Abstract
We previously discovered that the ubiquitin protease Ubp10/Dot4p is important for telomeric silencing through its interaction with Sir4p. However, the mechanism of Ubp10p action was unknown. We now provide evidence that Ubp10p removes ubiquitin from histone H2B; cells with UBP10 deleted have increased steady-state levels of H2B ubiquitination. As a consequence, ubp10Δ cells also have increased steady-state levels of histone H3 Lys4 and Lys79 methylation. Consistent with its role in silencing, Ubp10p is preferentially localized to silent chromatin where its ubiquitin protease activity maintains low levels of H3 Lys4 and Lys79 methylation to allow optimal Sir protein binding to telomeres and global telomeric silencing. The ubiquitin protease Ubp8p has also been shown to remove ubiquitin from H2B, and ubp8Δ cells have increased steady-state levels of H2B ubiquitination similar to those in ubp10Δ cells. Unlike ubp10Δ cells, however, ubp8Δ cells do not have increased steady-state levels of H3 Lys4 and Lys79 methylation, nor is telomeric silencing affected. Despite their separate functions in silencing and SAGA-mediated transcription, respectively, deletion of both UBP10 and UBP8 results in a synergistic increase in the steady-state levels of H2B ubiquitination and in the number of genes with altered expression, indicating that Ubp10p and Ubp8p likely overlap in some of their target chromatin regions. We propose that Ubp10p and Ubp8p are the only ubiquitin proteases that normally remove monoubiquitin from histone H2B and, while there are regions of the genome to which each is specifically targeted, both combine to regulate the global balance of H2B ubiquitination.
Posttranslational modifications of nucleosomal histones play pivotal regulatory roles in all aspects of eukaryotic chromosome dynamics: replication, recombination, repair, segregation, and gene expression. These modifications include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (71). Many of these modifications often occur together within the same histone or nucleosome or within neighboring nucleosomes, creating distinct chromatin domains (71). Particular combinations of residue-specific modifications typically correlate with specific functional consequences for the modified chromatin domains (71).
In Saccharomyces cerevisiae, chromatin-mediated silencing at telomeres and the HM loci is particularly sensitive to histone acetylation and methylation, which appear to disrupt the ability of the silencing proteins Sir3p and Sir4p to bind to, or assemble along, the nucleosome fiber. For example, tethering of histone acetylases to silent domains increases histone acetylation within these domains and blocks the propagation of silencing (16). Similarly, overexpression of a histone methylase greatly increases global histone methylation and disrupts silencing (85, 92). In vitro, fragments of Sir3p bind unacetylated and unmethylated histone peptides better than those that are acetylated or methylated (13, 81), demonstrating the preference of Sir3p for unmodified histones. Because of this preference, histone acetylation and methylation appear to be excluded from silent chromatin (5-7, 63, 79, 88, 90).
In genetic screens looking for factors that compromise telomeric silencing, we previously identified a number of genes that encode protein modification enzymes, such as DOT1, DOT4, and RAD6 (37, 85). Dot1p is a histone methyltransferase that methylates Lys79 of histone H3 in the context of the nucleosome (24, 49, 65, 92). H3 Lys79 methylation is primarily associated with active chromatin—approximately 90% of histone H3 in yeast is methylated at this site (92)—and the modification is absent from silent chromatin loci (63). Loss of DOT1 completely abrogates Lys79 methylation, which results in a reduction of Sir protein binding to the telomeres and a concomitant loss of telomeric silencing (92). However, loss of DOT1 also results in increased Sir protein binding to subtelomeric regions (92). We have hypothesized that this reflects a promiscuous binding of Sir3p to nucleosomes outside of silent regions due to the absence of H3 Lys79 methylation (92). Because Sir3p is limiting in the cell (11, 87, 89), such promiscuous binding reduces the effective amount of Sir3p available for the normally silenced loci (93).
Methylation also occurs on Lys4 of histone H3 by action of the Set1 protein (8, 45, 62, 69, 77). Lys4 methylation is similar to Lys79 methylation: it is normally associated with active chromatin (5, 82), and Sir protein binding is reduced in silent regions when Lys4 methylation is abolished (81). Simultaneous loss of H3 Lys79 and Lys4 methylation by deletion of DOT1 and SET1 synergistically reduces Sir protein binding to the telomeres (64), indicating that these two modifications function together to prevent the promiscuous binding of Sir proteins to active chromatin.
The ubiquitin-conjugating enzyme Rad6p is also required for telomeric silencing (37). Rad6p functions with Bre1p, its cognate ubiquitin-protein ligase (39, 95), to attach ubiquitin to Lys123 of histone H2B (76). Over 25 years ago it was found that ubiquitinated H2B and H2A are associated with transcriptionally active chromatin in higher eukaryotes (54, 68). Consistent with this, components of the Paf1 transcriptional elongation complex are required for H2B ubiquitination in S. cerevisiae (46, 64, 96). Although its precise role in transcription is unclear, H2B ubiquitination is required in vivo for H3 Lys4 and Lys79 methylation (9, 20, 67, 91). It is proposed that the ubiquitin moiety on H2B may recruit proteasomal ATPases to prepare the chromatin for Set1p and Dot1p activity (22). Because loss of H3 Lys4 and Lys79 methylation disrupts silencing (81, 92), the requirement of Rad6p function for telomeric silencing is likely to be indirect and due to its role in ubiquitinating H2B as a precursor to establishing both H3 Lys4 and Lys79 methylation in active chromatin.
Taking all of this together, it is clear that silencing requires both the addition of histone acetylation and methylation in active chromatin and the removal or prevention of these histone modifications in silent regions. Removal of histone acetylation in silent regions is carried out by the silencing protein and histone deacetylase Sir2p (40, 50, 86). Although a demethylase that targets H3 Lys4 mono- and dimethylation has been identified in mammals (83), no homologous lysine demethylases are known in S. cerevisiae. Thus, it is not known if H3 Lys4 and Lys79 methylation can be directly removed from silent regions if established. Instead, removal of ubiquitin from H2B by ubiquitin proteases prior to the establishment of H3 Lys4 and Lys79 methylation may be one mechanism for limiting these H3 modifications in silent regions. Of the 17 known and putative ubiquitin proteases in S. cerevisiae (2), Ubp8p has been shown to regulate the levels of ubiquitinated H2B as part of the transcriptional activator complex SAGA (18, 33, 80), indicating that removal of ubiquitin from H2B does occur. However, Ubp8p is not known to function in silencing.
We previously discovered that the ubiquitin protease Dot4p (now known as Ubp10p) is important for silencing in vivo (43). The absence of Dot4p ubiquitin protease activity, either by deletion of DOT4 or mutation of the catalytic residue, results in reduced silencing, especially at telomeres (43, 85). By two-hybrid analysis, we found that Dot4p interacts with the silencing protein Sir4p (43), and we proposed that Dot4p acts at, or is part of, silent chromatin. However, we had not identified any in vivo substrates of Dot4p. Given the association of Dot4p with silent chromatin and the role of H2B ubiquitination in H3 Lys4 and Lys79 methylation, we investigated whether Dot4p's ubiquitin protease activity regulates the levels of H2B ubiquitination to limit H3 Lys4 and Lys79 methylation in silent regions, as has been previously proposed (78).
MATERIALS AND METHODS
Reagents.
General chemical and enzymatic reagents were obtained from New England Biolabs (Beverley, MA), Sigma (St. Louis, MO), Fisher (Pittsburgh, PA) and Pierce (Rockford, IL). Nitrocellulose (Protran; pore size of 0.2 μM) was obtained from Schleicher and Schuell (Keene, NH). The anti-Myc and anti-FLAG antibodies were obtained from Sigma and used at 1:10,000 dilutions for Western blotting or 1:500 dilutions for chromatin immunoprecipitations (ChIP). Anti-monomethyl, -dimethyl, and -trimethyl Lys4 histone H3 antibodies were obtained from Upstate (Waltham, MA) and used at 1:5,000 dilutions for Western blotting and 1:500 dilutions for ChIP. Anti-dimethyl Lys79 histone H3 antibody was made as previously described (92) and used at 1:2,000 dilutions for Western blotting and at 1:100 dilutions for ChIP. Purified anti-Sir2p polyclonal antibodies against the N-terminal 19 residues of Sir2p were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and used at a 1:500 dilution for ChIP. Mouse anti-Sir3p monoclonal antibodies were made as previously described (92), and used at 1:3 dilutions for ChIP. Vistra Green, ECL chemiluminescence immunodetection reagents, sheep anti-mouse horseradish peroxidase-conjugated antiserum, and donkey anti-rabbit horseradish peroxidase-conjugated antiserum were obtained from Amersham Biosciences (Piscataway, NJ); the antisera were used at 1:2,000 dilutions. IRDye800-conjugated goat anti-mouse and anti-rabbit antibodies were obtained from Rockland Immunochemicals (Gilbertsville, PA). Protein G Dynabeads were obtained from Dynal (Brown Deer, WI).
Recombinant DNA, molecular cloning, and yeast strains.
Standard molecular biology techniques were used. Plasmids and yeast strains used in this study are described in Table 1. Yeast deletion alleles were made by PCR amplification of the appropriate knockout construct, followed by transformation into yeast cells using standard yeast transformation techniques (http://www.fhcrc.org/labs/gottschling).
TABLE 1.
Plasmids, yeast strains, and oligonucleotides used in this study
| Plasmid, strain, or oligonucleotide | Genotype or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pJH23 | HIS3 CEN HTA1-HTB1 | 35 |
| pRS314-FLAG/HTB1 | TRP1 CEN FLAG-HTB1 | 76 |
| pRG422 | 1.4-kb BamHI-SacI FLAG-HTB1 fragment from pRS314-FLAG/HTB1 used to replace the corresponding region in pJH23 | This study |
| pRS406 | URA3 INT | 84 |
| pRG490 | 2.7-kb SacII-XhoI HTA1-FLAG-HTB1 fragment from pRG422 inserted between SacII-XhoI sites in pRS406 | This study |
| pRS306 | URA3 INT | 84 |
| pdot4-1-MT6 | pBluescript KS+ ubp10C371S-MT6 | 43 |
| pRS306-str4-1-MT6 | 3.8-kb BamHI-SfuI ubp10C371S-MT6 fragment from pdot4-1-MT6 inserted between the BamHI-HindIII (blunted) sites in pRS306 | This study |
| pdot4-5-MT6 | pBluescript KS+ ubp10Δ94-250-MT6 | 43 |
| pRS306-str4-5-MT6 | 3.3-kb BamHI-SfuI ubp10Δ94-250-MT6 fragment from pdot4-1-MT6 inserted between the BamHI-HindIII (blunted) sites in pRS306 | This study |
| pDot4-MT6 | pBluescript KS+ UBP10-MT6 | 43 |
| pRG637 | 850-bp HindIII-BsrGI wild-type fragment from pDOT4-MT6 used to replace the mutant region in pRS306-str4-1-MT6 | This study |
| pRS404 | TRP1 INT | 84 |
| pRG716 | 3.9-kb XhoI-SacII ubp10C371S-MT6 fragment from pRS306-str4-1-MT6 inserted between XhoI-SacII sites in pRS404 | This study |
| pRG717 | 3.5-kb XhoI-SacII ubp10Δ94-250-MT6 fragment from pRS306-str4-5-MT6 inserted between XhoI-SacII sites in pRS404 | This study |
| pRG718 | 3.9-kb XhoI-SacII UBP10-MT6 fragment from pRG637 inserted between XhoI-SacII sites in pRS404 | This study |
| pRS414 | TRP1 CEN | 84 |
| Yeast strains | ||
| AR120 | HMLa MATaHMRacdc7-1 bar1 trp1-289 ura3-52 leu2-3,112 his6 | Walt Fangman |
| UCC4861 | AR120 CDC7 | This study |
| UCC6195 | UCC4861 CDC7 HTAI-FLAG-HTB1 | This study |
| UCC6196 | UCC6195 sir2Δ::LEU2 | This study |
| UCC6197 | UCC6195 sir3Δ::LEU2 | This study |
| UCC6198 | UCC6195 sir4Δ::LEU2 | This study |
| UCC6199 | UCC6195 ubp10Δ::KanMX | This study |
| UCC6184 | UCC6199 ubp10Δ::KanMX::ubp10C371S-MT6::URA3 | This study |
| UCC6185 | UCC6199 ubp10Δ::KanMX::ubp10Δ94-250-MT6::URA3 | This study |
| UCC6186 | UCC6199 ubp10Δ::KanMX::UBP10-MT6::URA3 | This study |
| UCC4825 | MATaade2Δ::hisG ura3Δ0 ADE2-TEL-VR | 43 |
| UCC4857 | UCC4825 ubp10Δ::KanMX | 43 |
| UCC4870 | UCC4825 ubp10C371S-MT6 | 43 |
| UCC4896 | UCC4825 ubp10Δ94-250-MT6 | 43 |
| UCC4836 | UCC4825 sir4Δ::KanMX | This study |
| UCC7315 | MATalys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pCS1 | This study |
| UCC6286 | UCC7315 sir4Δ::KanMX | This study |
| UCC6288 | UCC7315 ubp8Δ::KanMX | This study |
| UCC6357 | UCC7315 ubp10Δ::NatMX ubp8Δ::KanMX | This study |
| UCC6361 | UCC7315 ubp10Δ::NatMX | This study |
| UCC6389 | MATalys2Δ0 trp1D63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 htaI-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 | This study |
| UCC6390 | UCC6389 ubp10Δ::NatMX | This study |
| UCC6391 | UCC6389 sir4Δ::KanMX | This study |
| UCC6392 | UCC6389 ubp8Δ::KanMX | This study |
| UCC6393 | UCC6389 ubp10Δ::NatMX ubp8Δ::KanMX | This study |
| UCC6163 | UCC6389 rad6Δ::KanMX | This study |
| UCC6394 | MATα lys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 | This study |
| UCC6395 | UCC6394 ubp10Δ::NatMX | This study |
| UCC6396 | UCC6394 sir4Δ::KanMX | This study |
| UCC6397 | UCC6394 ubp8Δ::KanMX | This study |
| UCC6398 | UCC6394 ubp10Δ::NatMX ubp8Δ::KanMX | This study |
| UCC6406 | UCC6390 ubp10Δ::NatMX::ubp10C371S-MT6::TRP1 | This study |
| UCC6407 | UCC6390 ubp10Δ::NatMX::ubp10Δ94-250-MT6::TRP1 | This study |
| UCC6408 | UCC6390 ubp10Δ::NatMX::UBP10-MT6::TRP1 | This study |
| UCC6475 | UCC6408 sir2Δ::HygMX | This study |
| UCC6477 | UCC6408 sir3Δ::HygMX | This study |
| UCC6479 | UCC6408 sir4Δ::HygMX | This study |
| UCC6422 | UCC6389 ppr1Δ::LYS2 | This study |
| UCC6423 | UCC6390 ppr1Δ::LYS2 | This study |
| UCC6424 | UCC6391 ppr1Δ::LYS2 | This study |
| UCC6425 | UCC6392 ppr1Δ::LYS2 | This study |
| UCC6426 | UCC6393 ppr1Δ::LYS2 | This study |
| UCC6432 | UCC6389 TRP1 | This study |
| UCC6435 | UCC6390 TRP1 | This study |
| Oligonucleotides | ||
| STR4RS+ (ubp10Δ 5′) | AAT CCG TCC TAT TGT CAT ATC ACA ATC ACA GAC TGA TTG TAC TGA GAG TGC ACC | 43 |
| STR4RS− (ubp10Δ 3′) | TCC AGG AAT ATC GAG TTT TTT CAT TTG GTG AAC CTG TGC GGT ATT TCA CAC CG | 43 |
| oRG255 (ubp8Δ 5′) | CTT CGG TCC TCG TCG TCC TAC TTG AAA CCC TGC TTT TTT TAT TTG TTA TTA ATA ATT CTG TGC GGT ATT TCA CAC CGC | This study |
| oRG256 (ubp8Δ 3′) | TAG CTT TTT CTT CTT TTT TGT TTT ATT ATT ATT GTT GAA TGC TAT TTG CTG AAT CAC AGA TTG TAC TGA GAG TGC ACC | This study |
| SIR4KO1 (sir4Δ 5′) | CAA CCC ACA ATA CCA AAA AAG CGA AGA AAA CAG CCA GAT TGT ACT GAG AGT GCA CC | This study |
| SIR4KO2 (sir4Δ 3′) | CAC TTC GTT ACT GGT CTT TTG TAG AAT GAT AAA AAG CTG TGC GGT ATT TCA CAC CG | This study |
| GAL1-1 | GAA GAA GTG ATT GTA CCT GAG | 92 |
| GAL1-2 | ACC TTT CCG GTG CAA GTT TC | 92 |
| ACT1-1 | CCA ATT GCT CGA GAG ATT TC | 92 |
| ACT1-2 | CAT GAT ACC TTG GTG TCT TG | 92 |
| oRG58 (SAN1 5′) | GCC CCT ACG CAC AAC CGC | This study |
| oRG59 (SAN1 3′) | GGA CGT GTT TTC GGA TGG G | This study |
| 5HMR | GAG AAT AAG CGC AGG TAC TCC | 92 |
| 5HMR-2 | TCT TGA GCG GTG AGC CTC TG | 92 |
| VIR-1 | CAG GCA GTC CTT TCT ATT TC | 92 |
| VIR-2 | GCT TGT TAA CTC TCC GAC AG | 92 |
ChIP assays.
ChIP assays were performed similar to those previously described (92), with some changes. Yeast extract-peptone-dextrose (YEPD; 200 ml) cultures were grown to a cell density of 2 × 107 cells/ml and subjected to cross-linking with 1% formaldehyde. Cell lysates were prepared, and lysates were sonicated eight times for 30 s using a Fisher Scientific sonic dismembrator model 60. Sonication resulted in an average fragment size of 500 to 800 bp (data not shown). Clarified lysates were stored at −80°C until required.
For the Ubp10p-Myc ChIPs, cross-linking was performed differently. Cells were first harvested by centrifugation, washed twice in phosphate-buffered saline, and resuspended in 50 ml of phosphate-buffered saline. Dimethyl adipimidate was added to a final concentration of 10 mM to improve cross-linking (48), and the cells were incubated at room temperature for 45 min. Formaldehyde was added to a final concentration of 1%, and the cultures were incubated at room temperature for an additional 15 min. Samples were prepared as described above.
Initial multiplex PCR amplifications were performed on various concentrations of immunoprecipitates and total lysates to determine the concentration required for amplification in the linear range. Final multiplex PCR amplifications were performed using a single concentration previously determined to be in the linear amplification range. PCR products were resolved on 2% agarose (1× Tris-acetate-EDTA) gels. Gels were stained with Vistra Green and analyzed using a PhosphorImager (Molecular Dynamics) and ImageJ software. Oligonucleotide sequences for PCR amplifications are listed in Table 1.
Western analyses.
Whole-cell lysates were prepared as previously described (26), with some changes. Cultures were grown in YEPD or yeast complete (YC) medium at 30°C to the desired optical density (2 × 107 cells/ml for logarithmically growing cells, overnight saturation for diauxic cells, and 7-day saturation for stationary cells), and equal numbers of cells (1.6 × 108) were harvested by centrifugation. Cells were lysed in 300 μl of SUME (8 M urea, 1% sodium dodecyl sulfate, 10 mM morpholinepropanesulfonic acid, pH 6.8, 10 mM EDTA) containing 0.01% bromophenol blue and 10 mM phenylmethylsulfonyl fluoride by mechanical shearing using acid-washed glass beads. For immunoblotting, 5- to 20-μl samples of the cellular lysates were resolved on 16% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, and the proteins were transferred to nitrocellulose and immunoblotted with the appropriate antibody.
Transcript DNA microarrays.
Transcript DNA microarrays similar to those previously described (23) were performed in triplicate. Dye-reversal experiments were performed to identify sequence-specific dye biases. Statistical analysis of microarray data was performed as previously described (17), using a Bayesian t statistic derived for microarray analyses (3) and a false discovery rate methodology (4) to account for multiple testing. The entire normalized data set is in Table S1 in the supplemental material; the National Center for Biotechnology Information Gene Expression Omnibus accession numbers for the series of individual data sets are GSE2329 (ubp10Δ and ubp8Δ series) and GSE2330 (ubp10C371S and ubp10Δ94-250 series).
RESULTS
In our earlier work, we found that loss of DOT4 results in reduced steady-state levels of HA epitope-tagged Sir4p, and we speculated that Dot4p might control Sir4p stability, perhaps by regulating its ubiquitin-dependent degradation (43). However, in subsequent testing we found that the HA epitope tag (or the T7- or Myc-epitope tag) added to the N terminus of Sir4p results in aberrant degradation of Sir4p (data not shown). Using new antibodies, we found that untagged Sir4p is normally stable and not subject to ubiquitination (27). We also found that the steady-state levels of untagged wild-type Sir4p are unchanged in dot4Δ cells (data not shown). Thus, the alteration of Sir4p stability by loss of DOT4 is an artifact of epitope tagging the protein and not a normal mode of regulation. As a result, we turned our attention to other potential substrates of Dot4p.
(Concurrent with our initial discovery (85), DOT4 was also found by homology searches for ubiquitin proteases and named UBP10 (36). Because UBP10 is now the standard name for the gene in the Saccharomyces Genome Database (http://db.yeastgenome.org/cgi-bin/locus.pl?locus = ubp10), we will henceforth use UBP10 rather than DOT4. By doing so, we have renamed the previously constructed dot4 alleles, dot4-1 and dot4-5 (43), with the more descriptive ubp10 nomenclature, ubp10C371S and ubp10Δ94-250, respectively.)
Ubp10/Dot4p negatively regulates histone H2B ubiquitination in vivo.
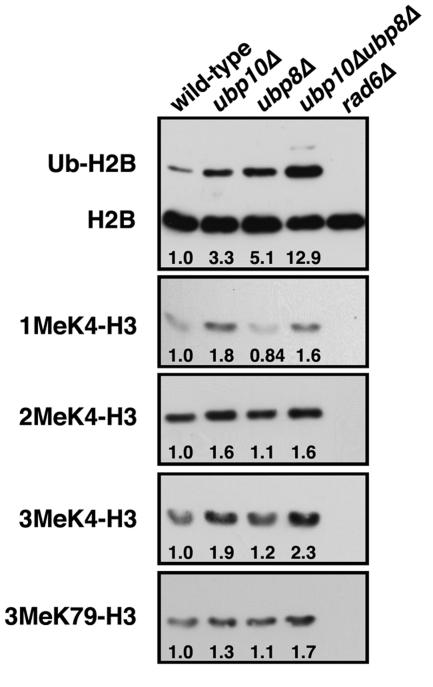
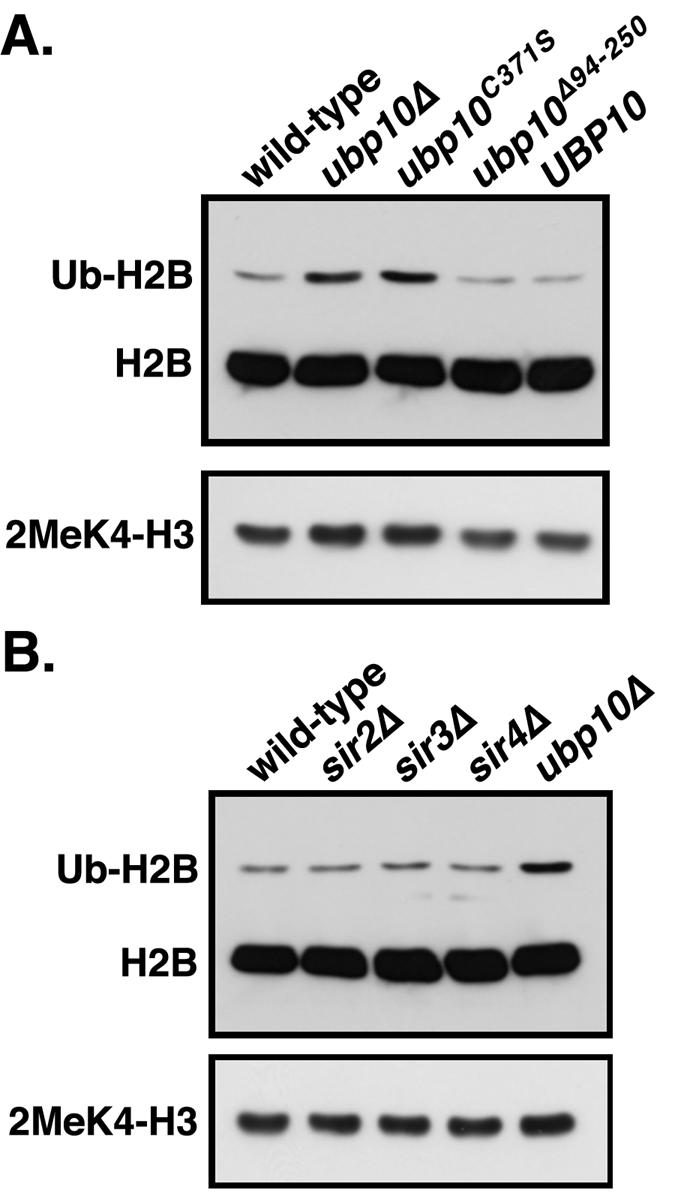
With Sir4p eliminated as a substrate of Ubp10p, we turned our attention to histone H2B due to the fact that H2B ubiquitination is required for histone H3 Lys4 and Lys79 methylation (9, 20, 67, 91), both of which antagonize Sir protein binding to chromatin (64, 81, 92). To determine if Ubp10p targets ubiquitinated H2B, we examined the steady-state levels of H2B ubiquitination in ubp10Δ cells. Compared to wild-type cells, ubp10Δ cells have approximately threefold higher steady-state levels of H2B ubiquitination (Fig. 1), supporting the idea that ubiquitinated H2B is a bona fide substrate of Ubp10p. In fact, the increased steady-state levels of H2B ubiquitination in ubp10Δ cells are similar to the approximately fivefold higher levels observed in ubp8Δ cells (Fig. 1); Ubp8p is a ubiquitin protease recently shown to regulate H2B ubiquitination (18, 33). Deletion of both UBP10 and UBP8 results in a further increase in the steady-state levels of H2B ubiquitination that is greater than that seen in either ubp10Δ or ubp8Δ cells (Fig. 1). These results indicate that Ubp10p and Ubp8p act upon separate populations of ubiquitinated H2B in part, which is consistent with their distinct roles in silencing and SAGA-mediated transcription, respectively (33, 43).
FIG. 1.
Ubp10p negatively regulates histone H2B ubiquitination and H3 Lys4 and Lys79 methylation levels. Global steady-state H2B ubiquitination levels from whole-cell lysates were assayed using anti-FLAG antibodies that recognize the FLAG epitope placed on the N terminus of H2B (76). Global steady-state H3 Lys4 and Lys79 methylation levels were also assayed using antibodies specific for mono-, di-, and trimethylated Lys4 forms of H3 as well as an antibody generated against the dimethylated Lys79 form of H3 (92). Whole-cell lysates were derived from strains UCC6369 (wild type), UCC6390 (ubp10Δ), UCC6392 (ubp8Δ), UCC6393 (ubp10Δ ubp8Δ), and UCC6163 (rad6Δ) that were harvested during log-phase growth. rad6Δ cells served as a negative control for H2B lacking ubiquitin. Changes in levels (n-fold) of H2B ubiquitination and H3 Lys4 and Lys79 methylation for each mutant strain relative to the wild-type strain are shown below each corresponding lane and are the average of three independent experiments. Quantitation was performed using National Institutes of Health ImageJ software.
Ubp10/Dot4p negatively regulates global histone H3 Lys4 and Lys79 methylation.
The previously documented dependence of histone H3 Lys4 and Lys79 methylation on H2B ubiquitination led us to examine if loss of UBP10 or UBP8 altered H3 methylation. Using specific antibodies, we examined steady-state levels of H3 Lys4 mono-, di- or trimethylation or H3 Lys79 dimethylation. Although the changes were often modest, ubp10Δ cells did have reproducibly higher steady-state levels of all forms of H3 Lys4 methylation and of H3 Lys79 methylation compared to wild-type cells (Fig. 1, and see Fig. 6 and 7). In contrast, we observed no detectable increase in H3 Lys4 or Lys79 methylation in ubp8Δ cells (Fig. 1), indicating that increased H2B ubiquitination alone is not sufficient for increased H3 methylation. In ubp10Δ ubp8Δ cells, H3 Lys4 and Lys79 methylation are also increased similar to ubp10Δ cells (Fig. 1B). Altogether, the higher steady-state levels of H2B ubiquitination in ubp10Δ cells likely present an increased opportunity for H3 Lys4 and Lys79 methylation.
FIG. 6.
Loss of silencing does not affect global H2B ubiquitination levels. (A) Loss of Ubp10p catalytic activity but not loss of Sir4p binding affects H2B ubiquitination and H3 Lys4 methylation. Global steady-state H2B ubiquitination and H3 Lys4 dimethylation levels from whole-cell lysates derived from strains UCC6195 (wild-type UBP10), UCC6199 (ubp10Δ), UCC6184 (ubp10C371S-6Myc), UCC6185 (ubp10Δ94-250-6Myc), and UCC6186 (UBP10-6Myc) were assayed using anti-FLAG antibodies (FLAG-tagged H2B) or antibodies that specifically recognized H3 Lys4 dimethylation. (B) Loss of silencing does not affect H2B ubiquitination or H3 Lys4 methylation. Global steady-state H2B ubiquitination and H3 Lys4 dimethylation levels were assayed as described in panel A using strains UCC6195 (UBP10), UCC6196 (sir2Δ), UCC6197 (sir3Δ), UCC6198 (sir4Δ), and UCC6199 (ubp10Δ).
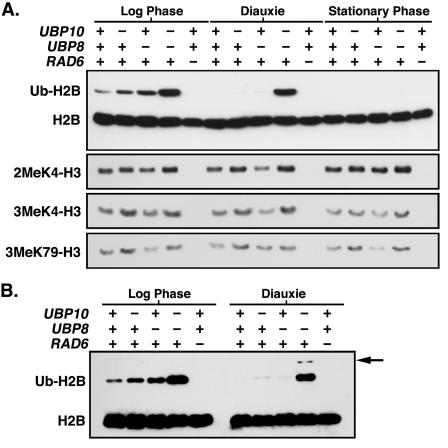
FIG. 7.
Ubp10p and Ubp8p overlap in their target chromatin regions. (A) Global steady-state levels of H2B ubiquitination and H3 Lys4 and Lys79 methylation from whole-cell lysates were assayed as described in the legend of Fig. 1. Whole-cell lysates were derived from strains UCC6369 (wild type), UCC6390 (ubp10Δ), UCC6392 (ubp8Δ), UCC6393 (ubp10Δ ubp8Δ), and UCC6163 (rad6Δ) that were harvested during log-phase growth, diauxie (overnight growth to saturation), and stationary phase (7-day saturation). (B) Some H2B ubiquitination persists in ubp10Δ and ubp8Δ cells in diauxie. H2B ubiquitination was assayed as described in the legend of Fig. 1. Arrow indicates where diubiquitinated form of H2B would run as expected in ubp10Δ ubp8Δ cells.
A recent study found modest decreases in the steady-state levels of H3 Lys4 di- and trimethylation but a 10-fold increase in the steady-state levels H3 Lys4 monomethylation in ubp8Δ cells (18). However, we do not see much change in the levels of any form of H3 Lys4 methylation in ubp8Δ cells (Fig. 1). One possibility for the difference is that H3 Lys4 methylation was analyzed in acid-extracted histones in the previous study (18), whereas we examined H3 Lys4 methylation in whole yeast cell extracts (see Materials and Methods). Perhaps there exists a soluble, nonnucleosomal pool of Lys4-methylated histone H3 that is detected by the whole-cell extract but not by the differential extraction method. If so, the extraction method would reveal information about the state of H3 Lys4 methylation exclusively in the nucleosome. Alternatively, the various forms of Lys4-methylated histone H3 could have different solubilities that affect recovery during the extraction procedure. Whatever the reason, our results in whole-cell extracts are reproducible and show little change in global H3 Lys4 methylation in the absence of UBP8 (Fig. 1; see Fig. 7), which is consistent with very little role for Ubp8p in global transcription (see Fig. 8).
FIG. 8.
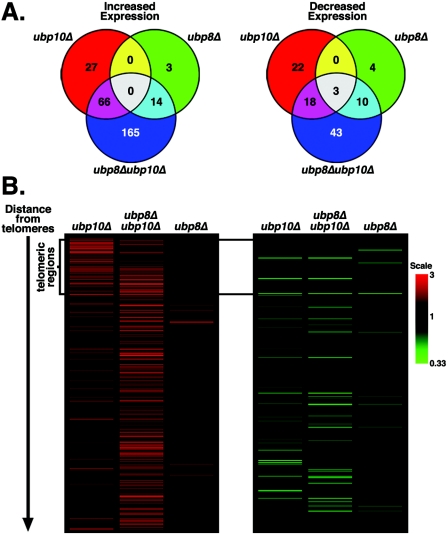
Loss of UBP10 and UBP8 has synergistic transcriptional effects primarily in active chromatin. Transcript array analysis using strains UCC6389 (wild type), UCC6390 (ubp10Δ), UCC6392 (ubp8Δ), and UCC6393 (ubp10Δ ubp8Δ) was performed as described in the legend of Fig. 2. (A) Venn diagrams show the degree of overlap in expression changes between ubp10Δ, ubp8Δ, and ubp10Δ ubp8Δ cells. (B) Expression changes in ubp10Δ ubp8Δ cells are primarily in nontelomeric regions. Positional cluster analysis shows expression changes of genes based on distance from telomeres. Only genes that were expressed greater than or less than 1.5-fold in at least one of the strains are shown (total genes shown is 368). Genes located in telomeric regions are marked on the left. See Table S1 in the supplemental material for the entire normalized data set.
Ubp10/Dot4p is enriched at silenced loci.
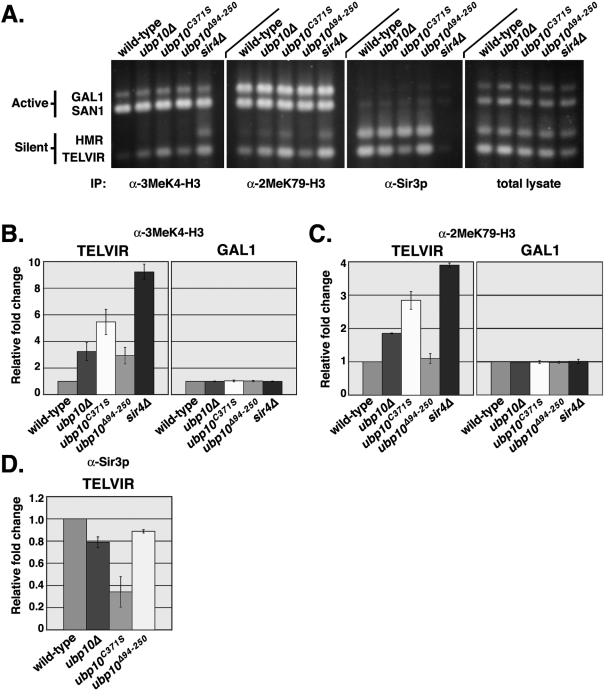
Negative regulation of H2B ubiquitination would explain the role of Ubp10p in silencing, given that H2B ubiquitination is required for histone H3 Lys4 and Lys79 methylation (9, 20, 67, 91), both of which antagonize Sir protein binding to chromatin (64, 81, 92). Because Ubp10p is involved in silencing telomeric gene expression through a direct interaction with Sir4p (43, 85), it seemed likely that Ubp10p is localized specifically to silent chromatin to reverse H2B ubiquitination. By chromatin immunoprecipitation, we found that Ubp10p is preferentially localized to the silent telomere VIR and the silent mating locus HMRa compared to active chromatin regions such as GAL1 and SAN1 (Fig. 2).
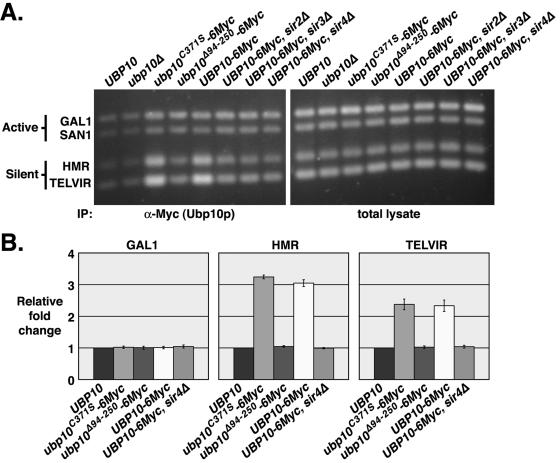
FIG. 2.
Ubp10p is preferentially localized to silent chromatin. In vivo cross-linking analysis was performed using strains UCC6389 (untagged UBP10), UCC6390 (ubp10Δ), UCC6408 (UBP10-6Myc), UCC6406 (ubp10C371S-6Myc), UCC6407 (ubp10Δ94-250-6Myc), UCC6475 (UBP10-6Myc, sir2Δ), UCC6477 (UBP10-6Myc, sir3Δ), and UCC6479 (UBP10-6Myc, sir4Δ). A ChIP assay of Ubp10 proteins was performed using anti-Myc antibodies specific for the six-Myc tag fused to the C termini of the wild-type Ubp10, mutant Ubp10C371S, and mutant Ubp10Δ94-255 proteins. ChIP was also performed using untagged wild-type Ubp10p as a control. Multiplex PCR amplifications were performed to assess Ubp10p binding at the silent domains of telomere VIR and HMRa, the active SAN1 gene, and the repressed GAL1 gene. DNA of the total lysate was amplified as a control. (A) Representative example of Vistra Green-stained PCR amplifications. (B) Quantitative analysis of data in shown in panel A. Values represent the ratio of immunoprecipitate to total lysate for the query gene normalized to the ratio of immunoprecipitate to total lysate for SAN1. All values are the averages of at least two independent experiments. Error bars represent the standard deviation.
Previously, we made a number of ubp10/dot4 mutant alleles that are compromised for telomeric silencing (43). In particular, we constructed two alleles: ubp10C371S (originally named dot4-1), which renders Ubp10p nonfunctional by substitution of the active site Cys371 with Ser, and ubp10Δ94-250 (originally named dot4-5), which is catalytically active but can no longer bind Sir4p due to deletion of the Sir4p-binding region spanned by residues 94 to 250. We found that the Ubp10C371S protein is preferentially localized to the silent regions at telomere VIR and HMRa, similar to wild-type Ubp10p (Fig. 2), whereas the Ubp10Δ94-250 protein has no preference for silent loci (Fig. 2). Loss of Ubp10p preferential localization to silent regions was also observed when the silencing genes SIR2, SIR3, or SIR4 were deleted. Thus, Ubp10p is enriched at silent chromatin via its interaction with Sir4p, and Ubp10p localization is independent of its ubiquitin protease activity.
Loss of Ubp10/Dot4p function and targeting disrupts global silencing of telomeric genes.
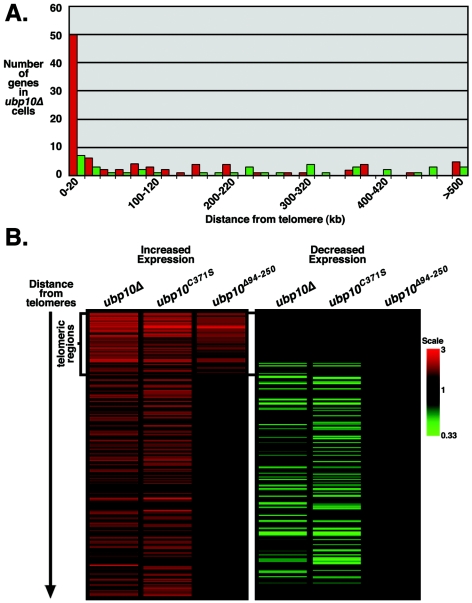
We next determined if Ubp10p's function in gene repression is similarly biased toward silent regions by examining global gene expression in ubp10 mutant cells using DNA microarrays. To avoid secondary effects due to the slow-growth phenotype of ubp10Δ and ubp10C371S cells in synthetic (YC) medium (43), we grew cells used for the expression analysis in rich (YEPD) medium. After applying rigorous statistical criteria (see Materials and Methods), we found that 90 genes have increased expression by 1.5-fold or greater in ubp10Δ cells, and 50 of these genes are located within 20 kbp of the telomeres (Fig. 3A and see Table S1 in the supplemental material). Because less than 5% of all genes are located within 20 kbp of the telomere, the enrichment of telomeric genes in the ubp10Δ expression profile indicates that the role of Ubp10p in gene expression is biased toward regulation of telomeric silencing. We also found that 40 genes have decreased expression by at least 1.5-fold in ubp10Δ cells; 7 of these genes are located within 20 kbp of the telomeres (Fig. 3A; see Table S1 in the supplemental material). As expected, the gene expression profile of ubp10C371S cells significantly overlaps that of ubp10Δ cells (Fig. 3B and see Table S1 in the supplemental material), indicating that Ubp10p's ubiquitin protease activity is largely responsible for its role in gene regulation near telomeres and elsewhere in the genome.
FIG. 3.
Loss of UBP10 primarily affects the expression of telomeric regions. Transcript array analysis was performed using strains UCC6389 (wild type) and UCC6390 (ubp10Δ) for panel A and strains UCC6432 (wild type), UCC6435 (ubp10Δ), UCC6406 (ubp10C371S), and UCC6407 (ubp10Δ94-250) for panel B. Cells were grown to log phase in rich medium (YEPD), and transcripts were isolated and analyzed. (A) Increased expression in ubp10Δ cells shows a telomeric bias. Number of genes increased in ubp10Δ cells plotted by position from the telomere. Histograms represent 20-kb increments from the telomeres. Red histograms represent genes with increased expression; green histograms represent genes with decreased expression. (B) ubp10Δ94-250 cells are specifically defective in telomeric silencing. Positional cluster analysis shows expression changes of genes based on distance from telomeres. Only genes expressed greater than or less than 1.5-fold in at least one of the strains are shown (total number of genes shown is 222). Genes located within 20 kbp of their respective telomeres are marked on the left as “telomeric regions.” See Table S1 in the supplemental material for the entire normalized data set.
The gene-expression profile of ubp10Δ94-250 cells contrasts with that from ubp10Δ and ubp10C371S cells in one notable way. Only telomeric genes have increased expression in ubp10Δ94-250 cells; none of the other nontelomeric genes with altered expression in ubp10Δ or ubp10C371S cells is affected in ubp10Δ94-250 cells (Fig. 3B and see Table S1 in the supplemental material), indicating that the Ubp10Δ94-250 protein, which has the Sir4p-binding region deleted, is specifically defective in telomeric silencing. These results also indicate that the nontelomeric gene expression changes in ubp10Δ and ubp10C371S cells are not an indirect result of losing telomeric gene silencing.
Ubp10/Dot4p reduces histone H3 Lys4 and Lys79 methylation at telomeric loci.
Ubp10p is preferentially localized to silent chromatin, important for repression of telomere-proximal genes, and negatively regulates H2B ubiquitination levels. It is therefore likely that the increased expression of telomere-proximal genes in the absence of Ubp10p function is the result of increased H2B ubiquitination within silent loci. To determine directly if this is the case, we required reagents (i.e., antibodies) that specifically recognize ubiquitinated H2B and can be used in chromatin immunoprecipitation experiments. Despite multiple efforts, we were unable to generate such reagents, and none are available elsewhere. Instead, we chose to use H3 Lys4 and Lys79 methylation as indirect markers of H2B ubiquitination because these modifications both require H2B ubiquitination and because antibodies specific for these modifications are readily available (9, 20, 67, 91).
For the ChIP assay, we used several loci indicative of different chromatin states including the repressed GAL1 locus, the actively transcribed SAN1 locus, the silent HMRa locus, and a silent region adjacent to telomere VIR. Under the growth conditions that we used for the assay, neither the repression of GAL1 nor the expression of SAN1 is altered by deletion or mutation of UBP10 (see Table S1 in the supplemental material). Thus, the GAL1 and SAN1 loci served as independent internal controls to judge the relative changes in H3 Lys4 and Lys79 methylation at the silent regions in HMRa and near telomere VIR.
Using antibodies that recognize H3 Lys4 trimethylation or Lys79 dimethylation, we found that both modifications are increased at the telomere-proximal position on telomere VIR in ubp10Δ cells (Fig. 4). We also found similar increases at a telomere-proximal position on telomere VIIL (data not shown). We could not detect an increase in Lys4 methylation at the silent mating-type loci HMRa in ubp10Δ cells but did detect a small increase in Lys79 methylation (Fig. 4). The lack of a significant increase in H3 Lys4 or Lys79 methylation at HMRa is consistent with the absence of transcription at the silent mating loci in ubp10Δ cells (see Table S1 in the supplemental material).
FIG. 4.
Loss of Ubp10p activity results in increased histone H3 Lys4 and Lys79 methylation and decreased Sir3p binding at telomeres. In vivo cross-linking analysis was performed using strains UCC4825 (wild type), UCC4857 (ubp10Δ), UCC4870 (ubp10C371S), UCC4836 (ubp10Δ94-250), and UCC4836 (sir4Δ). ChIP assays of H3 Lys4 and Lys79 methylation and Sir3p binding were performed using antibodies specific for H3 Lys4 trimethylation, H3 Lys79 dimethylation, or Sir3p. Multiplex PCR amplifications were performed to assess the degree of H3 Lys4 methylation, H3 Lys79 methylation, and Sir3p binding at the silent domains of telomeres VIR and HMRa, the active SAN1 gene, and the repressed GAL1 gene. DNA of the total lysate was amplified as a control. (A) Representative example of Vistra Green-stained PCR amplifications. (B to D) Quantitative analysis of data in panel A. Values for H3 Lys4 and Lys79 methylation represent the ratio of immunoprecipitate to total lysate for the query gene normalized to the ratio of immunoprecipitate to total lysate for SAN1. Values for Sir3p binding represent the ratio of immunoprecipitate to total lysate for telomere VIR normalized to the ratio of immunoprecipitate to total lysate for HMRa. All values are the averages of at least two independent experiments. Error bars represent the standard deviation. (B) Relative fold change of H3 Lys4 tri-methylation at telomere VIR and GAL1. (C) Relative increase in H3 Lys79 methylation at telomere VIR and GAL1. (D) Relative decrease in Sir3p binding at telomere VIR.
We also performed the ChIP analysis with either ubp10C371S or ubp10Δ94-250 cells. Interestingly, in ubp10C371S cells we saw a greater increase in H3 Lys4 and Lys79 methylation in the silent regions at telomeres VIR and HMRa, even though both ubp10Δ and ubp10C371S cells are equivalently deficient in Ubp10p catalytic activity (Fig. 4). Although ubp10Δ94-250 cells also have increased H3 Lys4 methylation at telomere VIR, the increases are less than in ubp10Δ cells (Fig. 4), which is consistent with a weaker silencing defect in ubp10Δ94-250 cells (see Table S1 in the supplemental material). We did not observe statistically significant increases in H3 Lys79 methylation at telomere VIR in ubp10Δ94-250 cells. However, is possible that the normally high levels of H3 Lys79 methylation in the genome (>90% of all nucleosomes) result in a higher background that masks small increases. Accordingly, increases in H3 K79 methylation at silent regions were consistently two- to threefold lower than increases in H3 Lys4 methylation (Fig. 4 and 5).
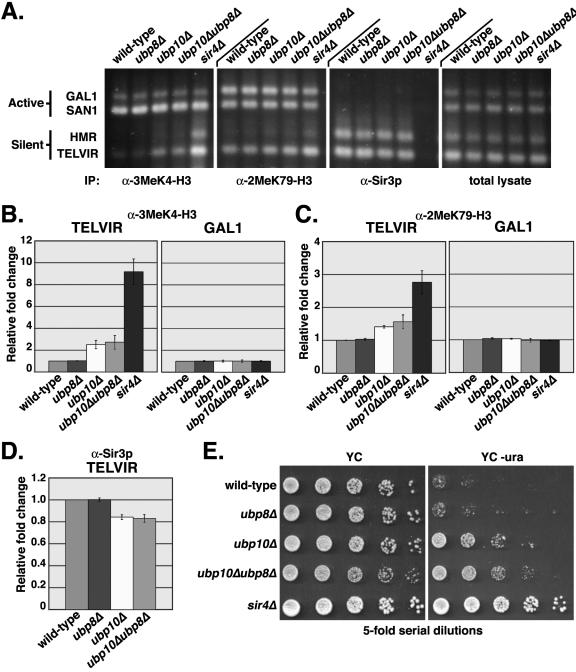
FIG. 5.
Loss of UBP8 does not result in increased histone H3 Lys4 and Lys79 methylation and decreased Sir3p binding at telomeres. In vivo cross-linking analysis was performed as described in the legend of Fig. 4 using stains UCC6389 (wild type), UCC6392 (ubp8Δ), UCC6390 (ubp10Δ), UCC6393 (ubp10Δ ubp8Δ), and UCC6391 (sir4Δ). (A) Representative example of Vistra Green-stained PCR amplifications. (B to D) Quantitative analysis of data in shown in panel A was performed identically as described in the legend of Fig. 4B to D. (B) Relative increase in H3 Lys4 trimethylation at telomere VIR and GAL1. (C) Relative increase in H3 Lys79 methylation at telomere VIR and GAL1. (D) Relative decrease in Sir3p binding at telomere VIR. (E) Ubp8p does not function in telomeric silencing. Overnight, saturated cultures of yeast strains UCC6422 (wild type), UCC6423 (ubp10Δ), UCC6424 (sir4Δ), UCC6425 (ubp8Δ), and UCC6426 (ubp10Δ ubp8Δ), which all carry the URA3 gene located near telomere VIIL (85), were serially diluted and spotted onto YC plates, with or without uracil. Cells were grown at 30°C for 3 days.
As expected, the greatest increases in H3 Lys4 and Lys79 methylation at telomeres VIR and HMRa occur in sir4Δ cells (Fig. 4), in which silencing is completely disrupted (Fig. 5E). Although loss of Ubp10p function at telomeres does not have as great an effect as loss of Sir4p function, all ubp10 mutations result in increased histone H3 modifications at silenced loci.
Increased H3 Lys4 and Lys79 methylation at telomere VIR in ubp10 mutant cells would be expected to result in reduced Sir protein binding. In ubp10Δ cells, we observed modest decreases in Sir3p binding to telomere VIR (Fig. 4) and telomere VIIL (data not shown), but there is no effect on Sir3p localization to HMRa (Fig. 4), which is consistent with the fact that ubp10Δ cells show little change in silencing at HMR (or HML) and are still able to mate with high efficiency (43, 85). As with the above H3 Lys4 and Lys79 methylation increases, the decreases in Sir3p localization at telomere VIR are far greater in ubp10C371S cells than in ubp10Δ cells (Fig. 4). One possible explanation for this difference is that the full-length, but catalytically inactive, Ubp10C371S protein has a longer association with Sir4p than wild-type Ubp10p, and this interferes with the binding of the other Sir proteins. Like ubp10Δ cells, ubp10Δ94-250 cells have modest reductions in Sir3p binding to telomeres. In all mutant ubp10 cells, changes in Sir2p binding to telomeres were similar to those observed for Sir3p, with no detectable change at the silent mating loci (data not shown).
While Sir2p and Sir3p binding in all ubp10 mutant cells can still be detected, Sir2p and Sir3p binding is eliminated in sir4Δ cells (Fig. 4 and data not shown), which is consistent with loss of SIR4 having a far greater effect on silencing than loss of UBP10 (85). Thus, it appears that Ubp10p activity is not required for Sir protein binding at silent loci but optimizes Sir protein association instead.
Ubp8p does not regulate Lys4 and Lys79 methylation within silent regions.
Loss of UBP10 results in increased H2B ubiquitination (Fig. 1), which is the likely cause for the increases in H3 Lys4 and Lys79 methylation in silent regions. Loss of UBP8 also results in increased H2B ubiquitination (Fig. 1), so we tested if similar increases in H3 Lys4 and Lys79 methylation occur at silent regions in ubp8Δ cells as they do in ubp10Δ cells. By ChIP analysis, we found no effect of losing UBP8 on H3 Lys4 or Lys79 methylation in silent regions, nor did we find a detectable change in Sir2p or Sir3p binding to the telomeres (Fig. 5A to D). Loss of UBP8 also has no effect on telomeric silencing (Fig. 5E; see Fig. 8). Ubp8p's lack of involvement in any aspect of silencing indicates that it is specifically Ubp10p's regulation of H2B ubiquitination that is required for silencing.
Loss of UBP8 has been previously reported to result in an increase in H3 Lys4 trimethylation at GAL1 under repressing conditions of growth in glucose (33). This is curious because H3 Lys4 trimethylation has been shown to occur specifically in the 5′ coding portion of active genes and is absent from inactive and repressed genes (66, 82). Consistent with GAL1 repression in glucose-grown cells, we find no noticeable change in H3 Lys4 trimethylation at GAL1 in the absence of UBP8 (Fig. 5A to C). The difference between our results and those from the earlier study might be explained by differences in the strains used in each study: Henry and colleagues used yeast strains derived from W303 (33), whereas we used yeast strains derived from S288c. In general, glucose repression appears to be less stringent in W303-derived strains than in S288c-derived strains (10). For GAL1 gene repression in particular, it has been shown that short-term GAL1 repression in W303-derived strains occurs more slowly and is less stringent than in S288c-derived strains (25). A low level of GAL1 expression in glucose-grown W303-derived strains, but not in S288c-derived strains, might explain the difference in the results.
Function of Ubp10/Dot4p is not restricted to silent loci.
The gene expression profiles of dot4 mutant cells (Fig. 3) indicated that Ubp10p activity is not restricted to the regulation of telomeric loci since the majority of total genes with altered expression in ubp10Δ cells (73 out of 130) are nontelomeric. The transcriptional effects at these loci are not the indirect result of derepression at telomeres because none of these genes shows altered expression in ubp10Δ94-250 cells, which are specifically defective in telomeric silencing (Fig. 3). Thus, Ubp10p likely has a direct effect on gene expression at these nontelomeric loci. However, it was not clear whether the expression of these nontelomeric loci is also regulated via modulation of H2B ubiquitination levels by Ubp10p. To address this issue, we examined total H2B ubiquitination and H3 Lys4 dimethylation levels in the various ubp10 alleles described above. Steady-state levels of both H2B ubiquitination and H3 Lys4 dimethylation are increased to the same level in ubp10Δ and ubp10C371S cells (Fig. 6A). Surprisingly, H2B ubiquitination and H3 Lys4 dimethylation levels in ubp10Δ94-250 cells are identical to wild-type UBP10 cells (Fig. 6A), even though silencing at telomeric loci is compromised in ubp10Δ94-250 cells. Because silent regions comprise approximately 10% of total chromatin (52, 55, 92), any increases in the steady-state levels of H2B ubiquitination that occur solely as a result of loss of silencing might not be detected by Western analysis. Supporting this idea, deletion of any individual SIR gene has no effect on the steady-state levels of H2B ubiquitination or H3 Lys4 dimethylation (Fig. 6B). Thus, the ubp10Δ94-250 allele reveals that Ubp10p activity is not restricted to silent regions and that Ubp10p also acts on nontelomeric regions to regulate H2B ubiquitination levels.
Ubp10/Dot4p and Ubp8p act on the same chromatin regions.
As shown above, ubp10Δ cells have higher steady-state levels of H2B ubiquitination than wild-type cells, and the levels are comparable to those in ubp8Δ cells (Fig. 1 and 7). Interestingly, deletion of both UBP10 and UBP8 results in a synergistic increase in H2B ubiquitination (Fig. 1 and 7). We estimate from dilution blotting and densitometric analyses of blots that 2 to 3% of total H2B is ubiquitinated in wild-type cells, 7 to 10% in ubp10Δ cells, 12 to 16% in ubp8Δ, and 35 to 40% in ubp10Δ ubp8Δ cells. An additive increase in the steady-state levels of H2B ubiquitination in ubp10Δ ubp8Δ cells would indicate that Ubp10p and Ubp8p exclusively act on separate regions of chromatin. However, the synergistic increase in H2B ubiquitination levels in ubp10Δ ubp8Δ cells indicates that Ubp10p and Ubp8p also act on overlapping regions of chromatin.
It was recently shown that as yeast cells enter diauxie or when glucose is depleted, H2B ubiquitination is no longer detectable (19). We examined wild-type cells in diauxie and similarly found that H2B ubiquitination completely disappears (Fig. 7A). H2B ubiquitination also disappears in either ubp10Δ or ubp8Δ cells during diauxie, though not completely as trace amounts of H2B ubiquitination could often be detected in both ubp10Δ and ubp8Δ cells with longer exposures (Fig. 7A and B). In ubp10Δ ubp8Δ cells, however, the majority of H2B ubiquitination originally detected in log phase is retained in diauxie (Fig. 7A). Although it is not known whether Rad6p-Bre1p ubiquitination activity is reduced as cells enter diauxie, maintenance of the high steady-state levels of H2B ubiquitination in ubp10Δ ubp8Δ cells indicates that Ubp10p and Ubp8p are the primary enzymes required for reduction of H2B ubiquitination during this time. The persistence of global H2B ubiquitination in ubp10Δ ubp8Δ cells during diauxie, but not in ubp10Δ or ubp8Δ cells, further supports the idea that Ubp10p and Ubp8p overlap in many of the chromatin regions that they modify.
The H2B ubiquitination that is still present in ubp10Δ ubp8Δ cells during diauxie is eliminated as cells enter stationary phase (Fig. 7A). The loss of H2B ubiquitination in stationary phase is the likely the result of two things: Rad6p-Bre1p no longer ubiquitinates H2B and another mechanism eliminates ubiquitinated H2B. In ubp10Δ ubp8Δ cells we frequently observed an additional modified form of H2B with an increased molecular weight indicative of diubiquitination (Fig. 7B). This correlated with the slow loss of H2B ubiquitination in ubp10Δ ubp8Δ cells as they transited into stationary phase (Fig. 7A). We speculate that the persistence of monoubiquitin on H2B in ubp10Δ ubp8Δ cells increases the chance that H2B will become polyubiquitinated and ultimately removed by the proteasome, perhaps by degradation. It is not clear if this mode of H2B ubiquitination removal is normally operative in wild-type cells or if it is only a condition of losing both Ubp10p and Ubp8p function.
We also examined the steady-state levels of H3 Lys4 and Lys79 methylation in diauxic and stationary cells. In contrast to the increased steady-state levels of H2B ubiquitination that disappear in ubp10Δ cells during diauxie and stationary phase, the increased steady-state levels of H3 Lys4 and Lys79 methylation remain throughout all phases (Fig. 7A). Thus, the transient increase in H2B ubiquitination levels in ubp10Δ cells leads to relatively stable increases in H3 Lys4 and Lys79 methylation.
Ubp10/Dot4p has a role in gene expression beyond telomeric loci.
From the synergistic increase in H2B ubiquitination in ubp10Δ ubp8Δ cells (Fig. 1 and 7), it seemed that Ubp10p and Ubp8p might regulate H2B ubiquitination levels within some of the same chromatin regions. To examine this issue further, we examined gene expression profiles of ubp10Δ, ubp8Δ, and ubp10Δ ubp8Δ cells.
As stated above, 90 genes in ubp10Δ cells have increased expression of 1.5-fold or greater, with 50 of these located within 20 kbp of the telomeres (Fig. 3 and 8). Forty genes have reduced expression of at least 1.5-fold in ubp10Δ cells, with only 7 genes located within 20 kbp of the telomeres (Fig. 3 and 8).
By contrast, 17 genes have increased expression of 1.5-fold or greater in ubp8Δ cells, and none are located within 20 kbp of telomeres (Fig. 8), which is consistent with the fact that Ubp8p does not function in telomeric silencing (Fig. 5). Also in ubp8Δ cells, 17 genes have reduced expression of at least 1.5-fold, and only 4 of these genes are located within 20 kbp of the telomeres. Of the genes with altered expression in the ubp8Δ transcript profile, the majority (22 of a total of 34) have SAGA-dominated gene expression (38), as expected from Ubp8p function in SAGA-mediated transcription (18, 33, 80). It is worth noting that none of the 17 genes with increased expression in ubp8Δ cells overlaps with those affected in ubp10Δ cells, while just 3 genes with decreased expression in ubp8Δ cells are shared with ubp10Δ cells (Fig. 8A).
As expected, the majority of genes affected in ubp10Δ and ubp8Δ cells are also affected in ubp10Δ ubp8Δ cells (Fig. 8A). However, over 160 additional genes have increased expression of 1.5-fold or greater in ubp10Δ ubp8Δ cells compared to cells carrying either ubp10Δ or ubp8Δ alone. Of these additional genes, only 13 are located within 20 kbp of the telomeres (Fig. 8B; see also Table S1 in the supplemental material). The greater number of nontelomeric genes with increased expression in ubp10Δ ubp8Δ cells indicates a possible redundancy for Ubp10p and Ubp8p at these additional loci—either ubiquitin protease may be able to compensate in the absence of the other to maintain lower levels of transcription or repression at these loci. A similar situation was seen for genes that have decreased expression in ubp10Δ ubp8Δ cells. Over 40 additional genes have reduced expression of at least 1.5-fold in ubp10Δ ubp8Δ cells compared to cells carrying either ubp10Δ or ubp8Δ alone (Fig. 8A). Together, these data support the idea that Ubp10p and Ubp8p regulate H2B ubiquitination and gene expression at a shared set of loci in the genome.
DISCUSSION
We previously identified Ubp10/Dot4p as a ubiquitin protease whose enzymatic activity is required for optimal telomeric silencing (43, 85). However, we had not identified any ubiquitinated proteins that are Ubp10p substrates. Here we report that Ubp10p negatively regulates histone H2B ubiquitination.
Ubp10/Dot4p's role in telomeric silencing may be the removal of H2B ubiquitination.
Consistent with a role in silencing, we found that Ubp10p is targeted to silent regions by its Sir4p-interaction domain (Fig. 2), and this interaction facilitates telomeric silencing by preventing the accumulation of H3 Lys4 and Lys79 methylation (Fig. 4). Given Ubp10p's ubiquitin protease activity and the requirement of H2B ubiquitination for H3 Lys4 and Lys79 methylation (9, 20, 67, 91), we propose that Sir4p targets Ubp10p to silent chromatin to remove ubiquitin from H2B. This, in turn, would decrease the probability that H3 Lys4 and Lys79 become methylated within silenced regions.
The role for Ubp10p in silencing is best considered in light of the dynamic binding nature of chromatin proteins. Chromatin proteins continually associate and dissociate from chromatin, including the proteins considered to have relatively stable associations in vitro (72, 73). Indeed, Sir protein binding to silent chromatin is dynamic throughout the cell cycle, even in G1 and M when the genome is not being replicated (14, 51, 56). Temporary dissociation of Sir proteins from silent chromatin could leave these regions briefly accessible to the Rad6p-Bre1p complex and thereby susceptible to H2B ubiquitination. If fortuitous H2B ubiquitination persists for too long, this reversible histone mark could be converted into the more permanent marks of H3 Lys4 and Lys79 methylation, which would prevent the reassociation of Sir proteins and result in the loss of silencing. We postulate that Sir4p-dependent enrichment of Ubp10p at silent chromatin concentrates its ubiquitin protease activity to maintain silencing in the face of Sir protein dynamics.
While Ubp10p is localized to the silent mating region HMRa in a Sir4p-binding domain-dependent manner (Fig. 2), loss of UBP10 does not appreciably alter HM silencing as measured by silencing reporters placed at either HM loci (85), global transcript analysis (see Table S1 in the supplemental material), ChIP assay of H3 Lys4 methylation (Fig. 4), and quantitative mating assays of ubp10Δ cells (data not shown). HM silencing is inherently more stable than telomeric silencing (31), due to the significant stabilization of Sir protein association with chromatin by the binding of Sir1p to HM loci silencer regions (15). If this greater stability occurs by a reduction in Sir protein dissociation, it would readily explain the reduced reliance on Ubp10p action at the silent mating loci and maximal Sir protein binding to HMRa in the absence of UBP10 (Fig. 4).
Ubp10/Dot4p negatively regulates global H2B ubiquitination and H3 Lys4 and Lys79 methylation.
We found that loss of silencing, by deletion of any single SIR gene or the Sir4p-binding region in Ubp10p, does not result in higher steady-state levels of H2B ubiquitination or H3 Lys4 methylation (Fig. 6), despite the fact that these manipulations result in increases in H3 Lys4 methylation in silent regions (Fig. 4). Yet loss of Ubp10p catalytic activity, by deletion of UBP10 or mutation of the catalytic Cys residue, does result in higher steady-state levels of H2B ubiquitination and H3 Lys4 and Lys79 methylation (Fig. 6), indicating that Ubp10p activity is not limited to silent chromatin.
From these observations, it is likely that Ubp10p has a similar function elsewhere in the genome as it does in silent chromatin—to limit or prevent H3 Lys4 and Lys79 methylation by reversing H2B ubiquitination. However, there may be a significant difference in how Ubp10p activity is used. In silent chromatin, it is proposed that all forms of H3 Lys4 and Lys79 methylation will interfere with Sir protein binding (64, 81, 92), so Ubp10p may be recruited to prevent any methylation of H3 Lys4 and Lys79. Elsewhere in the genome, the degree of methylation coincides with different functional states. For example, H3 Lys4 dimethylation and Lys79 methylation are present throughout the genome (5, 63, 64, 66, 82, 92) and help prevent the promiscuous binding of Sir proteins (82, 92). By contrast, H3 Lys4 trimethylation specifically occurs in the 5′ coding portion of active genes and is absent from inactive genes (66, 82). It has been suggested that H3 Lys4 trimethylation facilitates gene transcription and possibly serves as a memory mark for recent transcriptional activity (66, 82). At inactive genes, H2B ubiquitination would have to persist long enough for some methylation of H3 Lys4 and Lys79 to occur but be eliminated before H3 Lys4 trimethylation occurs. Considering how loss of UBP10 affects the steady-state levels of H3 Lys4 methylation (Fig. 1 and 7), we propose that Ubp10p acts at inactive or repressed genes to facilitate this timely removal of ubiquitin from H2B. Consistent with this idea, the majority of nontelomeric genes with altered expression in ubp10Δ and ubp10Δ ubp8Δ cells have increased transcription (Fig. 3 and 8), indicating that Ubp10p may be involved in maintaining their inactivity or repression. This is not an indirect effect of losing silencing because the altered expression of these genes does not occur in ubp10Δ94-250 cells, which are specifically defective in telomeric silencing. Perhaps Ubp10p is targeted to inactive genes by repressive complexes, similar to recruitment by Sir4p to silent regions. If so, Ubp10p binding to repressors would have to be through a different domain than that used for binding to Sir4p. Alternatively, the loss of Ubp10p activity might indirectly affect transcription by alterations in the ubiquitination levels of other proteins in addition to H2B.
Ubp10/Dot4p and Ubp8p have separate and overlapping functions.
In addition to its role in silencing, we propose that Ubp10p's regulation of global H2B ubiquitination levels overlaps, in part, with that of Ubp8p. While loss of either UBP10 or UBP8 results in higher steady-state levels of H2B ubiquitination and functionally different phenotypic consequences for the cell (Fig. 1 and 5), the absence of both UBP10 and UBP8 together results in a synergistic increase in H2B ubiquitination levels and transcription (Fig. 7 and 8). Overlapping roles for Ubp10p and Ubp8p in gene regulation or other chromatin-related functions is further underscored by the persistence of ubiquitinated H2B in ubp10Δ ubp8Δ cells as they enter diauxie, which contrasts with the virtual disappearance of ubiquitinated H2B in ubp10Δ and ubp8Δ cells (Fig. 7). Of course, Ubp8p and Ubp10p are separable in some functions: Ubp8p functions in SAGA-mediated transcription (18, 33, 80), whereas Ubp10p functions in silencing (Fig. 5). Also, Ubp10p, but not Ubp8p, regulates steady-state H3 Lys4 and Lys79 methylation levels, even though both regulate steady-state H2B ubiquitination levels (Fig. 1). Despite these separable functions, the synergistic increases in steady-state H2B ubiquitination levels and transcription in ubp10Δ ubp8Δ cells underscore a potential overlapping role for these ubiquitin proteases.
The ubp10Δ94-250 allele demonstrates that Ubp10p regulates H2B ubiquitination outside of its defined role in silencing, but no such separation-of-function alleles that demonstrate a role for Ubp8p outside of the SAGA complex have been isolated. It was recently found that Sgf11p facilitates the interaction of Ubp8p with SAGA (41, 53, 74). Interestingly, while purification of Sgf11p resulted primarily in the copurification of SAGA components, other transcriptional regulators also copurified with Sgf11p (74), indicating that Sgf11p may link Ubp8p to other chromatin-related processes in addition to SAGA-mediated transcription. Taking this and our findings into consideration, we believe Ubp8p functions in other aspects of H2B deubiquitination in addition to its defined role in SAGA-mediated transcription.
Although it is not clear at this point how Ubp10p and Ubp8p regulate H2B ubiquitination at common loci, such an effect could be the result of nonspecific chromatin binding that is independent of their targeted recruitment by Sir complexes and SAGA, respectively. As is the case for a number of histone acetylases and deacetylases (44, 47, 75, 94), Ubp10p and Ubp8p may be able to remove H2B ubiquitination from chromatin throughout the genome without a high-affinity interaction. Some loci may be more susceptible to such “hit-and-run” activities, and alterations in H2B ubiquitination and gene expression are only revealed when both Ubp10p and Ubp8p are absent.
Multiple reasons for removal of ubiquitin from H2B.
The cell may modulate H2B ubiquitination levels by these ubiquitin proteases for a variety of reasons. Removal of ubiquitin from H2B may be used during gene repression to prevent H3 Lys4 and Lys79 methylation, similar to Ubp10p's role in silencing (43, 85). Deubiquitination of H2B may also facilitate transcription, as is the case for Ubp8p's role in SAGA-mediated transcription (18, 33, 80). There is evidence that deubiquitination of H2B may also occur during mitosis to allow chromatin compaction (12, 57, 61). Removal of ubiquitin from H2B may be important in halting transcription of mitotic genes as cells exit the cell cycle and may be the reason for deubiquitination of H2B as yeast cells transit into stationary phase (Fig. 7). Lastly, removal of the single ubiquitin moiety from ubiquitinated H2B may be required to prevent multiubiquitination and subsequent degradation of modified H2B by chromatin-associated proteasomes (29, 30, 60). These and other yet to be discovered outcomes of removing H2B ubiquitination likely contribute to the dynamic and flexible nature of chromatin.
Ubp10/Dot4p may have other roles beyond regulating H2B ubiquitination.
It is important to note that Ubp10p may target other ubiquitinated proteins in addition to histone H2B. For instance, Ubp10p also appears to regulate the steady-state levels of the general amino acid permease Gap1p (42). Because plasma membrane permeases are subject to regulated ubiquitination and endocytosis (34), Ubp10p may indirectly affect transcription of some genes as a result of altered nutrient transport. In fact, ubp10Δ cells have a slow-growth phenotype that is exacerbated in synthetic medium and by the presence of multiple amino acid and nucleotide auxotrophies (43). Thus, in the absence of de novo biosynthetic capabilities, ubp10Δ cells may be starved for essential nutrients due to impaired transport. Previous transcript analyses of ubp10Δ cells grown in minimal medium found a large number of stress response genes with increased expression, coincident with an increase in cellular oxidative damage (70). Nutrient starvation often results in a significant transcriptional stress response (28), which is similar to that observed in ubp10Δ cells grown in minimal medium (70). Furthermore, protracted amino acid starvation induces production of reactive oxygen species and increases apoptosis in wild-type cells (21), similar to what is observed in ubp10Δ cells (70). The increased expression of stress response genes is likely a secondary response to nutrient starvation in ubp10Δ cells. Deletion of any SIR gene can partially compensate for the slow-growth phenotype and the transcriptional stress response in ubp10Δ cells (43, 70). Because a number of subtelomeric genes encoding cell wall stress proteins are regulated by modulation of silencing (1), sirΔ suppression is also likely to be a secondary effect resulting from the increased expression of these proteins that allows sufficient cell wall structural changes to facilitate nutrient transport and partially alleviate the starvation of ubp10Δ cells.
Understanding the complete nature of Ubp10p action in the cell will require efforts aimed at identifying all of its substrates and target chromatin regions. Construction and analysis of separation-of-function alleles, as done here with ubp10Δ94-250, will help delineate Ubp10p action in active chromatin from that in silent chromatin. Because histone deubiquitination may be operative in many different species as a regulatory mode (12, 32, 58, 59, 61), we believe further understanding of the role of Ubp10p in modulating histone H2B ubiquitination will yield greater insight into this common axis of chromatin regulation.
Supplementary Material
Acknowledgments
We thank M. A. Osley for the FLAG-tagged H2B plasmids and J. Delrow for statistical analysis of the transcript microarray data. We thank H. Eisen and the members of the Gottschling lab for insightful discussions and critical reading of the manuscript.
R.G.G. is a Bristol Myers Squibb fellow of the Life Sciences Research Foundation. This work was supported by National Institutes of Health grant GM43893 to D.E.G.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan, and X. F. Zheng. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10:1295-1305. [DOI] [PubMed] [Google Scholar]
- 2.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 3.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 5.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein, M., R. E. Sobel, C. D. Allis, B. M. Turner, and J. R. Broach. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 10.Brown, T. A., and B. L. Trumpower. 1995. Strain-dependent variation in carbon source regulation of nucleus-encoded mitochondrial proteins of Saccharomyces cerevisiae. J. Bacteriol. 177:1380-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck, S. W., and D. Shore. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9:370-384. [DOI] [PubMed] [Google Scholar]
- 12.Cai, S. Y., R. W. Babbitt, and V. T. Marchesi. 1999. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc. Natl. Acad. Sci. USA 96:2828-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, T. H., and M. R. Gartenberg. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14:452-463. [PMC free article] [PubMed] [Google Scholar]
- 15.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531-541. [DOI] [PubMed] [Google Scholar]
- 16.Chiu, Y. H., Q. Yu, J. J. Sandmeier, and X. Bi. 2003. A targeted histone acetyltransferase can create a sizable region of hyperacetylated chromatin and counteract the propagation of transcriptionally silent chromatin. Genetics 165:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullen, P. J., W. Sabbagh, Jr., E. Graham, M. M. Irick, E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. Sprague, Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 19.Dong, L., and C. W. Xu. 2004. Carbohydrates induce mono-ubiquitination of H2B in yeast. J. Biol. Chem. 279:1577-1580. [DOI] [PubMed] [Google Scholar]
- 20.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 21.Eisler, H., K. U. Frohlich, and E. Heidenreich. 2004. Starvation for an essential amino acid induces apoptosis and oxidative stress in yeast. Exp. Cell. Res. 300:345-353. [DOI] [PubMed] [Google Scholar]
- 22.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 23.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 25.Frolova, E., M. Johnston, and J. Majors. 1999. Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulation by glucose and chromatin structure. Nucleic Acids Res. 27:1350-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner, R. G., and R. Y. Hampton. 1999. A “distributed degron” allows regulated entry into the ER degradation pathway. EMBO J. 18:5994-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Degradation-mediated protein quality control in the nucleus. Cell 120:803-815. [DOI] [PubMed] [Google Scholar]
- 28.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillette, T. G., F. Gonzalez, A. Delahodde, S. A. Johnston, and T. Kodadek. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. USA 101:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez, F., A. Delahodde, T. Kodadek, and S. A. Johnston. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548-550. [DOI] [PubMed] [Google Scholar]
- 31.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 32.Henchoz, S., F. De Rubertis, D. Pauli, and P. Spierer. 1996. The dose of a putative ubiquitin-specific protease affects position-effect variegation in Drosophila melanogaster. Mol. Cell. Biol. 16:5717-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 35.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 37.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 39.Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone, and H. D. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 40.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 41.Ingvarsdottir, K., N. J. Krogan, N. C. Emre, A. Wyce, N. J. Thompson, A. Emili, T. R. Hughes, J. F. Greenblatt, and S. L. Berger. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25:1162-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahana, A. 2001. The deubiquitinating enzyme Dot4p is involved in regulating nutrient uptake. Biochem. Biophys. Res. Commun. 282:916-920. [DOI] [PubMed] [Google Scholar]
- 43.Kahana, A., and D. E. Gottschling. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6608-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 46.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 47.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 48.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 49.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 50.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau, A., H. Blitzblau, and S. P. Bell. 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16:2935-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurenson, P., and J. Rine. 1992. Silencers, silencing, and heritable transcriptional states. Microbiol. Rev. 56:543-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, K. K., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levinger, L., and A. Varshavsky. 1982. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell 28:375-385. [DOI] [PubMed] [Google Scholar]
- 55.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 56.Martins-Taylor, K., M. L. Dula, and S. G. Holmes. 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168:65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsui, S. I., B. K. Seon, and A. A. Sandberg. 1979. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc. Natl. Acad. Sci. USA 76:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 59.Mimnaugh, E. G., G. Kayastha, N. B. McGovern, S. G. Hwang, M. G. Marcu, J. Trepel, S. Y. Cai, V. T. Marchesi, and L. Neckers. 2001. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 8:1182-1196. [DOI] [PubMed] [Google Scholar]
- 60.Morris, M. C., P. Kaiser, S. Rudyak, C. Baskerville, M. H. Watson, and S. I. Reed. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423:1009-1013. [DOI] [PubMed] [Google Scholar]
- 61.Mueller, R. D., H. Yasuda, C. L. Hatch, W. M. Bonner, and E. M. Bradbury. 1985. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J. Biol. Chem. 260:5147-5153. [PubMed] [Google Scholar]
- 62.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 65.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 67.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 68.Nickel, B. E., C. D. Allis, and J. R. Davie. 1989. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry 28:958-963. [DOI] [PubMed] [Google Scholar]
- 69.Noma, K., and S. I. Grewal. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orlandi, I., M. Bettiga, L. Alberghina, and M. Vai. 2004. Transcriptional profiling of ubp10 null mutant reveals altered subtelomeric gene expression and insurgence of oxidative stress response. J. Biol. Chem. 279:6414-6425. [DOI] [PubMed] [Google Scholar]
- 71.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 72.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 73.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 76.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 77.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 79.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos-Rosa, H., A. J. Bannister, P. M. Dehe, V. Geli, and T. Kouzarides. 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 279:47506-47512. [DOI] [PubMed] [Google Scholar]
- 82.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 83.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, and R. A. Casero. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 84.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith, J. S., C. B. Brachmann, L. Pillus, and J. D. Boeke. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149:1205-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 90.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 91.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 92.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 93.van Leeuwen, F., and D. E. Gottschling. 2002. Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 14:756-762. [DOI] [PubMed] [Google Scholar]
- 94.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 95.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 96.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.