Abstract
Ubiquitin is important for the release of human immunodeficiency virus type 1 (HIV-1) and several other retroviruses, but the functional significance of Gag ubiquitination is unknown. To address this problem, we decided to analyze Gag ubiquitination in detail. A low percentage of the HIV-1 p6 protein has previously been shown to be ubiquitinated, and published mutagenesis data suggested that Gag ubiquitination is largely lost upon mutation of the two lysine residues in p6. In this study, we show that Gag proteins lacking the p6 domain or the two lysine residues within p6 are ubiquitinated at levels comparable to those of the wild-type Gag protein. We detected monoubiquitinated forms of the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins in mature virus preparations. Protease digestion of Gag polyproteins extracted from immature virions indicated that ubiquitinated MA, CA, and possibly NC are as abundant as ubiquitinated p6. The HIV-1 late-domain motifs PTAP and LRSLF were not required for Gag ubiquitination, and mutation of the PTAP motif even resulted in an increase in the amount of Gag-Ub conjugates detected. Finally, at steady state, ubiquitinated Gag proteins were not enriched in either membrane-associated or virus-derived Gag fractions. In summary, these results indicate that HIV-1 Gag can be monoubiquitinated in all domains and that ubiquitination of lysine residues outside p6 may thus contribute to viral release and/or infectivity.
Retrovirus assembly is driven by the Gag polyprotein, which can form virus-like particles (VLPs) and bud from cells in the absence of other viral components (5, 32). During or after virus release, Gag is cleaved by the viral protease (PR) into the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, which are common to all retroviruses. Proteolytic processing of human immunodeficiency virus type 1 (HIV-1) Gag additionally yields the C-terminal p6 protein and two small spacer peptides (SP1 and SP2, which separate CA from NC and NC from p6, respectively) (Fig. 1). Retroviral assembly can be subdivided into the distinct stages of Gag membrane targeting, virus bud formation and induction of membrane curvature, and release of the newly assembled virus bud by a membrane fission event (5, 6, 20, 32). Severing the virus bud from the cell surface depends on viral late domains within Gag (6, 20). So far, three different motifs are known to have late-domain function: P(T/S)AP, YPXL/LXXLF, and PPXY. HIV-1 Gag carries a PTAP motif as well as an LRSLF motif within its C-terminal p6 domain but does not contain a PPXY motif. Other retroviruses, like Rous sarcoma virus (RSV), murine leukemia virus (MLV), Mason-Pfizer monkey virus (M-PMV), and human T-cell leukemia virus type 1, and also unrelated enveloped viruses, such as Ebola virus and vesicular stomatitis virus, have been found to mainly require a PPXY motif for budding, sometimes in combination with a P(T/S)AP motif (6, 20).
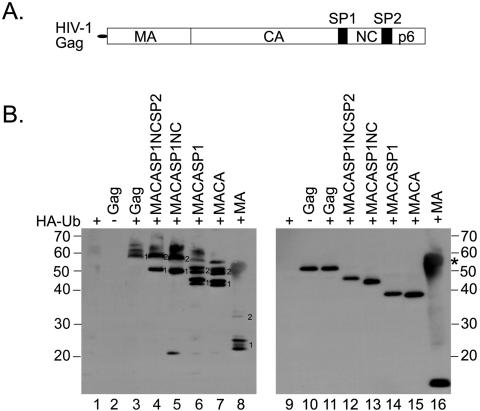
FIG. 1.
(A) Schematic representation of the HIV-1 Gag polyprotein. (B) Ubiquitination of C-terminally truncated Gag proteins. Truncated Gag proteins were expressed from pcDNA3-based Rev-independent Gag expression constructs. Proteins are designated by the domains that are present in the protein (panel A). The indicated proteins were expressed in the presence (+) or absence (−) of HA-Ub in 293T cells and, 48 h after transfection, immunoprecipitated with sheep anti-CA antiserum (lanes 1 to 7) or rabbit anti-MA antiserum (lane 8). The immunoprecipitates were examined by Western blotting with an anti-HA antibody to visualize Gag-HA-Ub conjugates (left). Proteins that are presumably modified by one or two ubiquitin moieties are marked 1 and 2, respectively. To visualize the recovered Gag proteins, the blot was reprobed with rabbit anti-MA antibodies (right). Since MA was immunoprecipitated with the same antibody, the heavy chain was also detected on the blot in this case (lane 16, marked by an asterisk). Molecular mass standards (in kDa) are shown at the side of each panel.
P(T/S)AP motifs function by recruiting the cellular ESCRT-I (endosomal complex required for transport I) complex through interaction with its component TSG101 (8, 19, 34). YPXL/LXXLF motifs function by binding to AIP1 (18, 30, 35). Both AIP1 and the ESCRT-I complex are components of a multiprotein network (initially described as class E vacuolar protein sorting proteins in yeast) that is required for the sorting of ubiquitinated membrane proteins into internal vesicles of the multivesicular body. AIP1 and ESCRT-I also interact with each other (18, 30, 35). The functional binding partners of PPXY motif late domains, on the other hand, appear to be ubiquitin (Ub) E3 ligases of the Nedd4 family (4, 11, 14, 33, 36). It is currently not clear, however, how the recruitment of these different proteins facilitates virus budding. Inserting a PPXY domain into the HIV-1 Gag protein, which does not normally contain this motif, induces ubiquitination of Gag (17, 29). Whether the function of Nedd4-like proteins in the release of viruses that contain a PPXY motif involves ubiquitination of Gag is not known.
There are several lines of evidence suggesting that Ub plays a role in the assembly of retroviruses, irrespective of the presence of a Ub ligase-recruiting PPXY motif. Different retroviruses (i.e., HIV, RSV, MLV, and simian immunodeficiency virus) incorporate unconjugated Ub into their virus particles at levels of up to 10% of Gag (22, 26). Furthermore, monoubiquitination of 2 to 5% of the virion-associated HIV-1 p6 and MLV p12 proteins has been reported (22). The equine infectious anemia virus (EIAV) p9 and mouse mammary tumor virus (MMTV) p8 and p14 proteins have also been shown to be ubiquitinated in the virus (23). Mutation of the target lysine residues in HIV-1 p6 and MLV p12 had no effect on virus release or replication, however, and did not result in a loss of incorporation of unconjugated Ub. Ubiquitination of HIV-1 Gag, on the other hand, was reported to be lost when the two lysine residues within p6 were altered (21). Several groups have shown that inhibition of the proteasome blocks release of HIV-1 and other retroviruses (MLV, M-PMV, HIV-2, simian immunodeficiency virus, and RSV), and this is possibly due to altered turnover of Ub (23, 25, 27). The proteasome inhibitor block to RSV release could be partially overcome by direct fusion of Gag with ubiquitin, suggesting that Gag ubiquitination might be important (25). However, there is no clear correlation between ubiquitination of Gag and sensitivity to proteasome inhibitors, since the Gag proteins of EIAV and MMTV are also ubiquitinated but insensitive to proteasome inhibition (23, 24). A role for ubiquitin in retrovirus release is also supported by the observation that overexpression of ubiquitin variants carrying mutations of residues important for ubiquitin function in endocytosis inhibited HIV-1 release (31). It remains unclear, however, whether the observed effects of proteasome inhibition or ubiquitin mutation are consequences of their interference with Gag ubiquitination or whether they may be due to effects on other ubiquitinated proteins. The functional consequences of Gag ubiquitination are also unknown. In vitro, the affinity of TSG101 for HIV-1 p6 is enhanced when p6 is expressed in C-terminal fusion with ubiquitin, suggesting that p6 ubiquitination might enhance the interaction between TSG101 and p6 (8).
The machinery that catalyzes Gag ubiquitination has not been identified. Since the PPXY motif recruits a Ub E3 ligase and can induce ubiquitination of HIV-1 Gag, which normally lacks a PPXY motif, it seems likely that ubiquitination of PPXY-containing Gag proteins is induced by Nedd4-like proteins (17, 31). It is unclear, however, how ubiquitination of Gag proteins lacking a PPXY motif is mediated and whether the P(T/S)AP motif is involved. Strack et al. reported that ubiquitination of an HIV-1 Gag-derived protein in which the entire NCSP2 domain had been replaced by the GCN4 leucine zipper was greatly stimulated when the p6 domain was present (29, 31). In contrast, Martin-Serrano et al. recently showed that the levels of Gag-Ub conjugates were increased when the PTAP motif was mutated (17).
In this paper, we analyze HIV-1 Gag ubiquitination in detail and show that Gag proteins lacking either lysine residues within p6 or the entire p6 domain are still ubiquitinated. In the mature virus, all major Gag domains (MA, CA, and NC) were found to be ubiquitinated at low levels. Our experiments also show that the HIV-1 PTAP and LRSLF motifs are not required for Gag ubiquitination and that, at steady state, ubiquitinated Gag proteins are not enriched in membrane- or virus-derived Gag fractions.
MATERIALS AND METHODS
Expression constructs and cloning procedures.
Hemagglutinin (HA)-Ub was expressed from a pBJ5-based HA-Ub expression vector (see Fig. 6) (31) or from pHA-Ub (see other figures). For the construction of pHA-Ub, the XhoI-EcoRI fragment of the pBJ5-based construct was subcloned into pcDNA3.1Zeo(−) (Invitrogen). pcDNA3-based Rev-independent Gag expression constructs were cloned by PCR amplification of the desired Gag portions with flanking BamHI and XbaI sites using pGag-EGFP (12) as a template. The PCR products were cut with BamHI and XbaI and cloned into the corresponding sites of pcDNA3. The late-domain mutations were introduced by PCR using wild-type (wt) or PTAP mutant (PTAP mutated to LIRL) pNL4-3 as a template (2, 13). The LRSLF motif was mutated in the wt or PTAP mutant context to ARSAA by PCR. For all late-domain mutants, PCR fragments were cut with ApaI and XbaI and cloned into the equivalent sites of pcDNA3Gag. The MA-green fluorescent protein (GFP) expression construct was kindly provided by T. Hope.
FIG. 6.
Ubiquitination of virus-derived HIV-1 Gag proteins. 293T cells were cotransfected with pΔR and pcDNA3 (lanes 1 and 3) or pΔR and an HA-Ub expression plasmid (lanes 2 and 4). Forty-eight hours after transfection, virions were pelleted through sucrose cushions, lysed in RIPA buffer, and immunoprecipitated with antiserum against MA (A), CA (B), or NC (C). Immunoprecipitates were resolved by SDS-PAGE and subjected to Western blotting with anti-HA antibodies (left) or HIV-1-specific antibodies (right). HA-Ub-modified proteins are identified at the sides of the left panels. Molecular mass standards (in kDa) are shown on the left side of each panel. Bands marked by an asterisk probably represent diubiquitinated versions of the respective HIV-1 proteins. LC indicates detection of the light chain of the antibody used for immunoprecipitation.
pΔR is based on the previously described pNLC4-3tr− (3), which contains all HIV-1 open reading frames under the control of the cytomegalovirus promoter. In contrast to pNLC4-3tr−, pΔR contains intact rev and tat open reading frames and carries a primer binding site deletion and a reverse transcriptase (RT) active-site mutation to render it noninfectious. To introduce the primer binding site deletion, pNLC4-3tr− was partially digested with EheI and subsequently cut with BssHII. The 5′ overhang of the BssHII site was filled with Klenow enzyme, and the vector was religated. The deletion (corresponding to nucleotides 640 to 711 of pNL4-3 [2]) was confirmed by sequencing. To repair the rev and tat reading frames, the mutated fragment was exchanged for the NL43 wt sequence using ApaI and NheI sites. Finally, RT was inactivated by two point mutations within its active site (D185D186 mutated to GA) by PCR. The mutated PCR product was reintroduced into the vector using SdaI and AgeI sites.
pΔR/PR(−) and the PTAP mutant pΔR/PP(−)/PR(−) were cloned by replacement of the wt ApaI-SdaI fragments with fragments from previously described mutated constructs (13, 16). For the construction of pΔR/p6(KR)/PR(−), the p6 mutant ApaI/SdaI fragment was subcloned from a previously described plasmid (21), and subsequently, the PR active-site mutation was introduced by PCR. All PCR-amplified inserts were confirmed by sequencing. Detailed cloning procedures and primer sequences are available on request.
Transfection, virion preparation, immunoprecipitation, and p24 ELISA.
293T cells were maintained and transfected using the calcium phosphate precipitation method as described previously (9). For cotransfections, the amount of total plasmid used for each sample was matched using empty vector (pcDNA3). VLPs were harvested by pelleting them through sucrose cushions as previously described for M-PMV (9), or virus was pelleted through 300 μl sucrose in a TLA45 rotor (119,000 × g; 30 min; 4°C) (see Fig. 3). For immunoprecipitation, cells were lysed on ice with RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 5 mM N-ethyl-maleimide [NEM], and protease inhibitors). The lysate was cleared by centrifugation (15,000 × g; 10 min; 4°C) and immunoprecipitated with protein A-sepharose (Amersham) and the indicated antiserum for 2 h at 4°C. In the case of NC, protein G-sepharose (Amersham) was used for immunoprecipitation. Immunoprecipitates were washed three times with RIPA buffer and once with TNE buffer (RIPA buffer without detergents), boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, and resolved by SDS-PAGE. In some cases (see Fig. 4 and 9), cells and/or virions were lysed at 95°C in a buffer containing 50 mM Tris [pH 8.0], 150 mM NaCl, and 1% SDS and boiled for 10 min at 95°C. Then, the lysate was adjusted to 1× RIPA buffer (by 10-fold dilution with 50 mM Tris [pH 8.0], 150 mM NaCl, 1.1% NP-40, 0.55% sodium deoxycholate), cleared by centrifugation (15,000 × g; 10 min; 4°C) and used for immunoprecipitation as described above. p24 enzyme-linked immunosorbent assay (ELISA) was carried out as described previously (15). For the detection of immature Gag proteins, samples were boiled in protein sample buffer before analysis by ELISA.
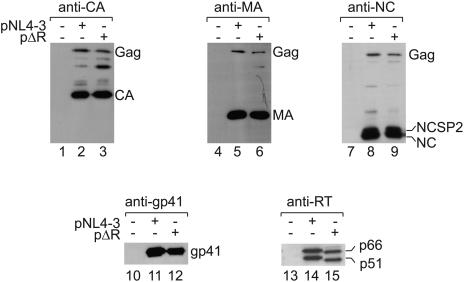
FIG. 3.
Comparison of the expression levels of viral proteins from pNL4-3 and pΔR. 293T cells were transfected with pcDNA3 (left lane in each blot), pNL4-3 (middle lane), or pΔR (right lane). Virions were pelleted through sucrose cushions and examined by Western blotting using anti-CA (top left), anti-MA (top middle), anti-NC (top right), anti-gp41 (lower left), or anti-RT (lower right) antibodies. The variant RT proteins expressed from pΔR travel slightly faster than the wild-type RT proteins (compare lane 15 to lane 14). HIV-1-specific proteins are identified on the right side of each panel.
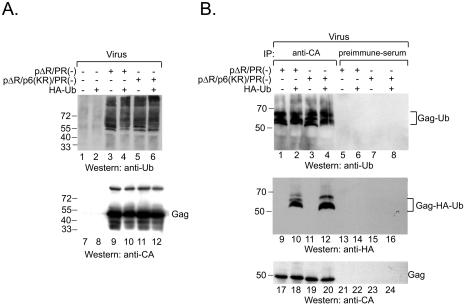
FIG. 4.
HIV-1 Gag lacking lysine residues in p6 is ubiquitinated. (A) The wild type (lanes 3, 4, 9, and 10) or p6 lysine mutant (lanes 5, 6, 11, and 12) pΔR/PR(−) was cotransfected with pHA-Ub as indicated (+). Virions were pelleted through sucrose cushions and directly analyzed by SDS-PAGE and Western blotting with an anti-Ub antibody (top). The blot was reprobed with anti-CA antibody (bottom). Molecular mass standards (in kDa) are shown at the side of each panel. (B) Transfection and virion preparation were carried out as described for panel A, but the virus pellets were resuspended in a buffer containing 1% SDS and boiled for 10 min. Subsequently, the buffer was adjusted to RIPA buffer and samples were split into two aliquots, one of which was immunoprecipitated (IP) with anti-CA antiserum (lanes 1 to 4, 9 to 12, and 17 to 20) while the other was immunoprecipitated with preimmune serum (lanes 5 to 8, 13 to 16, and 21 to 24). Gag samples were adjusted to equal amounts of Gag (as quantified by p24 ELISA), and the whole precipitates were analyzed in the case of the negative controls. Immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with anti-Ub antibodies (top). The membrane was stripped and consecutively reprobed with anti-HA (middle) and anti-CA (bottom) antibodies.
FIG. 9.
Ubiquitinated Gag is not enriched in the virus. 293T cells were cotransfected with pHA-Ub and ΔR/PR(−). Forty-eight hours after transfection, virions were pelleted through sucrose cushions and cells were detached from the culture dish and washed with PBS. Virus and cell pellets were boiled in a buffer containing 1% SDS for 10 min. The cell and virus lysates were adjusted to RIPA buffer, cleared, and immunoprecipitated with anti-CA antiserum. Samples were normalized for equal CA contents (by Western blotting with anti-CA antibody) (bottom) and analyzed by Western blotting with an anti-HA antibody (top).
In vitro PR cleavage.
Virions were pelleted through sucrose cushions as described above and resuspended in PR cleavage buffer (25 mM MES [morpholineethanesulfonic acid, pH 6.5], 250 mM NaCl, 50 μM ZnCl2, 10% glycerol, 1 mM dithiothreitol). Virions were lysed by adding Triton X-100 to a final concentration of 0.1%. HIV-1 PR was expressed and purified as described previously (15). Digestion was carried out using 120 nM PR for 1 hour at room temperature. In vitro processing was terminated by adding SDS-PAGE sample buffer and boiling the samples.
Membrane flotation assay.
Forty-eight hours after transfection, cells were detached with 10 mM EDTA in phosphate-buffered saline (PBS), washed once with PBS, and swollen on ice in hypotonic buffer (10 mM Tris [pH 8.0], 1 mM MgCl2, 5 mM NEM, and protease inhibitors) for 15 min. The cells were disrupted by Dounce homogenization and cleared by centrifugation (1,000 × g; 10 min; 4°C). The supernatant was adjusted to 40% OptiPrep (Progen), and 900 μl was placed at the bottom of a SW60 centrifuge tube and successively overlaid with 2.5 ml 28% Optiprep in TNE buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) and TNE buffer. The gradient was centrifuged at 165,000 × g in an SW60 rotor for 3 h at 4°C. The visible membrane-containing fraction and the bottom fraction containing soluble proteins were collected, diluted with RIPA buffer, and immunoprecipitated with anti-CA antibodies or analyzed directly by SDS-PAGE. All buffers used for flotation and immunoprecipitation contained 5 mM NEM.
Western blotting and antibodies.
Western blotting was performed using standard procedures with the indicated antibody dilutions. Rabbit anti-GFP, rabbit anti-CA (1:5,000), sheep anti-CA (1:5,000), rabbit anti-RT (1:2,000), and rabbit anti-MA (1:2,000) were raised against recombinant proteins. Goat anti-NC (1:5,000) was obtained from the AIDS Vaccine Program, SAIC-Frederick, Inc., Frederick, Md. Monoclonal rat anti-HA antibodies (1:500; clone 3F10) were purchased from Roche, mouse anti-GFP (1:1,000) from Santa Cruz, and anti-ubiquitin antibody (1:1,000; clone FK2) from Biomol. Chessie-8 anti-gp41 hybridoma supernatant was used undiluted (1). HIV-1 p24 hybridoma (183-H12-5C; 1:50-diluted culture supernatant) was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program.
RESULTS
Ubiquitination of Gag domains.
Previously published reports showed that the two lysine residues in the late-domain-containing p6 region of HIV-1 Gag are monoubiquitinated at low levels (21, 22). We noticed that the expression in 293T cells of several truncated Gag proteins lacking p6 yielded protein profiles that were strikingly similar to those of monoubiquitinated proteins, with a minor fraction of the protein shifted in apparent molecular mass by approximately 6 to 8 kDa (data not shown). To investigate whether subdomains of Gag can indeed serve as substrates for monoubiquitination, we coexpressed C-terminally truncated HIV-1 Gag proteins with HA-tagged ubiquitin (HA-Ub) (Fig. 1). Forty eight hours after transfection, Gag-derived proteins were immunoprecipitated with antiserum against CA or, in the case of MA, with antiserum against MA. Specifically precipitated proteins were then examined for modification by HA-Ub using Western blotting with an anti-HA antibody (Fig. 1B).
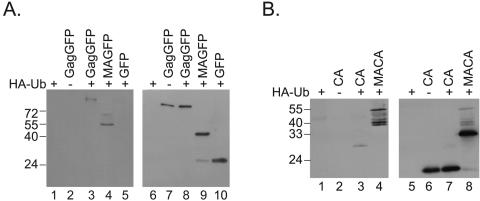
When HA-Ub or Gag was expressed alone, no signal for HA-Ub was detected in the precipitate (Fig. 1B, lanes 1 and 2). However, upon coexpression of full-length or C-terminally truncated Gag proteins with HA-Ub, modified Gag proteins were detected in each case (lanes 3 to 8). While the amounts of HA-Ub modified proteins seemed to be comparable for the truncated constructs, the signal for full-length ubiquitinated Gag was reproducibly weaker than that for ubiquitinated Gag proteins lacking p6 (compare lanes 4 to 7 to lane 3). The blotting membrane was reprobed with anti-MA antibodies to visualize the immunoprecipitated Gag proteins (right blot, lanes 9 to 16). The majority of the modified Gag proteins migrated with a 6- to 8-kDa difference relative to the unmodified protein or to each other, consistent with modification by one or two Ub molecules (Fig. 1B, left, bands marked 1 or 2). In the case of MA, MACA, or MACASP1, however, some of the bands detected by the anti-HA antibody were clearly not spaced at the expected intervals (Fig. 1B, lanes 6 to 8). These bands, therefore, might represent different isomers of mono- or diubiquitinated forms (different sites of Ub attachment), which can run differently in SDS-PAGE. Alternatively, the detection of double bands might be due to another covalent modification, such as phosphorylation, of the protein. Similar patterns of ubiquitination have been described previously (7). We conclude that C-terminally truncated Gag proteins containing at least the MA domain have the potential to become ubiquitinated when expressed in cell culture. We confirmed this result using an MA-GFP expression construct (Fig. 2A). In this experiment, ubiquitination of Gag-GFP and MA-GFP was observed (lanes 3, 4, 8, and 9), while no ubiquitinated GFP was detected (lanes 5 and 10), even though GFP contains 20 lysine residues.
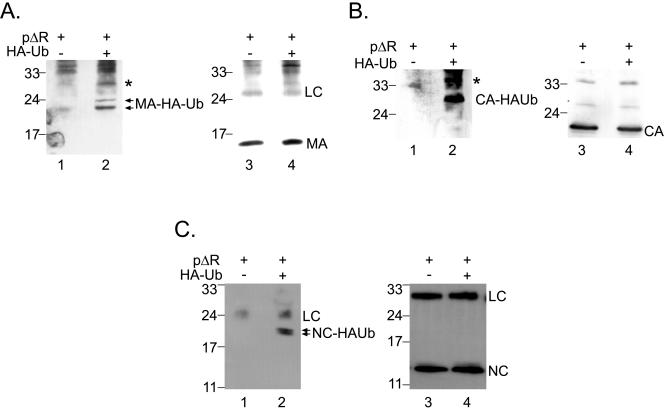
FIG. 2.
(A) Ubiquitination of MA-GFP. Gag-GFP, MA-GFP, or GFP alone was expressed in the presence (+) or absence (−) of HA-Ub as indicated. Cell lysates were immunoprecipitated with rabbit anti-GFP antiserum and probed for HA-Ub modification of precipitated proteins by Western blotting with anti-HA antibodies (left). The blot was reprobed using a monoclonal anti-GFP antibody (right). Molecular mass standards (in kDa) are shown on the left side. (B) Ubiquitination of CA. CA or MACA was expressed from pcDNA3-based, Rev-independent expression constructs in the presence (+) or absence (−) of HA-Ub as indicated. Immunoprecipitation was performed using rabbit anti-CA antiserum. HA-Ub-modified proteins were detected by Western blotting with an anti-HA antibody (left). The blot was reprobed with anti-CA antibodies (right). Molecular mass standards (in kDa) are shown on the left.
We next wanted to test whether Gag domains other than MA might also be ubiquitinated. For this purpose, we expressed CA or MACA in the presence or absence of HA-Ub (Fig. 2B). The cell extracts were immunoprecipitated with anti-CA antibodies, and bound material was examined for HA-Ub modification using Western blotting with an anti-HA antibody. This experiment showed that CA can also be monoubiquitinated in the absence of other Gag domains (Fig. 2B, lanes 3 and 7). However, the expression of MACA typically yielded much higher levels of ubiquitination than expression of CA alone (Fig. 2B, compare lane 4 to lane 3). Therefore, MA and CA have the potential for modification by ubiquitin when expressed alone, but there appears to be enhanced ubiquitination for MACA.
Ubiquitination of HIV-1 Gag proteins lacking lysine residues in p6.
We next determined whether our observation that domains other than p6 are ubiquitinated also holds true for the full-length Gag polyprotein expressed in the viral context. Since the biochemical analysis of ubiquitinated Gag proteins requires large quantities of particles, we constructed an HIV-1 expression construct giving rise to noninfectious VLPs in order to avoid taking biosafety measures for infectious HIV. This plasmid (pΔR) retains all open reading frames and expresses authentic viral transcripts in the correct ratios under the control of the cytomegalovirus promoter. It carries a primer binding site deletion, as well as two point mutations in the active site of the RT coding region, and the resulting virions are therefore released normally but are defective in reverse transcription and noninfectious. To test whether the expression of viral proteins from this construct exhibits patterns similar to expression from a wild-type provirus, 293T cells were transfected with empty vector (pcDNA3), the proviral plasmid pNL43 (2), or pΔR (Fig. 3). Virions were collected by centrifugation at 24 h after transfection and analyzed by Western blotting for the Gag cleavage products CA (lanes 1 to 3), MA (lanes 4 to 6), and NC (lanes 7 to 9); for the viral transmembrane glycoprotein gp41 (lanes 10 to 12); and for RT (lanes 13 to 15). From this analysis, as well as from electron microscopy, pulse-chase experiments, and mutagenesis data (late-domain mutations), we conclude that virus assembly following pΔR transfection is similar to pNL4-3 transfection.
Previously, Ott et al. reported that a small percentage of p6 is ubiquitinated both in the mature and in the immature virion and that Gag ubiquitination is lost upon mutation of the two lysine residues within p6 (21, 22). Since we observed that Gag domains other than p6 have the potential to be ubiquitinated, we wanted to test whether HIV-1 Gag proteins lacking lysine residues in p6 are still ubiquitinated. PR active-site-mutated variants of either pΔR [designated pΔR/PR(−)] or of a derivative in which both lysine codons in p6 were exchanged for arginine codons [pΔR/p6(KR)/PR(−)] were transfected into 293T cells, either together with pcDNA3 or with pHA-Ub (Fig. 4). Forty-eight hours after transfection, the particles were centrifuged through sucrose cushions and either directly analyzed by SDS-PAGE and Western blotting using antibodies against ubiquitin (Fig. 4A) or first subjected to immunoprecipitation with anti-CA antiserum (Fig. 4B) and then analyzed by Western blotting against ubiquitin. When virions were analyzed directly, Western blotting with anti-ubiquitin (Fig. 4A, top) or anti-HA (not shown) antibodies revealed that there was no significant difference in ubiquitinated products independent of the presence (Fig. 4A, lanes 3 and 4) or absence (Fig. 4A, lanes 5 and 6) of Lys residues in p6. Furthermore, ubiquitination of Gag polyproteins was not significantly altered when HA-Ub was coexpressed (Fig. 4A, compare lanes 3 and 5 with lanes 4 and 6). To confirm that particle-associated ubiquitinated proteins indeed represent modified Gag polyproteins, Gag products were immunoprecipitated from SDS-denatured concentrated particles and analyzed by immunoblotting. In the case of immunoprecipitation with anti-CA antiserum, the amounts of Gag were normalized by p24 ELISA before further analysis, while the whole samples were analyzed in the case of immunoprecipitation with preimmune serum. Immunoprecipitates were resolved by SDS-PAGE and subjected to Western blotting with anti-Ub (Fig. 4B, top) and anti-HA (Fig. 4B, middle) antibodies. This experiment confirmed that Gag polyproteins containing or lacking lysine residues in p6 were ubiquitinated at equal levels (Fig. 4B, top, compare lanes 1 and 2 with lanes 3 and 4), while the pattern of ubiquitinated products was slightly altered by the p6 mutation (Fig. 4B, top, compare lanes 3 and 1). All major products with apparent molecular masses between 55 and 72 kDa that were reactive with antibodies against ubiquitin were immunoprecipitated with antiserum against CA (Fig. 4B, lanes 1 to 4), but not preimmune serum (Fig. 4B, lanes 5 to 8). Furthermore, the pattern of Gag ubiquitination observed after coexpression of HA-Ub was virtually indistinguishable from that observed with endogenous ubiquitin (Fig. 4B, compare lane 2 to lane 1 and lane 4 to lane 3). Stripping and reprobing of the blotting membrane with an anti-HA antibody (Fig. 4B, middle) showed that the bands detected by Western blotting with anti-Ub antibodies could, as expected, also be detected with anti-HA antibodies. Reprobing the membranes from Fig. 4A and B with antiserum against CA (Fig. 4A and B, bottom) confirmed that comparable levels of Gag were loaded in each case. Similar results were obtained using cell-derived Gag proteins (data not shown). Therefore, we conclude that HIV-1 Gag proteins lacking lysine residues in p6 are still ubiquitinated.
Ubiquitination of Gag proteins carrying late-domain mutations.
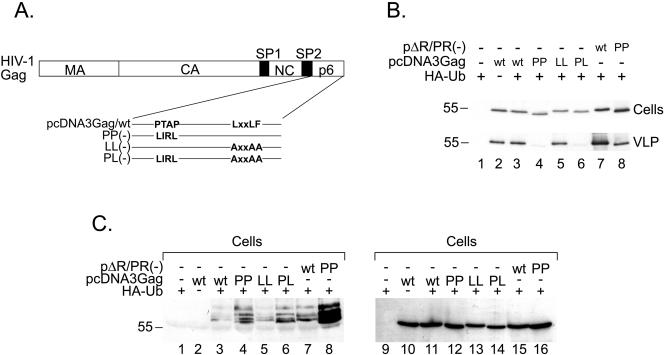
Previous publications suggested that ubiquitination of Gag may be linked to late-domain activity. Strack et al. showed that HIV-1 p6 can induce ubiquitination of a HIV-derived Gag protein with a heterologous dimerization motif (29, 31). As shown in Fig. 1, we observed ubiquitination of Gag domains in the absence of p6 and thus in the absence of the PTAP late domain and the LRSLF motif. Gag ubiquitination even seemed to be enhanced when the p6 domain was missing (Fig. 1). To address whether the late domain influences ubiquitination of Gag, we tested whether mutation of either motif would result in an alteration of the Gag ubiquitination pattern. For this, we cloned three Gag mutant constructs: pcDNA3Gag/PP(−), in which the PTAP motif was altered to LIRL; pcDNAGag/LL(−), in which the LRSLF motif was altered to ARSAA; and the PTAP and LRSLF double mutant pcDNA3Gag/PL(−) (Fig. 5A).
FIG. 5.
(A) Schematic representation of the PTAP and LRSLF mutant Gag proteins used. (B) 293T cells were cotransfected using (+) the indicated plasmids. Twenty hours after transfection, cells (top) and virions (bottom) were harvested and analyzed by Western blotting with an anti-CA antibody. Molecular mass standards (in kDa) are shown at the side of each panel. (C) The remaining cell extracts were immunoprecipitated with anti-CA antiserum and analyzed by SDS-PAGE and Western blotting with an anti-HA antibody (left). The blot was reprobed with an anti-CA antibody (right). Molecular mass standards (in kDa) are shown on the left side.
We and others (data not shown and reference 29) have observed that expressing high levels of Gag proteins in 293T cells can lead to late-domain-independent particle release. Therefore, we first wanted to establish that our analysis of Gag ubiquitination was performed under conditions for which a late-domain phenotype can be observed (Fig. 5B). Accordingly, 293T cells were cotransfected with the indicated plasmids and, less than 24 h after transfection, analyzed by Western blotting for intracellular Gag expression (Fig. 5B, top) and release of VLPs (bottom). For this experiment, expression vectors yielding Rev-independent production of wild-type and mutant HIV-1 Gag polyproteins (pcDNA3Gag derivatives), as well as the previously described plasmid pΔR and its PTAP-minus variant, were used. We observed that Gag proteins lacking a PTAP motif led to particle release at an efficiency of 30 to 40% compared to the respective wt proteins (Fig. 5B, compare lanes 4 to 3 and 8 to 7), confirming the late-domain phenotype. Release efficiencies were calculated as the ratio of virus-associated Gag to the sum of cell- and virus-associated Gag as quantified by ELISA (data not shown). Consistent with previous reports, altering the LRSLF motif did not result in reduced levels of virus release (compare lane 5 to lane 3) (30). Combined mutations of both motifs [pcDNA3Gag/PL(−)] also did not have an additive effect over altering the PTAP motif alone (compare lane 6 to lane 4).
Cell extracts from the same transfections were used to immunoprecipitate Gag proteins and analyze their levels of ubiquitination by detection of HA-Ub (Fig. 5C). None of the mutations introduced abolished Gag ubiquitination within cells (Fig. 5C, left) or VLPs (not shown). In contrast, inactivation of the PTAP motif even enhanced Gag ubiquitination both in the context of the Gag expression construct and in a viral context (compare Fig. 5C, lanes 4 to 3 and 8 to 7). Interestingly, the levels of ubiquitination of Gag expressed from pΔR/PR(−) were reproducibly higher than those of the Gag protein expressed from pcDNA3 in the absence of other viral components (compare lane 7 to lane 3). The Gag proteins derived from pcDNAGag are chimeras of the HXB2 and NL4-3 strains, while pΔR is based on the NL4-3 strain. The sequence differences between these two strains, therefore, could account for the differences observed. Another possibility is that viral proteins other than Gag might promote Gag ubiquitination. Reprobing the blot with an anti-CA antibody confirmed that the differences observed in Gag ubiquitination were not due to different amounts of immunoprecipitated Gag (Fig. 5C, right).
Detection of ubiquitinated Gag cleavage products in the mature virion.
Ott et al. previously reported that a small subset of p6 proteins in the mature virus are mono- or diubiquitinated (22). Since Gag proteins lacking p6 or lysine residues in p6 were ubiquitinated to an extent similar to that of wild-type Gag within VLP preparations (see above) and cells (data not shown), we tested if viral proteins other than p6 are ubiquitinated within the mature virus. For this, virions released from 293T cells transfected with pΔR alone or cotransfected with pΔR and an HA-Ub expression plasmid were collected 48 h after transfection and immunoprecipitated with antiserum against MA, CA, or NC (Fig. 6). Immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with anti-HA antibodies (Fig. 6A to C, left). No signal for HA-Ub was detected when supernatant from control cells expressing only HA-Ub was used (data not shown). In the case of immunoprecipitation with anti-MA antibodies (Fig. 6A), three bands were detected upon cotransfection with HA-Ub (lane 2) that were not present in the control (lane 1). The sizes of the two lower bands are consistent with modification of MA by one ubiquitin moiety, while the upper band most probably represents diubiquitinated MA. The ubiquitination pattern of virus-associated MA was similar to the pattern of MA ubiquitination obtained upon expression of MA alone (compare Fig. 1B, lane 8). The blot was reprobed with antiserum against MA to confirm that similar amounts of MA were immunoprecipitated in both cases (Fig. 6A, lanes 3 and 4). Thus, a subset of the MA proteins in the virus are ubiquitinated.
When virus lysates were immunoprecipitated with anti-CA antiserum, bands for mono- and possibly diubiquitinated CA were detected (Fig. 6B, lane 2) that were not present in the control immunoprecipitation (lane 1), showing that virus-derived CA is also ubiquitinated at low levels. Reprobing the blot confirmed that similar amounts of CA were loaded in each case (Fig. 6B, lanes 3 and 4). This blot shows two additional anti-CA-reactive bands migrating above CA whose identities are currently unclear. Immunoprecipitation of NC from virus lysates and reprobing with anti-HA revealed that NC is also ubiquitinated at low levels (Fig. 6C). In this case, two modified products were detected that probably both represent monoubiquitinated NC. Western blotting for NC confirmed again that similar amounts were immunoprecipitated in both cases (Fig. 5C, lanes 3 and 4). We therefore conclude that, in addition to ubiquitinated p6, small amounts of mono- and sometimes diubiquitinated species of the viral MA, CA, and NC proteins are present in mature HIV-1 particles.
Several domains are ubiquitinated in Gag polyproteins derived from immature virions.
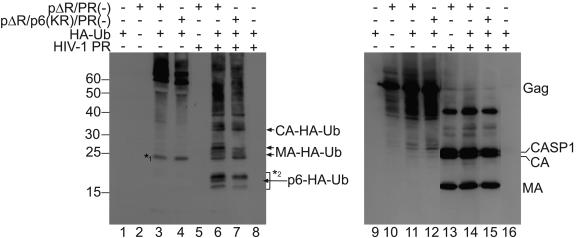
Having established that a subset of the MA, CA, and NC proteins (as well as the p6 protein) in mature HIV-1 particles are ubiquitinated, we wanted to address whether one site of ubiquitin attachment is predominant. To this end, we used Gag polyproteins from PR-negative virus, which were digested with exogenously added HIV-1 PR. For this, we expressed either HA-Ub alone or pΔR/PR(−) alone or coexpressed wild-type or p6 lysine mutant pΔR/PR(−) plasmids with HA-Ub. Forty-eight hours after transfection, virions were centrifuged through sucrose cushions and resuspended in PR cleavage buffer. The samples were adjusted to 0.1% Triton X-100, and each sample was divided into two aliquots, one of which was digested with recombinant HIV-1 PR while the other served as a control. All samples were resolved by SDS-PAGE and analyzed by Western blotting with an anti-HA antibody (Fig. 7, lanes 1 to 8), followed by reprobing the blot with a mixture of anti-CA and anti-MA antisera to confirm Gag cleavage (Fig. 7, lanes 9 to 16). When Gag was cleaved by PR, ubiquitinated Gag disappeared and several smaller ubiquitinated bands were detected (Fig. 7, lanes 6 to 7). We assigned the bands marked with MA-HA-Ub and CA-HA-Ub using immunoprecipitated HA-Ub-conjugated MA and CA expressed from the pcDNA3 constructs described above as markers (not shown). The band labeled with p6-HA-Ub was lost upon mutation of the two lysine residues in p6, and therefore most likely represents monoubiquitinated p6 (compare lane 7 to lane 6). There were at least two Ub conjugates that traveled lower than ubiquitinated MA and were not lost upon mutation of the two lysine residues in p6 (lanes 6 and 7). These bands, therefore, might represent monoubiquitinated NC or diubiquitinated SP2, which would also be expected to migrate between 15 and 20 kDa. We conclude that MA, CA, and at least one other Gag domain are ubiquitinated at levels comparable to p6.
FIG. 7.
Digestion of ubiquitinated Gag by exogenously added HIV-1 PR. 293T cells were transfected with either pHA-Ub alone or pΔR/PR(−) alone or cotransfected with pHA-Ub and pΔR/PR(−) or pΔR/p6(KR)/PR(−) as indicated. Forty-eight hours after transfection, virions were pelleted through sucrose cushions and resuspended in PR cleavage buffer. The virions were lysed by adjusting the buffer to 0.1% Triton and divided into two aliquots. While one aliquot was left untreated (lanes 1 to 4 and 9 to 12), the second aliquot was treated with 120 nM HIV-1 PR for 1 hour (lanes 5 to 8 and 13 to 16). After protease digestion, samples were analyzed by SDS-PAGE and Western blotting with anti-HA antibodies (left). The blot was reprobed with a mixture of anti-CA and anti-MA antisera to confirm Gag cleavage. The band marked by *1 represents a form of Ub that is detected in the absence of Gag cleavage. The bands marked by *2 represent ubiquitinated Gag cleavage products other than p6, MA, and CA, possibly NC or SP2.
Ubiquitinated Gag is not enriched in membrane or virus fractions.
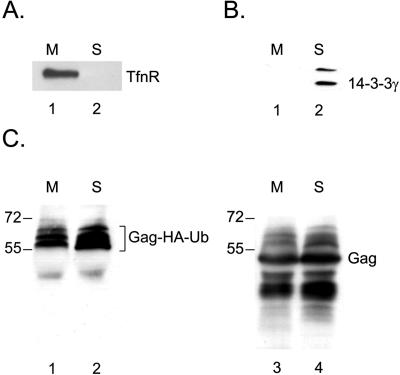
Several lines of evidence point to a role of ubiquitin in the late stages of virus release (20). If ubiquitination of Gag were to take place predominantly in the late stages of virus assembly, ubiquitinated Gag species might be enriched in membrane or virus fractions over soluble or cell-associated fractions. To test if ubiquitinated Gag is enriched in membrane fractions, we cotransfected 293T cells with pHA-Ub and pΔR/PR(−). Forty-eight hours after transfection, cells were detached from the culture dish, washed with PBS, swollen in hypotonic buffer, and disrupted by Dounce homogenization. Nuclei, as well as unbroken cells, were removed by centrifugation. The resulting lysate was subjected to membrane flotation on an Optiprep gradient, and after centrifugation, the visible membrane fraction and the bottom fraction of the gradient were collected. To confirm the quality of our fractionation, a small portion of each fraction was examined for the presence of cellular markers for membrane (transferrin receptor [TfnR]) or soluble (14-3-3γ) fractions (Fig. 8A and B). As expected, TfnR was detected exclusively in the membrane (M) fraction (Fig. 8A), and 14-3-3γ was found only in the soluble (S) fraction (Fig. 8B). Using this procedure, unmyristoylated (G2A mutant) Gag was recovered exclusively from the bottom fraction, confirming that flotation of Gag on this gradient depends on its membrane association (data not shown). The membrane and soluble protein fractions were diluted with RIPA buffer and immunoprecipitated for Gag. The precipitates were normalized for comparable Gag contents (Gag ratio as in Fig. 8C, right) and subjected to Western blotting with anti-HA antibodies (Fig. 8C, left). As the intensities of the signals for ubiquitinated Gag species were comparable to the amounts of Gag present in each fraction (Fig. 8C, compare lanes 1 and 2 with 3 and 4), there appears to be no enrichment of ubiquitinated Gag in either the membrane-associated or soluble protein fraction.
FIG. 8.
Ubiquitinated Gag is not enriched in membrane fractions. 293T cells were cotransfected with pHA-Ub and pΔR/PR(−) and, 48 h after transfection, subjected to membrane flotation. The visible membrane (M) and soluble (S) fractions were collected. A small part of each fraction was analyzed by Western blotting for the fractionation markers TfnR (A) and 14-3-3γ (B). The remaining part of each fraction was immunoprecipitated for the CA protein and analyzed by Western blotting using anti-HA antibodies (C, lanes 1 and 2). The blot was reprobed with monoclonal anti-CA antibodies (C, lanes 3 and 4) to confirm that similar amounts of Gag were analyzed.
We next wanted to test whether ubiquitinated Gag is enriched in virus preparations compared to cell-associated ubiquitinated Gag. In similar experiments, it has been shown that free ubiquitin, as well as cyclophilin A and TSG101, are enriched in virus fractions (10, 26). For this, 293T cells cotransfected with pHA-Ub and pΔR/PR(−) were detached using EDTA and washed with PBS, followed by lysis in a buffer containing 1% SDS at 95°C. The samples were boiled for 10 min, and the buffer was adjusted to RIPA buffer. We chose this procedure to inactivate deubiquitinating activity present in cell lysates and therefore to be able to accurately determine the cellular levels of ubiquitinated Gag. VLPs were prepared from the media by pelleting them through sucrose cushions, and the virus pellets were treated as described for the cell fraction. Both samples were cleared by high-speed centrifugation and immunoprecipitated with anti-CA antiserum. The precipitates were normalized for their Gag contents (Fig. 9, bottom) and examined for modification of Gag by HA-Ub (Fig. 9, top). This experiment clearly indicated that the ubiquitinated pool of Gag protein is not larger in the virus fraction than in the cell.
DISCUSSION
In this study, we showed that the HIV-1 Gag-derived proteins MA, CA, and NC, in addition to p6, are ubiquitinated at low levels. Previously published data suggested that ubiquitination of Gag is lost upon mutation of the two ubiquitin acceptor lysine residues within p6 (21). The discrepancy between that study and our observation that HIV-1 Gag lacking lysine residues in p6 or lacking all of p6 is still ubiquitinated could be due to several reasons. We used very large amounts of virus or immunoprecipitated Gag proteins for our analysis. This might have resulted in a higher sensitivity of detection of Ub conjugates. In addition, we mainly used coexpressed HA-tagged Ub, which further improved the sensitivity of detection of Gag-Ub conjugates. However, we were also able to detect ubiquitination of Gag polyproteins lacking lysine residues in p6 using antibodies against Ub, making this explanation rather unlikely. We reproducibly observed, on the other hand, that one of the bands detected for ubiquitinated Gag was lost when the p6 lysine residues were altered, and it is possible that this was the modified product mainly detected in the previous study.
So far, ubiquitination has been reported only for Gag domains that also carry known late-domain motifs (with the exception of MMTV, for which no late-domain sequences have yet been identified). Our observation that other Gag domains are ubiquitinated at levels very similar to that of the HIV-1 late-motif-containing p6 domain suggests that Gag ubiquitination, at least in the case of the P(T/S)AP motif, is not limited to the Gag domains containing the late-domain motif(s). We addressed a potential contribution of the PTAP and LRSLF motifs to Gag ubiquitination. The presence of these motifs was clearly not required for Gag ubiquitination, as Gag proteins lacking p6 or carrying mutations in the PTAP or LRSLF motifs were still ubiquitinated. In fact, upon PTAP mutation or p6 deletion, Gag ubiquitination was enhanced compared to wild-type Gag. This might reflect the recruitment of a Ub hydrolase by the PTAP motif (the action of such a hydrolase could also account for the large amount of free Ub in the virion), or alternatively, it could be the result of an alteration in Gag localization or the delay in release of late-domain mutant Gag. Our results are in agreement with a recent report by Martin-Serrano et al., who also observed that the PTAP motif results in decreased levels of Gag ubiquitination (17).
The identity of the ubiquitin ligase(s) responsible for HIV-1 Gag modification and its potential binding site(s) within Gag (if any) are unknown. We found p6 to be dispensable for Gag ubiquitination and showed that the MA and CA domains are monoubiquitinated independently of each other when expressed alone. Ubiquitination was enhanced, however, when MACA was expressed compared to either MA or CA alone. Conceivably, monoubiquitination of various Gag domains may represent a “nonspecific” modification of proteins that contain many exposed lysine residues, although this has not been reported for other proteins and was not observed for GFP in this study. Alternatively, there may be two specific ligase binding sites in the MA and CA domains of HIV-1 Gag or the two domains may act in a cooperative manner to recruit the ubiquitination machinery. Thus, a yet-undefined Ub-ligase-recruiting motif might be present in HIV-1 Gag (e.g., in MA and/or CA) or Gag ubiquitination might be induced or regulated by another protein. Interestingly, the levels of Gag ubiquitination were enhanced when a full-length HIV-1 expression plasmid was used rather than a Gag expression vector. While this may be due to strain-specific differences, it may also suggest that other viral proteins modify Gag ubiquitination.
The Gag proteins of several retroviruses have been found to be ubiquitinated, but it is still unknown whether Gag ubiquitination is functionally relevant and whether there is a difference between PPXY late-domain-containing viruses (capable of recruiting an E3 ubiquitin ligase of the Nedd4 family) and viruses lacking a PPXY motif (e.g., HIV-1). Interestingly, the levels of ubiquitination of virus-associated HIV p6 and MLV p12 (which contains a PPPY motif) are similar, indicating that HIV-1 Gag may be sufficient to induce ubiquitination (22). On the other hand, ubiquitination of the Gag proteins of PPXY-containing viruses may also be nonspecific. There is the possibility that Gag ubiquitination represents a bystander effect and recruitment of the ubiquitination machinery serves a different purpose (e.g., ubiquitination of a cellular protein). Alternatively, the overall level of Gag ubiquitination might be functionally relevant while no specific ubiquitination site is needed. This could explain why ubiquitination occurs in different Gag domains, as observed in this study, and would suggest that the various sites of modification may compensate for each other. A third possibility would be that one or several specific ubiquitination sites within HIV-1 Gag are relevant for Gag function. Interestingly, lysine residues in the C terminus of RSV MA have recently been described as important for RSV release (28). These lysine residues are in the region directly upstream of the RSV PPPY late domain. While Spidel et al. (28) have not formally shown that the lysine residues are indeed ubiquitinated and that the loss of ubiquitination is the reason for the observed budding defect, this explanation seems likely, since the addition of extra lysine residues at other locations within Gag could compensate for the loss of lysine residues in MA. Our data show that all major domains of HIV-1 Gag are ubiquitinated, and functionally relevant modified lysine residues may thus be found in MA, CA, NC (SP2), and/or p6. Ott et al. reported that the lysine residues in the p6 domain are dispensable for HIV-1 release and infectivity (21). On the other hand, Strack et al. showed that the HIV-1 PTAP late domain and determinants located within the NCSP2 region of HIV-1 Gag cooperate in promoting virus release (31). Given that NC (and possibly SP2) is indeed ubiquitinated, one may suggest that ubiquitination of the HIV-1 Gag domain adjacent to the late motif may be functionally relevant, similar to what had been observed for RSV. This could be the basis for the reported cooperativity between NCSP2 and the PTAP motif.
We did not observe an obvious accumulation of ubiquitinated Gag proteins at membranes or inside the virus, which would argue against Gag ubiquitination being a late event. However, interpretation of these results is clearly complicated if one takes into account that a deubiquitinating enzyme might act at the site of virus release and therefore in membrane fractions, as suggested by the accumulation of ubiquitinated Gag in a PTAP mutant. If ubiquitination of Gag is a transient and short-lived event, comparisons of the steady-state levels of ubiquitinated Gag should be interpreted with caution.
Acknowledgments
We thank Eric Freed, Tom Hope, Marylin Resh, and David Ott for expression constructs and Barbara Müller and Oliver Fackler for helpful comments on the manuscript. The anti-NC antiserum was a gift from the AIDS Vaccine Program, SAIC-Frederick, Inc., Frederick, Md.; HIV-1 PR was a gift from Jan Konvalinka.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (SFB638, project A9). E.G. is the recipient of a stipend from the Boehringer Ingelheim Fonds.
REFERENCES
- 1.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohne, J., and H. G. Krausslich. 2004. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 563:113-118. [DOI] [PubMed] [Google Scholar]
- 4.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 6.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman, M. H., J. Loureiro, H. L. Ploegh, and D. Tortorella. 2003. Ubiquitinylation of the cytosolic domain of a type I membrane protein is not required to initiate its dislocation from the endoplasmic reticulum. J. Biol. Chem. 278:34804-34811. [DOI] [PubMed] [Google Scholar]
- 8.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 9.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammarstedt, M., and H. Garoff. 2004. Passive and active inclusion of host proteins in human immunodeficiency virus type 1 gag particles during budding at the plasma membrane. J. Virol. 78:5686-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konvalinka, J., M. A. Litterst, R. Welker, H. Kottler, F. Rippmann, A. M. Heuser, and H. G. Krausslich. 1995. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J. Virol. 69:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krausslich, H. G. 1991. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc. Natl. Acad. Sci. USA 88:3213-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, P. D. Bieniasz, and A. Yaravoy. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 20.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 21.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 22.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 27.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spidel, J. L., R. C. Craven, C. B. Wilson, A. Patnaik, H. Wang, L. M. Mansky, and J. W. Wills. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 31.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 33.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 34.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 36.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]