HSC-F cells, a T-cell line from a cynomolgus monkey, and found that both CV1- and HSC-F-TRIM5αs could inhibit CD4-dependent HIV-1 infection, as well as vesicular stomatitis virus glycoprotein-mediated infection. CV1-TRIM5α could also inhibit SIVmac infection, whereas HSC-F-TRIM5α could not. In the SPRY (B30.2) domain of CV1-TRIM5α, there was a 20-amino-acid duplication that was not present in HSC-F-TRIM5α. A chimeric TRIM5α containing 37 amino acid residues from CV1-TRIM5α, which spanned the 20-amino-acid duplication, in the background of HSC-F-TRIM5α fully gained the ability to inhibit SIVmac infection. Conversely, the mutant CV1-TRIM5α lacking the 20-amino-acid duplication completely lost the ability to restrict SIVmac infection. These findings clearly indicated that a specific region of 37 amino acid residues in the SPRY domain of CV1-TRIM5α contained a determinant of species-specific restriction of SIVmac.
Human immunodeficiency virus type 1 (HIV-1) is thought to have been introduced into the human population from chimpanzees (9) and shows a very narrow host range limited only to humans and chimpanzees. HIV-1 does not experimentally infect Old World monkeys, such as rhesus and cynomolgus monkeys, and fails to replicate in activated CD4-positive T lymphocytes obtained from these monkeys (13, 31). In contrast, simian immunodeficiency virus (SIV) isolated from a macaque monkey (SIVmac) can replicate well in rhesus (13, 31) and cynomolgus monkeys (2, 3). The restricted host range of HIV-1 has greatly hampered its use in animal experiments and, hence, caused difficulty in developing prophylactic vaccines against HIV-1 infection.
Several studies have suggested that the block of HIV-1 replication in Old World monkey cells occurred at a postentry step (7, 13, 31) and appeared to result from a failure to initiate reverse transcription (13). The block was still observed when CD4-negative monkey cells were infected with HIV-1 pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G) and was overridden by a high multiplicity of infection (MOI) with VSV-G-pseudotyped virus or virus-like particles lacking genomic RNA (5, 10, 16, 19). Importantly, resistance against HIV-1 infection was shown to be dominant in heterokaryons between human and Old World monkey cells, suggesting the presence of inhibitory factor(s) against HIV-1 infection in Old World monkey cells (19). Studies on chimera of HIV-1 and SIVmac have suggested that restriction determinants lie within the HIV-1 P24 capsid protein (CA) (11, 23, 24, 29).
Recently, the screening of a rhesus monkey cDNA library identified tripartite motif 5α (TRIM5α), a component of cytoplasmic bodies, as a factor that confers resistance to HIV-1 infection (33). Shortly after, TRIM5α of African green monkey, another Old World monkey, was also shown to restrict HIV-1 infection, whereas human TRIM5α was reported to restrict N-tropic murine leukemia virus (12, 14, 25, 36).
An African green monkey kidney cell line, CV1, was shown to be highly resistant to SIVmac infection, as well as to HIV-1 infection. We analyzed TRIM5α of CV1 cells and HSC-F cells, a T-cell line from a cynomolgus monkey, and report here that the ability of CV1-TRIM5α to suppress SIVmac infection was determined by a small region composed of 37 amino acid residues in the SPRY (B30.2) domain of CV1-TRIM5α.
MATERIALS AND METHODS
Cloning and expression of TRIM5α.
TRIM5α cDNA was amplified by reverse transcription-PCR from the human T-cell line MT4, cynomolgus monkey T-cell line HSC-F (2, 3), and African green monkey cell lines CV1 and Vero by using 5′-GCGGCCGCTACTATGGCTTCTGG-3′ as a forward primer and 5′-GAATTCTCAAGAGCTTGGTGA-3′ as a reverse primer. Amplified products were then cloned into the vector pCR-2.1TOPO (Invitrogen), and the nucleotide sequence of 10 clones for each TRIM5α was determined.
The entire coding regions of selected clones were transferred to pcDNA3.1 (Invitrogen) by using NotI and EcoRI sites, which were introduced by primers used in the PCR step. Hamster TK-ts13 cells (4) were transfected with pcDNA3.1 carrying TRIM5α cDNA and cultured in the presence of 0.75 mg of G418 (Gibco)/ml for 14 days. The colonies obtained were examined for the expression of TRIM5α by using the TaqMan PCR method according to the manufacturer's instructions (Applied Biosystems). Sequences of the probe and primers used to specifically detect each TRIM5α were as follows: MT4-TRIM5, forward primer (5′-AACCTGGAGAAGGAGGAGGAAGAC-3′), reverse primer (5′-CTGGGTCTGCTGCACCAT-3′), and probe (5′-FAM-TCAGTTTCAGAGTTCG-TAMRA-3′); HSC-F-TRIM5, forward primer (5′-AACCTGGAGAAGGAGAAAGAAGAC-3′), reverse primer (5′-CTGGGTCTGCTGCACCAT-3′), and probe (5′-FAM-TTCGTTTCAGACTTTG-TAMRA-3′); and CV1-TRIM5, forward primer (5′-AACCTGGAGAAGGAGGAAGAAGA-3′), reverse primer (5′-CTGGGTCTGCTGCACCAT-3′), and probe (5′-FAM-TCCGTTTCAGACTTCG-TAMRA-3′). These primers amplify the coiled-coil region of TRIM5 genes. Plasmid DNA used for transfection served as a standard to determine the number of copies of TRIM5α transcripts. The parental TK-ts13 cells were totally negative for the primate TRIM5α expression. Clones expressing each TRIM5α at comparable levels (ca. 4 to 6 × 107 copies/μg of total RNA) were used for subsequent experiments.
To generate CV1-TRIM5α and HSC-F-TRIM5α cDNAs carrying a hemagglutinin (HA) tag (YPYDVPDYAA) at their C termini, cloned CV1-TRIM5α and HSC-F-TRIM5α cDNAs in pcDNA3.1 were used as templates for PCR amplification with a primer containing a nucleotide sequence corresponding to the HA tag fused with the C-terminal portion of TRIM5α. The C-terminal portion of TRIM5α fused with the HA tag (BamHI to NotI) and the N-terminal portion of TRIM5α (NotI to BamHI) were assembled on a pCEP4 vector (Invitrogen). To generate chimeric TRIM5α HSC-F+60tag, the 182-bp SphI-BamHI fragment of HSC-F-TRIM5α-tag was replaced with the corresponding 242-bp SphI-BamHI fragment of CV1-TRIM5α in the background of HSC-F-TRIM5α-tag. Conversely, the 242-bp SphI-BamHI fragment of CV1-TRIM5α was replaced with the 182-bp SphI-BamHI fragment of HSC-F-TRIM5α-tag in the background of CV1-TRIM5α-tag to generate CV1-60tag. PCR-based mutagenesis of HSC-F-TRIM5α-tag was performed to generate HSC-delete-tag, which possessed the 5′-proximal 84 bp of the SphI-BamHI fragment of CV1-TRIM5α in the background of HSC-F-TRIM5α-tag, and HSC-insert-tag, which possessed 3′-proximal 158 bp of the SphI-BamHI fragment of CV1-TRIM5α in the background of HSC-F-TRIM5α-tag. Similarly, CV1-delete-tag, which possessed the 3′-proximal 98 bp of the SphI-BamHI fragment of HSC-F-TRIM5α in the background of CV1-TRIM5α-tag, was generated by a PCR-based mutagenesis of CV1-TRIM5α-tag. The entire coding sequences of these TRIM5α-tags were then transferred to the NotI site of pSeV18+b(+). Recombinant Sendai viruses (SeVs) carrying various TRIM5α-tags were recovered according to a previously described method (32). The viruses passaged a second time in embryonated chicken eggs were used as stock for all experiments. The wild-type Z strain of SeV served as a control in all of the experiments.
To establish human cell lines which constitutively express primate TRIM5αs or their chimeras, human osteosarcoma C143 cells were transfected with pCEP4 containing cDNA of CV1-TRIM5α-tag, HSC-F-TRIM5α-tag, CV1-60tag, or HSC-F+60tag, and cells were cultured in the presence of 0.3 mg of hygromycin B (Gibco)/ml for 14 days.
Immunoprecipitation and Western blot analysis.
When we performed Western blot analysis of cells expressing HA-tagged TRIM5α proteins, we consistently observed nonspecific binding of anti-HA antibody to a protein that comigrated with HSC-F-TRIM5α. Therefore, we analyzed the expression of each HA-tagged TRIM5α protein in the hygromycin B-resistant C143 cells or MT4 cells infected with recombinant SeVs by immunoprecipitation, followed by Western blot analysis as described previously (20) to reduce nonspecific background. Briefly, cell lysate was first adsorbed with protein A-agarose before the addition of anti-HA antibody to avoid nonspecific protein binding to protein A-agarose. TRIM5α proteins in the cell lysate were then precipitated with anti-HA high-affinity rat monoclonal antibody (Roche) by using a protein A-immunoprecipitation kit (Roche). Precipitated materials were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 4 to 12% NuPAGE Bis-Tris gel (Invitrogen). Proteins in the gel were then electronically transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore). Blots were blocked and probed with anti-HA antibody overnight at 4°C and then incubated with peroxidase-conjugated anti-rat immunoglobulin G (American Qualex) and developed by using the Immun-Star HRP chemiluminescence kit (Bio-Rad). Visualized image was recorded by LAS1000 (Fuji) and quantified by ImageGauge (Fuji). At least three independent experiments were performed, and the means and standard deviations (SD) for the data were calculated.
Viruses and HIV-1 lentivirus vector.
VSV-G-pseudo typed HIV-1-NL43, SIVmac239, or HIV-2-GH123 was prepared by transfection of 293T cells with a combination of pMD.G (17, 18) and pNL432 (1), pBRmac239 (15), or pGH123 (30), respectively. HIV-1 vector expressing green fluorescence protein (GFP) was prepared as described previously (17, 18). Two days after transfection, culture supernatants of 293T cells were collected and assayed for reverse transcriptase activity using a reverse transcriptase colorimetric assay (Roche).
Viral infection.
Assays for the HIV-1 vector expressing GFP were performed in 24-well plates containing 4 × 104 Tk-ts13-derived target cells. Serially diluted VSV-pseudotyped HIV-1 vectors encoding GFP were inoculated, and infected cells were enumerated by using a flow cytometer (FACScan; Becton Dickinson) 40 h after infection. For VSV-pseudotyped HIV-1, SIVmac239, and HIV-2 infection assays, we inoculated viruses containing 1 ng of reverse transcriptase into 4 × 104 C143 cells. For CD4-dependent infection assays, 2.5 × 105 MT4 cells were infected with SeV expressing CV1-TRIM5α-tag, HSC-F-TRIM5α-tag, or the parental Z strain of SeV at a MOI of 10 PFU per cell, followed by incubation at 37°C for 9 h. Cells were then superinfected with 30 ng of p24 of an X4 HIV-1 strain, NL43, or 30 ng of p27 of SIVmac239. The culture supernatants were collected periodically, and the level of p24 or p27 was measured by using a RETROtek antigen ELISA kit (ZeptoMetrix).
Data deposition.
The sequences described here have been deposited in the GenBank database under accession numbers AB210050 to AB210052.
RESULTS
Variation in TRIM5α.
We cloned TRIM5α cDNA from the human T-cell line MT4, cynomolgus monkey T-cell line HSC-F, and African green monkey kidney cell lines CV1 and Vero. The predicted amino acid sequences of TRIM5αs are compared in Fig. 1A and B.
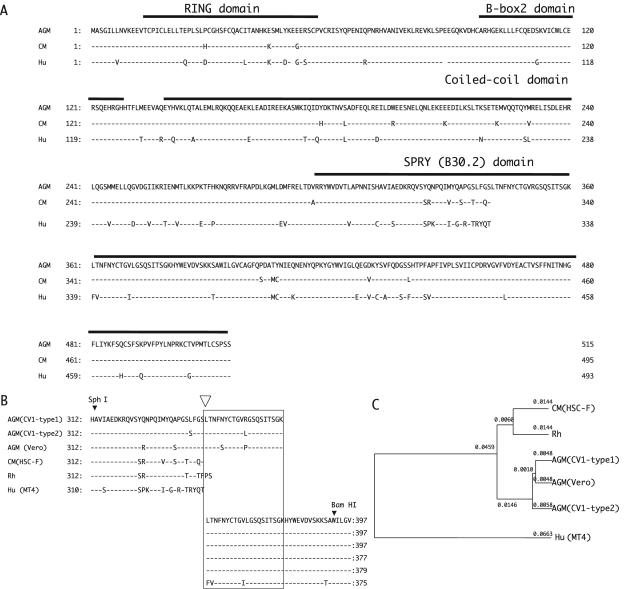
FIG. 1.
(A) Alignment of amino acid sequences of African green monkey (AGM, CV1-TRIM5α-type1), cynomolgus monkey (CM, HSC-F-TRIM5α), and human TRIM5α (Hu, MT4-TRIM5α) predicted from the sequences of the cDNAs used in the present study with key domains indicated. (B) Alignment of amino acid sequences of the highly variable region within the SPRY (B30.2) domain of TRIM5α. The rhesus monkey TRIM5α (Rh) sequence published by Stremlau et al. (33) was added. A box indicates the 20-amino-acid duplication within African green monkey TRIM5α. An open arrowhead denotes the recombination point in chimeric TRIM5αs, HSC-delete-tag, HSC-insert-tag, and CV1-delete-tag (see Fig. 4A). (C) Phylogenetic tree of various TRIM5α sequences produced by the UPGMA (unweighted pair-group method with arithmetic averages) method.
Human TRIM5α from MT4 cells differed at an amino acid position 249 (G249D) from the previously published sequence (33) and was designated MT4-TRIM5α. The cynomolgus monkey TRIM5α from HSC-F (HSC-F-TRIM5α) was two amino acids shorter than the rhesus monkey TRIM5α (33) and two amino acids longer than the human TRIM5α. All 10 clones derived from Vero cells had the same sequence as the previously published one (36). On the other hand, we found at least two distinct TRIM5α sequences in CV1 cells. The two major TRIM5α sequences obtained from CV1 were designated CV1-TRIM5α-type1 and CV1-TRIM5α-type2, and five amino acids were found to differ between the two sequences (I259V, L337S, R351L, G359R, and G438S). Among 10 cDNA clones obtained from CV1, there were four type 1 clones and four type 2 clones. The remaining two clones were most likely chimeric artifacts. Two recently published sequences of TRIM5α from CV1 cells (AY593973 and AY625002) showed differences at three positions—L7V, I259V, and G438S—and have S, L, and R at positions 337, 351, and 359, respectively (14, 36), whereas both the type 1 and 2 clones had leucines at the seventh position. All sequences obtained from Vero and CV1 contained a 20-amino-acid duplication within the SPRY domain, which was not observed in human MT4 and cynomolgus monkey HSC-F (Fig. 1B).
The phylogenetic tree of various TRIM5α sequences showed that cynomolgus and rhesus monkey TRIM5αs are similar to each other, a finding consistent with the fact that these two monkeys belong to the genus Macaca (Fig. 1C).
African green monkey and cynomolgus monkey TRIM5α inhibit HIV-1 infection in nonprimate cells.
We first sought to determine whether or not each TRIM5α can inhibit HIV-1 infection in the context of nonprimate cells because human and primate cells express endogenous TRIM5α that could complicate a functional analysis of TRIM5α-mediated restriction. The hamster cell line TK-ts13 was used, because it is very susceptible to a VSV-G-pseudotyped, HIV-1-based GFP-expressing lentivirus vector, HIV-1-GFP. Cell clones stably expressing MT4-, CV1-, and HSC-F-TRIM5α were selected according to the method described in Materials and Methods. The levels of expression of TRIM5α were determined by using a real-time PCR, and cells expressing comparable amounts of TRIM5α (4 × 106 to 6 × 106 copies/μg of total RNA) were used for subsequent study. Restriction can be quantified by comparing the percentage of GFP-positive cells with or without TRIM5α.
As can be seen in Fig. 2A, MT4-TRIM5α had a very weak anti-HIV-1 effect (∼1.2-fold), a finding consistent with a previous study (33). In contrast, restriction of HIV-1 was clearly evident over a wide range of initial MOIs in cells expressing HSC-F-TRIM5α, CV1-TRIM5α-type1, and CV1-TRIM5α-type2 (ca. 5- to 10-fold). There was no significant difference between CV1-TRIM5α-type1 and type2; therefore, we chose CV1-TRIM5α-type1 to be representative of CV1-TRIM5α in the subsequent experiments. We obtained the same results as described above when we used an HA-tagged version of TRIM5α (data not shown).
FIG. 2.
(A) TK-ts13 cell clones expressing MT4-TRIM5α (□), CV1-TRIM5α-type1 (•), CV1-TRIM5α-type2 (▪), HSC-F-TRIM5α (▴), or empty vector (○) were exposed to the indicated GFP-expressing HIV-1 vector. GFP-positive cells were counted with a flow cytometer. Data typical of at least three independent clones for each TRIM5α are shown. (B) Lysates of MT4 cells infected with recombinant SeV expressing CV1-TRIM5α-tag, HSC-F-TRIM5α-tag, or the parental Z strain were immunoprecipitated by anti-HA antibody. Resultant immunoprecipitates were visualized by Western blotting with an antibody to HA. A representative result of four independent experiments is shown. (C) MT4 cells were mock infected (○), or infected with SeV expressing CV1-TRIM5α-tag (▪), HSC-F-TRIM5α-tag (▵), or the parental Z strain (•). At 9 h after infection, cells were inoculated with an HIV-1 strain, NL43, and culture supernatants were periodically assayed for levels of p24. The datum points are means for triplicate samples with the SD.
African green monkey and cynomolgus monkey TRIM5α inhibit CD4-dependent HIV-1 infection in human cells.
To test the restriction properties of TRIM5α in CD4-dependent HIV-1 infection, we constructed a recombinant SeV expressing TRIM5α fused with the HA tag in the C-terminal of HSC-F-TRIM5α or CV1-TRIM5α (HSC-F-TRIM5α-tag SeV or CV1-TRIM5α-tag SeV). Human T-cell line MT4 cells were first infected with the SeV expressing TRIM5α-tag (Fig. 2B), incubated at 37°C for 9 h, and then infected with an X4-tropic HIV-1 strain NL43. As can be seen in Fig. 2C, both HSC-F-TRIM5α-tag and CV1-TRIM5α-tag completely inhibited HIV-1 replication, whereas MT4 cells infected with SeV empty vector fully supported HIV-1 replication.
Distinct patterns of restriction for SIVmac among nonhuman primate TRIM5α.
In African green monkey CV1 cells, both HIV-1 and SIVmac239 were restricted, whereas only HIV-1 was restricted in cynomolgus monkey HSC-F cells (Fig. 3A). Therefore, we examined whether or not CV1-TRIM5α also could inhibit the replication of SIVmac. In MT4 cells infected with recombinant SeV expressing HSC-F-TRIM5α-tag, the replication of SIVmac239 was not suppressed at all (Fig. 3B), indicating that the HSC-F-TRIM5α showed a similar specificity to rhesus monkey TRIM5α (33). In MT4 cells infected with recombinant SeV expressing CV1-TRIM5α-tag, in contrast, the replication of SIVmac239 was completely suppressed. These results suggested that the distinct sensitivity of African green monkey and cynomolgus monkey cells to HIV-1 and SIVmac infection was, at least partly, determined by TRIM5α.
FIG. 3.
(A) MT4 (○), HSC-F (▵), or CV1 (▪) cells were infected with VSV-pseudotyped NL43 or VSV-pseudotyped SIVmac239, and culture supernatants were periodically assayed for levels of p24 or p27. The datum points are means for triplicate samples with the SD. (B) MT4 cells were mock infected (○) or infected with SeV expressing CV1-TRIM5α-tag (▪) or HSC-F-TRIM5α-tag (▵). At 9 h after infection, cells were inoculated with SIVmac239, and culture supernatants were periodically assayed for levels of p27. The datum points are means for triplicate samples with the SD.
A small region of 37 amino acid residues in the SPRY domain of CV1-TRIM5α determines SIVmac restriction.
A comparison of the human and nonhuman primate TRIM5α sequences showed the presence of a highly variable region in the N-terminal portion of the SPRY domain (Fig. 1A and B). In this region, CV1 and Vero TRIM5α had a 20-amino-acid repetition that was totally absent in HSC-F-TRIM5α. These findings prompted us to test whether this highly variable region of TRIM5α determined the species-specific inhibition of SIVmac infection. We constructed chimeric TRIM5αs from HSC-F-TRIM5α-tag and CV1-TRIM5α-tag by using SphI and BamHI restriction enzyme digestion (Fig. 4A). HSC-F+60tag contained the 242-bp fragment of CV1-TRIM5α in the background of HSC-F-TRIM5α-tag. The reciprocal chimera, CV1-60tag, contained a 182-bp fragment of HSC-F-TRIM5α in the background of the CV1-TRIM5α-tag. In this fragment, the differences between CV1 and HSC-F TRIM5α, including the 20-amino-acid repetition, were located in a small region of 37 amino acid residues (Fig. 1B). Human osteosarcoma C143 cells stably expressing various TRIM5αs (Fig. 4B) were infected with VSV-G-pseudotyped HIV-1 NL43, and levels of P24 in the culture supernatants were assayed periodically. As expected, both chimeric TRIM5αs and parental TRIM5αs clearly inhibited the replication of HIV-1 NL43 (Fig. 4C). When these cells were infected with VSV-G-pseudotyped SIVmac239, the parental CV1-TRIM5α-tag could also inhibit the replication of SIVmac239, whereas CV1-60tag, which contained the 182-bp fragment of HSC-F-TRIM5α in the background of CV1-TRIM5α-tag, could not. Conversely, the parental HSC-F-TRIM5α-tag did not inhibit the replication of SIVmac239 at all, whereas HSC-F+60tag, which contained the 242-bp fragment of CV1-TRIM5α in the background of HSC-F-TRIM5α-tag, clearly inhibited SIVmac239 (Fig. 4C). We obtained the same results as described above when we used SeVs to express parental TRIM5α-tag or chimeras (Fig. 4D). These results indicated that the determinant of the species-specific inhibition of SIVmac239 replication is located in 37 amino acid residues in the SPRY domain of CV1-TRIM5α.
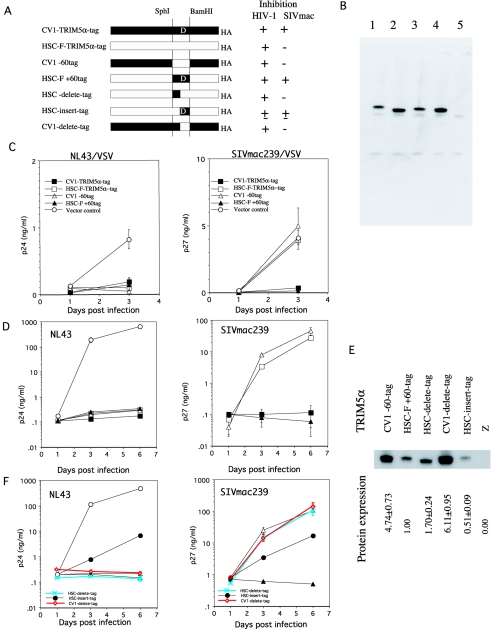
FIG. 4.
(A) Schematic representation of chimeric TRIM5α and summary of the results. Filled and open bars denote CV1 and HSC-F sequences, respectively. D, the CV-1-TRIM5α-specific 20-amino-acid duplication. The +, ±, and − symbols denote full, partial, and nosuppression, respectively. (B) Lysates of C143 cells expressing CV1-TRIM5α-tag (lane 1), HSC-F-TRIM5α-tag (lane 2), HSC-F+60tag (lane 3), CV1-60tag (lane 4), or empty vector (lane 5) were immunoprecipitated by using anti-HA antibody. The resultant immunoprecipitates were visualized by Western blotting with an antibody to HA. A representative result of three independent experiments is shown. (C) C143 cells expressing CV1-TRIM5α-tag (▪), HSC-F-TRIM5α-tag (□), CV1-60tag (▵), HSC-F+60tag (▴), or empty vector (○) were infected with VSV-pseudotyped NL43 or SIVmac239, and culture supernatants were periodically assayed for levels of p24 or p27. The datum points are means for triplicate samples with the SD. (D) MT4 cells infected with SeV expressing CV1-TRIM5α-tag (▪), HSC-F-TRIM5α-tag (□), CV1-60tag (▵), HSC-F+60tag (▴), or empty vector (○) were infected with NL43 or SIVmac239, and culture supernatants were assayed for levels of p24 and p27. The datum points are means for triplicate samples with the SD. (E) Lysates of MT4 cells infected with recombinant SeVs expressing CV1-60tag, HSC-F+60tag, HSC-delete-tag, CV1-delete-tag, HSC-insert-tag, or parental Z strain were immunoprecipitated by using anti-HA antibody. Resultant immunoprecipitates were visualized by Western blotting with an antibody to HA. A representative result of three independent experiments is shown. The relative amounts of TRIM5α protein to that of HSC-F+60tag were calculated, and means and SD values of three independent experiments are shown. (F) MT4 cells infected with SeV expressing CV1-60tag (▵), HSC-F+60tag (▴), HSC-delete-tag (asterisks with blue lines), HSC-insert-tag (•), CV1-delete-tag (diamonds with red lines), or empty vector (○) were infected with NL43 or SIVmac239, and culture supernatants were assayed for levels of p24 and p27. The datum points are means for triplicate samples with the SD.
To narrow the SIVmac restriction determinant more precisely, we generated two more chimeric TRIM5αs (Fig. 4A). HSC-delete-tag contained a CV1-derived 17-amino-acid fragment without the 20-amino-acid duplication in the background of the HSC-TRIM5α-tag. HSC-insert-tag contained the HSC-F-derived 17-amino-acid fragment with the CV-1 derived 20-amino-acid duplication in the background of HSC-TRIM5α-tag. Recombinant SeVs expressing these chimeric TRIM5αs were generated and used in the subsequent experiments. Although the expression levels of mRNA of each chimericTRIM5α was virtually identical to each other (data not shown), we observed considerable variations in the levels of TRIM5α protein expression among chimeras (Fig. 4E). The HSC-delete-tag showed slightly higher levels of protein expression than those of HSC-F+60tag. However, HSC-delete-tag failed to inhibit SIVmac replication, whereas it restricted HIV-1 replication as completely as HSC-F+60tag did (Fig. 4F). These results clearly indicated that the CV1-derived 17-amino-acid region alone was not sufficient for SIVmac restriction. On the other hand, HSC-insert-tag partially restricted both HIV-1 and SIVmac, although this chimera showed lower levels of protein expression than other chimeras did (Fig. 4E and F). These results indicated that the CV1-specific 20-amino-acid duplication was important in SIVmac restriction.
To determine whether the CV1-specific 20-amino-acid duplication was indispensable for SIVmac restriction, we generated CV1-delete-tag, which lacked the 20-amino-acid duplication in the CV1-TRIM5α-tag (Fig. 4A). The protein expression level of CV1-delete-tag was comparable to that of CV1-60tag (Fig. 4E), and CV1-delete-tag inhibited HIV-1 replication as completely as CV1-60tag did. However, CV1-delete-tag was shown to lose the ability to inhibit SIVmac infection (Fig. 4F). Taken together, our data clearly indicated that the 20-amino-acid duplication of CV1-TRIM5α was necessary for SIVmac restriction and suggested that the adjacent 17-amino-acid region of CV1-TRIM5α was also necessary to fully restrict SIVmac infection.
HIV-2 GH123 is sensitive to cynomolgus monkey TRIM5α, as well as African green monkey TRIM5α.
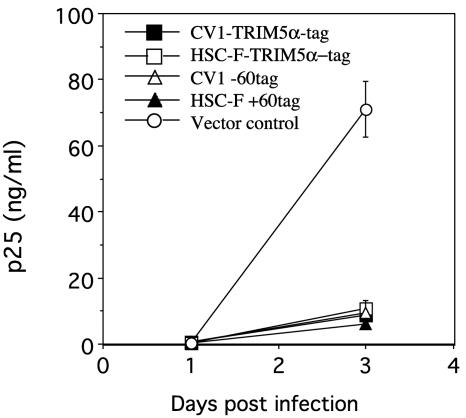
HIV-2 is closely related to SIVmac (9). We tested whether or not the sensitivity of HIV-2 to various TRIM5αs was similar to that of SIVmac239. C143 cells expressing CV1, HSC-F, and their chimeric TRIM5αs were infected with the VSV-G-pseudotyped HIV-2 strain GH123. Surprisingly, HSC-F-TRIM5α-tag inhibited HIV-2 replication as CV1-TRIM5α-tag had done (Fig. 5). Both chimeric TRIM5αs, CV1-60tag and HSC-F+60tag, also inhibited HIV-2 replication to a similar extent (Fig. 5). These results indicated that HIV-2 strain GH123 was sensitive to cynomolgus monkey TRIM5α despite its high level of sequence homology to SIVmac239.
FIG. 5.
C143 cells expressing CV1-TRIM5α-tag (▪), HSC-F-TRIM5α-tag (□), CV1-60tag (▵), HSC-F+60tag (▴), or empty vector (○) were infected with VSV-pseudotyped GH123, and culture supernatants were periodically assayed for levels of p25. The datum points are means for triplicate samples with the SD.
DISCUSSION
In the present study, we showed that both cynomolgus and African green monkey TRIM5αs could inhibit HIV-1 infection. African green monkey TRIM5α could also inhibit SIVmac infection, whereas cynomolgus monkey TRIM5α could not. Experiments on chimeras of the cynomolgus and African green monkey TRIM5αs unequivocally demonstrated that a small region composed of 37 amino acid residues in the SPRY domain of African green monkey TRIM5α was responsible for restricting the SIVmac infection.
A previous study showed that rhesus monkey TRIM5γ, a splicing variant lacking the SPRY domain, did not suppress HIV-1 infection (33). In the case of TRIM7, the SPRY domain alone was sufficient for binding to its ligand glycogenin (38). Deletion of the entire SPRY domain from TRIM11 also abolished its ability to bind Humanin (21). Therefore, it is reasonable to assume that the variable N-terminal region of the SPRY domain of TRIM5α binds to HIV-1 or SIVmac CA protein. This assumption is consistent with the recent findings that in owl monkey cells, HIV-1 infection was restricted by a TRIM5-cyclophilin A fusion protein in which the SPRY domain was replaced with cyclophilin A, since cyclophilin A is a well-known ligand of HIV-1 CA protein (22, 28).
In the attempt to further narrow the SIVmac restriction determinant more precisely, we were able to demonstrate that the African green monkey-specific 20-amino-acid duplication was indispensable for SIVmac restriction and that the adjacent 17-amino-acid region of African green monkey alone was not sufficient. However, HSC-insert-tag carrying the cynomolgus monkey-derived 17-amino acid region with African green monkey-specific 20-amino-acid duplication showed low levels of protein expression and only partial suppression of HIV-1 and SIVmac replication. It is possible that an artificial combination of African green monkey-specific 20-amino-acid duplication with the cynomolgus monkey-derived 17-amino-acid region made TRIM5α molecules unstable. Further studies, including mutational analysis of the African green monkey-specific 17-amino-acid region, are necessary to determine the precise role of this region in SIVmac restriction.
Despite its close similarity to SIVmac, HIV-2 strain GH123 was restricted by cynomolgus monkey TRIM5α, as well as by African green monkey TRIM5α. Although both HIV-2 and SIVmac were considered to come from SIVsm (9), it is possible that HIV-2 has been replicating in the human population in the absence of TRIM5α restriction for a certain period and has lost its ability to escape from cynomolgus monkey TRIM5α. However, it has also been reported that there was a considerable degree of variation in the ability to grow in monkey cells among HIV-2 strains (6, 8, 26). Therefore, it is necessary to examine various HIV-2 strains for their sensitivity to human and monkey TRIM5αs before we can draw a definite conclusion. It would also be interesting to identify specific amino acid changes determining the sensitivity to cynomolgus monkey TRIM5α in viral CA proteins, since nearly 90% of the amino acid residues in SIVmac239 CA protein are conserved in HIV-2GH123.
In CV1 cells, the level of TRIM5 gene expression was ca. 3 × 106 copies/μg of total RNA, a level similar to that observed in other human cell lines examined (data not shown). Although HIV-1 infection was suppressed in hamster TK-ts13 or human C143 cells expressing CV1-TRIM5α, relatively high levels of TRIM5α (nearly 5 × 107 copies/μg of total RNA) appeared to be required for a level of suppression similar to that observed in CV1 cells. One possible explanation for this discrepancy is that certain molecules cooperating with TRIM5α also showed species specificity, and CV1-TRIM5α was not fully supported in hamster and human cells. Because TRIM5 gene products are suspected to be an E3 ubiquitin ligase (35), it is important to identify the E2 ubiquitin-conjugating enzyme interacting with TRIM5α. Alternatively, restriction factors other than TRIM5α may exist in CV1 cells, or certain molecules required for efficient lentivirus infection may be absent in CV1 cells.
After we submitted these findings for publication, small amino acid differences in the SPRY domain between human and rhesus monkey TRIM5αs were reported to determine HIV-1 restriction (27, 34, 37). Our findings are in good agreement with the results of these studies.
Acknowledgments
We thank Setsuko Bando for skillful technical assistance. HSC-F cells were kindly supplied by Hirofumi Akari. pGH123 was a gift from Akio Adachi.
This study was supported by grants from the Human Science Foundation; the Ministry of Education, Culture, Sports, Science, and Technology; and the Ministry of Health, Labour, and Welfare of Japan.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koeing, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., K. Mori, K. Terao, I. Otani, M. Fukasawa, R. Mukai, and Y. Yoshikawa. 1996. In vitro immortalization of Old World monkey T lymphocytes with Herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218:382-388. [DOI] [PubMed] [Google Scholar]
- 3.Akari, H., K. H. Nam, K. Mori, I. Otani, H. Shibata, A. Adachi, K. Terao, and Y. Yoshikawa. 1999. Effects of SIVmac infection on peripheral blood CD4+ CD8+ T lymphocytes in cynomolgus macaques. Clin. Immunol. 91:321-329. [DOI] [PubMed] [Google Scholar]
- 4.Alder, H., C. D. Chang, S. T. Chen, I. Beck, C. Y. Chang, and R. Baserga. 1989. Temporary complementation of temperature-sensitive mutants of the cell cycle by transfection with a wild-type or a mutant cDNA of ADP/ATP translocase. J. Cell Physiol. 141:90-96. [DOI] [PubMed] [Google Scholar]
- 5.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro, B. A., M. Nepomuceno, N. W. Lerche, J. W. Eichberg, and J. A. Levy. 1991. Persistent infection of baboons and rhesus monkeys with different strains of HIV-2. Virology 184:219-226. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita, M., A. Yoshida, A. Sakurai, J. Tatsuki, F. Ueno, H. Akari, and A. Adachi. 2003. Susceptibility of HVS-immortalized lymphocytic HSC-F cells to various strains and mutants of HIV/SIV. Int. J. Mol. Med. 11:641-644. [PubMed] [Google Scholar]
- 9.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 10.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himathongkham, S., and P. A. Luciw. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485-488. [DOI] [PubMed] [Google Scholar]
- 14.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 16.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi, H., M. Takahashi, F. H. Gage, and I. M. Verma. 1997. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc. Natl. Acad. Sci. USA 94:10319-10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama, E. E., Y. Tanaka, Y. Nagai, A. Iwamoto, and T. Shioda. 2004. A CCR2-V64I polymorphism affects stability of CCR2A isoform. AIDS 18:729-738. [DOI] [PubMed] [Google Scholar]
- 21.Niikura, T., Y. Hashimoto, H. Tajima, M. Ishizaka, Y. Yamagishi, M. Kawasumi, M. Nawa, K. Terashita, S. Aiso, and I. Nishimoto. 2003. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur. J. Neurosci. 17:1150-1158. [DOI] [PubMed] [Google Scholar]
- 22.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putkonen, P., B. Bottiger, K. Warstedt, R. Thorstensson, J. Albert, and G. Biberfeld. 1989. Experimental infection of cynomolgus monkeys (Macaca fascicularis) with HIV-2. J. Acquir. Immune. Defic. Syndr. 2:366-373. [PubMed] [Google Scholar]
- 27.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 29.Shibata, R., M. Kawamura, H. Sakai, M. Hayami, A. Ishimoto, and A. Adachi. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65-:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata, R., T. Miura, M. Hayami, K. Ogawa, H. Sakai, T. Kiyomasu, A. Ishimoto, and A. Adachi. 1990. Mutational analysis of the human immunodeficiency virus type 2 (HIV-2) genome in relation to HIV-1 and simian immunodeficiency virus SIV (AGM). J. Virol. 64:742-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata, R., H. Sakai, M. Kawamura, K. Tokunaga, and A. Adachi. 1995. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt. 11):2723-2730. [DOI] [PubMed] [Google Scholar]
- 32.Shioda, T., E. E. Nakayama, Y. Tanaka, X. Xin, H. Liu, A. Kawana-Tachikawa, A. Kato, Y. Sakai, Y. Nagai, and A. Iwamoto. 2001. Naturally occurring deletional mutation in the C-terminal cytoplasmic tail of CCR5 affects surface trafficking of CCR5. J. Virol. 75:3462-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 34.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, L., L. Yang, P. K. Moitra, K. Hashimoto, P. Rallabhandi, S. Kaul, G. Meroni, J. P. Jensen, A. M. Weissman, and P. D'Arpa. 2003. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp. Cell Res. 288:84-93. [DOI] [PubMed] [Google Scholar]
- 36.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 38.Zhai, L., A. Dietrich, A. V. Skurat, and P. J. Roach. 2004. Structure-function analysis of GNIP, the glycogenin-interacting protein. Arch. Biochem. Biophys. 421:236-242. [DOI] [PubMed] [Google Scholar]