Abstract
The molecular mechanisms that regulate nuclear NF-κB to determine whether the stimulation of this pathway has a pro- or antiapoptotic effect on cells have yet to be fully defined. Nuclear compartmentalization is increasingly recognized as an important mechanism for regulating the activity of transcription-related proteins and modulating cell growth and death. We have investigated whether such compartmentalization serves as a mechanism for regulating NF-κB transcriptional activity. We demonstrate that the RelA component of NF-κB is sequestered in the nucleolus in response to the proapoptotic NF-κB stimuli aspirin, serum withdrawal, and UV-C radiation. In contrast, RelA is excluded from the nucleolus in response to the cytokines tumor necrosis factor and TRAIL. We identify an N-terminal motif of RelA that is essential for the nucleolar localization of the protein and show that deleting this motif inhibits the translocation of RelA from the nucleoplasm to the nucleolus. We demonstrate that the nucleolar accumulation of RelA is paralleled by a decrease in basal levels of NF-κB transcriptional activity and by apoptosis. Furthermore, we show that the retention of RelA in the nucleoplasm inhibits this decrease in NF-κB-driven transcription and blocks apoptosis induced by aspirin and UV-C radiation. This work identifies a novel cellular mechanism for regulating NF-κB-driven transcription and apoptosis, involving the nucleolar sequestration of a key NF-κB subunit. These data contribute to the understanding of the complexities of NF-κB function and have considerable relevance to cancer prevention and therapy.
NF-κB is a ubiquitously expressed, inducible transcription factor that regulates the transcription of a diverse array of genes (1, 30). There are five known members of the mammalian NF-κB/Rel family which can dimerize through their Rel homology domain to form a variety of different transcription factor complexes (45). Although all members of the NF-κB family can bind DNA, only RelA, c-rel, and RelB have transactivating functions. The most abundant form of NF-κB is a heterodimer of the p50 and RelA polypeptides. In most cell types, this complex exists in the cytoplasm bound to a family of IκB inhibitory proteins. Following cellular stimulation by specific inducers, IκB is phosphorylated by the IκB kinase (IKK) complex at serines 32 and 36 and then degraded by the 26S proteosome (20). Subsequently, NF-κB translocates to the nucleus, where it regulates the transcription of target genes.
It is now well established that NF-κB plays an essential role in controlling cellular growth properties and apoptotic cell death (21, 31). However, the cellular consequences of activating the NF-κB pathway are complex, since the nuclear translocation of NF-κB can have both pro- and antiapoptotic effects (14). For instance, the induction of the NF-κB pathway by cytokines such as tumor necrosis factor (TNF) results in the nuclear translocation of NF-κB complexes that are antiapoptotic and promote cell growth (2, 46). In keeping with an antiapoptotic role for NF-κB, inappropriate activation of the NF-κB pathway has been shown to contribute to tumor formation in a number of cancer types (21), while increased DNA binding of NF-κB is associated with tumor resistance to chemotherapy (44). In contrast, when stimulated by agents such as UV radiation and serum withdrawal, the nuclear translocation of NF-κB can promote cell death (5, 25, 39). In this lab, we are interested in the mechanism by which aspirin mediates the apoptosis of colon cancer cells. In previously published work, we reported that in the absence of additional cytokines, aspirin activates the NF-κB pathway and that NF-κB complexes induced by aspirin are also proapoptotic (13, 42). Consistent with the notion of differing cellular responses to NF-κB nuclear translocation, NF-κB can activate the transcription of both proapoptotic and antiapoptotic genes (30). However, the mechanisms that regulate nuclear NF-κB to determine the spectrum of genes which are activated/repressed by a specific signal are complex, and the downstream effects on cell growth and death have yet to be fully defined (8).
The compartmentalization of transcription-associated proteins within nuclear bodies is increasingly recognized as an important mechanism for regulating gene expression, cell proliferation, and apoptosis (3, 7). For example, the activity of the p53 tumor suppressor gene is regulated by the nucleolar sequestration of its negative regulator, MDM2 (37, 47), and c-myc-induced progression through the cell cycle is inhibited by the sequestration of the protein in the nucleolus (10). In terms of NF-κB, the nucleolar protein p14ARF has recently been shown to interact with RelA and inhibit NF-κB-driven transcription (32), while in two recent screens for NF-κB-interacting partners, the predominant proteins identified were the nucleolar proteins NFBP (43) and nucleophosmin/B23 (12). Furthermore, the NF-κB regulators NIK (NF-κB-inducing kinase) (4) and NRF (NF-κB repressing factor) (29) have very recently been shown to function through nucleolar shuttling. However, the regulation of NF-κB transcriptional activity and apoptosis by the nuclear organization of NF-κB proteins has yet to be examined.
Here we report that the RelA component of NF-κB translocates to the nucleolus following induction by the proapoptotic NF-κB stimuli aspirin, serum deprivation, and UV-C radiation. We identify a nucleolar localization signal (NoLS) at the N terminus of RelA and, using a dominant-negative mutant of RelA with a deletion of this nucleolar localization motif, we demonstrate that the nucleolar sequestration of RelA causes a reduction in basal levels of NF-κB transcriptional activity and apoptosis. These findings suggest a novel mechanism to regulate nuclear NF-κB activity and apoptosis involving the nucleolar compartmentalization of RelA.
MATERIALS AND METHODS
Cell culture and reagents.
SW480 and HRT18 colon cancer cell lines are available from the American/European Type Culture Collections, and immortalized rela null mouse embryo fibroblasts (MEFs) were kindly provided by R. T. Hay (University of St. Andrews). SW480 cells were maintained in L-15 medium, HRT18 in RPMI 1640, and MEFs in Dulbecco modified Eagle medium. All media were supplied by Gibco BRL and supplemented with penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% fetal calf serum (FCS). The generation of HRT18 cells expressing superrepressor IκBα (IκBSR or IκBs32-36) has been described previously (42). IκBSR-expressing clones were subcultured in RPMI under Geneticin (Gibco BRL, Paisley, United Kingdom) selection and then grown in the absence of Geneticin during aspirin treatment. Aspirin (Sigma) was solubilized in water using 10 N NaOH, and then the pH was adjusted to 7.0. For aspirin treatment, cells were plated at a density of 1 × 105 per 50-ml flask, grown until 60 to 80% confluent, and then treated continuously with aspirin in either 0.5% FCS (SW480 and HRT 18 cells) or 10% FCS (MEFs) for a further 16 h or 72 h or as specified below. To study the effects of inhibitors on the response to aspirin, cells were preincubated in low-serum medium containing 50 μM MG132 (Calbiochem), 20 nM leptomycin B, 200 μM pyrrolidine dithiocarbamate (PDTC; Sigma), 0.5 μg/ml actinomycin D (Sigma), or 10 μM cycloheximide (Sigma) for 1 h prior to the addition of aspirin (5 mM) for a further 16 h. Cells were treated with TNF and TRAIL (R&D Systems) in low-serum medium as described above for the times specified below. For serum-deprivation experiments, cells were grown overnight in medium containing 10% serum. This medium was then replaced with either 10% or 0% FCS-containing medium, and cells were incubated for a further 96 h. For UV-C irradiation, cells (seeded in six-well plates) were washed in phosphate-buffered saline, exposed to UV-C irradiation (40 J/m2) using a Stratalinker instrument (Stratagene), and then recovered in original medium for the times specified below.
Plasmids.
The 3enhancer CONA (3x κB ConA-Luc) and human immunodeficiency virus (HIV)-long terminal repeat (LTR) wild-type (WT) and κB-deleted reporter constructs were supplied by R. T. Hay (University of St. Andrews) and have been described elsewhere (34). The pCMV-β-galactosidase plasmid is commercially available (Promega). The green fluorescent protein (GFP)-RelA expression construct was a kind gift from E. Qwarnstrom (University of Sheffield) (6). To generate GFP-tagged deletion mutants, RelA fragments corresponding to amino acids (aa) 1 to 311, 290 to 511, 105 to 511, and 50 to 511 were PCR amplified using GFP-RelA as a template. PCR fragments were cloned in fusion with GFP in the pEGFP-C1 (Clonetech) vector. The QuikChange site-directed mutagenesis kit (Stratagene), along with primers made using QuikChange primer design, was used to generate the RelA construct with a deletion of amino acids 27 to 30 (RelAΔ27-30) and the K28/R mutation. The sequences of all constructs were confirmed by the sequencing of plasmid DNA on an ABI3700 genetic analyzer.
Antibodies.
The following primary antibodies were used: sheep polyclonal anti-IκBα (gift from R. T. Hay [University of St. Andrews]), sheep polyclonal anti-Cu/ZnSOD (The Binding Site), rabbit polyclonal anti-p65 (Santa Cruz), mouse monoclonal anti-p65 (Santa Cruz), mouse monoclonal anti-nucleolin (Santa Cruz), goat polyclonal anti-fibrillarin (Santa Cruz), mouse monoclonal anti-Ki-67 (Santa Cruz), mouse monoclonal anti-pk TAG (TCS Cellworks), rabbit polyclonal anti-GFP (Santa Cruz), and goat polyclonal anti-caspase-3 (R&D Systems).
Transfections and reporter assays.
For the analysis of NF-κB-driven transcription, cells were transfected with the indicated reporter plasmids using Lipofectin (SW480 and HRT18 cells) or Lipofectamine 2000 (MEFs), as described by the manufacturer (Gibco BRL). Cells were grown for 24 h and then treated with aspirin (0 to 10 mM) for the times specified below. Luciferase and β-galactosidase activities were measured in cell extracts using a luciferase reporter assay kit (Promega) and a β-galactosidase assay kit (Promega) as per the manufacturer's instructions. The relative luciferase activity was calculated as the units of luciferase activity per unit of β-galactosidase activity. All results shown are the means of triplicates of at least three independent experiments. To analyze the effects of aspirin and UV-C radiation on the cellular localization of wild-type and mutant RelA, SW480 cells were transfected with GFP-RelA expression constructs as described above. Transfected cells were recovered for 24 h and then treated with aspirin for a further 16 h or recovered for 8 h after UV-C irradiation (40 J/m2). The localization of GFP-tagged proteins was analyzed in adherent cells using an Axiovert 100 inverted microscope (Zeiss) at an excitation wavelength of 488 nm. Images were captured and processed using IPLab 3.6. To determine the effects of N-terminal deletions of RelA on NF-κB-driven transcription in SW480 cells, cells were transfected with either GFP-RelA or RelAΔ27-30 along with 3xκB ConA-luc as described above. The total adherent cell population was harvested, and then cells expressing GFP-tagged protein were isolated from nonexpressing cells using a fluorescence-activated cell sorter (Becton Dickinson). Luciferase assays were performed on lysates from GFP-expressing cells, and the relative luciferase activity was calculated as the level of luciferase activity per GFP-tagged cell (as determined during cell sorting).
Western blot analysis.
Following aspirin treatment, cytoplasmic and nuclear extracts were prepared from adherent cells as previously described (35, 42). To examine the expression levels of GFP-tagged RelA proteins, whole-cell extracts (using lysis buffer consisting of 50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 0.5% NP-40, 10% glycerol, 0.1 mM EDTA, 0.5 mM dithiothreitol, protease inhibitors) were prepared from transfected cells. Bradford assays (Bio-Rad) were used to measure protein content. Extracts (20 μg) were resolved on 10% sodium dodecyl sulfate-polyacrylamide gels, and immunoblotting was performed by standard procedures. Membranes were probed with anti-Cu/ZnSOD or anti-actin as a control for protein loading. Electrophoretic mobility shift assays (EMSAs) were performed on nuclear extracts as previously described by us (42).
Immunocytochemical staining.
SW480 cells were grown on sterilized coverslips until 60 to 80% confluent and then treated with aspirin in low-serum (0.5% FCS) medium as specified below. Cells were fixed in 1:1 methanol:acetone at −20°C for 20 min, and then immunocytochemistry was performed using standard procedures. Primary antibodies were diluted in 1.5% donkey serum as follows: p65 (rabbit polyclonal or mouse monoclonal) (1:100), nucleolin (C23) (1:100), fibrillarin (1:100), and Ki-67(1:50). Coverslips were mounted in Vectastain (Vector Laboratories) containing DAPI (4′,6′-diamido-2-phenylindole) and cells analyzed using fluorescent microscopy. The permeabilization of cells prior to fixation was carried out as previously described (22a). Fluorescent microscopy was performed with a Zeiss Axioplan microscope with a 63× Plan Neofluor objective and a Chroma 83000 filter set. Each channel was recorded independently, and pseudocolor images were superimposed. Images were captured using in-house scripts written for IPLab Spectrum 3.6.
Isolation of nucleoli.
Nucleoli were isolated from 6 × 108 SW480 cells as described elsewhere (http://www.lamondlab.com/f5nucleolarprotocol.htm). Nucleoli were resuspended in phosphate-buffered saline, and the protein contents of all cell fractions were analyzed using Bradford assays (Bio-Rad). All steps were carried out at 4°C. Western blot analysis was performed as described above.
Viability and apoptosis assays.
Staining for cell surface phosphatidylserine was used as a marker for apoptosis and was carried out using an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Oncogene Research Products) as per the manufacturer's instructions. The percentage of apoptotic cells within the cell population was determined by microscopy. At least 250 cells from multiple fields of view were counted for each sample. To analyze the effects of the GFP-RelA deletion mutants on the modulation of cell viability, cells were transiently transfected with the GFP-tagged expression constructs and then stimulated as described above. Phase-contrast images of randomly selected fields of adherent cells were captured using an Axiovert 100 inverted microscope (Zeiss), and then the total number of cells was determined using hemocytometric counts. To examine the effects of RelA cellular distribution on aspirin- and UV-C-induced apoptosis, cells were transfected with the GFP-RelA wild-type and deletion mutants and treated as described above. Apoptotic cells were detected using an annexin V-biotin kit (Oncogene) with a streptavidin-Texas Red conjugate (Oncogene) as per the manufacturer's instructions. The numbers of nonapoptotic (bright-field channel), apoptotic (Texas Red channel), transfected (FITC channel), and transfected apoptotic cells were determined by microscopy, and the percentages of transfected cells, apoptotic cells, and transfected apoptotic cells within the cell population were calculated. At least 250 transfected cells were examined for annexin V binding in each sample. All results presented are the means of at least three experiments ± standard errors (SE).
RESULTS
Nucleolar localization of RelA.
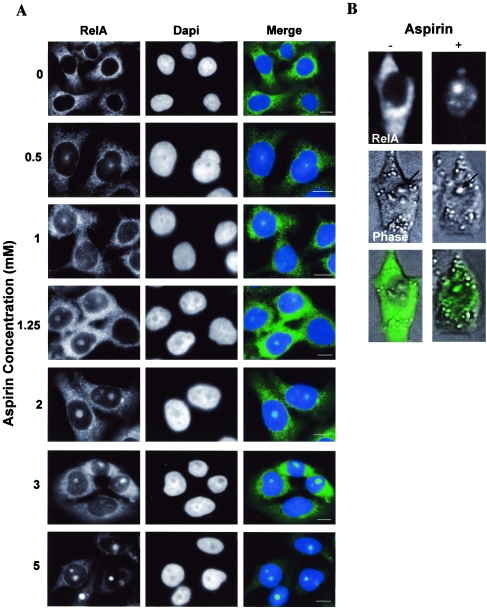
We set out to determine whether NF-κB effects on cell growth and death were regulated at the nuclear level by the compartmentalization of NF-κB proteins. We previously reported that the stimulation of the NF-κB pathway by aspirin induces nuclear translocation of NF-κB complexes that promote apoptosis (13, 42), and so we focused on aspirin as a proapoptotic NF-κB stimulus. Using immunocytochemical analysis, we first examined the subnuclear distribution of endogenous NF-κB proteins in aspirin-treated SW480 colon cancer cells. Our previous studies indicated that the NF-κB complex induced by aspirin consisted predominantly of RelA (42), and so we examined this critical component of NF-κB. We found that prior to induction by aspirin, RelA was present mainly in the cytoplasm, with a small amount of nucleoplasmic staining (Fig. 1A). However, in response to aspirin, RelA accumulated in the nucleoplasm and, more interestingly, became compartmentalized into distinct nuclear bodies (Fig. 1A). Cells showing nuclear foci of RelA were observed at aspirin concentrations as low as 500 μM, which is comparable to the salicylate levels that we measured in plasma samples from subjects given standard analgesic doses of aspirin (42). Furthermore, the number of cells showing these foci increased in a dose-dependent manner (Fig. 1A and data not shown). To confirm that the observed nuclear accumulations of RelA were not an artifact of the staining procedure, SW480 cells that had been transfected with a GFP-RelA expression construct were analyzed. Using live-cell imaging, we found that the GFP-tagged RelA showed the same subcellular localization that we demonstrated for endogenous protein (Fig. 1B). GFP-tagged RelA was localized mainly to the cytoplasm and, to a much lesser degree, in the nucleoplasm, prior to aspirin exposure. However, in response to aspirin, there was a dramatic relocalization of RelA from the cytoplasm/nucleoplasm to distinct subnuclear bodies. Aspirin had no effect on the distribution of GFP alone (see below). These data establish that RelA accumulates in distinct subnuclear bodies in response to aspirin.
FIG. 1.
RelA accumulates in distinct nuclear bodies following aspirin treatment. (A) Immunomicrograph (magnification, ×63) showing the subcellular distributions of RelA in control and aspirin-treated (0.5 to 2 mM for 72 h or 3 to 5 mM for 16 h) SW480 cells. Nuclear accumulations of RelA are observed at pharmacologically relevant doses. Nuclei are stained with DAPI. Bars, 10 μm. (B) SW480 cells were transiently transfected with a GFP-RelA expression vector 24 h prior to aspirin (0 [−] or 5 [+] mM for 16 h) stimulation. Fluorescent and phase-contrast images (magnification, ×40) of adherent cells were captured using an Axiovert 100 inverted microscope. Arrows indicate nucleoli.
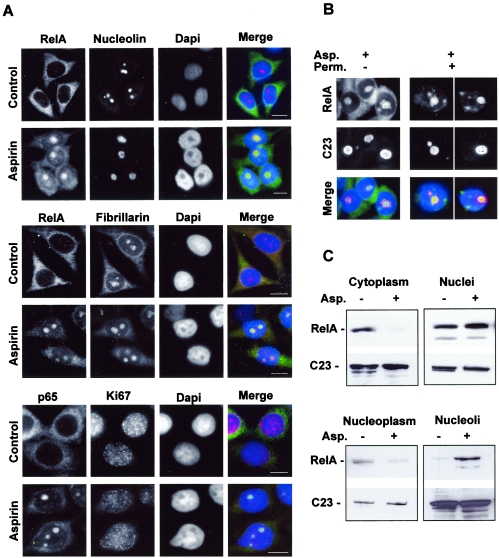
The eukaryotic nucleus contains a number of well-defined structures, including splicing speckles, Cajal bodies, chromatin territories, and nucleoli (3, 7, 24). Since the nuclear foci of RelA colocalized with dense, refractile nuclear bodies, as shown by phase microscopy (Fig. 1B), and with areas unstained by DAPI (Fig. 1A), as shown by immunocytochemical analysis, we considered it likely that the RelA was accumulating within nucleoli. This was confirmed by immunocytochemical analysis demonstrating that aspirin-induced nuclear RelA colocalizes with the nucleolar proteins nucleolin (C23) and fibrillarin (Fig. 2A). Ki-67 is a proliferation marker whose presence in the nucleolus is cell cycle dependent (11). We found that Ki-67 was mainly nucleoplasmic in aspirin-treated cells and showed minimal colocalization with RelA (Fig. 2A), suggesting that the nucleolar accumulation of RelA is a protein-specific event and not a general consequence of changes in proliferation. The permeabilization of cells prior to immunocytochemical analysis confirmed that RelA was bound within the nucleolus after aspirin treatment (Fig. 2B).
FIG. 2.
RelA localizes in the nucleolus in response to aspirin. (A) Immunocytochemical staining (magnification, ×63) showing that RelA colocalizes with the nucleolar proteins nucleolin and fibrillarin, but not with Ki67, when induced by aspirin. SW480 cells were unstimulated (control) or stimulated with aspirin (5 mM, 16 h) and then fixed, and immunocytochemistry was performed using the specified antibodies. DNA is stained by DAPI. Bars, 10 μm. (B) RelA is held within nucleolar bodies after aspirin treatment. The immunomicrographs (magnification, ×63) show the cellular localization of RelA in aspirin (Asp.)-treated (5 mM, 16 h) SW480 cells with (+) and without (−) permeabilization (Perm.) prior to fixation and immunocytochemical analysis. Nucleolin (C23) staining depicts nucleoli. Merged image shows DAPI-stained DNA (blue). (C) Aspirin induces a decrease in nucleoplasmic and an increase in nucleolar levels of RelA. Nontreated and aspirin (Asp.)-treated (5 mM, 16 h) SW480 cells were fractionated using sucrose gradients. Western blot analysis was then performed on protein extracted from the specified cell fractions using anti-RelA followed by anti-nucleolin (C23) antibodies.
To further confirm that aspirin mediates the nucleolar accumulation of RelA, nucleoli were purified from untreated and aspirin-treated SW480 cells. Western blot analysis of cell fractions revealed substantially increased levels of RelA in nucleoli from aspirin-treated cells compared to nucleoli purified from untreated controls (Fig. 2C). Furthermore, the increase in nucleolar RelA was associated with decreased levels of the protein in the cytoplasm and, importantly, in the nucleoplasm (Fig. 2C). Aspirin had no significant effect on nucleolar or nucleoplasmic levels of nucleolin (C23) (Fig. 2C). The purity of isolated nucleoli was confirmed using lamin B (a nuclear protein that does not accumulate in nucleoli), which showed minimal staining in the purified nucleolar fractions (data not shown). Taken together, these cell-imaging and molecular studies confirm that RelA localizes to the nucleolus in response to aspirin.
Specificity of stimuli causing nucleolar localization of RelA.
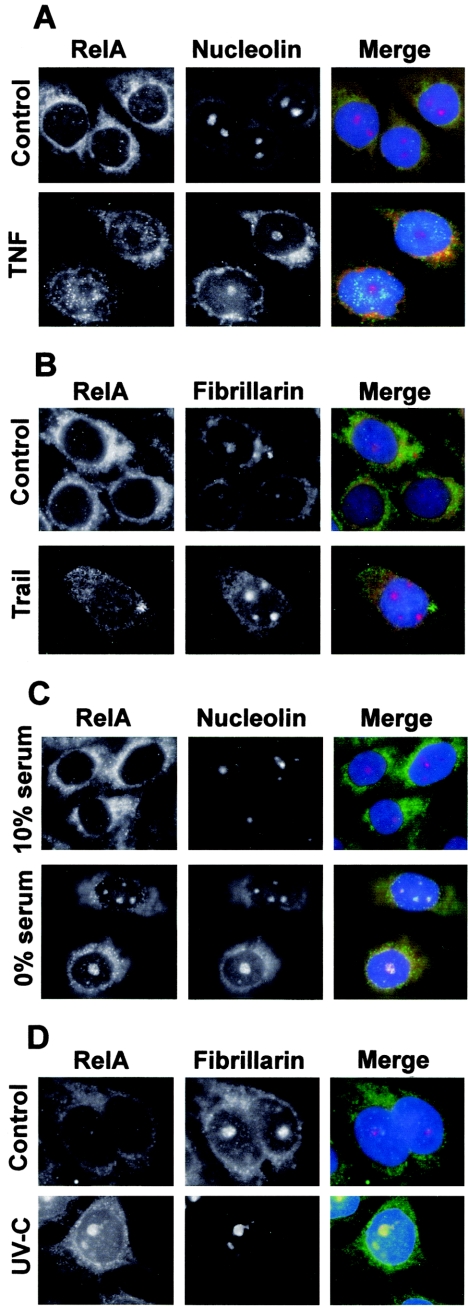
We wished to determine whether other NF-κB stimuli, particularly those known to be proapoptotic, had effects similar to that of aspirin on the nuclear distribution of RelA. Using immunocytochemical analysis, we examined the distribution of RelA in response to the proapoptotic NF-κB stimuli serum withdrawal, UV-C, and TRAIL and to the antiapoptotic signal TNF. Following exposure to TNF and TRAIL, we found that RelA was concentrated within the nucleoplasm as expected, but there was no detectable nucleolar staining for RelA (Fig. 3A and B). In contrast, serum deprivation (27) and UV-C (40 J/m2) radiation (22) resulted in RelA colocalization with the nucleolar proteins nucleolin and fibrillarin, thereby confirming that RelA also localizes to the nucleolus following exposure to these atypical activators of the NF-κB pathway. These data demonstrate clear differences in the nuclear distributions of RelA, depending upon the nature of the inducer. The data also suggest the intriguing possibility that the nucleolar sequestration of RelA is a general mechanism for regulating NF-κB activity and apoptosis.
FIG.3.
Specificity of stimuli causing nucleolar localization of RelA. (A) SW480 cells were unstimulated (control) or stimulated with TNF (10 ng/ml, 1 h) and then fixed, and immunocytochemistry was performed using specified antibodies. Nucleoli are stained with nucleolin. (B) Immunocytochemistry was performed on SW480 cells either untreated or treated with TRAIL (10 ng/ml) for 16 h. Fibrillarin was used to identify nucleoli. (C) SW480 cells were seeded in medium containing 10% serum and grown for 16 h. Medium was then replaced by fresh medium containing either 10% or 0% serum, and cells were grown for a further 96 h. Following fixation, immunocytochemistry was performed using antibodies to RelA and nucleolin as described above. (D) Immunocytochemistry, using anti-RelA and anti-nucleolin antibodies, was performed on SW480 cells 5 h after mock or UV-C (40 J/m2) irradiation. In all merged panels, DNA is stained by DAPI and appears blue.
Mode of accumulation of RelA within nucleoli.
To begin to elucidate the functional significance of these observations, we wished to understand the mechanism by which RelA is targeted to the nucleolus. We are particularly interested in the proapoptotic effects of aspirin and so used this agent as a model system for examining nucleolar targeting of RelA.
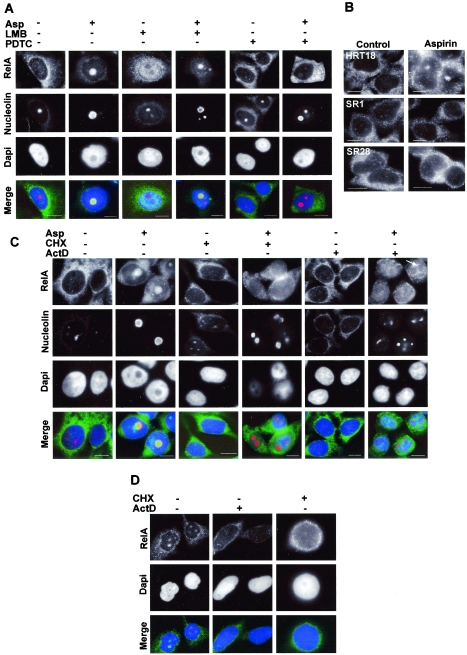
It is now recognized that RelA shuttles continuously between the cytoplasm and the nucleus of resting cells, utilizing the Crm1p nuclear export pathway (33). Since this export pathway is also used to export ribosomes from the nucleolus to the cytoplasm, we speculated that RelA exits the nucleus via the nucleolus and that nucleolar accumulation of RelA is caused by the disruption of Crm1p-mediated nuclear export. We postulated that if this were the case, then nucleolar accumulation of RelA would be apparent in the presence of the Crm1p inhibitor, leptomycin B. However, we found that in the presence of leptomycin B, RelA accumulated in the nucleoplasm and was excluded from nucleoli (Fig. 4A). Furthermore, leptomycin B had no effect on aspirin-induced nucleolar accumulation of RelA (Fig. 4A). These data indicate that the nucleolar localization of RelA is independent of Crm1p-mediated nuclear export.
FIG.4.
Mode of nucleolar accumulation of RelA. (A) Immunocytochemical staining showing localization of RelA in SW480 cells treated with aspirin (Asp; 5 mM for 16 h) in the presence (+) and absence (−) of the inhibitor of nuclear export, leptomycin B (LMB), and of the inhibitor of IκB degradation, PDTC. Nucleoli are stained with nucleolin (C23), and nuclei are stained with DAPI. (B) Expression of superrepressor IκB inhibits nucleolar accumulation of RelA. Parental HRT18 cells and two clones expressing mutant IκB, resistant to aspirin-induced degradation (IκB SR1 and SR28) were untreated (control) or treated with 5 mM aspirin for 16 h. The cellular localization of RelA was determined using immunocytochemical staining. (C) Nucleolar accumulation of RelA requires de novo protein synthesis. The immunomicrographs show the cellular localization of RelA and nucleolin following treatment with aspirin (Asp.) (5 mM, 16 h) in the presence and absence of 0.5 μg/ml actinomycin D (ActD) or 10μM cycloheximide (CHX). The arrow indicates the exclusion of RelA from remaining nucleoli in actinomycin D-treated cells. (D) Immunomicrographs showing the cellular localization of RelA in cells exposed to 0% serum for 96 h in the presence or absence of actinomycin D (ActD) or cycloheximide (CHX) as described above. DNA is stained by DAPI and appears blue in the merged panels. Bars, 10 μm.
Next, we wished to establish whether translocation from the cytoplasm to the nucleus is required for the nucleolar accumulation of RelA or whether it is the nucleoplasmic pool of RelA that is targeted to translocate to the nucleolus. We initially treated SW480 cells with aspirin in the presence of the inhibitor of IκB phosphorylation and degradation, PDTC. We found that PDTC blocked both aspirin-induced degradation of IκB (data not shown) and the nucleolar accumulation of RelA (Fig. 4A). This was further confirmed using HRT18 colon cancer cells which we engineered to constitutively express a superrepressor form of IκB (IκBSR). This IκB is mutated at the critical serine phosphorylation sites (S32/S36) and is resistant to aspirin-induced phosphorylation and degradation (42). Clones IκBSR1 and IκBSR28 express levels of the mutant IκB protein that inhibit aspirin-induced (42) and TNF-induced (51) nuclear translocation of NF-κB. We found that IκBSR1 and 28 clones showed nucleoplasmic staining for RelA prior to aspirin treatment (Fig. 4B). This would suggest that the basal turnover of NF-κB in these cells is independent of IκB 32/36 phosphorylation (36). However, the IκBSR clones showed no nucleolar accumulation of RelA upon the induction of the NF-κB pathway by aspirin (Fig. 4B). In contrast, aspirin-mediated nucleolar accumulation of RelA was observed in parental HRT18 cells (Fig. 4B). These data suggest that there are two independent pools of RelA/NF-κB complexes in these cells: a basal pool and an inducible, cytoplasmic pool. The data clearly show that the induction of the inducible pool to the nucleus is required for the nucleolar translocation of basal complexes.
Because the accumulation of several proteins within the nucleolus is transcription dependent, we next determined whether the nucleolar localization of RelA was modified by actinomycin D. Using immunocytochemistry and with nucleolin as a nucleolar marker, we found that the treatment of cells with aspirin in the presence of actinomycin D resulted in RelA staining throughout the nucleoplasm, with exclusion from the remaining nucleoli (Fig. 4C). Treatment with aspirin in the presence of cycloheximide also prevented translocation of RelA to the nucleolus, causing it to accumulate in the nucleoplasm (Fig. 4C). Similar results were obtained when SW480 cells were serum deprived in the presence of actinomycin D or cycloheximide, in that the nucleolar translocation of RelA was abrogated (Fig. 4D). These data confirm that aspirin and serum deprivation induce the translocation of RelA from the cytoplasm to the nucleoplasm and suggest that synthesis of one or more proteins is required for RelA translocation from the nucleoplasm to the nucleolus.
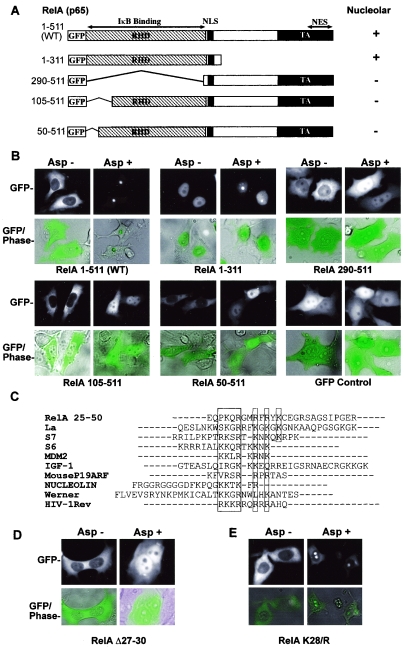
Identification of a RelA nucleolar localization domain.
To further understand the mechanism by which RelA translocates from the nucleoplasm to the nucleolus, we constructed a series of deletion mutants of RelA as GFP fusion proteins (Fig. 5A). The respective deletions were transiently transfected into SW480 cells and their subcellular distributions monitored in individual live cells. Prior to treatment, WT and deleted proteins were distributed within the cell as expected from published data (18). RelA containing the region between amino acids 1 and 311 (RelA 1-311) was predominantly nuclear, which has previously been described for RelA mutants lacking the nuclear export signal at the C terminus (Fig. 5B). RelA WT, 105-511, and 50-511 were localized within the cytoplasm, with a small amount of nucleoplasmic staining, while RelA 290-511 was distributed throughout the cell (Fig. 5B). Following exposure to aspirin, there was a decrease in cytoplasmic, and a corresponding increase in nuclear, levels of RelA in all cases, confirming the activation of the NF-κB pathway. However, there were distinct differences in the nuclear distributions of the GFP-tagged proteins, dependent upon the presence of the N terminus of the protein. We found that RelA WT and 1-311 accumulated in the nucleolus in response to aspirin (Fig. 5B). In contrast, mutants lacking the N-terminal 50 amino acids of the protein accumulated in the nucleoplasm and were excluded from the nucleolus (Fig. 5B). These data suggest that amino acids 1 to 50 of RelA contain the domain required for nucleolar localization. Aspirin had no effect on the cellular distribution of the GFP control (Fig. 5B).
FIG.5.
Aspirin-induced nucleolar translocation of RelA requires an N-terminal nucleolar localization motif. (A) Deletions of RelA were cloned into the PEGFP-C1 vector. RHD, Rel homology domain; NLS, nuclear localization signal; NES, nuclear export signal. The distributions of the respective deletions are shown. +, nucleolar; −, nucleoplasmic. (B) Deleting the N-terminal 50 aa of RelA blocks nucleolar translocation of the protein. SW480 cells were transfected with the specified GFP-tagged RelA expression constructs 24 h prior to treatment with 0 or 5 mM aspirin. The cellular distribution of GFP-tagged protein was determined in live/adherent cells using an Axiovert 100 inverted fluorescent microscope (magnification, ×40). (C) The N terminus of RelA contains a potential NoLS. Amino acids 1 to 50 of RelA were aligned to other published nucleolar localization signals (19, 47) using ClustalW. Each shaded area indicates a potential NoLS. (D) Deletion of aa 27 to 30 inhibits nucleolar translocation of RelA. SW480 cells were transfected with the RelA Δ27-30 expression construct 24 h prior to treatment with 0 or 5 mM aspirin. The cellular distribution of GFP-tagged protein was determined as described above. (E) Nucleolar localization of RelA does not involve posttranslational modification of lysine (K) 28. SW480 cells were transfected with a full-length GFP-RelA expression construct containing a K-to-R (arginine) mutation at residue 28 (RelA K28/R). The cellular distribution of this mutant was determined following aspirin treatment as described above.
For several proteins, it has been demonstrated that nucleolar localization is dependent upon the presence of a NoLS (19, 47). Sequence alignment of amino acids 1 to 50 of RelA with other published sequences known to mediate nucleolar localization revealed a potential NoLS at positions 27 to 38 of the protein. In particular, a motif (PKQR) at amino acids 27 to 30 appeared to be conserved among these sequences (Fig. 5C). Using site-directed mutagenesis, we generated a GFP-tagged RelA construct with a deletion of this motif (RelAΔ27-30). As shown in Fig. 5D, the RelAΔ27-30 mutant did not translocate to the nucleolus and consequently accumulated in the nucleoplasm.
RelA function is known to be modulated by acetylation (9) and ubiquitination (40), which both occur on lysine (K) residues. Since the motif we identified to be important for the nucleolar localization of RelA contained a lysine residue (K28), we postulated that this response may involve one of these modifications. However, we found that full-length RelA with a single K-to-R (arginine) mutation at residue 28 (GFP-RelA K28/R), which precludes acetylation or ubiquitination at this site, translocated to the nucleolus in response to aspirin in a fashion similar to that of the WT protein (Fig. 5E). This result is in keeping with aa 27 to 30 of RelA acting as an NoLS, since K and R residues are interchangeable within these motifs (47). Taken together, these data strongly suggest that RelA is actively transported from the nucleoplasm to the nucleolus and that there is a nucleolar localization domain located at the N terminus of the protein.
Nucleolar translocation of RelA is paralleled by a decrease in NF-κB-driven transcription.
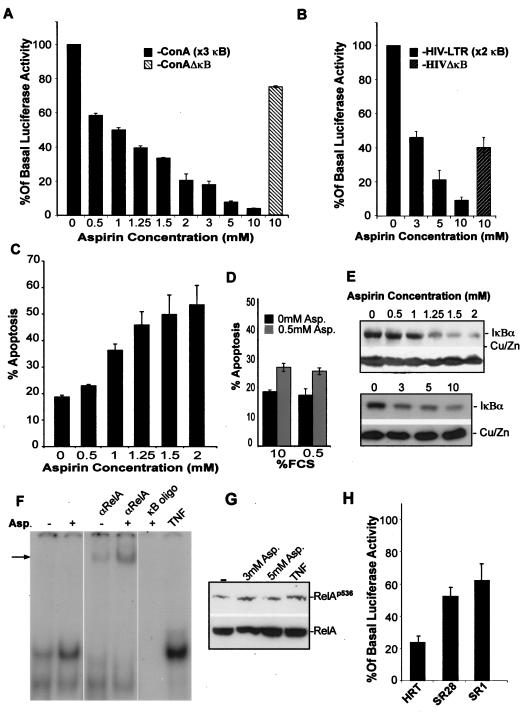
Having established that RelA accumulates in the nucleolus in response to specific proapoptotic NF-κB stimuli, we wished to determine whether this compartmentalization regulates nuclear NF-κB activity and apoptosis. To do this, we initially examined the effect of aspirin on NF-κB transcriptional activity using the HIV-LTR and 3enhancer-CON-A (3x κB ConA-luc) NF-κB-dependent luciferase reporter plasmids. We found that aspirin induced a significant decrease in basal NF-κB-driven luciferase activity in colon cancer cells (Fig. 6A and B). This decrease was apparent at 500 μM and was dependent upon both aspirin concentration and the number of κB sites in the luciferase construct (Fig. 6A and B). Furthermore, this decrease in NF-κB transcriptional activity paralleled the dose-dependent increase in the number of cells showing nucleolar foci of RelA that we observed (Fig. 1A). Aspirin (at the highest concentration used [10 mM]) had a minimal effect on the transcription from control plasmids with deleted κB sites compared to wild-type equivalents (Fig. 6A and B), demonstrating that the observed decrease is dependent upon NF-κB. Using annexin V-FITC staining of cell surface phosphatidylserine residues, we demonstrated that aspirin effects on the nuclear distribution of RelA and NF-κB transcriptional activity were also paralleled by the apoptosis of colon cancer cells (Fig. 6C). In the above experiments, cells were treated with aspirin and other NF-κB stimuli in conditions of low serum (0.5% FCS). Since total serum deprivation (0% FCS) had an effect similar to that of aspirin on RelA nuclear distribution (Fig. 3C), we considered whether the NF-κB and apoptotic responses to low-dose aspirin were enhanced by low-serum conditions. However, we found no difference in aspirin response when treatment occurred in 0.5% and 10% FCS media in terms of the nucleolar translocation of RelA, decreased NF-κB-driven transcription, and apoptosis (Fig. 6D and data not shown).
FIG. 6.
Nucleolar localization of RelA is paralleled by a decrease in NF-κB-driven transcription. (A and B) Pharmacologically relevant doses of aspirin repress NF-κB transcriptional activity. SW480 cells were transfected with 6 μg of wild-type (3x κB ConA and HIV-LTR) NF-κB-dependent luciferase reporter constructs or with equivalent plasmids with κB sites deleted (HIVΔκB, ConAΔκB) along with 3 μg of pCMVβ control plasmid. Twenty-four hours after transfection, cells were either unstimulated or stimulated with aspirin for a further 16 (3 to 10 mM) or 72 (0 to 2 mM) hours. Results were normalized using β-galactosidase activity and are presented as the percentages of relative luciferase activity compared to basal levels (untreated controls). (C) Aspirin induces apoptosis. Annexin V-FITC apoptosis assays were performed on aspirin-treated (0 to 2 mM, 72 h) SW480 cells. Apoptotic cells were visualized using fluorescent microscopy, and the percentages of apoptotic cells within the total cell populations were calculated. (D) Aspirin-induced apoptosis is not enhanced in low-serum conditions. Annexin V-FITC apoptosis assays were performed on SW480 cells treated with aspirin (Asp.) (0 or 0.5 mm, 72 h) in low-serum (0.5% FCS) or normal-serum (10% FCS) conditions. (E) Aspirin induces degradation of IκB. Immunoblot showing cytoplasmic levels of IκBα and control protein (Cu/ZnSOD) in SW480 cells treated with 0 to 2 mM aspirin for 72 h or 3 to 10 mM aspirin for 16 h. (F) Aspirin induces nuclear translocation of RelA/NF-κB complexes. SW480 cells were either unstimulated (−) or stimulated (+) with aspirin (5 mM for 16 h) or TNF (10 ng/ml, 1 h). Nuclear extracts were prepared and analyzed by EMSAs using a κB oligonucleotide. The incubation of extracts with the RelA antibody prior to the assay resulted in a complete shift of theaspirin-induced complex, confirming the complex consists predominantly of this component. The arrow indicates the supershifted band. The aspirin-induced complex was also competed out with 100× molar excess of unlabeled κB oligonucleotide. (G) Phosphorylation of RelA serine 536 in response to aspirin. SW480 cells were treated with aspirin (0 to 5 mM, 16 h) or TNF (10 ng, 1 h), and then immunoblot analysis was performed on total-cell extract. Blots were initially probed with a RelA phospho 536-specific antibody (Calbiochem) and then stripped and reprobed for total RelA. (H) Expression of superrepressor IκB blocks aspirin-mediated repression of NF-κB transcriptional activity. HRT18 parental cells and clones IκBSR1 and SR28 (which are resistant to aspirin-induced nuclear and nucleolar translocation of RelA) were transfected with the κB (HIV-LTR) and pCMV-β reporter constructs prior to a 16-h treatment with 0 or 5 mM aspirin. Results shown are normalized using β-galactosidase activity and are presented as described above.
Aspirin has previously been shown to inhibit TNF-induced NF-κB-driven transcription, and this has been attributed to the inhibition of IKK activity/degradation of IκB (23, 49, 50). To eliminate the possibility that the decrease in NF-κB-driven transcription we observed in response to this particular agent is caused by the inhibition of IκB degradation, we further examined the aspirin-mediated activation of the NF-κB pathway. Western blot analysis of cytoplasmic extracts and EMSAs performed on nuclear extracts demonstrated reduced levels of cytoplasmic IκB (Fig. 6E) and a strong induction of nuclear NF-κB (Fig. 6F) in response to aspirin. The aspirin-induced NF-κB complex had the same mobility as that induced by TNF, and supershift EMSAs confirmed that it consisted predominantly of the RelA transcriptional activator (Fig. 6F). Since the IκB promoter is recognized to be a target of NF-κB, we used the MG132 proteosome inhibitor to confirm that the reduction in cytoplasmic IκB levels was mediated by the proteosomal degradation of the protein (42; also data not shown). The phosphorylation of RelA at serine 536 is critical for the full activation of the protein (8). Western blot analysis, using phospho 536-specific antibodies, indicated that the aspirin-induced phosphorylation of RelA at this residue was comparable to that seen following treatment with TNF (Fig. 6G). These data confirm that the observed decrease in NF-κB transcriptional activity in response to aspirin is associated with IκB degradation and the nuclear translocation of RelA/NF-κB complexes, thereby providing strong evidence against the direct inhibition of IKK activity as the underlying cause of the aspirin effects. The upstream mechanisms by which aspirin activates the NF-κB pathway are currently subject to ongoing investigation in our lab and are outside the focus of this report.
Since we observed constitutive nuclear NF-κB activity in colon cancer cells, which decreased upon treatment with aspirin, we postulated that aspirin may (directly or indirectly) induce the repression of NF-κB-driven transcription within the nucleus independently of its effects on cytoplasmic NF-κB and the nuclear distribution of RelA. Therefore, we wished to test whether, like nucleolar accumulation of RelA, the cytoplasmic-to-nuclear translocation of NF-κB complexes was also required for this reduction in NF-κB-driven transcription. We found that aspirin mediated a 5.8-fold decrease in NF-κB transcriptional activity in parental HRT 18 cells, compared to a 1.9-fold decrease in cells expressing dominant-negative IκBα (IκBSR) (Fig. 6H). This diminished response was independent of basal transcription because, consistent with the basal nucleoplasmic staining for RelA (Fig. 4B), the IκBSR clones showed levels of basal NF-κB driven transcription similar to those of their parental counterparts (data not shown). These data indicate that the repression of NF-κB-driven transcription is also a consequence of IκB degradation and the nuclear translocation of NF-κB complexes.
On the basis of our observations so far, we hypothesized that the stimulation of the NF-κB pathway by particular signals promotes apoptosis by inducing the nucleolar sequestration of RelA and the inhibition of NF-κB-driven transcription. Next, we undertook a series of experiments to determine whether the nucleolar translocation of RelA is causally involved in the reduction of κB-driven transcription and the induction of apoptosis.
Temporal relationship of IκB degradation, nucleolar accumulation of RelA, repression of NF-κB activity, and apoptosis.
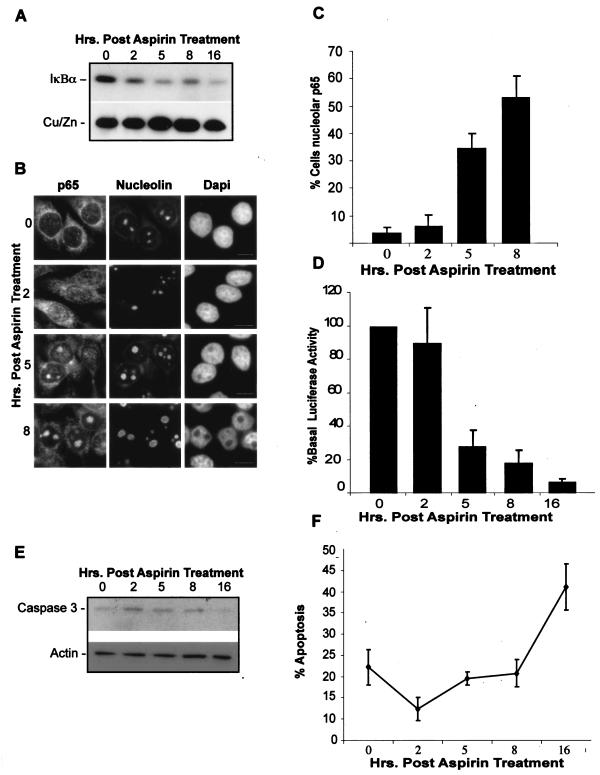
As a first step, we examined the kinetics of these responses. We found that aspirin-induced IκB degradation occurred within 2 hours of treatment (Fig. 7A) and was associated with an initial nucleoplasmic accumulation of RelA (Fig. 7B). This nucleoplasmic RelA had no effect on NF-κB-driven transcription. The percentage of cells showing nucleolar RelA increased progressively to a peak at 5 to 8 h (Fig. 7B and C), which coincided with the onset of NF-κB transcriptional repression (Fig. 7D). The nucleolar accumulation of RelA was not a consequence of cell death, since apoptosis, as determined by caspase-3 cleavage (Fig. 7E) and annexin V staining (Fig. 7F), was not observed until 16 h after aspirin treatment.
FIG. 7.
IκB degradation, nucleolar translocation of RelA, repression of NF-κB transcription, and apoptosis occur sequentially. (A) IκB degradation occurs 2 to 5 h after stimulation with aspirin. Cells were treated with aspirin (10 mM) for 0 to 16 h, and then cytoplasmic IκBα and control protein (CU/Zn SOD) levels were determined using Western blot analysis. (B and C) Nucleolar accumulation of RelA (p65) is apparent 5 to 8 h after aspirin stimulation. Immunocytochemistry was performed on SW480 cells treated as described above. DAPI staining depicts nuclei. Nucleoli are stained with nucleolin. Bars, 10 μm. The percentage of cells in the population showing nucleolar RelA (p65) was quantified. Data are the means of at least three experiments (± SE). (D) Decreased NF-κB-driven transcription occurs concurrently with nucleolar accumulation of RelA. SW480 cells were transfected with the NF-κB dependent 3xκB ConA-luc and the control pCMVβ reporter constructs. Twenty-four hours after transfection, cells were treated as described above. Values are presented as the percentages of luciferase activity at time zero and are the means (± SE) of three independent experiments after β-galactosidase normalization. (E) Aspirin-induced caspase-3 cleavage occurs 16 h after stimulation. SW480 cells were treated as described above and levels of uncleaved caspase-3 determined by Western blot analysis performed on cytoplasmic extracts. (F) Percentages of SW480 cells undergoing apoptosis, as determined by annexin V staining, following aspirin treatment as described above.
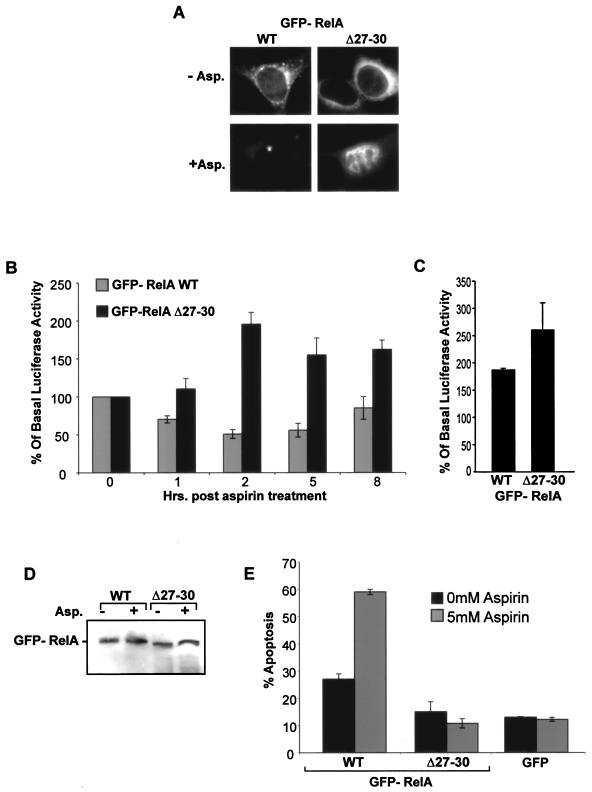
Inhibiting nucleolar translocation of RelA inhibits the aspirin-induced decrease in NF-κB transcriptional activity and apoptosis.
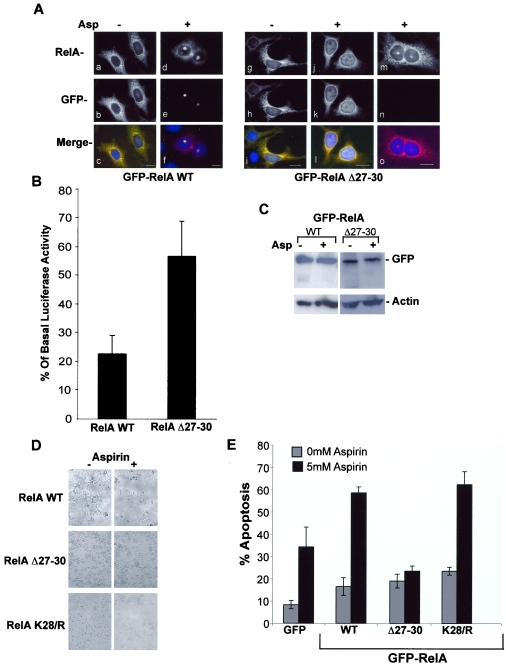
We next utilized the RelA NoLS deletion mutant (RelA Δ27-30) to directly test whether the nucleolar localization of RelA is causally involved in the repression of NF-κB-driven transcription and apoptotic cell death. We determined whether exogenously expressing RelAΔ27-30 modulates the distribution of endogenous protein. Using RelA immunocytochemistry performed on transfected SW480 cells, we found that, as expected, cells expressing RelA WT showed nuclear and nucleolar localization of RelA in response to aspirin (Fig. 8A). However, in cultures expressing RelA Δ27-30, which lacks the N-terminal NoLS, there was no nucleolar translocation of either mutant or endogenous protein in transfected cells. In contrast, adjacent nontransfected cells showed aspirin-mediated nucleolar accumulation of endogenous protein (Fig. 8A). These results suggest that mutants which are unable to translocate to the nucleolus act in a dominant-negative fashion to block the nucleolar translocation of endogenous RelA.
FIG.8.
Inhibiting nucleolar translocation of RelA blocks aspirin-induced repression of NF-κB-driven transcription and apoptosis. (A) Exogenally expressed RelA Δ27-30 blocks nucleolar translocation of endogenous RelA. Immunomicrographs (magnification, ×63) showing localization of all (RelA) or GFP-tagged (GFP) RelA in SW480 cells transfected with GFP-WT (a to f) or Δ27-30 (g to o) RelA and then treated with 0 (−) or 5 (+) mM aspirin (Asp) for 16 h. Panels m to o show RelA in nontransfected cells (confirmed by the lack of a GFP signal in panel n) adjacent to the RelA Δ27-30-transfected cells shown in panels j to l. Blue depicts DAPI staining of DNA. Bars, 10 μm. (B) Deletion of amino acids 27 to 30 of RelA inhibits aspirin effects on NF-κB-driven transcription. SW480 cells were transfected with the GFP-RelA WT or Δ27-30 expression vector, along with the 3x κB ConA-luc reporter plasmid. Following aspirin treatment (5 mM, 16 h), GFP-tagged cells were isolated using a fluorescence-activated cell sorter. Relative luciferase activity was calculated as the level of luciferase activity per GFP-expressing cell in the population. Results are presented as the reduction in relative luciferase activity compared to untreated controls and are the means of at least three experiments (± SE). The basal (12.3 × 106; SE, 3.2 × 106) levels of luciferase activity were comparable between cells expressing WT-RelA and those expressing Δ27-30-RelA (11 × 106; SE, 1.55 × 106). (C) Immunoblots showing GFP and control protein (Actin) expression levels in cells transfected with GFP-tagged WT or Δ27-30 RelA before and after aspirin (5 mM, 16 h) treatment. (D) Deletion of amino acids 27 to 30 of RelA inhibits aspirin effects on cell viability. SW480 cells were transfected with the indicated RelA expression vectors 24 h prior to stimulation with aspirin (5 mM, 16 h). Phase-contrast images of randomly selected fields are shown (magnification, ×10). (E) Deletion of amino acids 27 to 30 of RelA inhibits aspirin-induced apoptosis. SW480 cells were transfected with the specified vectors and treated with aspirin as described above. Annexin V-biotin staining, with a Texas Red-streptavidin conjugate, was used to identify apoptotic cells. The percentages of cells expressing GFP-tagged RelA undergoing apoptosis were determined by fluorescent microscopy in at least 250 transfected cells for each sample. The number of apoptotic cells in the nontransfected population was also determined. The results are the means of at least three independent experiments ± standard errors.
Using the dominant-negative NoLS deletion mutant, we initially examined the effects of inhibiting the nucleolar sequestration of RelA on the aspirin-induced repression of NF-κB-driven transcription. SW480 cells were transiently transfected with the GFP-RelA WT or Δ27-30 expression constructs along with the 3x κB ConA-Luc reporter vector. Following aspirin treatment, RelA-expressing cells were isolated from nontransfected cells using a fluorescence-activated cell sorter. The NF-κB reporter activity was then assayed in the GFP-RelA WT/Δ27-30-expressing population. Our data show that the aspirin-induced repression of NF-κB transcriptional activity was substantially abrogated in RelA Δ27-30-expressing cells (in which nucleolar translocation had been blocked) compared to those expressing the wild-type protein (RelA-WT) (Fig. 8B). The differences in responses could not be accounted for by differing basal levels of NF-κB-driven transcription, because these were comparable for cells expressing RelA-WT (12.3 × 106; SE, 3.2 × 106) and RelA Δ27-30 (11 × 106; SE, 1.55 × 106). The levels of expression of GFP-tagged protein were also comparable for wild-type and mutated RelA and remained unchanged by aspirin treatment (Fig. 8C).
To determine whether inhibiting RelA nucleolar translocation blocks the aspirin-induced growth inhibition of cancer cells, we initially examined aspirin effects on the viability of SW480 cell cultures transfected with GFP-RelA WT, K28/R, or the construct with the deletion of the NoLS, RelA Δ27-30. Aspirin induced a marked reduction in the number of viable colorectal cancer cells in cultures expressing GFP alone, GFP-RelA WT, or K28/R. However, this effect was inhibited in cultures expressing RelA Δ27-30 (data not shown). Phase-contrast microscopy confirmed the extensive cell death in cultures expressing RelA WT and RelA K28/R, which translocate to the nucleolus, while aspirin had a minimal effect on the numbers and morphology of cells within cultures expressing RelA Δ27-30, which is excluded from the nucleolus (Fig. 8E). Transfection efficiencies were similar for all expression vectors.
Next, we investigated the effects of expressing WT and mutated RelA on aspirin-induced apoptosis. Use of a biotin-annexin V assay, along with a streptavidin-Texas Red conjugate, allowed us to distinguish apoptotic and transfected cells and thus determine the percentages of cells undergoing apoptosis specifically in the transfected and nontransfected cell populations. We found that aspirin-induced apoptosis was inhibited in cells expressing RelA-Δ27-30 (Fig. 8F). In contrast, compared to the expression of GFP alone, apoptosis was enhanced in cells expressing RelA-WT and K28/R. Aspirin-induced apoptosis was observed to similar extents in all nontransfected cells (data not shown).
Amino acids 27 to 30 of RelA are essential for aspirin-induced repression of NF-κB-driven transcription and apoptosis.
To definitively establish whether the nucleolar targeting of RelA is absolutely required for aspirin effects on NF-κB-driven transcription and apoptosis, we utilized rela null MEFs transfected with either GFP, GFP-RelAWT, or -RelAΔ27-30. Aspirin effects on the localization of these GFP-tagged proteins were similar to those observed in colorectal cancer cell lines; both RelA WT and Δ27-30 translocated from the cytoplasm to the nucleus in response to aspirin. However, only RelA WT was able to translocate from the nucleoplasm to the nucleolus (Fig. 9A). The deletion of amino acids 27 to 30 abrogated this response, causing RelA to accumulate in the nucleoplasm (Fig. 9A). Aspirin had no effect on the localization of GFP alone (data not shown). Next, we examined aspirin effects on NF-κB-driven transcription in these cells using the 3x κB ConA-luc NF-κB-dependent reporter plasmid. We found that 2 to 5 h after aspirin (10 mM) treatment, there was a twofold repression in NF-κB transcriptional activity in cells expressing GFP-RelA WT (Fig. 9B). In marked contrast, there was a twofold activation of NF-κB-driven transcription in cells expressing GFP-RelA Δ27-30. WT and Δ27-30 RelA activated NF-κB-driven transcription in response to TNF to comparable degrees (Fig. 9C). Neither aspirin nor TNF had any effect on the equivalent NF-κB reporter plasmid with κB sites deleted (data not shown), confirming the specificity of these effects. Western blot analysis confirmed that WT and Δ27-30 RelA proteins were expressed at similar levels before and after aspirin treatment (Fig. 9D) and that the MEFs were indeed null for endogenous rela (data not shown). These data establish the crucial importance of aa 27 to 30 relative to aspirin effects on NF-κB transcriptional activity and also demonstrate that the differences in aspirin responses between the wild-type protein and the protein with the deletion of NoLS do not reflect a general difference in their DNA binding and transactivating potentials.
FIG. 9.
Amino acids 27 to 30 of RelA are essential for aspirin effects on NF-κB transcriptional activity and apoptosis in rela null MEFs. (A) rela null MEFs were transfected with 3 μg of GFP-RelA WT or -RelAΔ27-30. Cells were either unstimulated (−Asp.) or stimulated for 5 h with aspirin (10 mM) (+Asp.), and then the localization of GFP-tagged proteins was analyzed in fixed cells using fluorescent microscopy (magnification, ×63). (B and C) rela null MEFs were transfected with the above plasmids along with 3 μg of 3xκB ConA NF-κB dependent luciferase reporter constructs and 3 μg of pCMVβ control plasmid. Twenty-four hours after transfection, cells were either unstimulated, (B) stimulated with aspirin (10 mM) for the times specified, or (C) stimulated with TNF (10 ng/ml) for 5 h. Results were normalized using β-galactosidase activity and are presentedas the percentages of relative luciferase activity compared to basal levels (untreated controls). The basal luciferase values for cells expressing GFP-RelAWT (26.36 × 106; SE, 8.6 × 106) and those expressing GFP-RelA Δ27-30 (17.15 × 106; SE, 5.9 × 106) were not significantly different. (D) Western blot analysis showing expression levels of GFP-RelA WT and Δ27-30 in aspirin-treated and untreated rela null MEFs. (E) The NoLS of RelA is required for aspirin-induced apoptosis. SW480 cells were transfected with the specified vectors and treated with aspirin as shown. Annexin V-biotin staining, with a Texas Red-streptavidin conjugate, was used to identify apoptotic cells. The percentages of cells expressing GFP-tagged RelA undergoing apoptosis were determined by fluorescent microscopy in at least 250 transfected cells for each sample. The results are the means of at least three independent experiments ± standard errors.
We next used annexin V assays to investigate the apoptotic response of these cells to aspirin. We found that nontransfected rela null MEFs were resistant to aspirin-induced apoptosis but underwent extensive apoptosis in response to TNF (data not shown). However, when they were transfected with WT RelA, there was a decrease in the number of viable cells (data not shown) and a substantial increase in apoptosis in response to aspirin (Fig. 9E). This apoptotic response was blocked in cells expressing RelA with a deletion of the NoLS and in cells expressing GFP alone (Fig. 9E). These data confirm the importance of RelA, and in particular the NoLS of RelA (aa 27 to 30), in aspirin-induced apoptosis.
Inhibiting the nucleolar translocation of RelA inhibits UV-C-induced apoptosis.
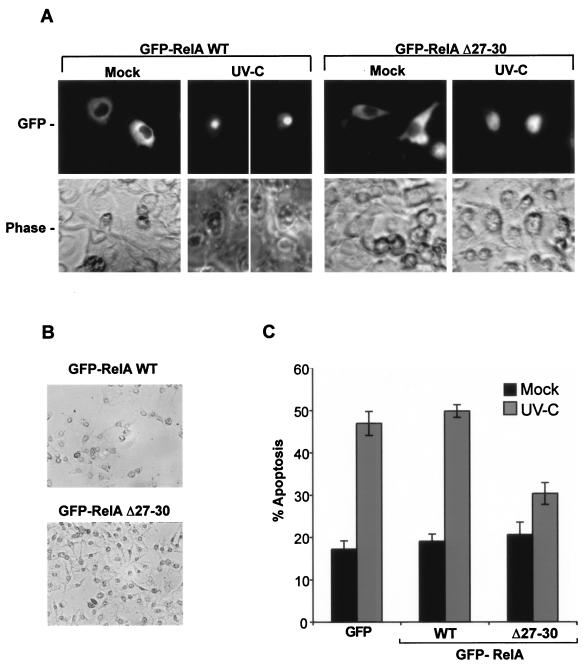
UV-C radiation is a recognized proapoptotic stimulus of NF-κB (5, 22) that, as we have shown, also targets RelA to the nucleolus (Fig. 3C). Having established the importance of the nucleolar translocation of RelA in aspirin-induced apoptosis, we next determined whether blocking this pathway also inhibited the proapoptotic effects of UV-C. First, we needed to know whether the region from aa 27 to 30 was involved in the UV-C-induced nucleolar translocation of RelA. SW480 cells were transfected with GFP, GFP-RelA WT, or GFP-RelAΔ27-30 and then mock or UV-C (40 J/m2) irradiated. Live-cell imaging was used to determine UV-C effects on the nuclear distribution of GFP-tagged proteins. Figure 10A demonstrates that GFP-RelA WT, in a manner similar to the aspirin response, accumulates in the nucleolus after UV-C treatment and that GFP-RelAΔ27-30 is excluded from the nucleolus, accumulating in the nucleoplasm. RelA WT and Δ27-30 remained cytoplasmic in all mock-irradiated cells, and UV-C had no effect on the distribution of GFP alone (data not shown). These data confirm the importance of aa 27 to 30 for the nucleolar targeting of RelA.
FIG. 10.
Nucleolar localization of RelA is required for UV-C-induced apoptosis. (A) Deleting aa 27 to 30 inhibits UV-C-induced nucleolar translocation of RelA. SW480 cells were transfected with 6 μg of GFP, GFP-RelA WT, or RelA Δ27-30 24 h prior to UV-C treatment (40 J/m2). The localization of GFP-tagged proteins was determined in live cells using an Axiovert 100 inverted fluorescent microscope (magnification, ×40) 8 h after treatment. (B) Expression of RelA Δ27-30 inhibits UV-C effects on cell viability. SW480 cells were transfected and UV-C treated as described above. Phase-contrast images (magnification, ×20) taken of randomly selected fields 24 h after treatment are shown. (C) Expression of RelA Δ27-30 inhibits UV-C effects on apoptosis. Annexin V apoptosis assays were performed on SW480 cells transfected with the specified GFP-tagged vectors 24 h after UV-C (40 J/m2) treatment. The percentages of cells expressing GFP-tagged protein undergoing apoptosis were determined by fluorescent microscopy in at least 250 transfected cells for each sample. The results are the means of at least three independent experiments ± standard errors.
Next, we examined the effects of expressing GFP-RelA WT and Δ27-30 on UV-C-induced changes in cell viability. We found that UV-C (40 J/m2) irradiation caused a marked reduction in the number of viable SW480 cells in cultures expressing GFP alone or GFP-RelAWT (data not shown). However, these effects were abrogated in cultures expressing RelAΔ27-30. Phase-contrast microscopy confirmed that UV-C mediates extensive cell loss in cultures expressing GFP-RelAWT while having a minimal effect on the numbers or morphology of cells in cultures expressing RelAΔ27-30 (Fig. 10B). To confirm that the observed cell loss was mediated by apoptosis and that Δ27-30 RelA blocked this apoptosis, we used biotin-conjugated annexin V apoptosis assays, along with streptavidin-linked Texas Red, to determine the percentage of transfected cells undergoing apoptosis. Figure 10D demonstrates that there are twofold increases in the percentages of cells undergoing apoptosis in cells expressing GFP alone or GFP-RelAWT in response to UV-C. However, this effect is blocked by the expression of the NoLS deletion mutant, RelAΔ27-30. Taken together with data presented above, these data provide compelling evidence that the nucleolar targeting of RelA is involved in the regulation of NF-κB and apoptosis in response to particular proapoptotic NF-κB stimuli.
DISCUSSION
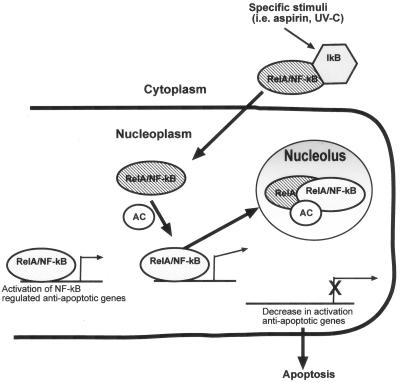
Mechanisms that regulate NF-κB activity within the nucleus are critical for determining the effect that stimulating this pathway has on downstream target genes and thus on the growth and death of cells (8). Here we demonstrated, using immunocytochemistry to detect endogenous protein, live-cell imaging, and biochemical assays, that the RelA component of NF-κB localizes to the nucleolus in response to aspirin and other proapoptotic NF-κB stimuli. Our analysis of the function of this subnuclear compartmentalization suggests a model for the regulation of nuclear NF-κB activity and the modulation of apoptosis (Fig. 11).
FIG. 11.
A model for the compartmental modulation of NF-κB transcriptional activity and apoptosis. In nonstimulated tumor cells, there are two pools of NF-κB, an inducible cytoplasmic pool and a basal pool that drives the transcription of antiapoptotic genes. Upon induction by aspirin, UV-C radiation, serum deprivation, and similar stimuli, inducible RelA/NF-κB complexes translocate into the nucleus, recruiting an additional cofactor (AC) to the basal pool. The induced RelA/cofactor complex does not activate transcription but complexes with basal RelA/NF-κB, and then all nuclear RelA translocates to the nucleolus. Once in the nucleolus, RelA is in a physical location different from that of its target promoters, resulting in a decrease in transcription of antiapoptotic genes and, consequently, apoptosis.
In addition to ribogenesis, it is increasingly apparent that the nucleolus has an important role in regulating cell cycle transition, cell proliferation, and apoptosis through the sequestration of key cell cycle and transcription factors (7, 38). Examples include the NF-κB coactivator E2F-1 (26), hypoxia-inducible factor-1 (15), and p53 (37, 47). Here we identify an N-terminal motif of the RelA component of NF-κB that targets the protein to the nucleolus. We demonstrate both in colorectal cancer cell lines and in mouse embryo fibroblasts that this nucleolar localization motif is required for aspirin-induced apoptosis. Furthermore, using RelA with a deletion of this motif, we demonstrate that the nucleolar translocation of RelA is required for UV-C induced apoptosis. Therefore, we conclude that the nucleolus also plays a role in regulating NF-κB effects on cell growth and death.
Our model suggests that the mechanism by which the nucleolar targeting of RelA mediates apoptosis is by sequestering RelA away from target promoters, thus causing a decrease in the transcription of NF-κB-dependent, antiapoptotic genes. Our conclusion that the compartmentalization of RelA within the nucleolus modulates NF-κB-driven transcription is based on the following observations. First, dose-response (Fig. 1 and 6) and kinetic (Fig. 7) studies show a relationship between the nucleolar accumulation of RelA and a reduction in NF-κB-driven transcription. Second, cell fractionation studies indicate that the decrease in the basal pool of RelA in the nucleoplasm (where target promoters are located) and the corresponding increase in nucleolar RelA in response to aspirin are associated with a reduction in NF-κB transcriptional activity (Fig. 2C and 6A). Finally, and most convincingly, blocking the nucleolar localization of RelA with the dominant-negative NoLS deletion mutant (RelA Δ27-30) reversed the effects of aspirin on basal NF-κB transcriptional activity in each of the cellular models studied (Fig. 8 and 9).
In order to be fully active, nuclear RelA is required to undergo a number of posttranslational modifications, principally phosphorylation and acetylation (8). Therefore, we considered the possibility that the reductions in NF-κB-driven transcription and apoptosis were caused by the induction of transcriptionally inactive RelA rather than by nucleolar translocation of basal RelA per se. However, the NoLS deletion mutant of RelA had the same established phosphorylation and acetylation sites as the wild-type GFP-tagged protein. However, the RelA with the deletion activated, rather than repressed, NF-κB-driven transcription in response to aspirin and activated NF-κB transcriptional activity in response to TNF to a level comparable with that of wild-type protein. Furthermore, mutation of the lysine residue within this motif, which is a potential site for acetylation, did not affect the outcome of the aspirin treatment. Our data suggest that a cofactor(s) is required for the nucleolar translocation of RelA. Therefore, we also considered that it is the binding of this cofactor to basal RelA that causes it to become actively repressive at promoter sites, inducing apoptosis. However, our kinetic studies show that at 2 h, when aspirin-induced RelA is nucleoplasmic, there is no change in NF-κB transcriptional activity (Fig. 7). It is only during RelA nucleolar translocation (at 5 to 8 h) that aspirin mediates a decrease in NF-κB-driven transcription.
We did consider the possibility that the nucleolar sequestration of RelA and the reduction in NF-κB transcriptional activity were consequences rather than the causes of cell death. However, we observed extensive apoptosis of SW480 colon cancer cells in response to the activator of NF-κBdriven transcription, TRAIL (42, 51; also data not shown), but RelA was nucleoplasmic and remained excluded from the nucleolus (Fig. 3). Furthermore, our kinetic studies clearly demonstrated that aspirin-induced nucleolar localization of RelA and reduction in NF-κB transcriptional activity occurred before caspase-3 cleavage and also before any detectable increase in apoptotic cell populations. Finally, and most convincingly, aspirin- and UV-C-induced apoptosis was inhibited in cells expressing the RelAΔ27-30 mutant (Fig. 8, 9, and 10). Therefore, it is very unlikely that the observed responses are a result of apoptosis. Taken together, these data provide convincing evidence that diverting RelA from its gene targets in the nucleoplasm to the nucleolus can regulate nuclear NF-κB transcriptional activity and mediate apoptosis. These novel findings contribute to the growing literature on the importance of the nucleolus as a regulatory structure.
We observed nucleolar translocation of RelA in response to UV-C radiation and demonstrated that deleting the NoLS of RelA blocked UV-C-induced apoptosis. Our model would suggest that the mechanism for this apoptosis is a decrease in the transcription of NF-κB-dependent, antiapoptotic genes. This is of particular interest because Campbell et al. (5) recently demonstrated that UV-C-induced nuclear translocation of RelA/NF-κB complexes mediated the repression of the antiapoptotic Bcl-xl gene. However, they observed increased binding of RelA at the Bcl-xl promoter in response to UV-C and proposed a mechanism of active repression, whereby basal NF-κB complexes are converted from active to repressive. This is in contrast to the model that we propose, whereby induced RelA relocalizes basal RelA/NF-κB complexes to the nucleolus, sequestering them away from target promoters. Because of differences in experimental setups and outcomes, it is currently difficult to reconcile the differences between these two models. However, it is possible that in the complexities of the subcellular environment, both models might apply.
The degradation of IκB and the cytoplasmic-to-nuclear translocation of NF-κB/RelA complexes were absolutely required for nucleolar translocation of both induced and basal RelA (Fig. 4). Therefore, we propose that the NF-κB complexes induced by aspirin, UV-C radiation, and serum withdrawal recruit a specific cofactor(s) to the basal, promoter-bound complexes, which then transport all nuclear RelA to the nucleolus. This is supported by the kinetic studies demonstrating that induced RelA distributes throughout the nucleoplasm in basal complexes (Fig. 7) prior to localization in the nucleolus. Furthermore, in the absence of protein synthesis, RelA accumulates in the nucleoplasm in response to aspirin and serum deprivation, suggesting that an additional cofactor(s) is required for transport to the nucleolus. Although we do not know the nature of this cofactor at present, it is of particular interest that RelA has recently been found to interact with the nucleolar proteins NFBP (43) and nucleophosmin/B23 (12). Since the increased transcription of nucleophosmin has been identified as an early gene response to specific external stimuli (48), this is a particularly strong candidate. Another potential candidate is the nucleolar protein p14ARF, which has recently been shown to interact with RelA and inhibit NF-κB-driven transcription (32). We show here and elsewhere (42, 51) that NF-κB is rapidly induced by TNF and TRAIL and localizes in the nucleoplasm, whereas RelA localizes to the nucleolus in response to aspirin, serum deprivation, and UV-C radiation, which all induce NF-κB with much slower kinetics (5, 16, 42). It therefore seems possible that the kinetics of NF-κB induction determines the recruitment of the specific cofactors responsible for the subnuclear distribution of RelA and the downstream effects on cell growth and death. Defining the mechanism that determines the subnuclear distribution of translocated NF-κB complexes could have considerable relevance to the development of chemotherapeutic agents and also provide further insight into the complexities of the NF-κB pathway.
Our finding that aspirin activates the NF-κB pathway would appear to question a number of studies demonstrating that nonsteroidal anti-inflammatory drugs (NSAIDs) inhibit the activation of NF-κB (23, 49, 50). However, these studies examined only the very short-term effects of NSAIDs on the cytokine-induced activation of NF-κB. In the present study, we investigated the longer-term effects of aspirin alone on NF-κB signaling. Since aspirin mediates the apoptosis of cancer cells in vitro in the absence of additional cytokines, these experimental conditions are most relevant to the proapoptotic properties of the agent. The NSAID celecoxib (28) and the COX-2-specific inhibitor NS-398 (41) have also been shown to induce the degradation of IκB and the nuclear translocation of NF-κB in the absence of additional NF-κB stimulators. Whether activation of the NF-κB pathway is a primary effect of aspirin or secondary to its effect on another cellular pathway has yet to be established. However, we have demonstrated that nuclear and nucleolar translocations of RelA are absolutely required for aspirin-mediated apoptosis, indicating that it is this pathway that is ultimately responsible for the proapoptotic effects of the agent.
The constitutive activity of NF-κB has been shown to contribute to carcinogenesis in a number of cancer types (17). Therefore, in identifying a cellular mechanism to repress NF-κB-driven transcription, the results presented here may have general relevance to future rational drug design for cancer therapy. Furthermore, the mechanism of the apoptotic response induced by aspirin has particular relevance to understanding the mechanism by which NSAIDs protect against cancer. Gaining further understanding of how RelA is sequestered in the nucleolus may allow the design of small molecules which specifically target RelA to this compartment.
Acknowledgments
We gratefully acknowledge the gifts from Ron T. Hay (University of St. Andrews) of the IκBα antibody, the HIV-LTR and 3enhancer CON-A luciferase reporter constructs, and the rela null mouse embryo fibroblasts, and we thank him for critically reviewing this report. We also thank E. Quarnstrom (University of Sheffield) for kindly providing the RelA-GFP expression vector. P. Perry wrote the scripts for image capture, K. Reid was responsible for the isolation of nucleoli, and A. Sanderson provided help with cell sorting.
The work was supported by grants from the Scottish Executive Chief Scientist's Office (K/MRS/50/C2719 and CZB/4/41) and the AICR (02-330). L.A.S. is a Caledonian Research Fellow. M.G.D. is supported by Cancer Research United Kingdom Programme Grant funding (C348/A3758).
REFERENCES
- 1.Baeuerle, P. A. 1998. Pro-inflammatory signaling: last pieces in the NF-kappaB puzzle? Curr. Biol. 8:R19-R22. [DOI] [PubMed] [Google Scholar]
- 2.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 3.Belmont, A. 2003. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr. Opin. Cell Biol. 15:304-310. [DOI] [PubMed] [Google Scholar]
- 4.Birbach, A., S. T. Bailey, S. Ghosh, and J. A. Schmid. 2004. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-kappaB inducing kinase NIK. J. Cell Sci. 117:3615-3624. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, K. J., S. Rocha, and N. D. Perkins. 2004. Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol. Cell 13:853-865. [DOI] [PubMed] [Google Scholar]
- 6.Carlotti, F., R. Chapman, S. K. Dower, and E. E. Qwarnstrom. 1999. Activation of nuclear factor kappaB in single living cells. Dependence of nuclear translocation and anti-apoptotic function on EGFPRELA concentration. J. Biol. Chem. 274:37941-37949. [DOI] [PubMed] [Google Scholar]
- 7.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L. F., and W. C. Greene. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392-401. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L. F., Y. Mu, and W. C. Greene. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539-6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta, A., A. Nag, W. Pan, N. Hay, A. L. Gartel, O. Colamonici, Y. Mori, and P. Raychaudhuri. 2004. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 35:36698-36707. [DOI] [PubMed] [Google Scholar]
- 11.David-Pfeuty, T., and Y. Nouvian-Dooghe. 2002. Human p14(Arf): an exquisite sensor of morphological changes and of short-lived perturbations in cell cycle and in nucleolar function. Oncogene 21:6779-6790. [DOI] [PubMed] [Google Scholar]
- 12.Dhar, S. K., B. C. Lynn, C. Daosukho, and D. K. St. Clair. 2004. Identification of nucleophosmin as an NF-κB co-activator for the induction of the human SOD2 gene. J. Biol. Chem. 279:28209-28219. [DOI] [PMC free article] [PubMed]
- 13.Din, F. V., M. G. Dunlop, and L. A. Stark. 2004. Evidence for colorectal cancer cell specificity of aspirin effects on NFkB signalling and apoptosis. Br. J. Cancer 91:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, C., J. Yang, and J. F. Engelhardt. 2002. Temporal pattern of NFkappaB activation influences apoptotic cell fate in a stimuli-dependent fashion. J. Cell Sci. 115:4843-4853. [DOI] [PubMed] [Google Scholar]
- 15.Fatyol, K., and A. A. Szalay. 2001. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha) and inhibits HIF-1-mediated transcription. J. Biol. Chem. 276:28421-28429. [DOI] [PubMed] [Google Scholar]
- 16.Grimm, S., M. K. Bauer, P. A. Baeuerle, and K. Schulze-Osthoff. 1996. Bcl-2 down-regulates the activity of transcription factor NF-kappaB induced upon apoptosis. J. Cell Biol. 134:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwick, J. C., G. R. van den Brink, G. J. Offerhaus, S. J. van Deventer, and M. P. Peppelenbosch. 2001. NF-kappaB, p38 MAPK and JNK are highly expressed and active in the stroma of human colonic adenomatous polyps. Oncogene 20:819-827. [DOI] [PubMed] [Google Scholar]
- 18.Harhaj, E. W., and S. C. Sun. 1999. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol. Cell. Biol. 19:7088-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horke, S., K. Reumann, M. Schweizer, H. Will, and T. Heise. 2004. Nuclear trafficking of La protein depends on a newly identified NoLS and the ability to bind RNA. J. Biol. Chem. 279:50302-55039. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M. 1999. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 21.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 22.Kato, T., Jr., M. Delhase, A. Hoffmann, and M. Karin. 2003. CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol. Cell 12:829-839. [DOI] [PubMed] [Google Scholar]
- 22a.Kilbanov, S. A., H. M. O’Hagen, and M. Ljungman. 2001. Accumulation of soluble and nucleolar-associated p53 following cellular stress. J. Cell Sci. 114:1867-1873. [DOI] [PubMed] [Google Scholar]
- 23.Kopp, E., and S. Ghosh. 1994. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science 265:956-959. [DOI] [PubMed] [Google Scholar]
- 24.Leung, A. K., and A. I. Lamond. 2002. In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles. J. Cell Biol. 157:615-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, N., and M. Karin. 1998. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc. Natl. Acad. Sci. USA 95:13012-13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martelli, F., T. Hamilton, D. P. Silver, N. E. Sharpless, N. Bardeesy, M. Rokas, R. A. DePinho, D. M. Livingston, and S. R. Grossman. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mogi, M., N. Ozeki, H. Nakamura, and A. Togari. 2004. Dual roles for NF-kappaB activation in osteoblastic cells by serum deprivation: osteoblastic apoptosis and cell-cycle arrest. Bone 35:507-516. [DOI] [PubMed] [Google Scholar]
- 28.Niederberger, E., I. Tegeder, G. Vetter, A. Schmidtko, H. Schmidt, C. Euchenhofer, L. Brautigam, S. Grosch, and G. Geisslinger. 2001. Celecoxib loses its anti-inflammatory efficacy at high doses through activation of NF-kappaB. FASEB J. 15:1622-1624. [DOI] [PubMed] [Google Scholar]
- 29.Niedick, I., N. Froese, A. Oumard, P. P. Mueller, M. Nourbakhsh, H. Hauser, and M. Koster. 2004. Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J. Cell Sci. 117:3447-3458. [DOI] [PubMed] [Google Scholar]
- 30.Pahl, H. L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 31.Perkins, N. D. 2004. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol. 14:64-69. [DOI] [PubMed] [Google Scholar]
- 32.Rocha, S., K. J. Campbell, and N. D. Perkins. 2003. p53- and Mdm2-independent repression of NF-kappa B transactivation by the ARF tumor suppressor. Mol. Cell 12:15-25. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez, M. S., J. Thompson, R. T. Hay, and C. Dargemont. 1999. Nuclear retention of IkappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. J. Biol. Chem. 274:9108-9115. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, M. S., J. Wright, J. Thompson, D. Thomas, F. Baleux, J. L. Virelizier, R. T. Hay, and F. Arenzana-Seisdedos. 1996. Identification of lysine residues required for signal-induced ubiquitination and degradation of I kappa B-alpha in vivo. Oncogene 12:2425-2435. [PubMed] [Google Scholar]
- 35.Roff, M., J. Thompson, M. S. Rodriguez, J. M. Jacque, F. Baleux, F. Arenzana-Seisdedos, and R. T. Hay. 1996. Role of IkappaBalpha ubiquitination in signal-induced activation of NFkappaB in vivo. J. Biol. Chem. 271:7844-7850. [DOI] [PubMed] [Google Scholar]
- 36.Romieu-Mourez, R., E. Landesman-Bollag, D. C. Seldin, and G. E. Sonenshein. 2002. Protein kinase CK2 promotes aberrant activation of nuclear factor-kappaB, transformed phenotype, and survival of breast cancer cells. Cancer Res. 62:6770-6778. [PubMed] [Google Scholar]
- 37.Rubbi, C. P., and J. Milner. 2000. Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19:85-96. [DOI] [PubMed] [Google Scholar]
- 38.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 39.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-kappaB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 40.Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y. C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K. P. Lu. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12:1413-1426. [DOI] [PubMed] [Google Scholar]
- 41.Smartt, H. J., D. J. Elder, D. J. Hicks, N. A. Williams, and C. Paraskeva. 2003. Increased NF-kappaB DNA binding but not transcriptional activity during apoptosis induced by the COX-2-selective inhibitor NS-398 in colorectal carcinoma cells. Br. J. Cancer 89:1358-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark, L. A., F. V. N. Din, R. M. Zwacka, and M. G. Dunlop. 2001. Aspirin-induced activation of the NF-κB signalling pathway: a novel mechanism for aspirin-mediated apoptosis in colon cancer cells. FASEB J. 15:1273-1275. (First published 20 March 2001; 10.1096/fj.00-0529fje.) [PubMed] [Google Scholar]
- 43.Sweet, T., K. Khalili, B. E. Sawaya, and S. Amini. 2003. Identification of a novel protein from glial cells based on its ability to interact with NF-kappaB subunits. J. Cell. Biochem. 90:884-891. [DOI] [PubMed] [Google Scholar]
- 44.Tergaonkar, V., M. Pando, O. Vafa, G. Wahl, and I. Verma. 2002. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell 1:493-503. [DOI] [PubMed] [Google Scholar]
- 45.Thanos, D., and T. Maniatis. 1995. NF-kappa B: a lesson in family values. Cell 80:529-532. [DOI] [PubMed] [Google Scholar]
- 46.Wang, C. Y., J. C. J. Cusack, R. Liu, and A. S. J. Baldwin. 1999. Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat. Med. 5:412-417. [DOI] [PubMed] [Google Scholar]
- 47.Weber, J. D., M. L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, M. H., and B. Y. Yung. 2002. UV stimulation of nucleophosmin/B23 expression is an immediate-early gene response induced by damaged DNA. J. Biol. Chem. 277:48234-48240. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, Y., M. J. Yin, K. M. Lin, and R. B. Gaynor. 1999. Sulindac inhibits activation of the NF-kappaB pathway. J. Biol. Chem. 274:27307-27314. [DOI] [PubMed] [Google Scholar]
- 50.Yin, M. J., Y. Yamamoto, and R. B. Gaynor. 1998. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature 396:77-80. [DOI] [PubMed] [Google Scholar]
- 51.Zwacka, R. M., L. A. Stark, and M. G. Dunlop. 2000. NF-kappaB kinetics predetermines TNF-alpha sensitivity of colorectal cancer cells. J. Gene Med. 2:334-343. [DOI] [PubMed] [Google Scholar]