Abstract
Yeast (Saccharomyces cerevisiae) SWI/SNF is a prototype for a large family of ATP-dependent chromatin-remodeling enzymes that facilitate numerous DNA-mediated processes. Swi2/Snf2 is the catalytic subunit of SWI/SNF, and it is the founding member of a novel subfamily of the SF2 superfamily of DNA helicase/ATPases. Here we present a functional analysis of the diagnostic set of helicase/ATPase sequence motifs found within all Swi2p/Snf2p family members. Whereas many of these motifs play key roles in ATP binding and/or hydrolysis, we identify residues within conserved motif V that are specifically required to couple ATP hydrolysis to chromatin-remodeling activity. Interestingly, motif V of the human Swi2p/Snf2p homolog, Brg1p, has been shown to be a possible hot spot for mutational alterations associated with cancers.
In order to ensure faithful DNA replication, DNA repair, and gene expression, eukaryotic organisms must contend with the packaging of genomic DNA into chromatin. Dynamic changes in chromatin organization can be catalyzed by ATP-dependent chromatin-remodeling enzymes that use the energy derived from ATP hydrolysis to alter histone-DNA interactions (9, 28). All members of this class of remodeling enzyme contain a catalytic subunit that harbors a conserved ATPase domain related to yeast (Saccharomyces cerevisiae) Swi2/Snf2, the founding member of a subfamily of the SF2 superfamily of DNA and RNA helicases (7). Although no member of the Swi2/Snf2 family exhibits bona fide DNA helicase activity, all show DNA-stimulated ATPase activity that is required for chromatin-remodeling function in vitro and/or in vivo (6).
The diagnostic feature of Snf2/Swi2 ATPases is a set of seven ATPase/helicase sequence motifs (7). Structural studies of several bacterial SF2 family members indicate that this ATPase domain is assembled as two subdomains—a conserved N-terminal subdomain I (motifs I, Ia, II, and III) that is required for ATP binding and hydrolysis, and a C-terminal subdomain II (motifs IV to VI) which may play a role in energy transduction (4, 10). More biochemical information exists for motifs within the N-terminal subdomain I, due in part to the fact that it is found in a more diverse group of ATPases, which includes the recombination protein RecA (4). Residues within each of the seven Swi2/Snf2 ATPase/helicase motifs are essential for the functions of the yeast SWI/SNF chromatin-remodeling enzyme in vivo, and single amino acid changes within motif I (Walker A box) eliminate the ATPase activity of SWI/SNF in vitro (15, 24). In contrast, the biochemical roles for the remaining six Swi2/Snf2 ATPase motifs have not been previously investigated.
The ∼1 MDa yeast SWI/SNF complex was one of the first ATP-dependent chromatin-remodeling enzymes to be identified, and consequently many in vivo and in vitro studies have focused on this enzyme (28). SWI/SNF is required for expression of a subset of highly inducible yeast genes as well as for gene expression during late mitosis (14, 32). Furthermore, SWI/SNF can interact directly with a variety of gene-specific transcriptional activators, and these contacts can lead to the recruitment of SWI/SNF remodeling activity to target nucleosomes both in vivo and in vitro (23). How does SWI/SNF alter chromatin? In vitro assays have demonstrated that SWI/SNF-like enzymes can promote the accessibility of nucleases or DNA-binding transcription factors to nucleosomal sites. This enhanced accessibility of nucleosomal DNA may be due to the ATP-dependent movement of histone octamers in cis along the DNA, the transfer of histone octamers from one nucleosomal array to another (transfer in trans), the removal or replacement of nucleosomal histones, or the creation of alternative histone-DNA contacts that might include accessible loops of DNA on the nucleosomal surface (reviewed in reference 28). The degree to which each of these biochemical activities contributes to chromatin-remodeling reactions in vivo remains unclear.
We are interested in understanding how ATP hydrolysis is utilized during the process of chromatin remodeling. To this end we purified SWI/SNF complexes that harbor Swi2/Snf2 subunits containing amino acid substitutions within the conserved ATPase/helicase motifs. Swi2/Snf2 residues were identified that disrupt the various kinetic parameters for ATP hydrolysis (e.g., Km or kcat), and these studies are consistent with previous functional analyses of distantly related DNA or RNA helicases. In contrast, we find that alterations within conserved motif V have little to no effect on ATPase activity but cripple the remodeling activity of SWI/SNF. The data presented here suggest that motif V plays an important role in coupling the hydrolysis of ATP to the mechanism of chromatin remodeling. The evidence also suggests a model whereby residues within motif V play a role in nucleosomal substrate recognition.
MATERIALS AND METHODS
ATPase assays.
All ATPase assays were performed as previously described (20 mM Tris, pH 8.0, 5 mM MgCl2, 0.1 mg/ml bovine serum albumin, 5% glycerol, and 0.2 mM dithiothreitol) (16). DNA (208-11 template) or nucleosomal array was added to a final concentration of 13 nM. Reactions were conducted at 30°C. Rates of hydrolysis were measured by spotting time points on polyethyleneimine-cellulose and resolving released 32P-labeled phosphate from ATP by thin-layer chromatography in 750 mM potassium phosphate, pH 3.5. Analysis of hydrolysis rates was performed using a Molecular Dynamics PhosphorImager and Imagequant v1.2 (Amersham). For kinetic experiments, velocities were determined over time ranges that gave linear ATP hydrolysis rates for ATP concentrations (2 μM to 1 mM) for the various complexes. The kinetic parameters Km, Vmax, and kcat were determined by nonlinear fitting to the Michaelis-Menten equation using KaleidaGraph v3.6 (Synergy Software).
Generation of nucleosomal cruciform templates.
Nucleosomal templates for chromatin torsion assays were generated by salt dialysis as previously described (16). Briefly, 2 μg of 208-11 DNA (carrier DNA) and 0.5 μg of [α-32P]dCTP-labeled pXG540 was mixed with 1 μg of histones in TE buffer (10 mM Tris pH 7.5, 0.1 mM EDTA) containing 2 M NaCl. These amounts of histone octamers and DNA generate templates containing on average one nucleosome per 200 bp of DNA. Step dialysis with decreasing salt concentration was performed as described previously (16). Reconstitutions were verified by EcoRI digestion and native polyacrylamide gel electrophoresis (PAGE) analysis (16).
Torsion assays.
DNA or nucleosomal cruciform formation assays were performed in 30-μl reaction volumes containing 1× remodeling buffer (10 mM Tris, pH 8.0, 50 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, and 0.1 mg/ml bovine serum albumin), 3 mM ATP, 0.15 μg/ml endonuclease VII (Endo VII), 0.1 nM of [α-32P]dCTP pXG540 DNA or nucleosomal pXG540, and 1.5 nM SWI/SNF complex. Rates of cruciform extrusion were measured over 120 min at 25°C. Three-microliter time points were quenched and deproteinated by the addition of 2× Stop buffer (10 mM Tris, pH 8.0, 0.6% sodium dodecyl sulfate [SDS], 40 mM EDTA, 5% glycerol, and 0.1 mg/ml proteinase K) and incubated at 50°C for 20 min to deproteinate samples. Samples were then resolved on 4% 1× Tris-borate-EDTA (TBE) native acrylamide gels and imaged using a Molecular Dynamics PhosphorImager. The percentage of pXG540 fragment cut was determined by using Imagequant v1.2 (Amersham).
Reconstitution of “601” mononucleosomes.
A 343-bp DNA fragment was generated by digesting plasmid CP1024 with EcoRI and HindIII, gel purified, and labeled by Klenow fragment with [α-32P]dCTP. Labeled DNA fragments were used to generate mononucleosomes by oligonucleosome transfer using 2.5 μg of chicken oligonucleosomes (34) and 5.0 ng of labeled DNA fragment. Reconstitutions were performed by stepwise serial dilution as previously described (22). Nucleosome deposition was verified with native PAGE gels. For recombinant histone preparations, histones were expressed and purified as previously described (18). Recombinant intact (3.6 μg) or tailless (5.2 μg) octamers were mixed with 4.9 μg of unlabeled 208-11 DNA template and 90 ng [α-32P]dCTP-labeled 343-bp “601” DNA fragment. Mononucleosome reconstitutions were assembled by stepwise salt dialysis as previously described (16). Reconstitution of the labeled 601 DNA fragment was assayed by native PAGE.
Nucleosome mobilization assay.
Mononucleosomes (343 bp) were used to assay SWI/SNF-induced nucleosome mobilization. Thirty-microliter reaction volumes containing 0.3 ng (0.1 nM) labeled mononucleosome (130 ng total oligonucleosomes; 50 nM) were incubated at 30°C with 3 nM SWI/SNF complex and 3 mM ATP in 1× remodeling buffer for 90 min. Fractions were taken at various times and quenched with 20% glycerol and 600 ng/μl plasmid DNA and placed on ice. Fractions were resolved on 4% native TBE polyacrylamide gels for 90 min at 150 V. Gels were dried and imaged using a Molecular Dynamics PhosphorImager.
Exo III mapping of nucleosome boundaries.
Mononucleosomes for exonuclease III (Exo III) mapping were generated as follows. Plasmid CP1024 was digested with PstI, labeled with [γ-32P]dATP in a partial denaturing T4-PNK reaction mixture, and digested with EcoRI, and the resulting 326-bp DNA fragment was gel purified. This DNA fragment, labeled at the PstI end, was used to generate mononucleosomes by salt dialysis. Thirty-microliter reaction volumes containing 3 nM mononucleosomes or naked DNA were incubated with 3 nM SWI/SNF and 3 mM ATP in 1× remodeling buffer (see “Torsion assays” above) at 30°C. At each time point, 5 μl was withdrawn and incubated with 10 U apyrase for 2 min followed by 50 U of Exo III for an additional 2 min. Reactions were quenched in 2× Stop buffer (see “Torsion assays” above) and incubated at 50°C for 20 min to deproteinate samples. Deproteinated samples were boiled in an equal volume of formamide buffer (80% [wt/vol] deionized formamide, 10 mM EDTA, 1 mg/ml xylene cyanol FF, and 1 mg/ml bromophenol blue) and electrophoresed at 650 V for 55 min on 1× TBE 6% sequencing gels. Gels were dried and imaged using a Molecular Dynamics PhosphorImager (Amersham).
Mononucleosome restriction enzyme accessibility assays.
Thirty-microliter reaction volumes containing 0.3 ng (0.1 nM) mononucleosome (50 nM total nucleosomes) were incubated with 1.0 nM SWI/SNF complex, 3 mM ATP, and 40 U of restriction enzyme (HhaI or PmlI) in 1× remodeling buffer (see “Torsion assays” above) at 30°C. Time points (3 to 4 μl) were taken as indicated above and quenched in 2× Stop buffer (see “Torsion assays” above). Reactions were incubated at 50°C for 20 min to deproteinate samples. Deproteinated samples were resolved on 8% native TBE polyacrylamide gels, dried, and imaged using a Molecular Dynamics PhosphorImager (Amersham).
RESULTS
Purification of SWI/SNF complexes harboring an altered Swi2/Snf2 subunit.
Previously, we conducted a mutational analysis of the Swi2/Snf2 ATPase domain that led to the identification of a number of conserved residues that are essential for SWI/SNF function in vivo (as tested by the expression of several SWI/SNF-dependent genes) but have no effect on SWI/SNF complex assembly (24). Our goal in the current study was to use these mutant strains to purify SWI/SNF complexes containing altered Swi2/Snf2 subunits to investigate how the ATPase motifs contribute to the mechanism of chromatin remodeling. To facilitate purification of these defective SWI/SNF complexes, we used homologous recombination to introduce sequences that encode a tandem affinity purification (TAP) module at the extreme C terminus of the Swi2/Snf2 coding region. SWI/SNF complexes selected for purification include those that contain alterations in Swi2/Snf2 motif I (K798A), motif Ia (P824A), motif III (P932A and W935A), motif V (R1164A, ΔSTRAGGLG; residues 1162 to 1169), or motif VI (R1196K). We also included an alteration (K1088A) that substitutes a residue within a novel motif (IVb) found within the subdomain I and II spacer domain found only in the Swi2/Snf2 family (24). Diploid strains were then constructed by mating each mutant strain to a wild-type (WT) haploid strain so that an untagged version of SWI2 would complement the growth defect of the swi2 ATPase mutants (See Table 1 for the strain list). Since each SWI/SNF complex contains only one copy of the Swi2 ATPase subunit (27), affinity purification of the TAP-tagged copy of Swi2 selectively isolates the defective SWI/SNF complex. SWI/SNF complexes were purified by sequential chromatography on immunoglobulin G agarose and calmodulin agarose resins as previously reported (27), and SWI/SNF complex integrity was monitored by Western blot and SDS-PAGE/silver stain analyses (Fig. 1A and data not shown). The results confirm that each of the SWI/SNF complexes is of similar purity and that alterations within the Swi2/Snf2 ATPase domain have no effect on subunit composition or apparent stoichiometry.
TABLE 1.
Strains used in this study
| Namea | Relevant genotype |
|---|---|
| CY 296 | MATagal4Δ::leu2 leu2Δ1 his3Δ200 trp1Δ99 ura3Δ99 lys2Δ99 ade2− |
| CY 394 | MATα swi2Δ::HIS3 ura3::swi2(R1196A)-HA-His6HO-lacZ |
| CY 396 | MATα swi2Δ::HIS3 ura3::SWI2-HA-His6HO-lacZ |
| CY 397 | MATα swi2Δ::HIS3 ura3::swi2(K798A)-HA-His6HO-lacZ |
| CY 452 | MATα swi2Δ::HIS3 ura3::swi2(P824A)-HA-His6HO-lacZ |
| CY 453 | MATα swi2Δ::HIS3 ura3::swi2(P932A)-HA-His6HO-lacZ |
| CY 454 | MATα swi2Δ::HIS3 ura3::swi2(W935A)-HA-His6HO-lacZ |
| CY 455 | MATα swi2Δ::HIS3 ura3::swi2(R994A)-HA-His6HO-lacZ |
| CY 456 | MATα swi2Δ::HIS3 ura3::swi2(H1061A)-HA-His6HO-lacZ |
| CY 457 | MATα swi2Δ::HIS3 ura3::swi2(K1088A)-HA-His6HO-lacZ |
| CY 458 | MATα swi2Δ::HIS3 ura3::swi2(R1164A)-HA-His6HO-lacZ |
| CY 519 | MATα swi2Δ::HIS3 ura3::swi2(ΔSTRAGGLG)-HA-His6HO-lacZ |
| CY 943* | MATa/α swi2Δ::HIS3 ura3::swi2(K798A)-TAP HO-lacZ diploid from CY397 × CY296 |
| CY 944* | MATα swi2Δ::HIS3 ura3::swi2-TAP HO-lacZ created from CY396 |
| CY 1114* | MATa/α swi2Δ::HIS3 ura3::swi2(P824A)-TAP HO-lacZ diploid from CY452 × CY296 |
| CY 1115* | MATa/α swi2Δ::HIS3 ura3::swi2(P832A)-TAP HO-lacZ diploid from CY453 × CY296 |
| CY 1116* | MATa/α swi2Δ::HIS3 ura3::swi2(W935A)-TAP HO-lacZ diploid from CY454 × CY296 |
| CY 1117* | MATa/α swi2Δ::HIS3 ura3::swi2(K1088A)-TAP HO-lacZ diploid from CY457 × CY296 |
| CY 1118* | MATa/α swi2Δ::HIS3 ura3::swi2(R1164A)-TAP HO-lacZ diploid from CY458 × CY296 |
| CY 1119* | MATa/α swi2Δ::HIS3 ura3::swi2(R1196A)-TAP HO-lacZ diploid from CY394 × CY296 |
| CY 1120* | MATa/α swi2Δ::HIS3 ura3::swi2(ΔSTRAGGLG)-TAP HO-lacZ diploid from CY519 × CY296 |
| CY 1121* | MATa/α swi2Δ::HIS3 ura3::swi2(H1061A)-TAP HO-lacZ diploid from CY456 × CY296 |
Strains marked by asterisks were created for this study.
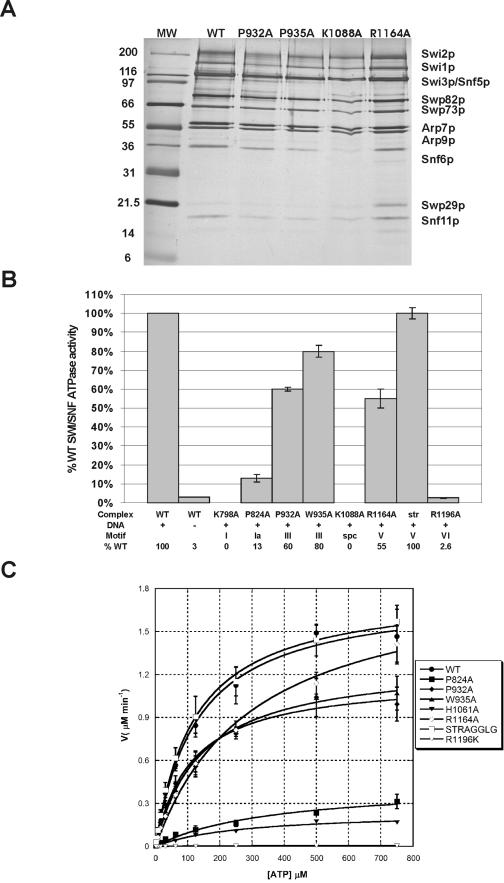
FIG.1.
ATPase activity of altered SWI/SNF complexes. (A) The indicated purified complexes were subjected to SDS-PAGE on 10% polyacrylamide gels and silver stained. (B) Assays for ATPase activity of altered SWI/SNF complexes performed as described in Materials and Methods. This graph represents numerous trials from two to four preparations of each of the various SWI/SNF complexes, normalized for protein concentration by Western blotting and silver staining. All experiments were conducted in the presence of dsDNA and ATP in excess of Km. The DNA sample labeled “WT −” lacks DNA. Error bars represent the standard deviations for the total trials for each individual complex. In the data table, “spc” denotes the spacer region in between motif III and IV in the ATPase domain. (C) Kinetic parameters of ATP hydrolysis for altered SWI/SNF complexes. For all concentrations of ATP, initial velocities were determined from multiple time courses over time ranges giving linear hydrolysis of ATP. Velocities were plotted as functions of ATP concentration and fitted to the Michaelis-Menten equation. Each error bar represents the standard deviation for at least three separate trials for each ATP concentration.
ATPase kinetics of defective SWI/SNF complexes.
Swi2p/Snf2p is a DNA-stimulated ATPase that shows greater than 30-fold stimulation of ATP hydrolysis in the presence of double-stranded DNA (dsDNA). For an initial characterization of the SWI/SNF complexes, dsDNA-stimulated ATPase activity was analyzed for each mutant at a single enzyme concentration (Fig. 1B). Enzyme concentrations for each preparation were calculated by Western blot analyses of serial dilutions using antibodies to the Arp9 subunit. For the >30 preparations of wild-type complex purified to date, we have observed a remarkable reproducibility in the correlation between Arp9 levels and ATPase activity. As a further control for variations in the number of active molecules of enzyme for different mutant complexes, all data shown are the results of assays performed with at least three to five independent preparations of enzyme.
As expected, many of the ATPase domain alterations lead to defects in ATP hydrolysis, and, in general, the magnitudes of the in vitro defects correlate well with the severities of the in vivo phenotypes (24). For instance, a substitution within motif I (K798A) eliminates ATPase activity, and this swi2K798A allele has a null phenotype in vivo (24). Likewise, amino acid substitutions within motif IVb (K1088A) and within motif VI (R1196K) lead to severe defects in the rates of ATP hydrolysis (0% and 3%, respectively, of the rate for the wild type) (Fig. 1B), and each of these mutants show major defects in vivo. A single amino acid change within motif III (W935A) yields an intermediate phenotype in vivo, and this also correlates well with a mild defect in ATP hydrolysis in vitro (∼80% of WT) (Table 2). In contrast, an alteration in motif V (ΔSTRAGGLG) shows no defect in ATP hydrolysis (100% of WT) (Fig. 1B), while this swi2ΔSTRAGGLG mutant has an in vivo phenotype that is equivalent to a complete deletion of SWI2 (24).
TABLE 2.
ATPase kinetic parameters for SWI/SNF complexes
| Parameter | Complex
|
||||||
|---|---|---|---|---|---|---|---|
| WT | P824A | P932A | W935A | R1164A | ΔSTRAGGLG | R1196K | |
| Km (μM) | 152 ± 18 | 406 ± 76 | 113 ± 13 | 172 ± 20 | 332 ± 35 | 125 ± 6 | 234 ± 45 |
| Vmax (μM/min) | 1.90 ± .07 | 0.46 ± .04 | 1.18 ± .04 | 1.41 ± .05 | 2.00 ± .08 | 1.81 ± .03 | 0.01 ± .001 |
| kcat (min−1) | 632 ± 23 | 154 ± 14 | 394 ± 13 | 471 ± 17 | 669 ± 27 | 604 ± 8 | 2 ± 0.5 |
| kcat/Km (M−1 s−1) | 2.5 × 108 | 6.0 × 107 | 2.1 × 108 | 1.6 × 108 | 1.2 × 108 | 2.9 × 108 | 5.1 × 105 |
Following initial ATPase characterization, we conducted a more thorough kinetic analysis of each SWI/SNF variant. The kinetic parameters for ATP hydrolysis, Km and kcat, were determined by nonlinear fitting to the Michaelis-Menten equation for a range of ATP concentrations (2 μM to 750 μM) in the presence of excess DNA cofactor (Fig. 1C). These results are summarized in Table 2. We note that the value of Km is relatively independent of enzyme concentration, and thus changes in the Km value are unlikely to be due to variations in the proportions of active SWI/SNF complexes. Two substitutions, K798A (motif I) and K1088A (motif IVb region), were not analyzed, since they do not show detectable levels of ATP hydrolysis. For the remaining six complexes, effects on both ATP binding (Km) and hydrolysis (kcat) were observed in this analysis. A single amino acid change within motif Ia (P824A) led to an increased Km and a decreased turnover rate (kcat), suggesting that this motif contributes to both ATP binding and hydrolysis. In contrast, alteration of motif VI primarily decreased the turnover rate (kcat was decreased 300- to 400-fold). Interestingly, the ATPase kinetic parameters for the ΔSTRAGGLG complex (motif V) were virtually identical to those for the wild-type complex, and the R1164A substitution within this motif yielded only a twofold increase in Km. Thus, alterations within motif V produce little to no defect in ATPase activity, even though these mutations cause dramatic phenotypes in vivo.
Swi2/Snf2 motif V is required to couple ATP hydrolysis to generation of superhelical torsion.
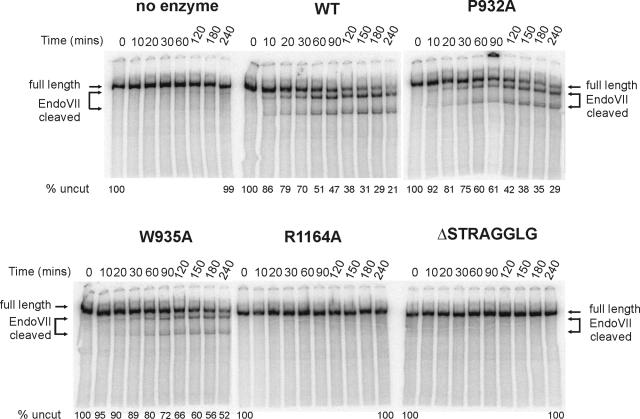
A hallmark of the Swi2p/Snf2p subfamily of ATPases is the ability to generate superhelical torsion in DNA or nucleosomal DNA substrates. This ATP-dependent activity has been suggested to represent the basic biomechanical force that is crucial for all chromatin-remodeling activities (11). We investigated how each ATPase domain alteration affected the generation of superhelical torsion by use of a cruciform extrusion assay (11). For these experiments, a linear 3.8-kb DNA fragment that contains an inverted [AT]34 was used as a template. When this DNA fragment is subjected to a reduction of superhelicity, the [AT]34 repeat is extruded as a cruciform that can be cleaved by the junction-resolving enzyme, T4 endonuclease VII (11). In the absence of remodeling enzyme, no cruciform is extruded, and consequently no T4 Endo VII products are detected (Fig. 2). In contrast, incubation with wild-type SWI/SNF and ATP leads to time-dependent formation of the DNA cruciform and subsequent cleavage by T4 Endo VII, generating two smaller DNA fragments (Fig. 2). As expected, complexes crippled for ATPase activity (K798A, P824A, K1088A, and R1196K) were unable to create levels of torsion in DNA that could be detected by this assay (11; also data not shown). Likewise, complexes that have intermediate ATPase defects also have corresponding intermediate defects in the rates of torsion generation (Fig. 2). Surprisingly, both alterations within motif V (R1164A and ΔSTRAGGLG) severely decrease the ability of SWI/SNF to generate torsion on DNA templates, even though these complexes are proficient at ATP hydrolysis, suggesting that residues within Swi2/Snf2 motif V may be required to couple ATP hydrolysis to chromatin remodeling.
FIG. 2.
Role of Swi2p ATPase motifs in generation of superhelical torsion. The ability to generate DNA superhelical torsion was tested on a linearized (AvaI-digested) pXG540 template which contains an inverted [AT]34 repeat. Reaction mixtures containing 0.1 nM of AvaI-linearized pXG540, 1.5 nM SWI/SNF (WT or variant), 3 mM ATP, and 0.15 μg/ml Endo VII were incubated for 4 h. Samples were taken at the denoted times, quenched, deproteinated and electrophoresed on 4% native PAGE gels (as described in Materials and Methods). The data shown are representative of multiple experiments.
SWI/SNF-dependent nucleosome remodeling requires Swi2/Snf2 motif V.
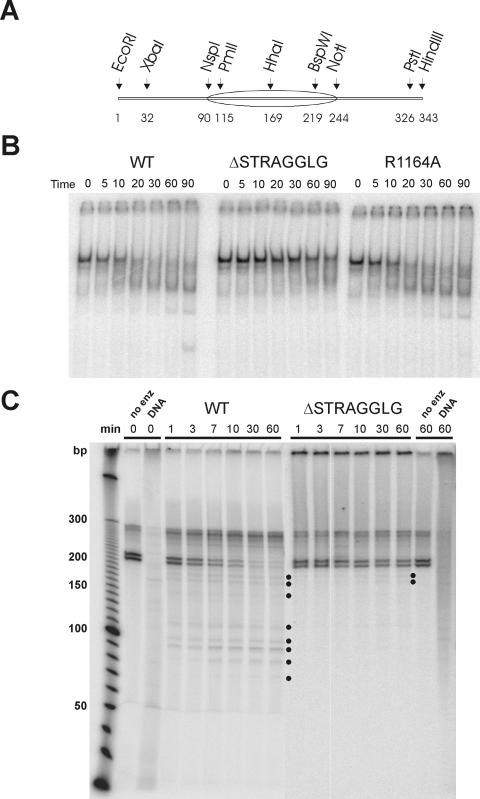
Since disrupting motif V has a major effect on the generation of topological stress, we tested whether alterations within motif V also affect the chromatin-remodeling activities of SWI/SNF. One of the characteristics of the SWI/SNF complex is the ability to mobilize positioned nucleosomes to the ends of short DNA fragments (31). We analyzed the ability of wild-type, R1164A, and ΔSTRAGGLG SWI/SNF complexes to mobilize a centrally positioned mononucleosome. The DNA template used for these studies was a 32P-end-labeled, 343-bp DNA fragment that contains a 601 nucleosome-positioning element located in the center (Fig. 3A). The 601 element was originally isolated in a screen to select for DNA sequences that bind nucleosomes very tightly (17), and unlike most other positioning sequences (e.g., 5S rRNA genes), reconstitution of a nucleosome onto the 601 element leads to one major translational position (Fig. 3A and C). Indeed, prior to incubation with SWI/SNF, >95% of the mononucleosomes migrate as a single species on native PAGE gels (Fig. 3B). The addition of wild-type SWI/SNF led to the ATP-dependent formation of a set of faster-migrating species, and after 90 min, a species that comigrates with free DNA was generated. The simplest interpretation of these results is that SWI/SNF action leads to a movement of the nucleosome in cis towards the ends of the DNA. In contrast, the ΔSTRAGGLG complex is almost completely defective for mobilization of the nucleosome (Fig. 3B).
FIG. 3.
Swi2p motif V is required for nucleosome mobilization by SWI/SNF. (A) Schematic of the 343-bp 601 DNA fragment. Numbers at the bottom indicate base pairs. (B) A representative native PAGE analysis of a 343-bp mononucleosome incubated with WT, ΔSTRAGGLG, or R1164A SWI/SNF complexes. Samples were taken from the reactions at indicated times, incubated with excess dsDNA and glycerol to remove SWI/SNF, and electrophoresed on 4% TBE PAGE gels. (C) Exo III mapping of nucleosome positions after SWI/SNF remodeling confirms mobilization of nucleosomes on the 601 substrate. A 3 nM concentration of DNA fragment (EcoRI-PstI) labeled at the PstI site was incubated with 3 nM SWI/SNF complex (WT or ΔSTRAGGLG) and 3 mM ATP in 1× remodeling buffer. At indicated times, apyrase was added to stop remodeling, which was followed by incubation with Exo III to identify nucleosome boundaries. After deproteination, samples were examined on DNA sequencing gels. WT, wild-type SWI/SNF; no enz, no remodeling enzyme; DNA, nucleosome-free DNA. Dots indicate novel Exo III stops generated after remodeling.
Although changes in nucleosome mobility in native PAGE are commonly interpreted as changes in nucleosome positions, it is also possible that SWI/SNF might generate altered nucleosomes that retain their positioning but contain exposed loops or bulges on the surface (1). Such products might also migrate faster than the initial mononucleosome substrate. To confirm that SWI/SNF was indeed mobilizing nucleosomes, we used exonuclease III to map nucleosome boundaries before and after the remodeling reactions. After incubation of the mononucleosomes with either wild-type or ΔSTRAGGLG SWI/SNF complexes, remodeling was terminated by the addition of apyrase to remove ATP, and the reactions were treated with Exo III. In the absence of chromatin remodeling, a set of two closely spaced Exo III stops that represents nucleosomes with boundaries near the NspI site (Fig. 3C; Fig. 3A shows a schematic of the DNA template) was observed. Wild-type SWI/SNF rapidly disrupts these nucleosome boundaries and generates a pattern of smaller digestion products (Fig. 3C; lanes labeled 1 to 60 min, WT). These data are consistent with the SWI/SNF-dependent movement of nucleosomes to one of the DNA ends. In contrast, significant disruption of the nucleosome boundary was not observed when the ΔSTRAGGLG complex was used during the time course (Fig. 3C; lanes labeled 1 to 60 min, ΔSTRAGGLG). In this case, even at extended reaction periods, only a few of the nucleosomes were moved ∼20 bp from their initial positions (Fig. 3C, 30 to 60 min time points, ΔSTRAGGLG, labeled with two dots). Thus, motif V is required to couple ATP hydrolysis to the nucleosome mobilization activity of SWI/SNF.
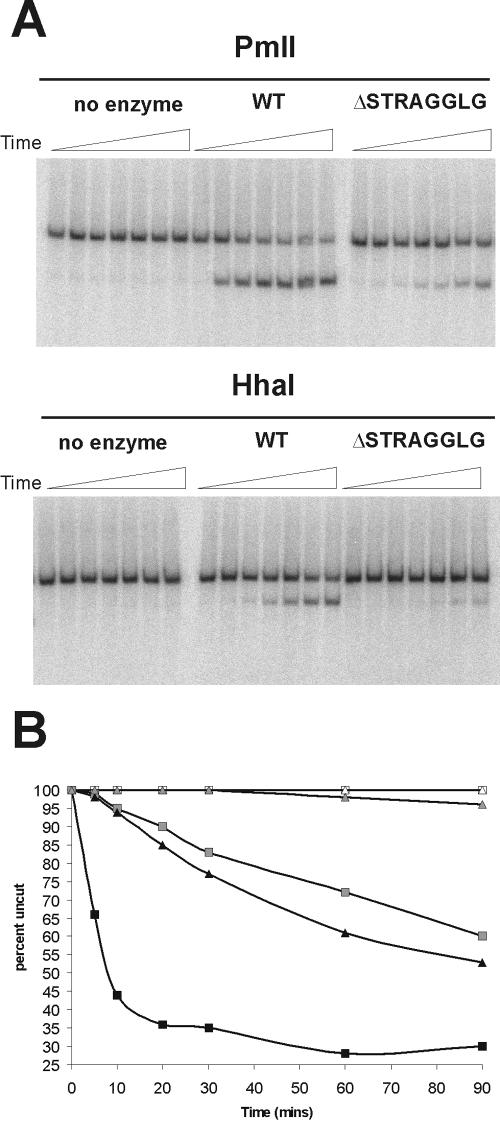
An additional hallmark of SWI/SNF action is the ATP-dependent increase in the restriction enzyme accessibility of mononucleosomes. In principle, this accessibility could result from either movement of nucleosomes or changes in histone-DNA contacts on the surface of the nucleosome. As the small deletion (ΔSTRAGGLG) within motif V cripples nucleosome mobilization, we tested whether there was also a defect in remodeling as assayed by analysis of restriction enzyme accessibility. On the 343-bp centrally positioned mononucleosome, a unique PmlI restriction site is located 25 bp from one DNA entry/exit site of the nucleosome, while a unique HhaI site is near the dyad axis of the positioned nucleosome (Fig. 3A). Remodeling by the wild-type SWI/SNF complex leads to rapid ATP-dependent accessibility of the PmlI site; 65% of the PmlI sites were accessible in 20 min (Fig. 4A and B). In contrast, the ΔSTRAGGLG complex showed drastically decreased activity; less than 10% of the template was cleaved in 20 min (Fig. 4A and B). Likewise, the ΔSTRAGGLG complex is unable to generate significant accessibility of the dyad axis, as illustrated by HhaI accessibility (∼5% accessibility in 90 min). Wild-type SWI/SNF, on the other hand, shows a linear increase in HhaI digestion kinetics that parallels the rate of appearance of the faster-migrating species in the mobility assays (50% complete within 90 min). Thus, these data indicate that the extent of nucleosome mobilization directly correlates with the kinetics of restriction enzyme accessibility and that an intact Swi2/Snf2 motif V is required to couple ATP hydrolysis to the chromatin-remodeling activities of SWI/SNF.
FIG. 4.
Swi2p motif V is required for enhanced restriction enzyme accessibility on mononucleosomes. (A) Reaction mixtures containing 1.0 nM wild-type SWI/SNF or ΔSTRAGGLG SWI/SNF were incubated with 130 ng (50 nM) total nucleosomes (0.1 nM 601 mononucleosomes), 3 mM ATP, and 40 U of restriction enzyme (HhaI or PmlI). Samples were removed from the reaction mixtures at given times (see panel B), quenched, and deproteinated. Samples were resolved on 8% TBE native PAGE gels. (B) Graphical representation of data from panel A. Reaction mixtures containing no enzyme (white symbols), WT (black symbols), and ΔSTRAGGLG (gray symbols) were incubated with either HhaI (triangles) or PmlI (squares).
Swi2/Snf2 motif V functionally interacts with nucleosomal histones.
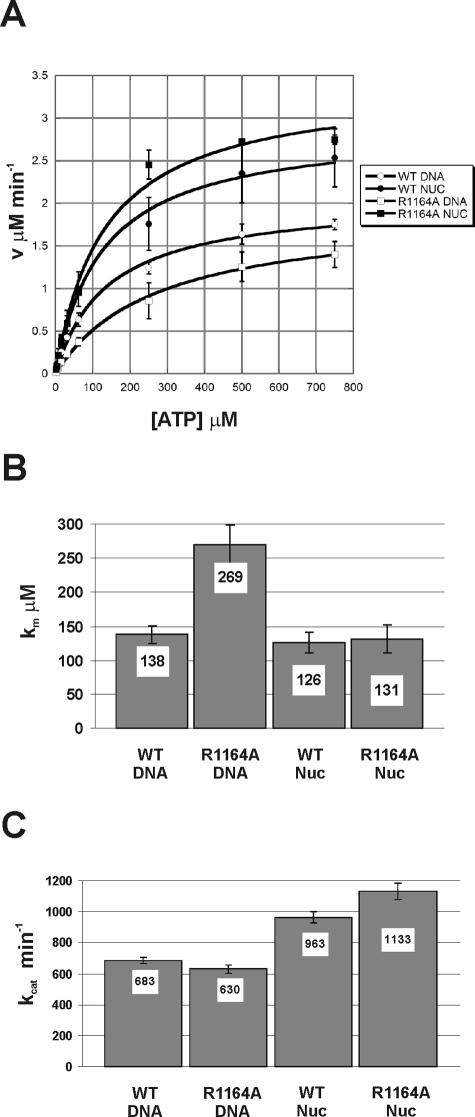
For the most part, the in vitro remodeling defects of Swi2/Snf2 alterations correlate well with the severities of the in vivo mutant phenotypes. For instance, Swi2/Snf2 alterations that eliminate ATPase activity and thus remodeling are strongly defective in vivo (e.g., K798A), and the eight-amino-acid deletion in motif V (ΔSTRAGGLG) eliminates remodeling activity in vitro and shows a null phenotype in vivo. However, the single amino acid substitution within motif V (R1164A) severely decreases ATP-dependent generation of DNA superhelical torsion, but the Swi2R1164A allele has only a modest defect in vivo. Furthermore, the R1164A complex shows a twofold decrease in ATPase activity, whereas the ΔSTRAGGLG deletion in this same motif has no effect on ATPase activity. Surprisingly, when nucleosomal DNA is used as the cofactor for ATPase activity, the ATPase activity of the R1164A complex is restored to wild-type levels (Fig. 5A). A detailed kinetic analysis of the R1164A ATPase assays indicates that nucleosomal DNA lowers the Km to wild-type levels (∼130 μM) (Fig. 5B). In the presence of either substrate, the kcat values for R1164A complex are nearly identical to that of the wild type (Fig. 5C). Interestingly, for both enzymes, the kcat values with nucleosomal DNA as a substrate were 50% higher then with DNA alone (Fig. 5C).
FIG. 5.
Nucleosomal substrates rescue the R1164A ATPase defect. ATPase activity of 4 nM WT or Swi2R1164A SWI/SNF complexes tested with 30 nM dsDNA or 30 nM nucleosomal DNA in a standard ATPase reaction buffer. (A) Hydrolysis rates were determined over time periods that gave linear hydrolysis and plotted as a function of ATP concentration and fitted to the Michaelis-Menten equation. Each error bar represents the standard deviation of at least three separate trials for each ATP concentration. (B) Km values with standard deviation calculated from the data shown in panel A. (C) kcat values for experiment shown in panel A.
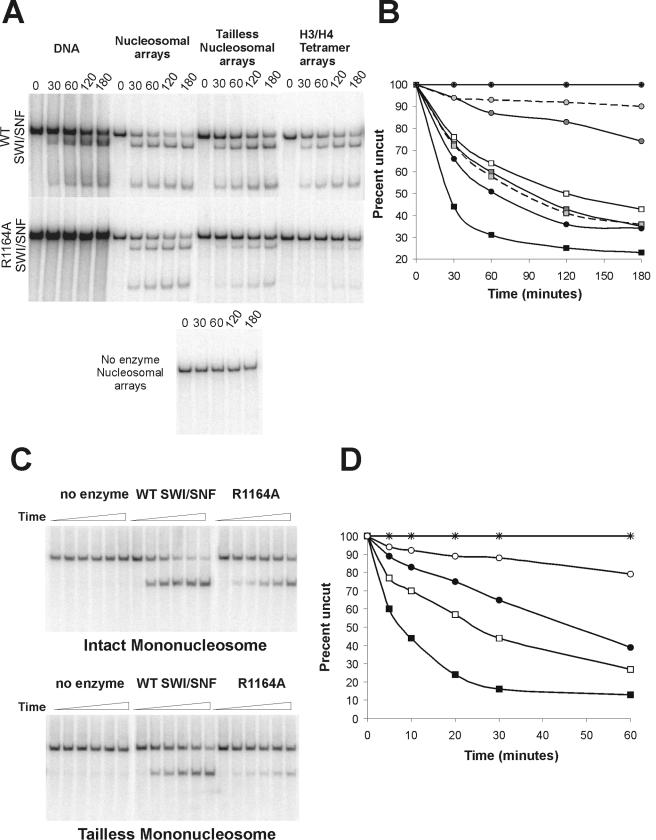
The ability of nucleosome assembly to alleviate the ATPase defect of the R1164A complex led us to analyze the remodeling properties of this complex in greater detail. First, we performed cruciform extrusion assays with nucleosomal substrates. Previous studies have shown that the ISWI and Mi-2 ATP-dependent remodeling enzymes require the presence of nucleosomes on the DNA template in order to produce superhelical torsion. In contrast, SWI/SNF is able to generate superhelical torsion with either nucleosomal or free DNA substrates (Fig. 6A and B and reference 11). We find however that the assembly of the cruciform substrate into a nucleosomal array creates a better substrate for SWI/SNF, as the wild-type enzyme shows a three- to fourfold increase in the rate of torsion generation compared to the free DNA substrate (Fig. 6A and B). Surprisingly, nucleosome assembly also restores the ability of the R1164A complex to generate superhelical torsion; in the presence of nucleosomes the R1164A complex exhibits only a twofold-slower rate of torsion generation compared to wild-type SWI/SNF (Fig. 6A and B). Notably, the ΔSTRAGGLG complex remains highly defective for torsion generation, consistent with the strong swi− phenotype of this mutant in vivo (data not shown). The ability of the R1164A complex to generate torsion on a nucleosomal substrate may explain why this complex functions well in the nucleosome mobilization assay (Fig. 3). Thus, the Swi2 R1164A substitution creates a SWI/SNF enzyme that requires nucleosomal structure for the generation of superhelical torsion, a novel requirement that was previously seen only in the ISWI and Mi-2 subfamilies of ATP-dependent remodeling enzymes.
FIG.6.
Assembly of intact nucleosomes alleviates the remodeling defects of the R1164A SWI/SNF complex. (A) A 1.5 nM concentration of SWI/SNF was incubated with 0.1 nM nucleosomal cruciform template or 0.1 nM DNA template, 0.15 μg/ml endonuclease VII, and 3 mM ATP. (B) Graphical representation of data from panel A. The rates of torsion generation by no enzyme (⧫) or by wild-type (squares) or R1164A (circles) complexes were assayed on DNA (open symbols) or chromatin templates containing intact octamers (black symbols), tailless octamers (dark gray symbols), or (H3-H4)2 tetramers (light gray symbols connected by dashed lines). (C) The abilities of wild-type and R1164A SWI/SNF complexes to create restriction enzyme accessibility (HhaI) were tested on mononucleosomes with intact and tailless octamers. SWI/SNF (1.0 nM WT or R1164A) was incubated with 1 nM mononucleosomes, 3 mM ATP, and 40 U HhaI. Samples were removed from the reaction mixtures at specific time points over 1 h and prepared as described in Materials and Methods. (D) Graphical representation of data from panel C. Wild-type SWI/SNF (squares) or R1164A (circles) reactions were incubated with either intact octamers (filled symbols) or tailless octamers (open symbols). Reactions containing no enzyme were also incubated with intact octamers (×) or tailless octamers (+).
Previous studies have shown that the nucleosome-dependent activities of the ISWI remodeling enzyme requires the histone N-terminal tail domains, and the histone H4 tail appears to play a key role (2, 5). In contrast, the nucleosome-stimulated activities of Mi-2 do not require the histone tail domains, but instead appear to require the histone H2A/H2B dimers (2). To investigate the key determinant that rescues R1164A activity, cruciform substrates were reconstituted with either recombinant histone octamers, recombinant histone H3/H4 tetramers, or recombinant histone octamers that lacked the histone N-terminal tail domains. These chromatin substrates were then used in superhelical torsion assays with either the wild-type or R1164A SWI/SNF complex (Fig. 6A and B). In the case of wild-type SWI/SNF, superhelical torsion was generated on all chromatin substrates, although torsion generation was much less on both the tailless and tetramer templates than on the intact nucleosomal templates (Fig. 6A and B). Interestingly, the levels of generation of torsion by SWI/SNF on the tailless and tetramer substrates were very similar to the levels of torsion generation on naked DNA. Likewise, the Swi2 R1164A complex showed very little ability to generate superhelical torsion on either the tailless or the H3/H4 tetramer templates, although activity was greater than the free DNA substrate. To investigate whether the defects observed on the tailless substrates reflect a role for a unique set of one or more N-terminal tails (e.g., the H2A/H2B tails), we also reconstituted templates with recombinant histones that lacked either the H3/H4 or the H2A/H2B tails. In both cases, torsion generation was equivalent to that of the intact nucleosomal substrates, indicating that a pair of histone tails is sufficient to provide an optimal substrate for both the wild-type and R1164A SWI/SNF complexes (data not shown). These data suggest that the histone tails and the histone H2A/H2B dimers contribute to the formation of an optimal nucleosomal substrate for both wild-type and R1164A SWI/SNF complexes.
Impaired remodeling of chromatin substrates lacking histone N termini.
The cruciform extrusion assays suggest that SWI/SNF prefers to interact with an intact nucleosome; however, it remains a possibility that the defective chromatin templates have differing capacities to constrain transient superhelical torsion generated by SWI/SNF. One prediction of the latter model is that removal of the histone tails should not influence SWI/SNF remodeling activity in other assays. To test this possibility, mononucleosomes were reconstituted using the same recombinant histone octamers used for the torsion assays, and SWI/SNF remodeling activity was monitored by the HhaI restriction enzyme accessibility assay. In the case of the WT SWI/SNF complex, the absence of the N-terminal tails leads to a threefold-slower rate of HhaI accessibility than for the intact mononucleosome substrate (50% cleavage at 24 min for tailless substrate versus 8 min for the intact substrate) (Fig. 6C and D). In the case of the R1164A enzyme, there is a much greater defect observed on tailless nucleosomes relative to the intact nucleosomes (sixfold or greater; ∼15% cleavage at 60 min for tailless substrate versus ∼10 min for intact substrate) (Fig. 6C and D). Thus, the results of these accessibility experiments correlate well with the cruciform extrusion assays, and they are consistent with the idea that the histone tails and/or the presence of an intact octamer play an important role in functional interactions with the Swi2/Snf2 ATPase domain.
DISCUSSION
In this study we characterized the ATPase kinetics and chromatin-remodeling activities of SWI/SNF complexes that harbor alterations within many of the diagnostic ATPase/helicase motifs of the Swi2p catalytic subunit. As expected, many of these Swi2 alterations lead to defects in ATP hydrolysis that result in SWI/SNF complexes crippled for various chromatin-remodeling activities. In contrast, we find that deletion of eight amino acids within motif V (ΔSTRAGGLG) does not alter the ATPase activity of SWI/SNF but that the disruption of motif V cripples the chromatin-remodeling activities of SWI/SNF. Notably, this altered complex maintains the capacity to interact with DNA, as the ATPase activity is still fully DNA stimulated. Thus, the ΔSTRAGGLG complex is specifically impaired in coupling the energy from ATP hydrolysis to the actual biomechanical force required for chromatin remodeling.
The specific role of motif V in canonical SF1 and SF2 helicases is not well understood. Motif V shows considerable sequence variability among helicase-like proteins. In NS3 and UvrB, residues within motif V interact with single-stranded DNA (12, 20, 35). In contrast, alterations within motif V of the Plum pox potyvirus cytoplasmic inclusion RNA helicase exhibit no oligonucleotide binding defect, but these mutants show reduced ATPase and helicase activities (8). Motif V within the PcrA helicase is a third example in which contacts between AMPPNP are mediated through residue E571 of motif V and residue H565 interacts with DNA (30). The differing roles for motif V among these enzymes have led to the hypothesis that motif V might be involved in coupling ATPase activity to the specific function of each individual enzyme. When the amino acid sequence of Swi2p is threaded onto the PcrA crystal structure (C. L. Smith and C. L. Peterson, unpublished observations), motif V within Swi2p encompasses a large, predicted loop that contains the amino acid sequence STRAGGLG. For the PcrA and NS3 helicases and the DNA repair protein CSB, the terminal glycine residue within this loop plays a key role(s) in vivo, and this residue makes important biochemical contacts with either DNA or ATP in vitro (4, 21). Recently, Thoma and colleagues have reported a crystal structure for the Swi2/Snf2-like ATPase domain from the zebra fish Rad54 protein (29). Notably, motif V is clearly visible as an exposed, flexible loop that protrudes into the predicted DNA binding cleft (29).
Among the more closely related Swi2/Snf2 homologs, the conservation of motif V is very high (85% or higher) (Table 3). The conservation of this motif is also high in the ISWI and Mi-2 subfamilies of ATP-dependent chromatin-remodeling enzymes (70 to 85%) (Table 3). In contrast, the Swi2p-like DNA repair proteins (e.g., ATRX, Rad54b, and ERCC) show only weak conservation within this portion of the ATPase domain (Table 3). Given that the motif V domain of zebra fish Rad54 forms the predicted flexible loop within the ATP binding cleft (29), the sequence divergence of motif V among the repair proteins may reflect altered functional properties for these Swi2-like enzymes. In this regard, it is intriguing to note that yeast Rad54 does not exhibit potent nucleosome mobilization activity, although this enzyme is quite active in superhelical torsion assays (13).
TABLE 3.
Motif V alignment within the SWI2/SNF2 ATPase family
| Name | Organism | Motif | Sequencea | % Identity |
|---|---|---|---|---|
| Snf2p | Saccharomyces cerevisiae | V | FILSTRAGGLGLNLQTADTVI | 100 |
| Brg1p | Tetrahymena thermophila | V | FILSTRAGGLGLNLOTADTVI | 100 |
| SPCC1620.14c | Schizosaccharomyces pombe | V | FMLSTRAGGLGLNLQTADTVI | 95 |
| Sth1p | Saccharomyces cerevisiae | V | FLLSTRAGGLGLNLQTADTVI | 95 |
| MG06388.4 | Magnaporthe grisea | V | FLLSTRAGGLGLNLQTADTVI | 95 |
| PSA-4 | Caenorhabditis elegans | V | FMLSTRAGGLGLNLQTADTVI | 95 |
| None (hypothetical protein) | Neurospora crassa | V | FLLSTRAGGLGLNLQTADTVI | 95 |
| SPCC830.01c | Schizosaccharomyces pombe | V | FMLSTRAGGLGLNLQTADTVI | 95 |
| BRAHMA | Caenorhabditis elegans | V | FMLSTRAGGLGLNLQTADTVI | 95 |
| SPAC1250.01 | Schizosaccharomyces pombe | V | FLLSTRAGGLGLNLQTADTVI | 95 |
| BRAHMA | Drosophila melanogaster | V | FLLSTRAGGLGLNLQTADTVV | 90 |
| ENSANGP | Anopheles gambiae | V | FLLSTRAGGLGLNLQTADTVV | 90 |
| SNF2-BETA | Homo sapiens | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| SMARCA4-1 | Homo sapiens | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| BRG1 | Homo sapiens | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| BRG1 | Gallus gallus | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| SMARCA4-2 | Homo sapiens | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| SMARCA4-1 | Mus musculus | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| SMARCA4-2 | Mus musculus | V | FLLSTRAGGLGLNLQSADTVI | 90 |
| BRM | Gallus gallus | V | FLLSTRAGGLGLNLQAADTVI | 90 |
| Hrp1 | Schizosaccharomyces pombe | V | FLLSTRAGGLGINLNTADTVI | 85 |
| HBRM | Homo sapiens | V | FLLSTRAGGLGLNLQAADTVV | 85 |
| Mi-2b | Mus musculus | V | FILSTRAGGLGINLATADTVI | 85 |
| INO80 | Saccharomyces cerevisiae | V | FILSTRAGGLGINLTAADTVI | 81 |
| ISW1 | Saccharomyces cerevisiae | V | FLLTTRAGGLGINLTSADVVV | 71 |
| ATRX | Homo sapiens | V | FIISTKAGSLGINLVAANRVI | 62 |
| RAD54b | Gallus gallus | V | FLLSSKAGGVGLNLVGASHLI | 57 |
| ERCC-6 (CSB) | Homo sapiens | V | FLLTTRVGGLGVNLTGANRVV | 57 |
| Consensus SWI2 family | V | FLLSTRAGGLGLNLQTADTVI | ||
| Consensus SF2 residues (10) | V | +XTXXXXXG+o+Xo+DS |
For SF2 consensus residues, + represents hydrophobic residues, o represents hydrophilic residues, and X represents any residue.
Functional interactions between Swi2 motif V and the nucleosomal substrate.
One of the defining features of the SWI/SNF subfamily of chromatin-remodeling enzymes is the equal stimulation of ATPase activity by both free and nucleosomal DNA. In contrast, both the ISWI and Mi-2 subfamilies show a strong preference for nucleosomal DNA. These data have contributed to the view that SWI/SNF-type enzymes may exclusively recognize the DNA component of the nucleosome. Here we have provided several lines of evidence which suggest that SWI/SNF may preferentially interact with nucleosomal DNA and that motif V within the Swi2p catalytic subunit influences (directly or indirectly) this preferential interaction. We found that SWI/SNF generates superhelical torsion on nucleosomal templates preferentially compared to its activity on DNA substrates and that this enhanced activity on nucleosomes is sensitive to the presence of both the histone N-terminal tails and the H2A/H2B dimers. Importantly, this preference for a nucleosomal substrate does not reflect a peculiarity of the torsion assay, as we also find that SWI/SNF enhances the restriction enzyme accessibility of mononucleosomes that contain histone tails preferentially compared to its activity relative to substrates that lack histone tails. Furthermore, using a restriction enzyme accessibility assay, we previously showed that arrays of H3/H4 tetramers were inefficient substrates for SWI/SNF remodeling (3).
One of the more surprising results that we obtained in our analyses was the response of the R1164A enzyme to nucleosomal substrates. This single amino acid substitution within motif V leads to an approximately twofold decrease in ATPase activity when DNA was used as the nucleic acid cofactor, and this decreased activity correlates with an approximately twofold-lower apparent affinity for ATP (increased Km). However, when the same DNA was incorporated into nucleosomes, the ATPase activity (and Km value) was restored to wild-type levels. Likewise, the R1164A enzyme is unable to generate detectable levels of superhelical torsion on a free DNA substrate, but reconstitution of this DNA into a nucleosomal array almost completely alleviates this defect. We propose a model in which Swi2 R1164 plays a key role in orientating the motif V loop in response to nucleosomal DNA, thereby allowing the transduction of the energy released from ATP hydrolysis into the actual remodeling event. In this model, the motif V loop senses a nucleosome substrate that contains a full complement of intact histones, perhaps by monitoring the histone-DNA environment that is unique to the two adjacent gyres of nucleosomal DNA. This central role for Swi2 in nucleosome recognition is consistent with the observation that Swi2 is the sole SWI/SNF subunit that cross-links only to nucleosomal DNA in vitro (26).
An alternative model posits that the R1164A substitution alters the conformation of SWI/SNF or reduces the activity of the Swi2 motor in a way that can be compensated by the presentation of intact nucleosomes as a substrate. In this alternate view, there is no requirement for motif V to directly interact with nucleosomal DNA. However, it is intriguing to note that the recent crystal structure of the Swi2-like ATPase domain of zebra fish Rad54 shows that the motif V loop is positioned within the center of a cleft that is predicted to bind the DNA substrate (29).
Amino acid substitutions within motif V of hBRG1 (human Swi2p) are associated with cancers.
One of the human homologs of SWI2, hBRG1, has been the focus of numerous studies due to a connection to cancer in mammals (25). Recently, Medina and colleagues screened a number of lung cancer patients for mutations within the hBRG1 gene (19). They identified two types of alterations within hBRG1—a small insertion that occurred in one patient and mutations within the ATPase domain occurring in two patients. It is quite remarkable that both of these ATPase domain mutations alter residues within motif V of hBRG1 (Table 4). Similarly, Wong and colleagues also identified a single amino acid change within the BRG1 ATPase domain in a colon cancer cell line, HCT-116 (33). In this case, the codon for L1164 within motif V was changed to a proline codon (Table 4). Together, these data point to a novel, conserved role for motif V in the mechanism of chromatin remodeling by the SWI2/SNF2 family that may be especially key for the tumor suppressor roles of SWI/SNF.
TABLE 4.
Cancer mutations found in the ATPase motif V of hBRG1
| Isoform | Motif V sequencea | Note (reference) |
|---|---|---|
| WT BRG1 | FILSTRAGGLGLNLQTADTVIIFDS | WT Brg1 |
| L1164P | FILSTRAGGLGPNLQTADTVIIFDS | HCT-116, colon cancer cell line (33) |
| G1160R | FILSTRAGRLGLNLQTADTVIIFDS | G1160R lung carcinoma from patient (19) |
| S1176C | FILSTRAGGLGLNLQTADTVIIFDC | S1176C lung carcinoma from patient (19) |
Underlining indicates point mutations.
Acknowledgments
We thank Jonathan Widom for the gift of the 601 DNA plasmid and Tom Owen-Hughes for the gift of the endonuclease VII. We also thank Pedro Medina for communicating results prior to publication.
These studies were supported by an NIH grant (GM49650) to C.L.P.
REFERENCES
- 1.Aoyagi, S., G. Narlikar, C. Zheng, S. Sif, R. E. Kingston, and J. J. Hayes. 2002. Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions. Mol. Cell. Biol. 22:3653-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, L. A., C. Logie, E. Bonte, P. B. Becker, P. A. Wade, A. P. Wolffe, C. Wu, A. N. Imbalzano, and C. L. Peterson. 2000. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275:18864-18870. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, L. A., X. Shao, R. H. Ebright, and C. L. Peterson. 2000. Roles of the histone H2A-H2B dimers and the (H3/H4)2 tetramer in nucleosome remodeling by the SWI-SNF complex. J. Biol. Chem. 275:11545-11552. [DOI] [PubMed] [Google Scholar]
- 4.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 5.Clapier, C. R., G. Langst, D. F. Corona, P. B. Becker, and K. P. Nightingale. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 7.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez, A., H. S. Guo, P. Saenz, L. Simon-Buela, M. Gomez de Cedron, and J. A. Garcia. 1997. The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25:4474-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165-173. [DOI] [PubMed] [Google Scholar]
- 10.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 11.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. J. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, D. S., S. T. Kim, Q. Sun, and A. Sancar. 1995. Structure and function of the UvrB protein. J. Biol. Chem. 270:8319-8327. [DOI] [PubMed] [Google Scholar]
- 13.Jaskelioff, M., S. Van Komen, J. E. Krebs, P. Sung, and C. L. Peterson. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212-9218. [DOI] [PubMed] [Google Scholar]
- 14.Krebs, J. E., C. J. Fry, M. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 15.Laurent, B. C., I. Treich, and M. Carlson. 1993. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 7:583-591. [DOI] [PubMed] [Google Scholar]
- 16.Logie, C., and C. L. Peterson. 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304:726-741. [DOI] [PubMed] [Google Scholar]
- 17.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 18.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 19.Medina, P. P., J. Carretero, M. F. Fraga, M. Esteller, D. Sidransky, and M. Sanchez-Cespedes. 2004. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer 41:170-177. [DOI] [PubMed] [Google Scholar]
- 20.Moolenaar, G. F., R. Visse, M. Ortiz-Buysse, N. Goosen, and P. van de Putte. 1994. Helicase motifs V and VI of the Escherichia coli UvrB protein of the UvrABC endonuclease are essential for the formation of the preincision complex. J. Mol. Biol. 240:294-307. [DOI] [PubMed] [Google Scholar]
- 21.Muftuoglu, M., R. Selzer, J. Tuo, R. M. Brosh, Jr., and V. A. Bohr. 2002. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene 283:27-40. [DOI] [PubMed] [Google Scholar]
- 22.Owen-Hughes, T., R. T. Utley, D. J. Steger, J. M. West, S. John, J. Cote, K. M. Havas, and J. L. Workman. 1999. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol. Biol. 119:319-331. [DOI] [PubMed] [Google Scholar]
- 23.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 24.Richmond, E., and C. L. Peterson. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24:3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, C. W., and S. H. Orkin. 2004. The SWI/SNF complex—chromatin and cancer. Nat. Rev. Cancer 4:133-142. [DOI] [PubMed] [Google Scholar]
- 26.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636-12644. [DOI] [PubMed] [Google Scholar]
- 27.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10:141-145. [DOI] [PubMed] [Google Scholar]
- 28.Smith, C. L., and C. L. Peterson. 2004. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115-148. [DOI] [PubMed] [Google Scholar]
- 29.Thoma, N. H., B. K. Czyzewski, A. A. Alexeev, A. V. Mazin, S. C. Kowalczykowski, and N. P. Pavletich. 2005. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12:350-356. [DOI] [PubMed] [Google Scholar]
- 30.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 31.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 32.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 33.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 34.Yager, T. D., C. T. McMurray, and K. E. van Holde. 1989. Salt-induced release of DNA from nucleosome core particles. Biochemistry 28:2271-2281. [DOI] [PubMed] [Google Scholar]
- 35.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463-467. [DOI] [PubMed] [Google Scholar]