Abstract
Although several genes involved in mitochondrial function are direct Myc targets, the role of Myc in mitochondrial biogenesis has not been directly established. We determined the effects of ectopic Myc expression or the loss of Myc on mitochondrial biogenesis. Induction of Myc in P493-6 cells resulted in increased oxygen consumption and mitochondrial mass and function. Conversely, compared to wild-type Myc fibroblasts, Myc null rat fibroblasts have diminished mitochondrial mass and decreased number of normal mitochondria. Reconstitution of Myc expression in Myc null fibroblasts partially restored mitochondrial mass and function and normal-appearing mitochondria. Concordantly, we also observed in primary hepatocytes that acute deletion of floxed murine Myc by Cre recombinase resulted in diminished mitochondrial mass in primary hepatocytes. Our microarray analysis of genes responsive to Myc in human P493-6 B lymphocytes supports a role for Myc in mitochondrial biogenesis, since genes involved in mitochondrial structure and function are overrepresented among the Myc-induced genes. In addition to the known direct binding of Myc to many genes involved in mitochondrial structure and function, we found that Myc binds the TFAM gene, which encodes a key transcriptional regulator and mitochondrial DNA replication factor, both in P493-6 lymphocytes with high ectopic MYC expression and in serum-stimulated primary human 2091 fibroblasts with induced endogenous MYC. These observations support a pivotal role for Myc in regulating mitochondrial biogenesis.
MYC, which encodes a transcription factor, c-Myc (herein termed Myc), plays a central role in the regulation of cell size, cell proliferation, and apoptosis (6, 11, 28, 40, 41) through its regulation of RNA polymerase I-, II-, and III-dependent genes (2, 17, 18, 39). Deregulated expression of MYC contributes to at least 100,000 U.S. cancer deaths annually, but 20 years after its discovery as a proto-oncogene, its full spectrum of regulatory functions remains incompletely understood (8, 34). Myc dimerizes with Max and binds E boxes (5′-CACGTG-3′) to activate transcription (16). Myc also represses transcription by interfering with the function of other transcriptional activators, such as Miz-1, NF-Y, or Sp1 (23, 45). Myc regulates several major cellular functions, such as cell proliferation, cell adhesion, and cell size, which explains the diversity of Myc target genes found in a variety of metabolic pathways, including amino acid, nucleotide, lipid, and glucose metabolism (www.myccancergene.org) (48). While Myc has been linked to energy metabolism through its regulation of glycolysis (26), its role in mitochondrial biogenesis is less well understood. It is noteworthy that mitochondria are required for a number of important biochemical pathways, including heme synthesis, fatty acid oxidation, and amino acid and steroid metabolism, among many others. Hence, it stands to reason that in driving cellular replication, Myc could also directly or indirectly affect the generation of mitochondria to maintain a steady-state respiratory and metabolic capacity as cells traverse through the cell cycle (1).
Large-scale gene expression analysis in rat or human systems suggests that overexpression of Myc induces nuclearly encoded mitochondrial genes (7, 20, 31, 37, 38, 46). Genes that bind to Myc in Drosophila, rat, or human systems include those encoding mitochondrial proteins or those involved in mitochondrial biogenesis (15, 31, 33, 36, 38, 47). Collectively, these studies suggest that Myc could affect mitochondrial protein expression. Here, we sought to determine the effects of Myc on mitochondrial biogenesis in an inducible Myc-dependent human B-cell model of cell proliferation, a well-defined Rat1 fibroblast system that has been rendered Myc null through homologous recombination, and a model of conditional Myc knockout in primary murine hepatocytes (10, 30, 44). Not only did we observe that mitochondrial biogenesis depends on Myc, but we also found that among the Myc target genes most highly induced in the human B-cell system are those involved in mitochondrial biogenesis. Furthermore, we found that Myc directly regulates TFAM, which encodes a key factor involved in mitochondrial transcription and mitochondrial DNA (mtDNA) replication. These observations support a pivotal role for Myc in regulating mitochondrial biogenesis.
MATERIALS AND METHODS
Cell culture.
Cells were grown as previously reported. P493-6 cells are human B lymphocytes that express an EBNA2-estrogen receptor fusion protein and contain a tetracycline (Tet)-repressible human MYC construct (44). TGR1 (myc+/+), HO15 (myc−/−), and HO15-Myc (myc−/− + Myc; Myc was reconstituted in HO15) are rat fibroblasts (30). Human primary 2091 fibroblasts (American Type Culture Collection) were cultured, serum starved, and serum stimulated as described previously (47).
Primary hepatocyte preparation and adenoviral infection.
Primary murine hepatocytes were prepared and cultured on collagen-coated 35-mm plates as described for rat hepatocytes (32). One day after plating of 3 × 105 cells per plate, hepatocytes from floxed Myc mice were treated with either adenoviruses expressing Cre recombinase (a gift from F. Bunz and B. Vogelstein, Johns Hopkins University) and green fluorescent protein (GFP) or GFP alone (a gift from D. Johns, Johns Hopkins University) at approximately 5 × 108 PFU per plate (10, 42).
Microarray gene expression analysis.
P493-6 cells were treated in presence of 1 μM estradiol plus 0.1 μg/ml tetracycline (endogenous MYC) or 0.1 μg/ml tetracycline (low MYC) or in the absence of both (high ectopic MYC) for 72 h, and RNA was collected with an RNeasy minikit (QIAGEN). Hybridization on Affymetrix U133 Plus 2.0 chip and data analysis were performed in the Johns Hopkins Medical Institute microarray facility. The image data were processed with the statistical procedure Robust Multiarray Analysis to obtain gene expression signals (22), and statistically significant changes in gene expression was obtained with an empirical Bayes method using gamma-gamma or log normal-normal modeling (35).
PCR.
RNA isolated from tetracycline-treated or untreated P493-6 cells was first reverse transcribed to cDNA with TaqMan reverse transcription reagents (Applied Biosystems). Random hexamer primers were used for reverse transcription. Real-time PCR was performed with Sybr Green PCR core reagents (Applied Biosystems). Target gene primer pairs are listed in Table S1 in the supplemental material. The predeveloped TaqMan assay reagents (Applied Biosystems) 18S rRNA control kit was used in a real-time PCR procedure to detect 18S rRNA as an internal template control. cDNA transcribed with known concentrations of RNA was diluted at 10-fold series to generate a standard curve according to the cycle of threshold value obtained from real-time PCR. The quantity of cDNA of the tested gene was determined from the standard curve and normalized to the quantity of 18S rRNA.
Expression of human and mouse TFAM/Tfam and mouse Myc was measured with total RNA by using TaqMan one-step reverse transcription-PCR master mix reagents (Applied Biosystems). The primer and probe sequences are as follows: for human TFAM, 5′-AAGATTCCAAGAAGCTAAGGGTGA (forward), 5′-CAGAGTCAGACAGATTTTTCCAGTTT-3′ (reverse), and 6-carboxyfluorescein (FAM)-5′-TCACCGCAGGAAAAGCTGAAGACTGTAAAG-3′-6-carboxymethylrhodamine (TAMRA) (probe); for mouse Tfam, 5′-GATGGCGCTGTTCCGG-3′ (forward), 5′-TGGATAGCTACCCATGCTGGA-3′ (reverse), and FAM-5′-TCCCCTCGTCTATCAGTCTTGTCTGTATTC-3′-TAMRA (probe); and for mouse Myc, 5′-AGCCCCTAGTGCTGCATGA-3′ (forward), 5′-TCCACAGACACCACATCAATTTC-3′ (reverse), and FAM-5′-CAGCAGCGACTCTGAAGAAGAGCAAGAAGA-3′-TAMRA (probe). Rat Tfam was measured with Sybr Green core reagent from cDNA with primers 5′-AAAAATCTGTCTCATGATGAAAAGCAG-3′ (forward) and 3′-CTTCATTTCATTGTCATAACGAATTCTAT-3′ (reverse).
PCR conditions to assay floxed Myc versus the deletion allele and the primer sequences were as described previously (9, 10).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (4). Briefly, P493-6 cells were grown at 4 × 105 cells/ml either with or without 0.1 μg/ml tetracycline for 72 h. Human 2091 cells were serum starved and stimulated as described above. ChIP was performed with rabbit polyclonal anti-c-Myc sc-764X antibody (Santa Cruz Biotechnology). The human genomic sequence was downloaded from genome.ucsc.edu, and the E box was searched from 3 kb upstream of exon 1 to 7 kb downstream with the nucleic acids motif feature of OMIGA software. PCR primers are listed in Table S2 in the supplemental material. Real-time PCR was performed with Sybr Green PCR core reagents (Applied Biosystems). Absolute quantification was performed as described previously (47). The amount of DNA in the ChIP product was normalized to the amount of DNA in the total input chromatin.
Immunoblotting.
c-Myc was detected with monoclonal antibody 9E10. α-Tubulin (Ab-1) monoclonal mouse immunoglobulin G (Oncogene) was used to detect tubulin as a loading control.
Flow cytometric analysis.
Cells were incubated at 37°C with 5% CO2 for 30 min in the presence of 10 nM (P493-6 cells) or 100 nM (Rat1 fibroblasts) nonyl acridine orange (NAO) (Molecular Probes) and filtered and analyzed immediately with a Becton Dickinson FACScan flow cytometer.
Confocal and immunofluorescence microscopy.
P493-6 cells were incubated with MitoTracker Red CMX Ros (M7512; Molecular Probes) at 250 nM or 150 nM in presence of 10% or 0.25% fetal bovine serum for 15 min. Cells were then resuspended in fresh warm medium and cytospun onto slides. The slides were fixed, rinsed, and mounted. Images were collected with an UltraView (Perkin-Elmer) confocal microscope, and intensity measurements were performed with the signal intensity segmentation feature of IP lab software. Primary hepatocytes were stained with either MitoTracker Red (100 nM) or NAO (10 mM) as described previously (14, 19). Fluorescence microscopy was performed on living cells with an inverted Zeiss Axiovert 200 fluorescence microscope equipped with a digital camera.
Mitochondrial DNA copy measurement.
DNA was collected from whole cell lysates. Real-time PCR was performed for cytochrome c oxidase subunit 1 (COX1), peroxisome proliferative activated receptor gamma coactivator-related 1 (PPRC1), and cytokine-like protein C17. For primer sequences see Table S3 in the supplemental material. Absolute COX1 DNA copies were normalized to nuclear genes, PPRC1 or C17.
Measurement of total cellular oxygen consumption.
P493-6 cells were treated either with or without tetracycline as described above, and then cells were plated at 5 × 105 in the fluorescent dye-embedded 96-well microplate of the BD oxygen biosensor system (BD Biosciences). Cells were incubated for 4 h, and then results were read with a fluorescent microplate reader (Wallac Victor3V 1420 Multilabel Counter; Perkin-Elmer). The data were normalized according to the manufacturer's two-step normalization protocol (technical bulletin no. 448; BD Bioscience) and represented as normalized relative fluorescent units.
Electron microscopy.
Electron microscopy was performed in the microscopy core facility as described previously (47), and sections were viewed with a Philips CM 120 transmission electron microscope. Micrographs were obtained for 10 individual cells for each cell type. Micrographs were inspected, and mitochondrial morphology was determined.
RESULTS
Myc stimulates mitochondrial biogenesis.
Although gene expression and ChIP analyses indicate that Myc regulates genes involved in mitochondrial biogenesis, the effect of Myc on mitochondrial biogenesis has not been directly established. Hence, we sought to determine how Myc expression in the P493-6 human B-cell line influences mitochondrial mass and function. Myc is robustly induced in the tetracycline-responsive system without or with serum (Fig. 1A), as reported previously (43). Flow cytometry using NAO, whose staining of mitochondrial cardiolipin gives a measure of mitochondrial mass, revealed that Myc induction increases mitochondrial mass, although the induction is blunted in the absence of serum (Fig. 1B). Furthermore, quantitative fluorescence microscopy using Mitotracker Red, which stains functional mitochondria, corroborates the findings with NAO (Fig. 1C). These observations suggest that Myc can induce a slight increase in functional mitochondrial mass in the absence of cell proliferation. To further substantiate these findings, we measured mitochondrial DNA content by using real-time quantitative PCR and cellular oxygen consumption as indicators of mitochondrial mass and function, respectively. Both mitochondrial DNA and cellular oxygen consumption increased with Myc induction in the P493-6 B cells (Fig. 2). These observations support the notion that Myc induces mitochondrial biogenesis; however, whether Myc is necessary for mitochondrial biogenesis could not be discerned in the P493-6 system.
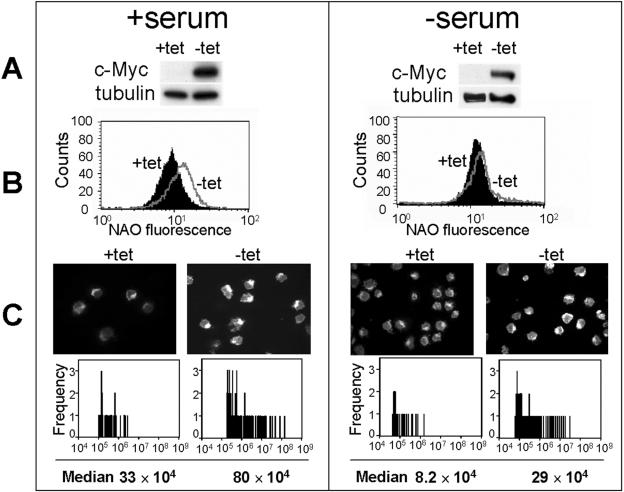
FIG. 1.
c-Myc increases mitochondrial mass and activity under high (10%; left panels)- or low (0.25%; right panels)-serum conditions. P493-6 cells were plated in presence (+tet, low Myc) or absence (-tet, high Myc) of tetracycline for 72 h in either high or low serum. The cells were collected as described in Materials and Methods. (A) c-Myc levels were determined by immunoblotting. Tubulin served as a loading control. (B) Fluorescence intensity of NAO staining (x axis) in cells as determined by flow cytometry. The y axis represents cell numbers. (C) Representative confocal micrographs of MitoTracker Red-stained cells are shown in the upper panels. The lower panels show the signal intensity distribution of MitoTracker Red staining as a measure of mitochondrial activity and content, using signal segmentation of the IP lab software. The median of relative signal intensity is also indicated. Exposure times for the confocal micrographs were kept constant between high-Myc and low-Myc conditions but were slightly different for high- versus low-serum conditions.
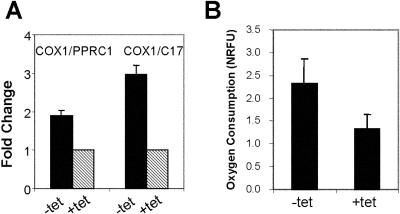
FIG. 2.
c-Myc induction increases mitochondrial DNA copies and cellular oxygen consumption. P493-6 cells were treated as described in the legend to Fig. 1 under high-serum conditions, and then cells were collected as described in Materials and Methods. (A) Quantitative real-time PCR was performed on the mitochondrial gene COX1, and the nuclear genes PPRC1 and C17 were used as internal controls. The fold changes were calculated relative to the DNA level in tetracycline-treated (+tet, low Myc) cells. Shown are averages from three independent experiments ± standard errors of the means. (B) Total cellular oxygen consumption, shown as averages from two independent experiments ± standard deviations. NRFU, normalized relative fluorescent units.
To determine whether Myc is necessary for normal mitochondrial biogenesis, we then studied whether depletion of Myc could influence mitochondrial content and structure in Rat1 fibroblasts that have been rendered Myc null by homologous recombination. Flow cytometric studies, as described above, reveal that Myc null cells have diminished mitochondrial mass and function (Fig. 3A and 3B). In fact, two NAO-stained Myc null populations were observed, corresponding to a larger group with markedly diminished mitochondrial mass and a smaller group with nearly normal mitochondrial mass. Although ectopic expression of Myc in the Myc null cells rescued their slowed growth (Fig. 3C), the recovery of mitochondrial mass was partial. We further examined the mitochondria with electron microscopy. Compared to wild-type fibroblasts, there is a unique Myc null cell population that contains abundant lysosome-like organelles and very few mitochondrial sections. A subpopulation of Myc null cells displays reduced mitochondrial sections. The normal mitochondrial sections reappear in Myc null fibroblasts rescued with ectopic Myc expression, although there appears to be a residual subpopulation with reduced normal mitochondrial sections (Fig. 4). These observations suggest that Myc is required for normal mitochondrial biogenesis, since chronic removal of Myc resulted in decreased mitochondrial mass.
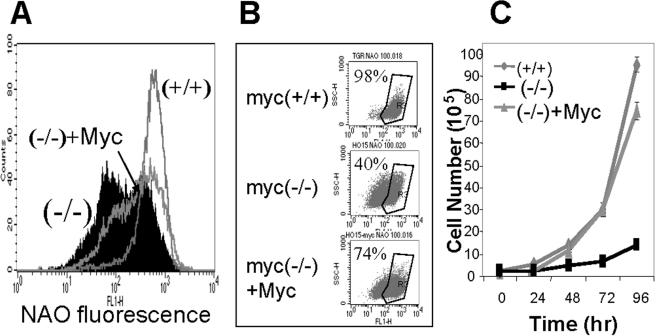
FIG. 3.
Mitochondrial mass is decreased in Myc null fibroblasts. (A and B) Log-phase-growing myc+/+, myc−/−, and myc−/− + Myc cells were stained with NAO. (A) The flow cytometric histogram shows the fluorescence intensity corresponding to mitochondrial mass. Myc+/+ cells have one signal peak at high fluorescence intensity, while myc−/− and myc−/− + Myc cells have two peaks. (B) The three small dot plots show the percentage of cells gated for higher fluorescence intensity. (C) Growth rates of myc+/+, myc−/−, and myc−/− + Myc fibroblasts.
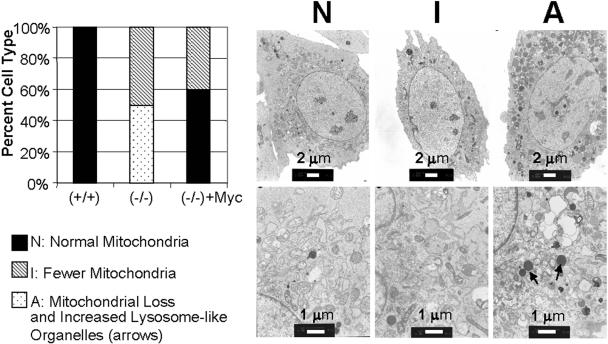
FIG. 4.
Ultrastructural analysis of mitochondria in rat fibroblasts. Left, quantitation of the types of cells found in fibroblasts. A total of 10 individual cells each from log-phase-growing myc+/+, myc−/−, and myc−/− + Myc cells were classified and are shown as the percentage displaying N, I, or A morphology. Right, electron micrographs of three major morphological categories: N, abundant, normal mitochondrial sections; I, fewer, normal mitochondrial sections; A, dramatic loss of mitochondrial sections with increased lysosome-like organelles. The lower panels are higher magnifications of the upper panels.
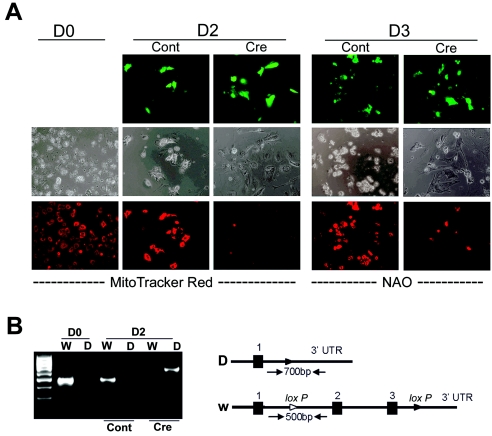
We sought to determine whether acute deletion of Myc from primary hepatocytes, which are rich in mitochondria, could affect mitochondrial mass. We obtained primary hepatocytes from homozygous mice that have floxed Myc alleles (10). Primary hepatocytes were treated with adenoviruses expressing either Cre recombinase and GFP or GFP alone. At 2 and 3 days after adenoviral treatment, hepatocytes were supravitally stained with MitoTracker Red or NAO and observed by fluorescence microscopy. As shown in Fig. 5A, the GFP fluorescence confirms adenoviral infections (top panels). Both MitoTracker Red and NAO displayed diminished staining in Cre recombinase-treated cells compared with controls (Fig. 5A). Genomic DNA samples harvested from untreated hepatocytes or from hepatocytes 2 days after adenoviral infection were assayed for the deleted or undeleted floxed Myc alleles (Fig. 5B). The controls treated with adenoviruses expressing only GFP retained the undeleted Myc configuration, whereas hepatocytes treated with Cre recombinase viruses had significant deletion of Myc and retained only a small fraction of the undeleted Myc alleles. Taken together our observations of the gain of Myc function in P493-6 and the loss of Myc in Rat1 cells and primary hepatocytes strongly support the hypothesis that Myc induces mitochondrial biogenesis.
FIG. 5.
Acute deletion of floxed Myc in isolated murine hepatocytes is associated with decreased stainable mitochondria. (A) Fluorescence and phase-contrast micrographs of cultured primary hepatocytes from homozygous mice with floxed Myc before (day 0) and 2 or 3 days after control (Cont) or Cre recombinase adenoviral infection. GFP fluorescence micrographs of adenovirally infected hepatocytes (top panels) at days 2 and 3 are shown. The same cells were stained with either MitoTracker Red (shown for day 2) or nonyl acridine orange (shown for day 3). Fluorescence micrographs of controls or Cre recombinase-treated cells were obtained with identical exposure times for each different fluorochrome (GFP, NAO, or MitoTracker Red). (B) Ethidium-stained agarose gel showing PCR-amplified (35 cycles) genomic DNA from untreated hepatocytes at day 0 or from control or Cre recombinase-treated hepatocytes at day 2. PCR primers used were specific for the undeleted (W) or deleted (D) floxed Myc alleles. W amplicon size, 500 bp; D amplicon size, 700 bp. Molecular markers are shown in the far left lane. UTR, untranslated region.
Myc induces genes involved in mitochondrial biogenesis.
We sought to determine how Myc expression in the P493-6 human B-lymphocyte cell line affects genes involved in mitochondrial function and biogenesis. The P493-6 cells have been engineered with two unique features: they contain an Epstein-Barr virus (EBV) genome with an estradiol-inducible EBNA2-estrogen receptor hormone binding domain (EBNA2-ER) fusion protein and a tetracycline-repressible human MYC allele (43, 44). EBNA2 is a key EBV transcriptional regulator necessary for EBV immortalization of human B cells. The phenotypes of P493-6 cells depend on the presence of estradiol and Tet. In the presence of Tet and absence of estradiol, P493-6 cells have a very low Myc expression and growth rate. However, addition of estradiol while Tet is present causes the P493-6 cells to express endogenous MYC, which is activated by EBNA2-ER, and to proliferate, resembling EBV-immortalized human lymphoblastoid cell lines. Ectopic MYC is induced in the absence of estradiol or tetracycline, causing the P493-6 cells to be tumorigenic in immunocompromised mice (R. Dinavahi and C. V. Dang, unpublished observation), resembling endemic Burkitt's lymphoma. This model provides a unique opportunity to study the classes of genes induced by the ectopic or endogenous expression of Myc.
Using oligonucleotide microarray analysis in four independent biological experiments, we found 2,679 genes (4,314 probe sets) that are responsive to ectopic Myc among a total of about 10,000 genes that are expressed in the P493-6 cells. About half (1,578) of the genes are induced, while the other half (1,101) are repressed. Among the 1,578 genes that are induced, expression analysis systematic explorer (EASE) gene ontology analysis revealed that 198 genes involved in the mitochondrion are overrepresented statistically out of 1,141 up-regulated genes that are able to be annotated (EASE score, 1.21E−45) (see Table S4 in the supplemental material) (21). Among the 1,101 down-regulated genes, 9 of them were annotated as associated with mitochondrial function. The names of the genes and fold changes are as follows: ALDH2, −2.47; ALDH6A1, −2.35; BCL2, −3.86; BNIP3L, −2.26; BCL2L1, −2.65; CPT1B, −2.00; COX4I2, −3.12; PSEN1, −1.93; and UCP2, −3.92. The up-regulated genes are grouped in relation to their participation in mitochondrial membrane, mitochondrial matrix, mitochondrial ribosomes, carboxylic acid metabolism, biosynthesis, electron transporter activity, oxidative phosphorylation, and mtDNA replication. Among these, the HSP60, cytochrome c (7, 20), sideroflexin 1, acetyl-coenzyme A acetyltransferase 1, isocitrate dehydrogenase (37), HSP10, and prohibitin (31, 37) genes have been reported to be induced by c-Myc. Cytochrome c, HSP60, PHB, PRDX3, and mSHMT have been shown to be bound by Myc by ChIP assays (15, 31, 33, 36, 47). To validate our microarray analysis, we selected 10 genes that are up-regulated and involved in mitochondrial DNA replication, oxidative phosphorylation, electron transport, carboxylic acid metabolism, or ATP synthesis to determine their expression by real-time PCR (see Table S5 in the supplemental material). The changes detected by microarray analysis highly correlate with the real-time PCR results.
We also examined the response of these genes to endogenous MYC via EBNA2-ER activation and found that mitochondrial genes are also robustly activated by endogenous MYC, as they are by ectopic MYC. In fact, 88% (175 of 198 annotated genes) of mitochondrion-related genes activated by ectopic MYC were also induced by endogenous MYC in the P493-6 system, and mitochondrion-related genes are overrepresented among genes responsive to endogenous MYC as determined by EASE analysis (data not shown). Since both endogenous and ectopic Myc could induce many mitochondrion-related genes, this analysis supports a major physiological role for Myc in mitochondrial biogenesis. While mitochondrion-related genes are overrepresented, other overrepresented groups of genes induced by Myc are also discernible through EASE analysis, including genes involved in metabolism, cell cycle regulation, and ribosome biogenesis (21).
Myc binds to TFAM.
Having observed that mitochondrion-related genes are overrepresented among Myc-responsive genes, we sought to determine whether Myc directly binds and regulates genes involved in mitochondrial function and biogenesis. The current Myc target gene database (www.myccancergene.org), with about 1,700 entries, contains 23 genes involved in mitochondrial function that are bound by Myc as determined by ChIP (Table 1). Ten of these are up-regulated in P493-6 cells as observed by microarray gene expression analysis (Table 1). In addition, global genomic mapping in Drosophila revealed that dMyc binds to six genes contributing to mitochondrial biogenesis, structure, and function (38). Among these, the TFAM and Tim10 human orthologs are also up-regulated by Myc in the P493-6 system, although their regulation by Myc in mammalian cells has not been studied.
TABLE 1.
Genes determined by ChIP to be Myc targets (www.myccancergene.org) and genes up-regulated by Myc in P493-6 cells as determined by microarray analysis
| Gene function | Gene determined by:
|
Fold inductionb | |
|---|---|---|---|
| ChIPa | Microarray | ||
| Oxidoreductase activity | NDUFA1 | ||
| NDUFA2 | |||
| NDUFB3 | |||
| NDUFB5 | |||
| NDUFS1 | NDUFS1 | 2.5 | |
| NDUFS6 | NDUFS6 | 2.2 | |
| NDUFV1 | |||
| PRDX3 | PRDX3 | 2.1 | |
| Ion transport | UCP1 | ||
| UCP3 | |||
| DNA metabolism | PHB | PHB | 5.0 |
| TFAM* | TFAM | 3.0 | |
| Protein metabolism | HSPD1 | HSPD1 | 5.3 |
| HSPE1 | HSPE1 | 5.3 | |
| TIMM17A | TIMM17A | 3.0 | |
| UQCRC2 | |||
| Amino acid metabolism | SHMT2 | SHMT2 | 3.1 |
| Tricarboxylic acid cycle | ACBP | ||
| DBI | |||
| Inner mitochondrial membrane | CYCS | CYCS | 3.4 |
| FCI-12 | |||
| TIMM10* | TIMM10 | 2.7 | |
| Other | NME1 | NME1 | 8.8 |
| MCFP | |||
| HAX1 | |||
The genes marked with an asterisk are genes bound by dMyc as determined by global genomic mapping in Drosophila.
Fold induction determined by microarray analysis between ectopic and low-Myc states.
Tfam is a key nuclearly encoded transcription factor, which translocates to mitochondria and activates mitochondrial transcription and mtDNA replication. In two independent experiments with P493-6 cells, TFAM was induced 2.8- and 3.0-fold by endogenous and ectopic MYC, respectively, as detected by microarray analysis. Furthermore, we observed a twofold induction of murine Tfam (relative to 18S rRNA) in livers treated with Myc adenoviruses compared with those treated with LacZ adenoviruses through tail vein injection (data not shown), which is a mouse model described previously (27). It is notable that rRNAs are also induced by Myc, and hence our values of Tfam induction may be underestimated (2, 17, 18).
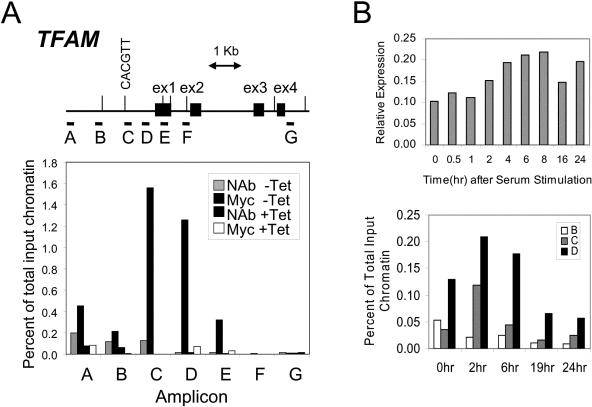
To further validate the role of Myc in the regulation of mitochondrial biogenesis, we sought to determine whether TFAM is a direct target of Myc in the P493-6 system. Using scanning chromatin immunoprecipitation assays and P493-6 cells, we found that Myc binds to TFAM at about 900 bp upstream of the transcription start site in the region of amplicons C and D (Fig. 6A). Amplicon C contains a 5′-CACGTT-3′ sequence that could be bound by Myc (3). Amplicon D has a palindromic sequence, 5′-CGCGCG-3′, that contains Myc/Max 5′-GCG-3′ half sites (3) and GC-rich regions with that are associated with dMyc in Drosophila cells (38). Neither amplicon contains phylogenetically conserved E boxes that are found in promoter and intron 1 regions (Fig. 6A). Both our gene expression and ChIP experiments support the direct regulation of TFAM by Myc.
FIG. 6.
c-Myc binds to TFAM in situ. (A) Human genomic sequence starting from 3 kb upstream of exon 1 to 7 kb downstream. Exons are represented by black boxes. The E boxes are indicated with vertical bars, and the E box in amplicon C is illustrated. Horizontal bars labeled A to G indicate the regions amplified for scanning ChIP analysis. P493-6 cells were plated in absence (−Tet) or presence (+Tet) of tetracycline as described in Materials and Methods, and then ChIP was performed with anti-c-Myc antibody (Myc −Tet or Myc +Tet). No-antibody control experiments (NAb −Tet or NAb +Tet) were performed at same time. −Tet corresponds to a high-Myc state, whereas +Tet represents a low-Myc state. Quantitative PCRs were performed. Shown are averages of triplicates. (B) Top, time-dependent expression of TFAM following serum stimulation of human 2091 primary fibroblasts. TFAM expression is shown as normalized expression relative to 18S rRNA. Mean values (with standard deviations of less than 5% of the mean) from triplicate real-time PCRs are shown. Bottom, chromatin immunoprecipitation assays showing time-dependent binding of Myc to TFAM following serum stimulation of human 2091 primary fibroblasts. Binding to amplicons B, C, and D, which are defined in panel A, is shown as a percentage of total input DNA.
To determine whether endogenous Myc could bind TFAM, we employed primary human 2091 fibroblasts as previously described (47). It has been known that MYC expression is induced maximally between 1 and 2 h after serum stimulation (47). Here we found a twofold increase in TFAM expression relative to 18S rRNA at 4 to 8 h after serum stimulation (Fig. 6B, top). ChIP experiments following serum stimulation reveal increased Myc binding to both amplicons C and D, with peak binding at 2 and 6 h after serum stimulation (Fig. 6B, bottom). No increased binding to the control amplicon B was seen. These experiments further confirm that Myc binds TFAM. Both our gene expression and ChIP experiments support the direct up-regulation of TFAM by Myc. These observations further support the notion that Myc affects mitochondrial biogenesis via transcriptional regulation.
We also sought to determine the dependence of Tfam expression on endogenous Myc, by measuring the expression of Tfam in rat fibroblasts rendered Myc null by homologous recombination and in primary murine hepatocytes acutely deprived of Myc through conditional floxed alleles. Chronic deprivation of Myc in rat cells resulted in decreased Tfam expression: 1.0-fold for myc+/+, 0.7-fold for myc−/−, and 0.93-fold for myc−/− + Myc. In contrast, acute deprivation of Myc in hepatocytes with more than a 10-fold reduction of Myc expression was not associated with a significant decrease in Tfam expression 2 days after Cre recombinase adenoviral infection. Although the Tfam mRNA half-life is unknown, our results suggest that Tfam expression in primary hepatocytes could be dependent on other factors that are codominant with Myc.
DISCUSSION
The Myc oncogenic transcription factor affects the expression of more than 10% of human genes and emerges as a master switch that couples metabolism with cell growth and proliferation. Since Myc is estimated to affect the expression of about 3,500 human genes, the major challenge is attaining an understanding of the biological context of Myc's regulatory function. A cascading effect of Myc's function may be envisioned through its regulation of other transcription factors and their cofactors, which in turn regulate genes that are involved in specific cellular processes. The spectrum of genes regulated by Myc includes those involved in mitochondrial function; however, whether Myc directly affects mitochondrial biogenesis has not been well established. Three previous studies, one with PRDX3 (47) (a mitochondrial peroxiredoxin), one with serine hydroxymethyltransferase (36) (a major source of the one-carbon unit), and one with cytochrome c (33), suggest that Myc may directly influence mitochondrial homeostasis, cell metabolism, and apoptosis, although none directly addressed whether Myc could affect the production of mitochondria in mammalian cells. In this report, we provide evidence both by in vitro and in vivo ectopic expression of Myc and by studying Myc null fibroblasts and acute deletion of floxed murine Myc in primary hepatocytes that Myc affects mitochondrial biogenesis, presumably through its direct regulation of genes involved in mitochondrial biogenesis.
The use of the P493-6 system allowed us to identify a set of Myc-responsive genes that are involved in mitochondrial biogenesis. In fact, among the Myc-responsive genes, the group annotated as being related to mitochondrial structure and function ranks among the highest that are overrepresented by EASE analysis. Particularly intriguing is that Tfam, a key mitochondrial transcriptional regulator and mtDNA replication factor, is Myc responsive and, as we demonstrated in this report, a direct target of Myc in the P493-6 and primary human fibroblast systems. Tfam overexpression appears sufficient to induce mitochondrial transcription but not mtDNA replication in vitro (29). In transgenic mice, however, overexpression of human TFAM is sufficient to increase mtDNA copy number (12). On the other hand, reduction of TFAM by RNA interference in HeLa cells or gene deletion in the mouse reduces mtDNA copy number, indicating that Tfam is necessary for mtDNA replication and mitochondrial biogenesis (12, 24). It is notable that our in silico work to identify transcription factors that cooperate with Myc reveals nuclear respiratory factor NRF1 as one among six predicted Myc collaborators in cis-regulatory modules (13). NRF1 is a major transcriptional regulator of mitochondrial biogenesis and recently has been thought to collaborate with E2F, although TFAM was found bound by NRF1 but not E2F (5, 25). In contrast, our studies reveal that Myc is able to directly bind TFAM, suggesting that Myc has a distinct nonoverlapping function with E2F in inducing mitochondrion-related genes. Although Myc binds the TFAM promoter region, this area lacks canonical E boxes and resembles Myc binding regions of the glycolytic TPI and GAPD genes that lack canonical Myc binding sites (26). It is notable that while chronic deletion of Myc in rat fibroblasts resulted in decreased Tfam expression, the acute deletion of Myc from primary mouse hepatocytes was not associated with diminished Tfam expression. With the caveat that the half-life of the Tfam mRNA is not known, these observations suggest that the dependence of Tfam expression on endogenous Myc could be tissue type dependent or that endogenous Myc could contribute to Tfam expression but not be required. The decrease in hepatocyte mitochondrial mass following acute removal of Myc is hence likely to depend on other Myc target genes involved in mitochondrial biogenesis. Notwithstanding the uncertainty surrounding the sufficiency and necessity of Myc in regulating Tfam, together with previous identification of direct Myc target genes involved in mitochondrial function or biogenesis, our current findings firmly support Myc's role in mitochondrial biogenesis.
Our functional studies verify that upon Myc induction, P493-6 cells increase their consumption of oxygen, mitochondrial mass and function, and mtDNA content. Removal of Myc from the Rat1 fibroblasts, conversely, resulted in cells with diminished numbers of normal mitochondrial sections and evidence of dysmorphic, presumably incompletely assembled mitochondria. Although we do not understand the bimodal distribution of Myc null cells according to nonylacridine orange staining, we speculate that the proliferative compartment of these cells, compared with the majority of cells that are resting, may attain a higher mitochondrial mass independent of Myc. Reconstitution of Myc in the Myc null cells, however, not only partially rescues mitochondrial mass in these knockout cells but also increases the number of morphologically normal mitochondrial sections as determined by electron microscopy. These observations suggest that Myc increases the propensity of these cells to generate functional mitochondria. In addition, acute deletion of floxed murine Myc by Cre recombinase resulted in diminished mitochondrial mass in primary hepatocytes. In summary, our findings demonstrate a role for Myc in regulating genes involved in mitochondrial structure and function and in mitochondrial biogenesis and further establish that Myc is a master switch that couples cellular metabolic needs to cell growth and proliferation.
Supplementary Material
Acknowledgments
We thank D. Eick for P493-6 cells, J. Sedivy for TGR and HO15 cells, and P. Puigserver for comments on the manuscript. We thank D. Murphy for kind instruction on sample preparation and data analysis for confocal microscopy and electron microscopy, Y. Ko for technical assistance with the stimulated cellular oxygen consumption assay, and L. Blosser for the flow cytometry assay. We thank Francisco Martinez Murillo and Chunfa Jie for microarray hybridization and data analysis.
This work was supported by NIH/NCI grants CA52497, CA57341, and CA09159 and the Training Program in Human Genetics and Molecular Biology. J. Kim is a Howard Hughes Medical Institute predoctoral fellow. C. Dang is the Johns Hopkins Family Professor in Oncology Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front Biosci. 3:d250-d268. [DOI] [PubMed] [Google Scholar]
- 2.Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shiue, K. Fatyol, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, L. G. Larsson, and A. P. Wright. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell. Biol. 7:303-310. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell, T. K., J. Huang, A. Ma, L. Kretzner, F. W. Alt, R. N. Eisenman, and H. Weintraub. 1993. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 13:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 6.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 7.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang, C. V. 1999. c-Myc targets genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Alboran, I. M., E. Baena, and A. C. Martinez. 2004. c-Myc-deficient B lymphocytes are resistant to spontaneous and induced cell death. Cell Death Differ. 11:61-68. [DOI] [PubMed] [Google Scholar]
- 10.de Alboran, I. M., R. C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R. A. DePinho, and F. W. Alt. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 11.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 12.Ekstrand, M. I., M. Falkenberg, A. Rantanen, C. B. Park, M. Gaspari, K. Hultenby, P. Rustin, C. M. Gustafsson, and N. G. Larsson. 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13:935-944. [DOI] [PubMed] [Google Scholar]
- 13.Elkon, R., K. I. Zeller, C. Linhart, C. V. Dang, R. Shamir, and Y. Shiloh. 2004. In silico identification of transcriptional regulators associated with c-Myc. Nucleic Acids Res. 32:4955-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elmore, S. P., Y. Nishimura, T. Qian, B. Herman, and J. J. Lemasters. 2004. Discrimination of depolarized from polarized mitochondria by confocal fluorescence resonance energy transfer. Arch. Biochem. Biophys. 422:145-152. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell. Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 17.Grandori, C., N. Gomez-Roman, Z. A. Felton-Edkins, C. Ngouenet, D. A. Galloway, R. N. Eisenman, and R. J. White. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell. Biol. 7:311-318. [DOI] [PubMed] [Google Scholar]
- 18.Grewal, S. S., L. Li, A. Orian, R. N. Eisenman, and B. A. Edgar. 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell. Biol. 7:295-302. [DOI] [PubMed] [Google Scholar]
- 19.Guidot, D. M. 1998. Endotoxin pretreatment in vivo increases the mitochondrial respiratory capacity in rat hepatocytes. Arch. Biochem. Biophys. 354:9-17. [DOI] [PubMed] [Google Scholar]
- 20.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 60:5922-5928. [PubMed] [Google Scholar]
- 21.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 23.Izumi, H., C. Molander, L. Z. Penn, A. Ishisaki, K. Kohno, and K. Funa. 2001. Mechanism for the transcriptional repression by c-Myc on PDGF beta-receptor. J. Cell Sci. 114:1533-1544. [DOI] [PubMed] [Google Scholar]
- 24.Kanki, T., K. Ohgaki, M. Gaspari, C. M. Gustafsson, A. Fukuoh, N. Sasaki, N. Hamasaki, and D. Kang. 2004. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 24:9823-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. W., K. I. Zeller, Y. Wang, A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 24:5923-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S., Q. Li, C. V. Dang, and L. A. Lee. 2000. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl. Acad. Sci. USA 97:11198-11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutz, W., J. Leon, and M. Eilers. 2002. Contributions of Myc to tumorigenesis. Biochim. Biophys. Acta 1602:61-71. [DOI] [PubMed] [Google Scholar]
- 29.Maniura-Weber, K., S. Goffart, H. L. Garstka, J. Montoya, and R. J. Wiesner. 2004. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 32:6015-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 31.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mezey, E., L. Rennie-Tankersley, and J. J. Potter. 2001. Liver alcohol dehydrogenase is degraded by the ubiquitin-proteasome pathway. Biochem. Biophys. Res. Commun. 285:644-648. [DOI] [PubMed] [Google Scholar]
- 33.Morrish, F., C. Giedt, and D. Hockenbery. 2003. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 17:240-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18:3004-3016. [DOI] [PubMed] [Google Scholar]
- 35.Newton, M. A., C. M. Kendziorski, C. S. Richmond, F. R. Blattner, and K. W. Tsui. 2001. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comput. Biol. 8:37-52. [DOI] [PubMed] [Google Scholar]
- 36.Nikiforov, M. A., S. Chandriani, B. O'Connell, O. Petrenko, I. Kotenko, A. Beavis, J. M. Sedivy, and M. D. Cole. 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 22:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 38.Orian, A., B. van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oskarsson, T., and A. Trumpp. 2005. The Myc trilogy: lord of RNA polymerases. Nat. Cell. Biol. 7:215-217. [DOI] [PubMed] [Google Scholar]
- 40.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 41.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 42.Rajagopalan, H., P. V. Jallepalli, C. Rago, V. E. Velculescu, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature 428:77-81. [DOI] [PubMed] [Google Scholar]
- 43.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuhmacher, M., M. S. Staege, A. Pajic, A. Polack, U. H. Weidle, G. W. Bornkamm, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9:1255-1258. [DOI] [PubMed] [Google Scholar]
- 45.Wanzel, M., S. Herold, and M. Eilers. 2003. Transcriptional repression by Myc. Trends Cell Biol. 13:146-150. [DOI] [PubMed] [Google Scholar]
- 46.Watson, J. D., S. K. Oster, M. Shago, F. Khosravi, and L. Z. Penn. 2002. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 277:36921-36930. [DOI] [PubMed] [Google Scholar]
- 47.Wonsey, D. R., K. I. Zeller, and C. V. Dang. 2002. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. USA 99:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.