Abstract
Retroviral late (L) domains present within Gag act in conjunction with cellular proteins to efficiently release virions from the surface of the cell. Three different critical core sequences have been identified as required elements for L-domain function: PPPY, PTAP (also PSAP), and YPDL, with different retroviruses utilizing one or two of these core sequences. The human immunodeficiency virus type 1 (HIV-1) L domain is centered around a PTAP sequence in the p6 region of Gag. To assess the ability of heterologous L-domain sequences to be functionally interchanged for those in full-length HIV-1, we produced a series of constructs that replaced PTAP-containing p6Gag sequences with those of PPPY- or YPDL-based L domains. While previous studies had found that L domains are interchangeable in other retroviruses, most of the sequences introduced into p6Gag failed to substitute for PTAP-mediated L-domain function. One exception was the 11-amino-acid p2b sequence of Rous sarcoma virus (RSV) Gag, which could fully restore HIV-1 budding, while a PPPPY sequence exchange alone did not. This suggests that the RSV L domain consists of more than simply its core L-domain sequence. The HIV-p2b chimera was as infectious as the wild type, produced normal virions, and was sensitive to proteasome inhibitors. These results show that L-domain sequences are not necessarily interchangeable. Thus, HIV-1 Gag might have a more stringent requirement for L-domain function than the other retroviruses previously studied.
Human immunodeficiency virus type 1 (HIV-1) assembly takes place in association with membranes (53) and is clearly observed at the plasma membrane in most cell types (11), although in macrophages assembly occurs primarily at internal membranous structures (42, 46). Assembly is a process that is driven by Gag, the primary structural protein of the virion and the only viral protein strictly required for particle formation (53). Gag is a polyprotein that consists of the mature internal structural proteins, minimally matrix (MA), capsid (CA), and nucleocapsid (NC), that are produced from this precursor by viral protease processing (53). (The organization of HIV-1 Gag and the mature Gag cleavage products are presented on the NL4-3 diagram in Fig. 1.) For HIV-1, processing begins during assembly and concludes sometime after the virion buds from the cell (22, 23).
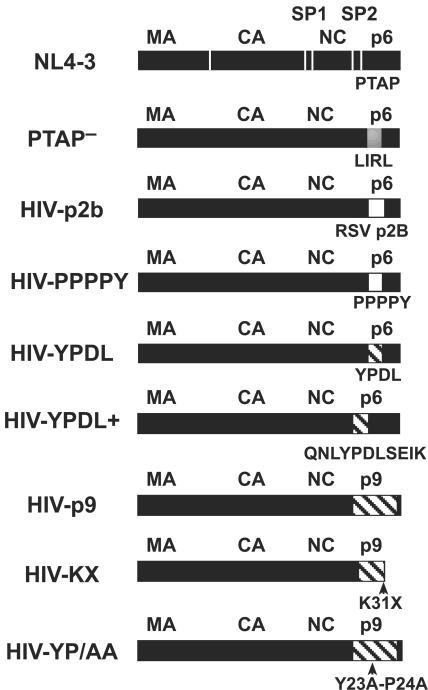
FIG. 1.
DNA constructs. Diagrams of the various Gags from the full-length HIV-1 constructs used in this study are presented with HIV-1 sequences denoted in black, RSV in white, and EIAV in hatched shading. Sequence changes are indicated below each diagram. Protease cleavage sites that produce the mature Gag proteins are denoted in the NL4-3 diagram by white lines in Gag.
While retroviruses can assemble and form lollipop-like budding structures driven by Gag interactions, late (L)-domain sequences within Gag act in concert with various members of the class E vacuolar protein-sorting (VPS) pathway to efficiently release these budding virions from the cell (reviewed in references 5, 9, and 34). L domains interact with their cellular partners after Gag assembly begins (41), apparently catalyzing the fusion of membranes that is required for pinching off the distended viral buds to release particles (5, 9, 34). Without these interactions, viruses remain tethered to the plasma membrane, arrested at this rate-limiting budding step (7, 15, 62). While the exact mechanism that provides this release function remains unknown, it is clear that HIV-1 and equine anemia virus (EAIV) L domains interact with the ESCRT I/III (endosomal sorting complex required for transport I/III) pathway during assembly and budding (6, 8, 10, 12, 13, 31-33, 35, 43, 49, 51, 54, 56, 57).
Wills et al. initially discovered that the p2b region of Rous sarcoma virus (RSV) Gag, located between MA and CA, was important for efficient release of virus particles (58) and later coined the term “L domain” (38). The essential core sequence in p2b is PPPY (58, 59), a sequence that was later observed in and found to be part of the L domain for the Gag proteins of Mason-Pfizer monkey virus (16, 60), murine leukemia virus (MuLV) (62), and human T-cell leukemia virus type 1 (19, 26). This PPPY sequence can bind members of the HECT family of ubiquitin ligases and appears to functionally interact with several family members: LDI-1 for RSV (24, 55); WWP1, WWP2, and Itch for MuLV (29); BUL1 for Mason-Pfizer monkey virus (61); and Nedd4.1 and WWP1 for human T-cell leukemia virus type 1 (3, 19). The L domain of HIV-1 is centered around a PTAP core sequence (7, 21, 28) found in the N-terminal region of p6Gag, the C-terminal protein of Pr55Gag (Fig. 1). This core sequence binds Tsg101 and works in concert with other members of the ESCRT pathway to promote virus budding (6, 8, 10, 12, 13, 32, 33, 35, 43, 56, 57). In addition to the PTAP-based L domain, there is a secondary sequence in p6Gag, L41XXLF46, that binds AIP1/Alix in the cell, which in turn appears to recruit ESCRT III proteins to assist in HIV-1 Gag release (28, 51). A third core L-domain sequence, YPDL, within the p9Gag protein of EIAV Gag (apparently the p6Gag homolog for EIAV), is required for its L-domain activity (4, 27, 44). The p9Gag protein can bind the AP-2 adapter protein complex (45) and recently has been found to bind AIP1 and, ultimately, the ESCRT III complex to mediate particle release (31, 51, 57).
One interesting clue to the nature of viral budding is that L domains can be interchanged between different viruses: HIV p6Gag, EIAV p9Gag, or a portion of p6Gag that includes PTAP can provide L-domain function for RSV Gag (38). Also, the PPPY sequence in MuLV can be complemented with p2b or p6Gag sequences (62). Similar results were found for EIAV budding: the defect observed by the removal of the p9Gag sequence with its YPDL sequence from EIAV Gag was complemented by p6Gag or p2b sequences (38, 44, 49, 62). In addition to being interchangeable, L domains can act in a positionally independent manner, thereby functioning as true modular domains apart from the remainder of Gag (27, 38, 44, 49, 62). Thus, the various types of Gag appear to exploit different cellular pathways for release with the route taken dictated by the particular type of L domain present. This concept is supported by the ability of Gag L-domain mutants to be rescued in trans by coexpression of Gag proteins with heterologous L domains (28, 30, 58).
While the L-domain core sequences are required for function, it is not clear if these four amino acid sequences are entirely sufficient for function, especially when placed within heterologous Gag sequences. Mutating individual sequences flanking the core domain has essentially no effect on L-domain function, indicating that these flanking sequences are not strictly required (21, 38, 44, 59). However, experimental evidence suggests that these core L-domain sequences may not be sufficient for function in heterologous contexts (52).
Although the exchangeability of L domains has been established in several retroviruses, less has been done in the HIV-1 context (38, 44, 62). Recently, HIV-1 Gag with a complete p6Gag deletion was not rescued by coexpression of Gag containing several portions of PPPY-based heterologous L-domain sequences (30). To examine the interchangeability of various L-domain-associated sequences within full-length HIV-1, we tested L-domain sequences from EIAV p9Gag and RSV p2b for their ability to provide L-domain function in an HIV-1 proviral context. Our results show that only the RSV p2b sequence could provide L-domain function for HIV-1, suggesting that heterologous L-domain sequences can have limited ability to promote release of HIV-1.
MATERIALS AND METHODS
DNA mutagenesis.
The pNL4-3 infectious molecular clone of HIV-1 (1) was used for these studies and altered by site-directed mutagenesis using PCR-based methods, either by a single amplification with a mutagenic primer method or by two rounds of amplification using the overlap extension procedure (20). The PTAP− mutant (21), carrying a PTAP-to-LIRL change in p6Gag (amino acids 7 to 10), was provided by Eric Freed (Drug Resistance Program, NCI—Frederick, Frederick, MD). Mutations introduced into p6Gag by this method are as follows: HIV-PPPPY, exchange of p6Gag amino acids 6 to 10 (EPTAP) for RSV p2b amino acids 5 to 9 (PPPPY); HIV-p2b, exchange of p6Gag amino acids 3 to 13 (SRPEPTAPPEE) for RSV p2b (TASAPPPPYVG); HIV-YPDL, exchange of p6Gag amino acids 7 to 10 (PTAP) for EIAV p9Gag amino acids 23 to 26 (YPDL); HIV-p9, exchange of p6Gag amino acids 4 to 36 for EAIV amino acids 4 to 45 of p9Gag; HIV-KX, the HIV-p9 construct with a K31 to a nonsense codon; HIV-YP/AA, the HIV-p9 construct with the Y28P29 of p9Gag mutated to AA; HIV-YPDL+, exchange of p6Gag amino acids 4 to 14 (RPEPTAPPEES) for EIAV p9Gag amino acids 20 to 30 (QNLYPDLSEIK); and HIV-p2b/L45P, which contains an L45-to-P mutation in the remaining p6Gag sequence of the HIV-p2b construct. After construction, the region of DNA that was PCR amplified was sequenced to confirm the mutation and to rule out the possibility of any additional changes introduced during the mutagenesis process.
Cell culture methods.
293T human embryonic kidney cells and HeLa-CD4-LTR-lacZ (HCLZ) cell lines were cultured in Dulbecco's modified Eagle's medium, while H9 T-cell leukemia cells were cultured in RPMI 1640. All media were supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 100 U per ml penicillin, and 100 μg per ml streptomycin. All cell culture products were obtained from Invitrogen Inc. (Carlsbad, CA). Transient transfections of 293T cells were carried out using a calcium phosphate mammalian cell transfection (17). Viral protein production was measured by reverse transcriptase assay on cell culture supernatants as previously described (14). The HIV-1 infection assays using HCLZ cells as a lacZ-Tat transcomplementation reporter for HIV-1 infection were carried out as previously described (14). The cells were infected with dilutions of transfection supernatants, and the assay was developed for β-galactosidase activity by 5-bromo-4-chloro-3-indolyl-β-galactoside staining 48 h postinfection. Positive-staining cells (those colored blue from infection) were observed by light microscopy and counted to score infection events. Viral replication assays were carried out by infecting 1 × 106 H9 cells with dilutions of virions in a 24-well plate (Costar Corporation, Cambridge, MA), and clarified samples were taken routinely and monitored for the presence of virus by reverse transcriptase activity. All HIV-1 infections were carried out in the presence of 2 μg per ml Polybrene.
Protein analysis.
Virions were isolated by centrifugation through a 20% sucrose pad in an SW28 rotor at 112,000 × g at 4°C for 1 h. Immunoblot analysis was performed by lysing equal volumes of virion preparations with lysis buffer (125 mM Tris-HCl, pH 7.5, 4% [wt/vol] sodium dodecyl sulfate [SDS], and 1.8% [vol/vol] β-mercaptoethanol) and separating the proteins on a 4 to 20% Tris-glycine gel (Invitrogen). Proteins were electroblotted onto 0.45-μm-pore-size polyvinylidene difluoride membrane (Invitrogen) using a semidry technique (25). Primary goat antisera against p24CA and p7NC were obtained from the AIDS Vaccine Program (National Cancer Institute—Frederick, Frederick, MD). Proteins were detected by development with horseradish peroxidase-conjugated anti-goat secondary antibody (Biochain Institute, Hayward CA) and the Immun-star horseradish peroxidase substrate kit (Bio-Rad, Hercules, CA) on LumiFilm, Roche Applied Science (Indianapolis, IN). Metabolic labeling of transfected cells with [35S]Met and [35S]Cys was carried out as previously described (47). Briefly, cells were first Met/Cys starved for 1 h before being labeled with Promix, a mixture of 150 μCi of Met and 60 μCi of Cys (Amersham Biosciences, Piscataway, NJ). Virions were isolated by sucrose density centrifugation as described above. Gag was immunoprecipitated from viral and cellular lysates with goat antibodies to p24CA and p7NC and Ezview protein G beads (Sigma). Precipitates were separated on a 10% Tris-glycine SDS-polyacrylamide gel electrophoresis (PAGE) gel (Invitrogen) and analyzed with a PMI phosphorimager (Bio-Rad). Signal values were normalized for the relative amount of Met and Cys in the observed products. For proteasome inhibitor studies, 293T cells were transfected 24 h before study. Cells were pretreated with medium containing 10 μM of MG-132 (z-Leu-Leu-Leu-CHO, or zLLL) and lactacystin (Bio-Mol, Plymouth Meeting, PA) for 10 min (immunoblot) or 1 h (during the starvation phase of metabolic labeling) before changing the medium with fresh inhibitor-containing medium for either an 8-h virus collection phase (immunoblot) as previously described (36) or a 6-h metabolic radiolabeling as described above.
RESULTS
To investigate the interchangeability of L domains within HIV-1, we produced several full-length HIV-1 constructs that replaced portions of p6Gag sequences in the NL4-3 proviral clone with heterologous sequences from either RSV or EIAV L-domain regions to examine them for release-promoting function. The different mutants examined are presented in Fig. 1 and can be classified as either PPPY- or YPDL-based chimeras. Since p6Gag and protease genes overlap, the sequence alterations made in this region were made without introduction of nonsense codons in the pol reading frame to maintain protease expression and potential infectivity. Two PPPY-based chimeras were produced with RSV sequences. The HIV-p2b construct had the SRPEPTAPPEE p6Gag sequence in NL4-3 replaced with the complete 11-amino-acid p2b protein sequence (TASAPPPPYVG) that has been shown to provide L-domain function for other Gag proteins (62). To determine if simply the core RSV sequence is sufficient, the HIV-PPPPY construct was produced by replacing the EPTAP p6Gag sequence with the PPPPY RSV core L-domain motif. Five YPDL-based chimeras were made with EIAV sequences. The HIV-YPDL construct was produced by replacing the p6Gag PTAP sequence with the YPDL L-domain core sequence. HIV-YPDL+ was constructed by replacing the RPEPTAPPEES sequence with QNLYPDLSEIK, a p9Gag sequence that previously had been shown to be sufficient for L-domain function (44). The HIV-p9 construct was produced by replacing most of the p6Gag sequence, amino acids 4 to 36, with amino acids 4 to 45 of p9Gag. This portion of p9Gag has been shown to be sufficient for L-domain and other replicative functions in EIAV Gag (4). The N- and C-terminal p6Gag sequences were retained in the HIV-p9 construct to maintain the SP2 (p1Gag)-p6Gag cleavage site (GNF∧LQS) in the Gag frame and the preprotease leader (p6Pol)-protease cleavage site in the pol frame, as well as the LXXLF AIP1 binding motif in p6Gag (amino acids 45 to 49). Since the central region of p6Gag is dispensable for HIV-1 replication in vitro (2), this p9Gag-for-p6Gag substitution should not affect HIV-1 Gag functions other than release. Two additional mutants were produced from this HIV-p9Gag construct. The HIV-KX construct is an HIV-p9 derivative that contains a lysine-to-nonsense codon change at p9Gag position 31. In EIAV Gag, this mutation does not affect L-domain function in EIAV (4) but removes a ubiquitin-like sequence that has been proposed to assist EIAV release (39). The HIV-YP/AA construct contains a mutation in the HIV-p9Gag core L-domain sequence, YPDL to AADL, that destroys L-domain function in EIAV (44).
Virion production of mutants.
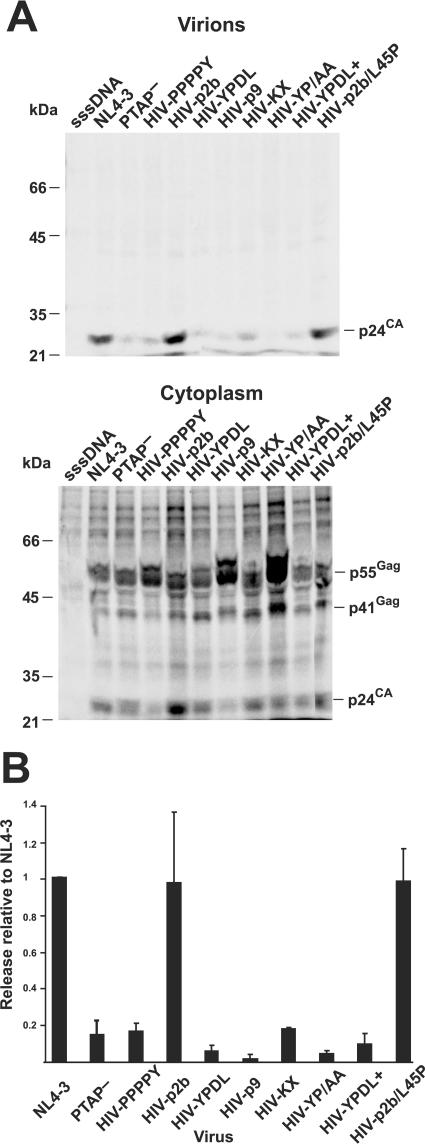
Virions were expressed by transfection of the proviral constructs into 293T cells, and virions were isolated out of 48-h cell culture supernatants. Equal volumes of the preparations were examined by p24CA immunoblot analysis. The intensities of the bands in the PTAP− mutant, all the EIAV sequence exchange chimeras, and the HIV-PPPPY mutant samples were considerably lower than that of the NL4-3 wild-type sample bands (Fig. 2). However, the bands in the HIV-p2b sample had only slightly less intensity than those in the wild-type sample. Values from the reverse transcription assays of supernatants from transfected cells mirrored the immunoblot results (data not shown). None of the viruses had severe protease processing defects (Fig. 2). Wild-type preparations contained unprocessed Gag, Pr55Gag, possibly due to the larger amount of virus present in the sample. While all of the mutants processed Gag, PTAP−, HIV-YPDL, HIV-KX, and HIV-YPDL+ had slight defects in processing, as evidenced by the increased presence of a p41Gag partially processed precursor relative to their respective p24CA levels.
FIG. 2.
Immunoblot analysis of virion preparations. A p24CA immunoblot of equal volumes of virions isolated from transfection supernatants is presented. Samples are identified above their respective lanes. Positions of the molecular mass standards are indicated at left, and reactive bands are identified at right. sssDNA, virus preparation isolated from a sheared salmon sperm DNA transfection.
To examine the budding of these mutants further, the release of virions was examined by metabolically labeling Gag in transfected cell cultures with [35S]Met/Cys for 6 h and then immunoprecipitation of Gag from lysates of virion and cytoplasmic preparations with anti-p24CA and anti-p7NC sera. Similar to the immunoblot results, the data showed that nearly all of the chimeric viruses were released at a much lower level than wild type (Fig. 3, Table 1, virions relative to NL4-3). In contrast, the HIV-p2b construct produced nearly wild-type levels of virions. To normalize for potential differences in protein expression, the total amounts of labeled Gag precipitated from the virion preparations and the cell lysates were quantified by phosphorimager analysis (Fig. 3A) and the release factor was calculated: i.e., the amount of virions released in 6 h (Gag pelletable by density centrifugation)/total Gag (pelletable Gag and Gag in cell lysates). (Typical results are provided in Table 1.) Release factors relative to the wild-type NL4-3 construct from three independent experiments revealed that the PTAP− mutant released particles at a 10-fold-lower level (Fig. 3B), a release value similar to that observed by others (15, 21). The core motif exchanges, HIV-PPPY and HIV-YPDL, both had similarly low release values. Therefore, these short core sequences themselves were insufficient to provide L-domain function in this context. Similarly, the HIV-YPDL+ virions failed to release efficiently; therefore, this sequence does not function in our HIV-1 Gag chimera, even though it can provide L-domain function to EIAV (27, 44). The release factors for the HIV-p9 and HIV-YP/AA constructs were considerably lower than those for the wild type and somewhat less than those for the PTAP− clone. While the HIV-K31X construct also failed to bud efficiently, it released better than HIV-p9, which suggests sequences in the C-terminal region of p9Gag might suppress HIV-1 budding. Taken together, our results show that the EIAV L-domain sequences and the PPPPY core sequence failed to function as L domains in these HIV-1 constructs.
FIG. 3.
[35S]Met/Cys metabolic radiolabeling and immunoprecipitation. (A) Phosphorimages of SDS-PAGE gels from a typical radioimmunoprecipitation analysis of metabolically labeled Gag from virion and cytoplasmic lysates are presented. The lysates examined are indicated above each image, and samples are identified above their respective lanes. Positions of the molecular mass standards are indicated at left, and reactive bands are identified at right. sssDNA, virus preparation isolated from a sheared salmon sperm DNA transfection. (B) Graphical results of release factors relative to wild-type virus from three independent experiments are presented.
TABLE 1.
Immunoprecipitation of [35S]Met-Cys-labeled Gag
| Virus | Amt of virionsa | Amt of virions relative to NL4-3 | Release factorb | Release relative to NL4-3 |
|---|---|---|---|---|
| NL4-3 | 1.9 × 104 | 1 | 0.33 | 1 |
| PTAP− | 1.3 × 103 | 0.07 | 0.031 | 0.09 |
| HIV-PPPPY | 2.6 × 103 | 0.1 | 0.064 | 0.2 |
| HIV-p2b | 2.7 × 104 | 1 | 0.27 | 0.8 |
| HIV-YPDL | 1.4 × 103 | 0.07 | 0.025 | 0.07 |
| HIV-p9 | 7.7 × 102 | 0.04 | 0.014 | 0.04 |
| HIV-KX | 4.3 × 103 | 0.2 | 0.067 | 0.2 |
| HIV-YP/AA | 1.9 × 102 | 0.01 | 0.0014 | 0.004 |
| HIV-YPDL+ | 1.4 × 103 | 0.07 | 0.035 | 0.1 |
| HIV-p2b/L45P | 2.5 × 104 | 1 | 0.32 | 1 |
Phosphorimager quantitation of SDS-PAGE gel analysis of Gag immunoprecipitations using p24CA and p7NC antisera in arbitrary phosphorimager pixel units subtracted for local background.
Relative amount of units in particle preparations versus total amount of Gag recovered from particles and cell lysates after 6 h of labeling.
In contrast to most of the HIV chimeras, the HIV-p2b construct budded almost as efficiently as the wild type (Table 1, Fig. 3), revealing that the complete p2b sequence can replace the HIV-1 L-domain function, even though the PPPPY sequences alone could not. Recently, it has been found that an L41RSLF46 motif in the C-terminal region of p6Gag binds AIP1 and provides additional L-domain function, although the contribution of this sequence to budding appears to be weaker than that of PTAP (31, 51). Also, this sequence can be deleted with a modest decrease in viral budding (21). The HIV-1 L domain might rely on both of these signals for complete function (28). To test the importance of this sequence in the context of HIV-p2b, we altered a critical AIP1-binding residue in p6Gag, L45 to P, to produce HIV-p2b/L45P. This virus was released as efficiently as the HIV-p2b construct in both immunoblot and radioimmunoprecipitation analyses (Table 1, Fig. 2 and 3), showing that an intact AIP binding site was not contributing to the release of HIV-p2b. Thus, the p2b sequence alone appears to be sufficient for L-domain function in this context.
Infectivity of mutants.
To determine if these L-domain exchanges affected other aspects of HIV-1 replication, we examined the infectivity of NL4-3, PTAP−, HIV-p2b, HIV-KX, and HIV-PPPPY in a single-round Tat lacZ complementation assay. The results with transfected cell supernatants revealed that the titer of HIV-p2b was equivalent to that of the wild type (typical results presented in Table 2). Thus, HIV-p2b has wild-type HIV-1 properties. Consistent with this conclusion, HIV-p2b displayed a normal virion morphology by electron microscopy (data not shown). In contrast, the titers of PTAP−, HIV-KX, and HIV-PPPPY were considerably lower than that of wild-type, consistent with their lower release factors (Table 1 and Fig. 3B). Therefore, these L-domain chimeras maintained a degree of infectivity that roughly mirrors their release rate. Replication assays with H9 T cells revealed that the HIV-p2b mutant end-point dilution values paralleled the infectivity results (typical results presented in Table 2). The HIV-KX mutant replicated in H9 cells at a much lower level than the wild type, and the PTAP− and HIV-PPPPY virions showed little capacity to replicate in H9 cells after 5 weeks in culture.
TABLE 2.
Infectivity and replication of HIV chimeras
| Virion | Titer of BCFUa | Titer relative to NL4-3 | H9 replication (TCID/ml)b |
|---|---|---|---|
| sssDNAc | 1 | 3 × 10−6 | <1 |
| NL4-3 | 2.9 × 105 | 1 | ≥106 |
| PTAP− | 1.9 × 103 | 7 × 10−3 | <1 |
| HIV-p2b | 3.1 × 105 | 1.1 | ≥106 |
| HIV-KX | 1.0 × 103 | 3 × 10−3 | ≤101 |
| HIV-PPPPY | 3.4 × 102 | 1 × 10−3 | <1 |
Titer of blue cell-forming units (BCFU) per ml of clarified transfection supernatant.
Tissue culture infective dose (TCID) per ml as measured by infecting H9 cells with 10-fold dilutions of clarified transfection supernatant. The highest dilution tested was 10−7.
sssDNA, virus preparation from a sheared salmon sperm DNA transfection.
HIV-p2b is sensitive to proteasome inhibitors.
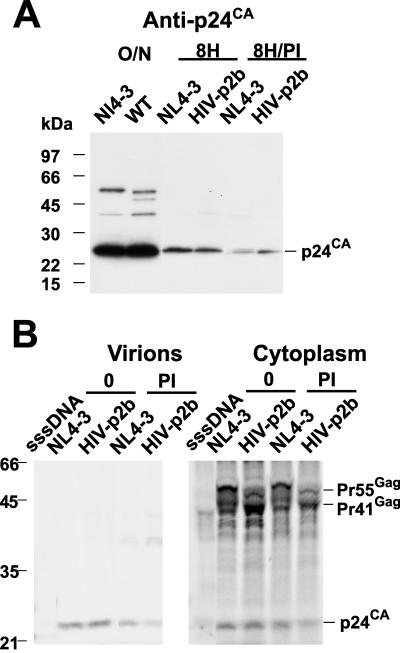
We previously have found that the budding of HIV-1 is reduced by proteasome inhibitors (48). However, not all retroviruses are sensitive to proteasome inhibitors: those that use either PPPY or PTAP for L-domain function are sensitive to proteasome inhibitors, while EIAV, which uses a YPDL L domain, or mouse mammary tumor virus, which uses an as yet undefined L domain, release normally under inhibitor treatment (36, 37, 40, 41, 49, 50). Interestingly, rhabdoviruses employing a PPPY L-domain motif are also sensitive to proteasome inhibitors (18). To determine if the release of the HIV-p2b chimera was altered by proteasome inhibitor treatment, we examined p24CA immunoblots of virion preparations produced during 8 h from cells expressing NL4-3 or HIV-p2b in the presence or absence of 10 μM MG-132 and lactacystin. These conditions do not have adverse effects on protein synthesis over this time period (48). The results showed that the proteasome inhibitor-treated cells produced noticeably less HIV, either wild type or HIV-p2b, than the untreated cultures (Fig. 4A). Electronic image capture and signal quantitation analysis of the immunoblot showed that the amount of HIV-p2b virus produced from proteasome inhibitor-treated cells was threefold lower than that of the untreated sample (Table 3). To confirm this observation, we metabolically labeled cells expressing these two viruses and determined the release factor with and without the inhibitors. The results showed that the budding of HIV-p2b was decreased approximately fourfold by the proteasome inhibitors (Fig. 4B). Thus, HIV-p2b maintains a requirement for active proteasomes as predicted (37, 49).
FIG. 4.
Proteasome inhibitor studies. (A) Immunoblot of cells treated with or without proteasome inhibitors for 8 h is presented. Samples are identified above their respective lanes. O/N, virion preparations prepared from an 18-h harvest of supernatants from transfected cultures. Positions of the molecular mass standards are indicated at left, and reactive bands are identified at right. (B) Phosphorimages of an SDS-PAGE gel from radioimmunoprecipitation analysis of metabolically labeled Gag from virion and cytoplasmic lysates are presented. Cells were labeled for 6 h in the presence or absence of proteasome inhibitors (MG-132 and lactacystin) as indicated. The lysates examined are indicated above each image, and samples are identified above their respective lanes. Positions of the molecular mass standards are indicated at left, and reactive bands are identified at right. sssDNA, virus preparation isolated from a sheared salmon sperm DNA transfection.
TABLE 3.
Proteasome inhibitor sensitivity of HIV-p2b
| Virusa | Value by:
|
|||||
|---|---|---|---|---|---|---|
| Immunoblotting
|
[35S]Met/Cys radioimmunoprecipitation
|
|||||
| Virionb | Value vs untreated | Virionc | Value vs NL4-3 | Release factord | Release relative to untreated | |
| NL4-3 | 1.0 × 103 | 1 | 1.4 × 104 | 1 | 0.23 | 1 |
| HIV-p2b | 7.3 × 102 | 1 | 2.1 × 104 | 1 | 0.38 | 1 |
| WT + PI | 1.7 × 102 | 0.2 | 7.7 × 103 | 0.5 | 0.16 | 0.68 |
| HIV-p2b + PI | 2.1 × 102 | 0.3 | 2.5 × 103 | 0.2 | 0.09 | 0.24 |
WT, wild type; PI, proteasome inhibitor treatment.
Arbitrary imager units by Versa-Doc image capture.
Phosphorimager quantitation of SDS-PAGE analysis of Gag immunoprecipitations using p24CA and p7NC antisera in arbritrary phosphorimager pixel units subtracted for local background.
Relative units in particle preparations versus total amount of Gag recovered from particles and cell lysates after 6 h of labeling.
DISCUSSION
We show here that only the p2b sequence of RSV was able to provide L-domain function for HIV-1 out of the seven chimeras tested in our constructs. The other heterologous sequences that we transferred into Gag, the RSV PPPPY core motif or EIAV p9Gag sequences, were unable to functionally replace the PTAP sequence. Thus, they do not function as L domains in these chimeric constructions (i.e., these sequences are not sufficient to mediate efficient release in this context). Other groups have found that the complete HIV-1 L domain appears to involve other sequences in addition to its PTAP sequence, the LXXLF motif (28, 51, 57), and sequences within the NC-SP2 (p1) region (52). It is important to note that our constructs maintained these sequences, so failure to complement the PTAP function seems to arise more from fundamental differences between the exchanged sequences.
One important caveat to this study is that the failure of heterologous sequences to provide L-domain function could be due to subtle factors in the constructs: e.g., the extent of the foreign sequence introduced, the amount of native sequence removed, or the region in the provirus chosen for the sequence exchange. To minimize these complications, the heterologous sequences were placed in the L-domain region of HIV-1 Gag, while maintaining important p6Gag sequences: e.g., pol and the AIP1-binding sequences. We also introduced L-domain sequences that can function in other Gag proteins. Additionally, our data showed that these mutant viruses were infectious and that Gag was processed. Thus, the L-domain-deficient virions have no gross defects in other Gag functions. Despite these precautions and results, we cannot eliminate the possibility that these sequences could not provide L-domain function in other HIV-1 contexts.
The failure of the EIAV constructs to supply PTAP function might be due to a difference in factors used by these two L domains. The EIAV YPDL sequence interacts with AIP1 and enters the ESCRT pathway by associating with the ESCRT III complex (31, 51, 57). In contrast, the PTAP sequence in HIV-1 Gag interacts with Tsg101 and enters the pathway at the ESCRT I stage (6, 8, 10, 12, 13, 32, 33, 35, 43, 56, 57), two steps upstream of ESCRT III (5, 34). Thus, these EAIV sequences might not function in HIV-1 due to a requirement for HIV-1 Gag targeting to VPS components found in the pathway before ESCRT III (likely present in ESCRT I and/or II). Consistent with this idea, EIAV can use the HIV-1 pathway: either PTAP-containing sequences (49) or those of the Vps28 protein, a component of the ESCRT I complex, can provide L-domain function when introduced into EIAV Gag (54). Proteasome inhibitor studies also suggest that the cellular mechanism used for EIAV release might be fundamentally different from HIV-1 and the PPPY L-domain-containing retroviruses (36, 37, 39).
While the PPPY sequence has been shown to be required for L-domain function in p2b (59), this core motif alone was insufficient to provide L-domain function in our HIV-PPPPY chimera. This implies that the p2b sequences flanking PPPPY in p2b are also required for L-domain function in this HIV-1 context. This is consistent with the previous interpretation that core L-domain sequences are required but not sufficient for function (52).
Currently, the cellular pathway for candidate retroviral PPPY-binding proteins in budding is not as well defined as the PTAP-mediated pathway. Experiments have functionally linked MuLV budding with the VPS system (29), though not directly with the ESCRT complexes. Thus it is likely that HIV-p2b enters the VPS system the same way as MuLV and presumably RSV. Since the AIP1 site was not required for efficient virus release, a direct interaction between HIV-p2b and ESCRT III does not appear to be responsible for L-domain function in this chimera.
As predicted based on our previous findings (36, 37, 40, 48-50), HIV-p2b budding was reduced by proteasome inhibitors, consistent with this type of L domain requiring proteasome function (37). This agrees with results showing that EIAV proteasome inhibitor sensitivity is dictated by its L domain (49). Taken together, these results confirm a linkage between proteasome activity and PTAP- or PPPY-based L-domain function, likely due to disruption of important VPS components that interact with these particular domains (36, 37, 40, 48-50).
Previously, Martin-Serrano et al. found that short PPPY-containing sequences from either Ebola virus or MuLV were unable to provide L-domain function in the context of full-length HIV-1 (30). There are significant differences between RSV p2b and the Ebola virus or MuLV PPPY-containing sequences examined that could be responsible for the difference between our results and theirs. In the same report, trans-complementation experiments that coexpressed a proviral clone that contained a p6Gag deletion with Gag proteins carrying heterologous L-domain substitutions found that EIAV p9Gag but not RSV p2b could rescue the budding of the p6Gag-deleted provirus in trans (30). The discrepancy between those results and our results using proviral constructs may be due to experimental approaches. Similar to our findings, Shehu-Xhilaga et al. mentioned that EIAV sequences were unable to provide L-domain function for HIV-1 p6Gag proviral chimeras (49). Despite these differences, our data support the general conclusion of Martin-Serrano that heterologous L domains are not simply interchangeable in the HIV-1 context, consistent with a previous proposal that the HIV-1 L domain is more complex than other viruses (52).
While many of the chimeric virions budded poorly, these L-domain-deficient virions still could undergo at least one round of replication: the infectivity of the mutants was roughly proportional to the amount of virus released. Therefore, the L domain is not strictly required for other Gag functions. Thus, the L-domain sequences appear to truly act like domains, functioning apart from the other functions of Gag.
The limited interchangeability of L domains and HIV-1 Gag we observe appears to underlie an important aspect of HIV-1 budding that is not yet clear. Understanding the complex interactions between retroviruses and cellular machinery is important, and our results point to important differences between HIV-1 and other retroviral budding pathways. Future studies should assist our understanding of this process.
Acknowledgments
We thank Eric Freed for the gift of the PTAP− mutant and Robert Gorelick for assistance in the preparation of the manuscript.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Adachi, A., S. Koenig, H. E. Gendelman, D. Daugherty, S. Gattoni-Celli, A. S. Fauci, and M. A. Martin. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleiber, G., S. Peters, R. Martinez, D. Cmarko, P. Meylan, and A. Telenti. 2004. The central region of human immunodeficiency virus type 1 p6 protein (Gag residues S14-I31) is dispensable for the virus in vitro. J. Gen. Virol. 85:921-927. [DOI] [PubMed] [Google Scholar]
- 3.Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 9.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 11.Gelderblom, H. R. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5:617-638. [PubMed] [Google Scholar]
- 12.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77:9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelick, R. J., S. M. Nigida, Jr., J. W. Bess, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H.-G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 18.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 21.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, A. H., and R. Swanstrom. 1991. HIV-1 gag proteins are processed in two cellular compartments. Proc. Natl. Acad. Sci. USA 88:4528-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 26.Le Blanc, I., M.-C. Prévost, M.-C. Dokhélar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., and P. D. Bieniasz. 2003. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 77:12373-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 35.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patnaik, A., and J. W. Wills. 2002. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76:2789-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G. P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 47.Rudner, L., S. Nydegger, L. V. Coren, K. Nagashima, M. Thali, and D. E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 79:4055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shehu-Xhilaga, M., S. Ablan, D. G. Demirov, C. Chen, R. C. Montelaro, and E. O. Freed. 2004. Late domain-dependent inhibition of equine infectious anemia virus budding. J. Virol. 78:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 52.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 54.Tanzi, G. O., A. J. Piefer, and P. Bates. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 77:8440-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 78:13943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 58.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xaing, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xaing, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76Gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]