Abstract
eRF3 is a GTPase associated with eRF1 in a complex that mediates translation termination in eukaryotes. In mammals, two genes encode two distinct forms of eRF3, eRF3a and eRF3b, which differ in their N-terminal domains. Both bind eRF1 and stimulate its release activity in vitro. However, whether both proteins can function as termination factors in vivo has not been determined. In this study, we used short interfering RNAs to examine the effect of eRF3a and eRF3b depletion on translation termination efficiency in human cells. By measuring the readthrough at a premature nonsense codon in a reporter mRNA, we found that eRF3a silencing induced an important increase in readthrough whereas eRF3b silencing had no significant effect. We also found that eRF3a depletion reduced the intracellular level of eRF1 protein by affecting its stability. In addition, we showed that eRF3b overexpression alleviated the effect of eRF3a silencing on readthrough and on eRF1 cellular levels. These results suggest that eRF3a is the major factor acting in translation termination in mammals and clearly demonstrate that eRF3b can substitute for eRF3a in this function. Finally, our data indicate that the expression level of eRF3a controls the formation of the termination complex by modulating eRF1 protein stability.
In eukaryotes, two release factors, eRF1 and eRF3, are required to complete protein synthesis. These translation termination factors associate in a complex which binds to the elongating ribosome when a stop codon enters the A site. eRF1 recognizes all three stop codons by direct interaction at the decoding A site (5, 10) and activates the peptidyltransferase center, which triggers the hydrolysis of the peptidyl-tRNA, generating a free full-sized polypeptide. eRF3 is a GTPase that stimulates eRF1 activity in a GTP-dependent manner (40). eRF3 alone can bind GTP, but its GTPase activity requires the presence of both eRF1 and ribosomes, which may play the role of a composite GTPase-activating protein (11, 12). The eRF3 GTP-bound form interacts with eRF1 in vitro and in vivo to constitute the active translation termination complex (12, 36, 40). In the yeast Saccharomyces cerevisiae, eRF1 and eRF3 are encoded by the essential genes SUP45 and SUP35, respectively. Mutations in either of these genes cause an omnipotent suppressor phenotype and the ability to enhance nonsense suppression by a weak suppressor tRNA (reviewed in reference 18). Recently, it has been shown in S. cerevisiae that eRF3 GTPase activity facilitates stop codon decoding by eRF1 (33).
The C-terminal regions of eRF3 proteins are highly conserved through evolution and carry the four canonical GTP-binding motifs of the GTPase superfamily (2). This domain is essential for translation termination and interaction with eRF1. The N-terminal region varies in both length and sequence among species. In yeast, it is neither essential for cell viability nor required for termination but is responsible for prion-like [PSI+] factor formation (29, 31). Otherwise, this domain participates in the interaction with eRF1 (15, 30) and is involved in eRF3 binding to the poly(A)-binding protein, PABP (14). In vivo, eRF3 interacts simultaneously with eRF1 and PABP when the latter is bound to the translation initiation factor eIF-4F (37). This complex mediates ribosome recycling and ensures the coupling between termination and initiation of translation. Moreover, in yeast, eRF3-PABP interaction couples translation to mRNA decay (17). Recently, the crystal structure of Schizosaccharomyces pombe N-terminally truncated eRF3 revealed a strong overall similarity with the elongation factors EF-Tu and eEF-1A, but also local structural changes that affect nucleotide and Mg2+ binding to eRF3 (23). Indeed, eRF3 has negligible affinity for GDP at physiological Mg2+ concentration, implying that the GDP-to-GTP transition of eRF3 would not require a guanine exchange factor. In addition, the interaction domain with eRF1 was localized close to the eRF3 C terminus, and it was shown that eRF3 N-terminal extension can block this domain, potentially regulating the interaction between the two factors (23).
Two distinct genes encoding eRF3 were identified in the human, mouse, and rat genomes, but not in the recently available chicken genome. These genes, called GSPT1/eRF3a and GSPT2/eRF3b (15, 16, 19), are located on human chromosomes 16 and X, respectively. The eRF3a gene contains several intronic sequences, whereas the eRF3b gene has none. At the mRNA level, eRF3a and eRF3b differ in tissue distribution and in expression during cell cycle progression (15, 16). eRF3b mRNA is poorly expressed in most mouse tissues tested except the brain, whereas eRF3a mRNA is abundant in all tissues, and its level varies during the cell cycle. The encoded proteins, eRF3a and eRF3b, share 87% identity, most of the differences being concentrated in their N-terminal domains. Both proteins bind to eRF1 and stimulate eRF1 release activity in vitro, suggesting that both can play a role in translation termination (15, 19). However, in yeast, the interaction of mouse eRF3a with eRF1 in the two-hybrid system is much less efficient than that of mouse eRF3b (15), and mouse eRF3b, but not eRF3a, can substitute for yeast eRF3 (25). Moreover, in human cells, the overexpression of eRF3b, but not that of eRF3a, has an antisuppressor effect (19, 26). These results call into question the role of eRF3a, and finally, whether both proteins can function as translation termination factors in vivo has not been clearly determined.
In the present work, we first analyzed the distributions of the eRF3a and eRF3b proteins in various human cell lines and mouse brain, and second, we investigated the roles of eRF3a and eRF3b in translation termination. For this purpose, we silenced the expression of each gene in cultured human cells, using short interfering RNAs (siRNAs). The effect of silencing on translation termination efficiency was examined by measuring the readthrough at a premature nonsense codon in a reporter mRNA. Our results show that eRF3a is present in all cell lines tested, whereas eRF3b is found only in mouse brain. In human 293 cells, we found that eRF3a silencing induces a significant increase in readthrough, whereas eRF3b silencing has no detectable effect. We also show that both eRF3a and eRF3b overexpression alleviates the effect of eRF3a silencing on readthrough efficiency. In addition, eRF3a depletion reduces the intracellular level of eRF1 protein by affecting its stability. Thus, in contrast with the literature on eRF3a and eRF3b, these results suggest that eRF3a is the major factor acting in translation termination in mammalian cells and that its expression level influences eRF1 protein stability, regulating the formation of the translation termination complex. We also demonstrate that eRF3b, which seems to be poorly expressed in mammalian cells, can substitute for eRF3a in the translation termination process.
MATERIALS AND METHODS
Cell lines.
The human cell lines 293 (ATCC CRL-1573), HeLa (ATCC CCL-2), Hs27 (ATCC CRL-1634), HepG2 (ATCC HB-8065), T-47D (ATCC HTB-6133), and HuH7 were cultured in Dulbecco modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 units/ml penicillin (hereafter referred to as complete DMEM). Pellets of WERI-Rb (ATCC HTB-169), U-2 OS (ATCC HTB-96), ARPE (ATCC CRL-2302), and glioblastoma cells (FOG cells) were a generous gift of Michel Kress (27). Pellets of human normal keratinocytes (SKP4 cells) were generously donated by Thierry Magnaldo (28).
Antibodies.
The antibodies against human eRF3a, eRF3b, and eRF1; mouse eRF3a and eRF3b; and yeast eRF3 were produced by Eurogentec (Belgium). Rabbits were immunized with synthetic peptides derived from the protein sequence. Antibodies were then obtained by affinity purification of the antisera using the corresponding immunogenic peptide: peptide GGRAAPVESSQEE of human eRF3a, peptide GKRMGRGAPVEPSRE of human eRF3b (Fig. 1A), a mixture of peptides VQQRLKLYNKVPPNG and EYQGGDDEFFDLDDY of human eRF1, peptide GGSSGDGRPPEESTQ of mouse eRF3a (mGSPT1), peptide SAPSGDGIAPAAMA of mouse eRF3b (mGSPT2), and a mixture of peptides MSDSNQGNNQQNYQ and LEKGTNRKSKKPPA of S. cerevisiae eRF3. The antibodies directed against Xenopus laevis eRF3 (XRF3) were described previously (26). The anti-α-tubulin (DM1A), anti-rabbit immunoglobulin G (IgG), and anti-mouse IgG peroxidase-linked antibodies were from Amersham Biosciences (England). The anti-Neo and anti β-galactosidase rabbit antibodies were from 5prime-3prime (France).
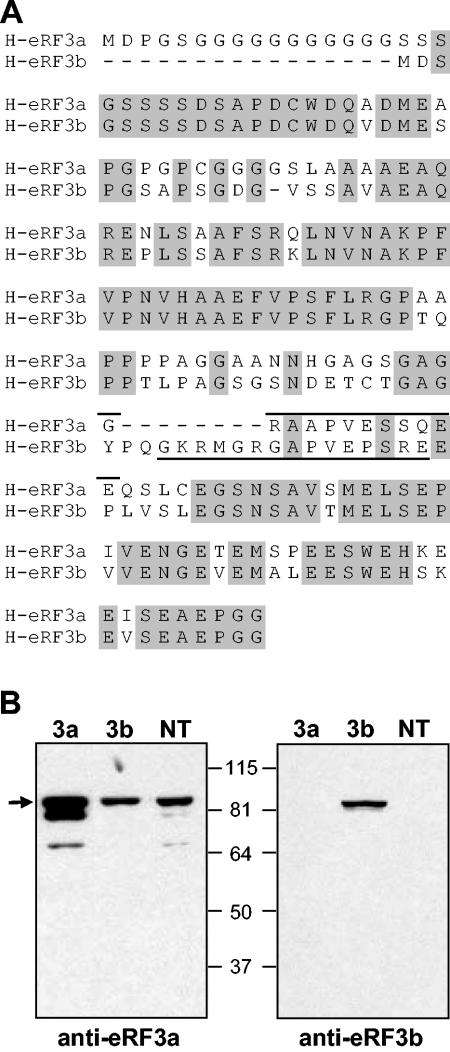
FIG. 1.
Specificity of anti-eRF3a and anti-eRF3b antibodies. (A) Alignment of the N-terminal sequences of human eRF3a and eRF3b; identical amino acids are shaded in grey. The sequences of the peptides used for immunization are indicated by solid lines above the sequence for eRF3a and below for eRF3b. (B) Extracts of 293 cells transfected with either plasmid pCMV-heRF3a (3a) or pCMV-heRF3b (3b) or not transfected (NT) and analyzed by Western blotting using anti-eRF3a (left) or anti-eRF3b antibodies (right). The arrow indicates the unprocessed form of eRF3a; molecular mass markers are indicated in kilodaltons.
Plasmid construction.
All DNA engineering was carried out using standard protocols (34). The plasmid pCMV-nls559C contains a lacZ gene in which the coding sequence is interrupted by a TAG stop codon. This plasmid was constructed in two steps: (i) a LacZ recombinant gene expressing a β-galactosidase hybrid protein targeted to nuclei (1) was inserted in the NheI-BamHI sites of the expression vector pBK-CMV (Stratagene), and (ii) the EcoRV-EcoRI fragment of the lacZ gene was replaced by the equivalent fragment of plasmid pRSV559C, which contained a TAG followed by a C at lacZ codon 600 (32). Plasmids pCMV-heRF3a, pCMV-heRF3b, pCMV-xeRF3, pCMV-heRF1 (26), and pCMV-ySUP35L contain the entire coding sequences of the human GSPT1 gene encoding eRF3a, the human GSPT2 gene encoding eRF3b, the Xenopus laevis eRF3 gene, the human eRF1 gene, and the Saccharomyces cerevisiae SUP35 gene encoding eRF3, respectively, inserted in pBK-CMV. Plasmids pCMV-heRF3a and pCMV-xeRF3 correspond to plasmid pBH35L and pBX35, respectively, which were described previously (21). The original plasmid encoding human eRF3b (GenBank accession no. AL522197) was generously donated by Genescope (Evry, France). Plasmid pCMV-ySUP35S is a derivative of pCMV-ySUP35L from which the NheI-HindIII restriction fragment extending from the 3′ end of the cytomegalovirus (CMV) promoter (NheI site) of pBK-CMV to the M domain sequence (HindIII site) of the S. cerevisiae SUP35 gene was removed. This resulted in the deletion of the region of the Sup35p open reading frame encoding the N and M domains (amino acids 1 to 253). Plasmid pCMV-heRF3b-GFP is a derivative of plasmid pEGFP-N1 (Clontech) in which a PCR-amplified fragment containing the entire sequence coding for human eRF3b and convenient restriction sites at the extremities was inserted in frame upstream of the enhanced green fluorescent protein gene. Plasmids pCMV-heRF3aNN and pCMV-heRF3bNN carry the double mutation S to N and T to N in the G1 motif GHVDAGKST of the GTPase domain (2) of eRF3a and eRF3b, respectively. These plasmids were obtained using the ExSite PCR-based site-directed mutagenesis kit (Stratagene), plasmids pCMV-heRF3a and pCMV-heRF3b as templates, and the appropriate oligonucleotides. For plasmids expressing small interfering RNA, pairs of oligonucleotides containing the appropriate 19-nucleotide-long target sequence in sense and reverse orientations and cohesive BglII and HindIII ends were annealed by incubation in 30 mM HEPES-KOH, pH 7.4, 100 mM potassium acetate, and 2 mM magnesium acetate for 4 min at 90°C and 10 min at 70°C, with slow cooling to room temperature. The annealed oligonucleotides were then inserted in the pSuper vector (3) linearized by BglII and HindIII. The siRNA-expressing constructs are summarized in Table 1.
TABLE 1.
eRF3a, eRF3b, and eRF1 sequences targeted by siRNAs
| Name of siRNA | Targeted sequence | Position | mRNA |
|---|---|---|---|
| 3a1 | 5′ GAGGAACAGUCAUUGUGUG 3′ | 606-624 | Human eRF3a (GenBank accession no. NM_002094) |
| 3a2 | 5′ CCUAAGUCUGUGGUUGCAC 3′ | 825-843 | |
| 3a9 | 5′ GAAUGUAAGGAGAAACUAG 3′ | 1353-1371 | |
| 3b1 | 5′ GGUUCCAAUUCAGCCGUUA 3′ | 547-565 | Human eRF3b (GenBank accession no. NM_018094) |
| 3b3 | 5′ UCUGUGAUCGUACCCUCAG 3′ | 761-769 | |
| 3b4 | 5′ GAACACUGGAGAAAUAUGA 3′ | 878-896 | |
| 1Y | 5′ CACAAGAAGGGUCUCAGUU 3′ | 1322-1340 | Human eRF1a |
Electroporation and transfection of human cells.
For electroporation, cells were plated the day before at 50% confluency in complete DMEM, and the evening before, the medium was exchanged for RPMI 1640 medium (Gibco) containing 0.1 mM dithiothreitol but no glutamine, fetal calf serum, or antibiotics. Immediately prior to electroporation, the cells were trypsinized, collected in phosphate-buffered saline (PBS; 10 mM phosphate buffer, pH 7.4, 140 mM NaCl), and resuspended at 1.2 × 107 cells/ml in RPMI medium containing 0.1 mM dithiothreitol. Four hundred microliters of the cell suspension was pipetted into a 4-mm electroporation cuvette, 10 μg of plasmid DNA was added, and the mixture was incubated for 10 min at room temperature with occasional gentle mixing. Electroporations were performed with a Gene Pulser II electroporation system (Bio-Rad) using a pulse of 300 V and 500 μF. The cells were then transferred into 10 ml of complete DMEM and plated. The medium was changed the day after electroporation, and cells were collected 2 to 4 days later by scraping them in 10 ml of PBS and pelleted in three aliquot fractions, which were used for β-galactosidase assays and Northern blot and Western blot analyses.
Transfection was carried out by the calcium phosphate coprecipitation method with 20 μg of DNA per 100-mm-diameter cell plate at 50% confluency. After overnight incubation, the medium and precipitate were removed and replaced with 10 ml of fresh medium. Following a further 24-h incubation, cells were collected by scraping them in 10 ml of PBS and pelleted.
To establish the 559C stable cell line, 1 μg of pCMV-nls559C was transfected in 293 cells. After overnight incubation, the cells were replated by diluting them 1:10 in complete DMEM. Clones were selected for 2 weeks in complete DMEM containing 800 μg/ml G418 (Roche) and screened for β-galactosidase activity by transfection of plasmid ptRNAam expressing a human UAG suppressor tRNA (26) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. Five positive clones, 559C-1 to 559C-5, were selected and stored frozen in liquid nitrogen. Subsequently, 559C stable cell lines were propagated in the absence of G418 selection for about 30 passages without observable loss of β-galactosidase activity.
RNA isolation and Northern blot analysis.
Total RNA was extracted as previously described (6). For Northern blot analysis, 20 μg of total RNA was subjected to electrophoresis in a 1% agarose-formaldehyde gel, blotted, and UV cross-linked onto a Hybond-N+ membrane (Amersham Biosciences). The membrane was sequentially hybridized in CHURCH solution (7) with randomly 32P-labeled fragments of human eRF3a (GSPT1) cDNA, human eRF1 cDNA, the lacZ gene of plasmid pCMV-nls559C, and human β-actin cDNA. Following hybridization, the membrane was washed under stringent conditions, exposed to X-ray film, stripped, and reprobed under the same conditions.
Western blot analysis.
Cell pellets were resuspended in 100 μl of PBS containing a 2× Complete EDTA-free cocktail of protease inhibitors (Roche), 1 μg/ml pepstatin, and 10 mM EDTA. The cells were lysed by sonication on ice and centrifuged for 5 min at 16,000 × g, and the supernatant was retained as cell extract. Mouse brain extracts were obtained as previously described (24). The protein concentrations of extracts were determined using the Micro BCA Protein Assay Reagent Kit (Pierce), with bovine serum albumin used as a standard. For each sample, 20 μg of total protein was loaded on an 8% polyacrylamide gel and subjected to electrophoresis. Proteins were subsequently electrotransfered onto a Hybond-C Extra membrane (Amersham Biosciences), and the membrane was blocked for 1 h in Tris-buffered saline (TBS)-Tween solution (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.2% Tween 20) containing 2% milk. The membrane was incubated overnight with primary antibodies at the appropriate dilution in TBS-Tween, washed five times for 10 min in TBS-Tween, and probed for 45 min with anti-rabbit IgG peroxidase-linked secondary antibodies at 1:30,000 dilution in TBS-Tween. The membrane was washed again five times in TBS-Tween and visualized by chemiluminescence and exposure to X-ray film.
Readthrough assay.
The pellets of electroporated cells were resuspended in 300 μl of 100 mM sodium phosphate buffer, pH 8. The cells were lysed by four cycles of freezing and thawing, and the cell lysates were centrifuged at 12,000 × g for 30 min at 4°C. The resulting supernatants were assayed for total proteins and β-galactosidase activity. For each sample, the total protein concentration was determined using the Micro BCA Protein Assay Reagent Kit (Pierce). The β-galactosidase assays were performed using the Luminescent Beta-galactosidase Detection Kit II (Clontech). Following the manufacturer's instuctions, β-galactosidase reactions were carried out in duplicate with 15 μl of extract and 200 μl of reaction buffer containing Galacton-Star substrate. After 1 h at room temperature, the β-galactosidase activity was measured as relative light units using a single-photon-counting program on a scintillation counter. The β-galactosidase activity was expressed as relative light units/μg of total protein, and the readthrough efficiencies were calculated as sample activity relative to negative control activity (negative controls are given in the figure legends).
Pulse-chase labeling of cell proteins and immunoprecipitation.
Three days after electroporation, cells were preincubated for 1 h in culture medium lacking methionine and cysteine and metabolically labeled for 2 h with 0.3 mCi of PRO-MIX L-[35S] in vitro Cell Labeling Mix (Amersham Biosciences) in 5 ml of culture medium lacking methionine and cysteine. The medium was removed, and the cell monolayer was washed with PBS and chased with complete DMEM. At various times postchase, cells were collected by scraping them in 10 ml of PBS and pelleted. The cells were lysed by sonication, and the extracts used for immunoprecipitation were prepared as described above for Western blot analysis. Immunoprecipitation was performed as described previously (39) with 2 × 107 cpm of radiolabeled cell extract and either 5 μl of anti-eRF1 antibodies or 2.5 μl of anti-α-tubulin antibodies. Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gels were fixed in 30% ethanol-10% acetic acid, dried, and developed by autoradiography. The protein bands of interest were quantified using the BAS1000 Fuji image plate program version 2.0.
RESULTS
Cellular and tissue expression of eRF3a and eRF3b.
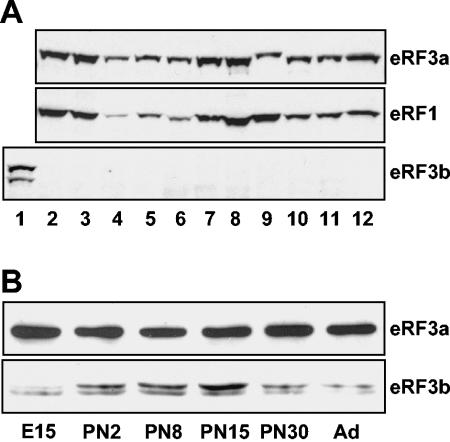
To investigate the roles of eRF3a and eRF3b in mammals, we first examined their cellular and tissue distributions using specific anti-peptide antibodies. Due to the very high similarities in the C-terminal domains, the peptides for rabbit immunization were chosen in the divergent regions of the N-terminal domains (Fig. 1A). The specificities of the antibodies were tested by Western blot analysis using extracts of human 293 cells overexpressing either eRF3a or eRF3b and of untransfected cells. As shown in Fig. 1B, antibodies directed against eRF3a recognized a single band of 84 kDa in untransfected and eRF3b-transfected cells. This band most likely corresponds to the endogenous form of eRF3a. Of the two lower bands detected in eRF3a-transfected cells (faintly visible in untransfected cells), the upper band of 80 kDa could correspond to the proteolytically processed form of eRF3a involved in apoptosis (13), and the faint band below to a degradation product. The fact that the same signal intensity was observed with anti-eRF3a antibodies for untransfected and eRF3b-overexpressing cells confirmed the specificity of these antibodies for eRF3a (Fig. 1B). With anti-eRF3b antibodies, a single band is observed in eRF3b-overexpressing cells only (Fig. 1B, right, lane 3b). The absence of signal in eRF3a-overexpressing cells confirmed the specificity of anti-eRF3b antibodies for eRF3b. Moreover, the absence of eRF3b signal in untransfected cells suggested that 293 cells contain a very low level of endogenous eRF3b, below the detection limit. Thus, the expression of eRF3a and eRF3b was analyzed in various human cell lines by Western blotting (Fig. 2A). eRF3a was expressed in all cell lines tested and, with the exception of U-2OS osteosarcoma cells (lane 9), its abundance was correlated with that of eRF1. In contrast, eRF3b was not detected in any of these cells. In addition, we were not able to detect eRF3b in various mouse tissues by Western blotting using specific antibodies directed against mouse eRF3b (data not shown). Since it has been reported that GSPT2 mRNA, encoding eRF3b, is more abundant in the brain than in the other tissues tested (15), we analyzed eRF3a and eRF3b expression in the mouse brain at various stages of development. As shown in Fig. 2B, eRF3b protein, barely detectable at the embryonic stage (embryonic day 15), increased after birth to reach a maximum at 15 days after birth. These results suggest that in mice, eRF3b expression is restricted to the brain and varies during development, whereas eRF3a seems to be ubiquitous and constantly expressed.
FIG. 2.
Cellular and tissue expression of eRF3a and eRF3b. (A) Western blot analysis of extracts from various human cell lines: 293 cells overexpressing human eRF3b (lane 1), untransfected 293 cells (lane 2), HeLa cells (lane 3), HuH7 hepatocarcinoma cells (lane 4), Hs27 foreskin fibroblasts (lane 5), HepG2 hepatocarcinoma cells (lane 6), T-47D breast cancer cells (lane 7), WERI-Rb retinoblastoma cells (lane 8), U-2OS osteosarcoma cells (lane 9), retinal pigmented epithelium ARPE (lane 10), SKP4 normal skin keratinocytes (lane 11), and glioblastoma FOG cells (lane 12). The membrane was revealed using anti-eRF3a, anti-eRF1, or anti-eRF3b antibodies as indicated. (B) Western blot analysis of mouse brain extracts at various stages of development, embryonic stage (embryonic day 15), postnatal stages (PN2 to PN30), and adult (Ad), using anti-mouse eRF3a or anti-mouse eRF3b antibodies as indicated.
Effect of siRNA targeted against eRF3a or eRF3b on readthrough of stop codon.
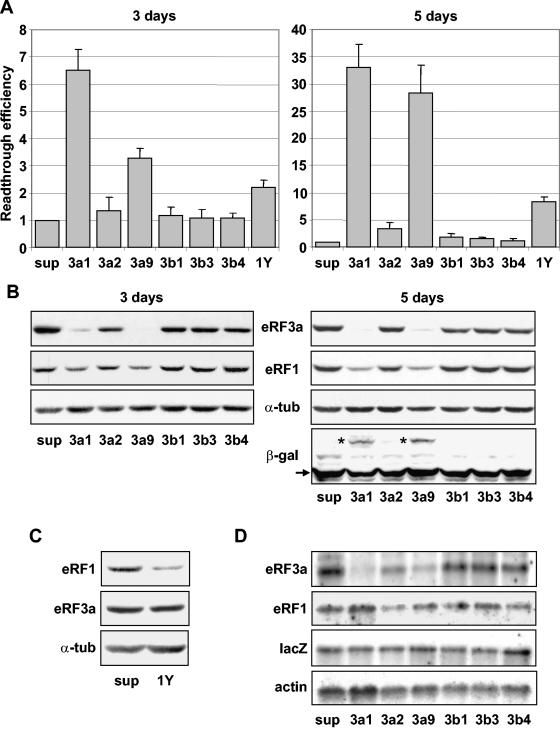
In yeast, depletion of the levels of release factors eRF1 and eRF3 causes a reduction in translation termination efficiency (33, 35). In mammalian cells, it has been shown that inhibition of eRF1 expression using siRNAs and antisense oligonucleotides enhances stop codon readthrough (4). Recently, an increase of stop codon readthrough was also obtained by silencing eRF3 (20). However, the specific effect of silencing either eRF3a or eRF3b was not analyzed. To investigate the involvement of eRF3a and eRF3b in translation termination, we analyzed the effects of siRNAs specifically targeting either eRF3a or eRF3b mRNAs on stop codon readthrough. For each mRNA, three plasmids directing the synthesis of three different siRNAs were constructed and transiently expressed in a modified 293 cell line stably expressing a lacZ reporter gene (the 559C cell line). This lacZ gene contained a premature nonsense TAG codon and encoded a truncated inactive β-galactosidase. The synthesis of full-length active β-galactosidase required readthrough of the stop codon. Thus, the level of β-galactosidase activity reflected the efficiency of stop codon readthrough. Parallel cultures of 559C cells were electroporated with the empty vector pSuper as a control or with plasmids expressing an siRNA directed against either eRF3a mRNA (si-3a1, si-3a2, and si-3a9), eRF3b mRNA (si-3b1, si-3b3, and si-3b4), or eRF1 mRNA (si-1Y). The β-galactosidase activity (relative light units/μg of protein) was measured in 3- or 5-day-postelectroporation cell extracts, and the relative readthrough efficiency was calculated by dividing the β-galactosidase activity in each sample by the β-galactosidase activity in the extract of control cells electroporated with pSuper. Three days after electroporation, a clear increase in readthrough was observed for cells expressing si-3a1 and si-3a9 (∼6-fold and ∼3-fold, respectively), whereas si-3a2, si-3b1, si-3b3, and si-3b4 had no effect (Fig. 3A). Expression of si-1Y targeting eRF1 mRNA and corresponding to si1187, described by Carnes et al. (4), increased readthrough ∼2-fold, a level in good agreement with the results reported by the authors. Five days after electroporation, readthrough increased greatly (∼30-fold) for si-3a1 and si-3a9 (Fig. 3A) and only moderately for si-1Y and si-3a2 (∼8-fold and ∼4-fold, respectively). We did not detect any increase when using si-3b1, si-3b3, and si-3b4 targeting eRF3b mRNA. The kinetics of readthrough levels induced by siRNAs targeting eRF3a 3 to 7 days postelectroporation showed that the readthrough level reached a maximum after 5 to 6 days and then decreased (data not shown). In comparison, the readthrough level induced by siRNA directed against eRF3 reported by Janzen and Geballe (20) was only two- to threefold higher than the control level. This difference from our results could be explained by the fact that these authors measured readthrough only 2 to 3 days after transfection.
FIG. 3.
Effects of siRNAs on stop codon readthrough. 559C cells stably expressing a lacZ gene containing a premature UAG stop codon were electroporated with the empty vector pSuper (sup) or with pSuper derivatives expressing siRNAs directed against eRF3a (3a1, 3a2, and 3a9), eRF3b (3b1, 3b3, and 3b4), or eRF1 (1Y). Cell extracts performed 3 or 5 days after electroporation, as indicated, were used for β-galactosidase assays and Western blot and Northern blot analyses. (A) Readthrough efficiencies of siRNAs were calculated by dividing the β-galactosidase activity in each sample by the β-galactosidase activity in pSuper-electroporated cell extract. The results were expressed as the mean of five experiments; the error bars show the standard errors of the mean. (B) Western blot analysis performed 3 or 5 days after electroporation of cells with the plasmids described above, using anti-eRF3a, anti-eRF1, anti-β-galactosidase (only shown for 5-day electroporation), or anti-α-tubulin antibodies as indicated. The positions of the truncated and full-length β-galactosidase are indicated by the arrow and asterisks, respectively. (C) Western blot analysis performed 5 days after electroporation of cells with pSuper (sup) or plasmid expressing siRNA directed against eRF1 (1Y), using anti-eRF1, anti-eRF3a, or anti-α-tubulin (α-tub) antibodies as indicated. (D) Northern blot analysis performed 5 days after electroporation of cells with the plasmids described above. The membrane was probed successively with eRF3a, eRF1, lacZ, and actin probes as indicated.
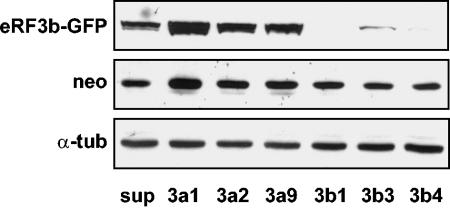
The efficiencies and specificities of the siRNAs for their targets were determined at the protein and mRNA levels by Western and Northern blot analyses, respectively. First, to test the efficiency of the eRF3a-targeted siRNAs, extracts from 3- and 5-day-postelectroporation cells were submitted to Western blot analysis using anti-eRF3a antibodies. The membranes were then reprobed sequentially with anti-eRF1, anti-β-galactosidase, and anti-α-tubulin antibodies as a loading control (a typical experiment is presented in Fig. 3B). A large decrease in eRF3a was observed for si-3a1- and si-3a9-expressing cells, in good correlation with the increase in readthrough levels. In comparison, si-3a2, which poorly affected readthrough, did not notably reduce eRF3a expression. As expected for specific siRNAs, si-3b1, si-3b3, and si-3b4, directed against eRF3b mRNA, did not change the expression level of eRF3a. The efficiency of eRF3b-targeted siRNAs and the absence of cross-response of eRF3a-targeted siRNAs on eRF3b expression were determined by Western blot analysis. Because endogenous eRF3b was not detectable in 293 cells, the cells were cotransfected with plasmid pCMV-heRF3b-GFP expressing an eRF3b-GFP fusion protein and plasmids expressing the siRNAs. The membrane was probed with anti-eRF3b antibodies, anti-neomycin antibodies as a control for the plasmid transfection level, and anti-α-tubulin as a loading control. The results show that siRNAs 3b1, 3b3, and 3b4 targeting eRF3b mRNA dramatically reduced the level of eRF3b-GFP protein, confirming their efficiency (Fig. 4). In contrast, siRNAs 3a1, 3a2, and 3a9 had no effect on eRF3b-GFP protein, demonstrating their specificity for eRF3a mRNA (Fig. 4). To verify the predicted interference of siRNAs with mRNA stability, the steady-state mRNA level was determined by Northern blot analysis for eRF3a and eRF1 mRNAs, with actin mRNA used as a loading control. As shown in Fig. 3D, the variations in eRF3a mRNA levels were similar in magnitude to those observed at the protein level: a strong reduction for si-3a1 and si-3a9, a moderate one for si-3a2, and no effect for siRNAs 3b1, 3b3, and 3b4.
FIG. 4.
Efficiencies of siRNAs targeting eRF3b mRNA. Extracts of 293 cells cotransfected with plasmid pCMV-heRF3b-GFP and either the empty vector pSuper (sup) or plasmids expressing siRNAs directed against eRF3a (3a1, 3a2, and 3a9) or eRF3b (3b1, 3b3, and 3b4) and analyzed by Western blotting using anti-eRF3b (eRF3b-GFP), anti-Neo (neo), or anti-α-tubulin (α-tub) antibodies as indicated.
In agreement with the results of the readthrough experiments presented above and with the results of Carnes et al. (4), a moderate decrease in eRF1 in si-1Y-electroporated cells was observed by Western blotting (only shown for 5-day-postelectroporation cells [Fig. 3C]). Surprisingly, in the Western blot experiments, we reproducibly observed a decrease in eRF1 levels in cells expressing si-3a1 and si-3a9. Only noticeable in 3-day-postelectroporation cells, this decrease was visibly enhanced 5 days postelectroporation, when the readthrough level reached its maximum (Fig. 3B). The decrease in the eRF1 protein level in si-3a1- and si-3a9-expressing cells was not due to a reduction in the eRF1 mRNA level, as confirmed by Northern blotting (Fig. 3D), and thus could not be attributed to cross-interference of these siRNAs with eRF1 mRNA.
In addition to its role in translation termination, eRF3 plays a role in other translation-coupled events, such as regulation of mRNA decay (17, 22) and recycling of ribosomes for translation initiation (8, 37). To determine whether these functions could influence the results of our readthrough assay, we examined the status of LacZ mRNA and of the encoded β-galactosidase in siRNA-expressing cells. Northern blot and Western blot analyses indicated that LacZ mRNA (Fig. 3D) and truncated β-galactosidase (Fig. 3B) levels remained unaffected by eRF3a depletion. Furthermore, the full-length β-galactosidase resulting from stop codon readthrough was clearly visible in extracts of si-3a1- and si-3a9-electroporated cells (Fig. 3B).
Altogether, these results show that (i) eRF3a depletion promotes stop codon readthrough, demonstrating the involvement of eRF3a in translation termination; (ii) eRF3b silencing has no effect on readthrough, likely due to the absence or small amount of this protein in 293 cells; and (iii) eRF3a depletion induces a reduction of the eRF1 protein level, which probably contributes to increased readthrough. Finally, our results suggest that in 293 cells, eRF3a is the factor associated with eRF1 in the translation termination complex. However, we cannot rule out the possibility that eRF3b might be involved in translation termination in other cell types.
eRF3b alleviates the effect of eRF3a depletion on stop codon readthrough.
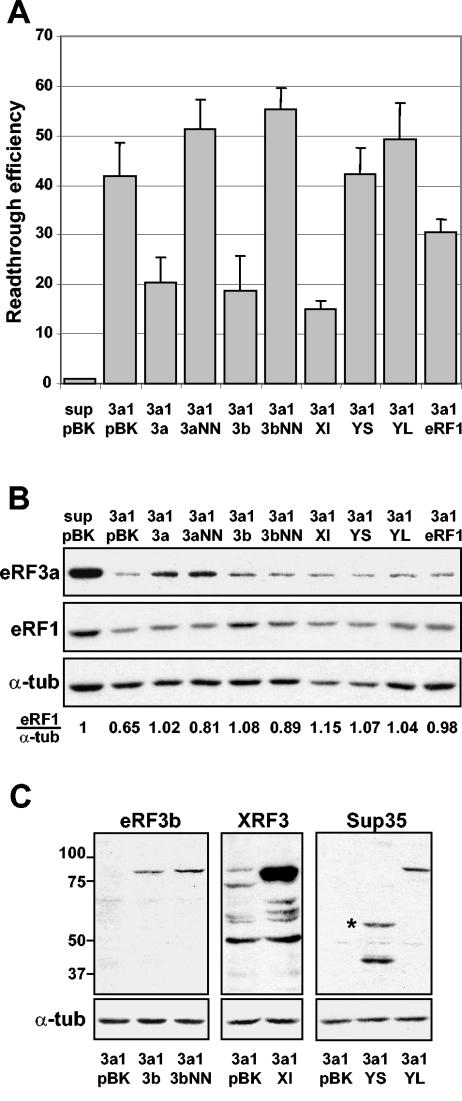
We next determined whether eRF3b could substitute for eRF3a in its role in translation termination and thus play the role of a release factor. For this purpose, eRF3a was depleted in 559C cells by electroporation of the plasmid expressing si-3a1, and 3 days later, the cells were reelectroporated with a set of pBK-CMV derivatives expressing either human eRF3a, human eRF3b, or their mutated forms carrying a double mutation in the G1 motif of the GTP-binding domain (see Materials and Methods). Three days after the second electroporation, the effect on stop codon readthrough was analyzed by measuring the β-galactosidase activity in cell extracts. Cells electroporated with plasmid pSuper and reelectroporated with plasmid pBK-CMV served as a negative control (Fig. 5A, lane sup pBK). Cells electroporated with a plasmid expressing si-3a1 and reelectroporated with plasmid pBK-CMV were used as the standard level for readthrough promoted by eRF3a depletion (Fig. 5A, lane 3a1 pBK). For each electroporation experiment, we verified the amounts of endogenous eRF3a and eRF1 (Fig. 5B) and of the overexpressed release factors (Fig. 5B, eRF3a and eRF1, and C, eRF3b) by Western blot analysis. As expected, eRF3a overexpression induced a clear decrease in readthrough up to half of the standard readthrough level of 3a1/pBK (Fig. 5A, lane 3a1 3a). Most interestingly, eRF3b overexpression also induced a reduction of readthrough that was as great as that produced by eRF3a overexpression (lane 3a1 3b). However, we did not observe any reduction with the GTPase mutated forms of eRF3a and eRF3b (lanes 3a1 3aNN and 3a1 3bNN). These results showed that eRF3b can substitute for eRF3a in the translation termination process and point out the importance of their GTPase activities. Moreover, when looking at eRF1 levels by Western blotting (Fig. 5B), we observed that overexpression of eRF3a and eRF3b rescued the defect of eRF1 generated by eRF3a depletion. This rescue was only partial with both GTPase mutated forms, as shown by the eRF1/α-tubulin ratio. Interestingly, this partial rescue did not induce a concomitant decrease in readthrough, suggesting a direct effect of eRF3a on termination efficiency.
FIG. 5.
Overexpression of various release factors in eRF3a-depleted cells. 559C cells were electroporated with the empty vector pSuper (sup) or with the plasmid expressing si-3a1 (3a1) and reelectroporated 3 days later with plasmid pBK-CMV (pBK) or pBK-CMV derivatives expressing various types of release factors: pCMV-heRF3a (3a), pCMV-heRF3aNN (3aNN), pCMV-heRF3b (3b), pCMV-heRF3bNN (3bNN), pCMV-xeRF3 (Xl), pCMV-ySUP35S (YS), pCMV-ySUP35L (YL), or pCMV-heRF1 (eRF1). Cell extracts performed 3 days after the second electroporation were used for β-galactosidase assays and Western blot analysis. (A) Readthrough efficiencies were calculated by dividing the β-galactosidase activity in each sample by the β-galactosidase activity in the extract of cells electroporated with pSuper and reelectroporated with pBK-CMV (sup pBK). Results were expressed as the mean of three experiments; the error bars show the standard errors of the mean. (B) Western blot analysis of cells electroporated either with the empty vectors, pSuper and pBK-CMV (sup pBK), as a negative control or with plasmid expressing si-3a1 and a set of pBK-CMV derivatives expressing various types of release factors (pBK, 3a, 3aNN, 3b, 3bNN, Xl, YS, YL, and eRF1), using anti-eRF3a, anti-eRF1, or anti-α-tubulin (α-tub) antibodies as indicated. The eRF1/α-tub ratio was calculated by densitometric analysis of the bands on the autoradiogram. This ratio was set to 1 for the sup/pBK control. (C) Control of the level of the overexpressed release factors in eRF3a-depleted cells: overexpression of eRF3b (3a1 3b), eRF3b GTPase mutated form (3a1 3bNN), X. laevis eRF3 (3a1 Xl), S. cerevisiae eRF3 short form (3a1 YS), and S. cerevisiae eRF3 long form (3a1 YL). For each Western blot experiment, extracts of eRF3a-depleted cells reelectroporated with plasmid pBK-CMV were used as a control (3a1 pBK). Anti-eRF3b, anti-X. laevis eRF3 (XRF3), anti-S. cerevisiae eRF3 (Sup35), or anti-α-tubulin antibodies were used as indicated for each panel. Molecular mass markers are indicated in kilodaltons. The anti-X. laevis eRF3 antibodies also recognized mammalian eRF3, and thus, the faint band of 84 kDa observed in blot XRF3, lane 3a1 pBK, corresponds to endogenous eRF3a. The short form of S. cerevisiae eRF3 is indicated with an asterisk.
The function of the eRF3 C-terminal domain carrying the GTPase activity in translation termination is well documented, whereas the function of the N-terminal extension remains mysterious. In S. cerevisiae, it was suggested that the eRF3 N-terminal domain was involved in binding to eRF1 and influenced the efficiency or specificity of termination (30). In addition, in the yeast two-hybrid system, deletion of the eRF3b N-terminal domain impaired its interaction with eRF1 (15). Recently, crystallographic studies of S. pombe eRF3 showed that the N-terminal domain might regulate its binding to eRF1 by masking the eRF1 binding site (23). Furthermore, Le Goff et al. (25) have shown that mouse eRF3b complemented a yeast S. cerevisiae eRF3 mutant, whereas eRF3a did not, except when its N-terminal sequence was deleted or structurally modified by fusion with an N-terminal tag sequence. These results pointed out the probable involvement of the N-terminal extension of eRF3 in the formation and the activity of the translation termination complex. Our next step was to determine whether eRF3 from other species, particularly from S. cerevisiae, which carries a highly divergent N-terminal extension, can substitute for eRF3a. For this purpose, X. laevis eRF3 (which is highly similar to human eRF3b) and two forms of S. cerevisiae eRF3, the entire protein and an N-terminally truncated form lacking the first 253 amino acids, were overexpressed in eRF3a-depleted cells. The level of overexpression was verified by Western blot analysis (Fig. 5C). As shown in Fig. 5A, X. laevis eRF3 alleviated the effect of eRF3a depletion at the same magnitude as did eRF3a or eRF3b (lane 3a1 Xl), demonstrating that X. laevis eRF3 can substitute for human eRF3a. Conversely, neither of the two forms of yeast eRF3 was able to substitute for eRF3a (lanes 3a1 YS and 3a1 YL). These results suggest that the C-terminal domain of eRF3, which is highly conserved among species (78% similarity between human and yeast), is not sufficient to confer on eRF3 its competence in translation termination and that the N-terminal domain is involved in the formation of an active translation termination complex. Furthermore, we have observed a slight but reproducible increase in the readthrough level, above the standard level of si-3a1/pBK cells, when overexpressing the entire S. cerevisiae eRF3 and the GTPase mutated forms of both eRF3a and eRF3b. One possible explanation is that these factors exert dominant-negative effects, i.e., they bind to eRF1 and sequester it in an inactive complex.
Finally, we examined how eRF1 overexpression affected the readthrough level in eRF3a-depleted cells. As shown in Fig. 5A, overexpressed eRF1 induced only a moderate decrease in the readthrough level compared to the standard readthrough level, from 42-fold to 30-fold (lane 3a1 eRF1). This reduction in the readthrough level was likely due to the restoration of the eRF1 protein level to almost normal (Fig. 5B). At this basal level of eRF1, the remaining 30-fold increase in readthrough was likely directly promoted by eRF3a depletion alone. However, these experiments did not allow us to decide whether an excess of eRF1 can rescue a defect of eRF3a.
eRF3a depletion induces eRF1 protein degradation.
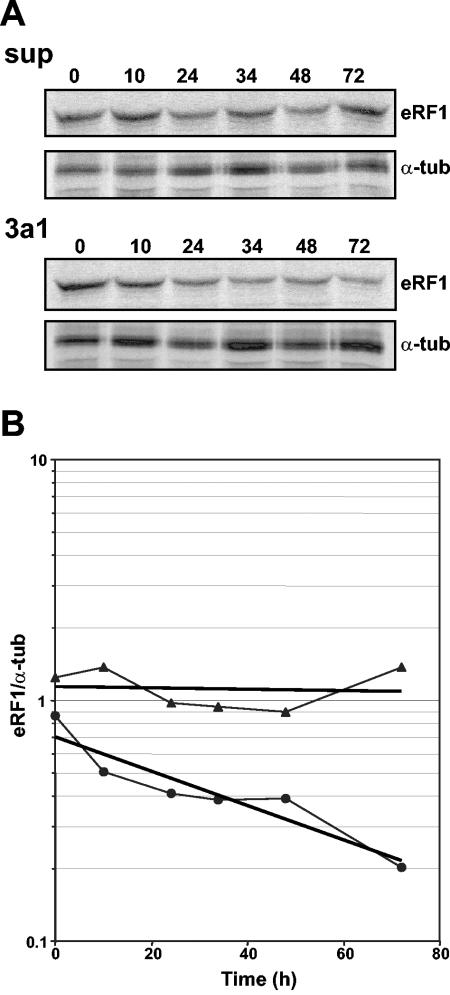
We have shown above that eRF3a depletion induced a reduction in the intracellular level of eRF1 protein (Fig. 3B) that was not due to a modification of the steady-state level of eRF1 mRNA (Fig. 3D). To further understand the mechanism involved in eRF1 reduction, we examined eRF1 protein stability in cells electroporated with si-3a1-expressing plasmid or pSuper as a control. For this purpose, 3 days after electroporation, cellular proteins were labeled by a 2-hour pulse with a mixture of [35S]methionine and [35S]cysteine and analyzed up to 72 h after being labeled by immunoprecipitation with anti-eRF1 and anti-α-tubulin antibodies (Fig. 6A). First, we observed that the amounts of eRF1 were almost the same for si-3a1-expressing cells and pSuper control cells after the 2-h labeling (Fig. 6B, time zero), suggesting that the translation of eRF1 was not affected by eRF3a depletion. Secondly, in si-3a1-expressing cells, there was a significant decrease of eRF1 protein as a function of time, which was not found in control cells (Fig. 6B). These results suggest that eRF1 is proteolytically degraded when not complexed with eRF3a, and they explain the reduction in eRF1 protein observed in eRF3a-depleted cells (Fig. 3B). However, with an eRF1 half-life of almost 40 h, this process seems to be very slow.
FIG. 6.
Stability of eRF1 protein in eRF3a-depleted cells. (A) 559C cells were labeled 3 days after electroporation with pSuper (sup) or si-3a1-expressing (3a1) plasmid by a 2-hour pulse with a mixture of [35S]methionine and [35S]cysteine, followed by a chase for various times as indicated above the lanes. Labeled proteins were immunoprecipitated with anti-eRF1 and anti-α-tubulin (α-tub) antibodies and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Dried gels were revealed by autoradiography. (B) eRF1 and α-tubulin bands were quantified using the BAS1000 Fuji image plate program version 2.0, and the eRF1/α-tubulin ratio at each time point was plotted on a semilogarithmic graph: eRF1/α-tubulin ratios in pSuper-electroporated cells (filled triangles); eRF1/α-tubulin ratios in si-3a1-expressing cells (filled circles). The regression line is presented for each curve.
DISCUSSION
In mammals, two genes encode two distinct eRF3 isoforms, eRF3a and eRF3b, which differ in their N-terminal domains. Both factors can bind eRF1 and have a GTPase activity which stimulates eRF1 release activity in vitro (15, 19, 40). However, most of the in vivo studies of mammalian eRF3 function in translation termination either were performed in yeast or did not discriminate between eRF3a and eRF3b, as in a recent report on eRF3 silencing in human cells (20). Intriguingly, it has been shown in yeast that mouse eRF3b, but not eRF3a, can substitute for yeast eRF3 (25). This result calls into question the roles of eRF3a and eRF3b in the translation termination process. The aim of this study was to gain insights into the differences between eRF3a and eRF3b functions during termination.
We have utilized specific antibodies directed against eRF3a or eRF3b to study their differential cellular expression. While eRF3a is expressed in all cell types tested with an abundance correlated with that of eRF1, eRF3b expression is restricted to brain tissue and varies during development. These results are in agreement with the tissue distribution of mouse eRF3a and eRF3b mRNAs reported by Hoshino et al. (15), showing by reverse transcription-PCR that eRF3b mRNA is present at lower levels than eRF3a mRNA in all tissues and is relatively enriched in the brain. Nevertheless, eRF3b cDNA was found in a wide variety of cDNA libraries, as shown by the numerous eRF3b sequences found in the GenBank EST collection. This could indicate that eRF3b protein may be expressed at a low level in all cell types, including 293 cells, but below the detection limits of our antibodies.
To determine the involvement of eRF3a and eRF3b in translation termination, we silenced eRF3a or eRF3b mRNA using siRNAs. Our results demonstrate unambiguously that eRF3a depletion induces a high level of stop codon readthrough whereas eRF3b silencing has no detectable effect. This suggests that in 293 cells, eRF3a plays a key role in translation termination efficiency and that eRF3b involvement, if any, was masked by the large amount of eRF3a. However, the demonstration that eRF3b can act as a release factor in vivo was obtained by overexpressing eRF3b in eRF3a-depleted cells (Fig. 5). Interestingly, the rescue of translation termination efficiency obtained with either eRF3a or eRF3b is roughly of the same magnitude. This suggests that eRF3b is as efficient as eRF3a in translation termination and that it must act in particular cell types or definite cellular states, which remain to be determined. As with eRF3a, we also observed a positive effect of eRF3b overexpression on the eRF1 level (Fig. 5B), confirming that eRF3b efficiently binds to eRF1 and restores its stability.
In addition, X. laevis eRF3, which is highly homologous to eRF3a and eRF3b in its C-terminal domain (∼96%) but closer to eRF3b than to eRF3a in its N-terminal extension, can also substitute for eRF3a. In contrast, neither the entire S. cerevisiae eRF3, which is highly divergent from human and Xenopus eRF3s in the N-terminal domain, nor the N-terminally truncated form of S. cerevisiae eRF3 carrying the conserved C-terminal domain only restores efficient termination (Fig. 5). However, in yeast, human eRF1 can complement yeast eRF1 mutants (38), showing that yeast eRF3 can associate with human eRF1 to form an active translation termination complex. Similarly, our results show that both forms of yeast eRF3 (entire and N-terminally truncated forms) rescue the eRF1 protein level (Fig. 5B), suggesting that they efficiently bind to human eRF1 but form an inactive complex. Interestingly, the overexpression of entire yeast eRF3 in eRF3a-depleted cells increases readthrough above the level obtained with si-3a1 alone, whereas the overexpression of the S. cerevisiae N-terminally truncated form does not. A possible explanation for this dominant-negative effect is that the long N-terminal extension of yeast eRF3 sequesters eRF1 in the inactive complex. These observations reinforce the previous suggestions (23, 25) that the N-terminal domain of eRF3 plays an important role in the formation of an active translation termination complex. The absence or improper conformation of the eRF3 N-terminal domain could possibly interfere either with the binding of the complex to the ribosome or with GTPase activity.
The same increase in readthrough above the level obtained with si-3a1 alone was found with eRF3a and eRF3b GTPase mutated forms, which carry a double mutation in the G1 motif of the GTPase domain (Fig. 5A). In addition, the results in Fig. 5B suggest that these mutated forms of eRF3 bind and stabilize eRF1, but less efficiently than wild-type eRF3a and eRF3b. The equivalent mutation in Ras GTPases (the S17N dominant-negative mutation) reduces the affinity for GTP, leading to the formation of a stable and inactive complex with the guanine exchange factor, but is insufficient to totally prevent the mutant form from binding GTP in vivo (9). As GTP binding seems to be required for eRF1-eRF3 association (22), we can speculate that our mutated forms of eRF3 bind GTP, though less efficiently than wild-type eRF3, and associate with eRF1 in an inactive complex.
In yeast, it has been reported recently that the reduction of the steady-state level of eRF3 does not modify eRF1 levels (33). In contrast, we show here that, in human cells, a depletion of eRF3a induces a significant reduction of the eRF1 level as observed by Western blotting (Fig. 3B). This reduction was further explained by a decrease in the eRF1 protein half-life (Fig. 6). In addition, overexpression of different forms of eRF3 alleviates eRF1 reduction (Fig. 5B). From these results, we hypothesize that eRF1 is proteolytically degraded when not complexed with eRF3. Thus, eRF1 degradation could allow the adjustment of its level to that of the eRF3 available for the formation of translation termination complexes, leading to the fine tuning of translation termination efficiency. This idea is reinforced when eRF3a and eRF1 expression levels in various human cell lines are compared, which shows that the abundance of eRF1 parallels that of eRF3a (Fig. 2A). However, the eRF1 protein seems to be very stable, and its mechanism of degradation is relatively slow. This is suggested by the pulse-chase experiment (Fig. 6) and the fact that, 5 days postelectroporation with si-3a1, only half of the eRF1 was degraded while eRF3a had almost completely disappeared (Fig. 3B). The high stability of eRF1 could explain the relatively inefficient silencing of eRF1 at the protein level in comparison with the almost complete knockdown of eRF3a. We also noticed that eRF1 depletion did not induce eRF3a reduction (Fig. 3C), suggesting that, due to its involvement in other cellular processes and association with factors such as PABP and Upf1, the intracellular amount of eRF3a is controlled by mechanisms other than termination complex formation.
In conclusion, in the present work, we have pointed out the importance of eRF3a in translation termination in mammalian cells: (i) eRF3a depletion induces an ∼30-fold increase in readthrough when a basal expression level of eRF1 is restored and (ii) eRF3a and eRF3b GTPase mutated forms and S. cerevisiae eRF3 overexpression in eRF3a-depleted cells restore eRF1 to a basal level without a concomitant decrease in readthrough. Both observations strongly suggest a direct effect of eRF3a on termination efficiency. Importantly, this work also gives prominence to the fact that eRF3a regulates the formation of the translation termination complex.
Acknowledgments
This work was supported by the Association Française contre les Myopathies. C.C. and W.V. held fellowships from the French Ministère de la Recherche et de l'Enseignement Supérieur (MENESR) and D.D. from the Association Française contre les Myopathies.
We thank Michel Philippe for critical reading of the manuscript.
REFERENCES
- 1.Bonnerot, C., D. Rocancourt, P. Briand, G. Grimber, and J. F. Nicolas. 1987. A beta-galactosidase hybrid protein targeted to nuclei as a marker for developmental studies. Proc. Natl. Acad. Sci. USA 84:6795-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 4.Carnes, J., M. Jacobson, L. Leinwand, and M. Yarus. 2003. Stop codon suppression via inhibition of eRF1 expression. RNA 9:648-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte, L., L. Frolova, L. Kisselev, and A. Favre. 2001. The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 268:2896-2904. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Church, D. M., C. J. Stotler, J. L. Rutter, J. R. Murrell, J. A. Trofatter, and A. J. Buckler. 1994. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat. Genet. 6:98-105. [DOI] [PubMed] [Google Scholar]
- 8.Cosson, B., N. Berkova, A. Couturier, S. Chabelskaya, M. Philippe, and G. Zhouravleva. 2002. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell 94:205-216. [DOI] [PubMed] [Google Scholar]
- 9.Feig, L. A. 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1:E25-E27. [DOI] [PubMed] [Google Scholar]
- 10.Frolova, L., X. Le Goff, H. H. Rasmussen, S. Cheperegin, G. Drugeon, M. Kress, I. Arman, A. L. Haenni, J. E. Celis, M. Philippe, J. Justesen, and L. Kisselev. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701-703. [DOI] [PubMed] [Google Scholar]
- 11.Frolova, L., X. Le Goff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334-341. [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova, L. Y., J. L. Simonsen, T. I. Merkulova, D. Y. Litvinov, P. M. Martensen, V. O. Rechinsky, J. H. Camonis, L. L. Kisselev, and J. Justesen. 1998. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 256:36-44. [DOI] [PubMed] [Google Scholar]
- 13.Hegde, R., S. M. Srinivasula, P. Datta, M. Madesh, R. Wassell, Z. Zhang, N. Cheong, J. Nejmeh, T. Fernandes-Alnemri, S. Hoshino, and E. S. Alnemri. 2003. The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J. Biol. Chem. 278:38699-38706. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino, S., M. Imai, T. Kobayashi, N. Uchida, and T. Katada. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274:16677-16680. [DOI] [PubMed] [Google Scholar]
- 15.Hoshino, S., M. Imai, M. Mizutani, Y. Kikuchi, F. Hanaoka, M. Ui, and T. Katada. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 273:22254-22259. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino, S., H. Miyazawa, T. Enomoto, F. Hanaoka, Y. Kikuchi, A. Kikuchi, and M. Ui. 1989. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 8:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosoda, N., T. Kobayashi, N. Uchida, Y. Funakoshi, Y. Kikuchi, S. Hoshino, and T. Katada. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278:38287-38291. [DOI] [PubMed] [Google Scholar]
- 18.Inge-Vechtomov, S. G., L. N. Mironova, and M. D. Ter-Avanesian. 1994. Ambiguity of translation: a eukaryotic version? Genetika 30:1022-1035. [PubMed] [Google Scholar]
- 19.Jakobsen, C. G., T. M. Segaard, O. Jean-Jean, L. Frolova, and J. Justesen. 2001. Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo. Mol. Biol. 35:672-681. [PubMed] [Google Scholar]
- 20.Janzen, D. M., and A. P. Geballe. 2004. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 32:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jean-Jean, O., X. Le Goff, and M. Philippe. 1996. Is there a human Ψ? C. R. Acad. Sci. Ser. III 319:487-492. [PubMed] [Google Scholar]
- 22.Kobayashi, T., Y. Funakoshi, S. Hoshino, and T. Katada. 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 279:45693-45700. [DOI] [PubMed] [Google Scholar]
- 23.Kong, C., K. Ito, M. A. Walsh, M. Wada, Y. Liu, S. Kumar, D. Barford, Y. Nakamura, and H. Song. 2004. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell 14:233-245. [DOI] [PubMed] [Google Scholar]
- 24.Larcher, J. C., L. Gasmi, W. Viranaicken, B. Edde, R. Bernard, I. Ginzburg, and P. Denoulet. 2004. Ilf3 and NF90 associate with the axonal targeting element of Tau mRNA. FASEB J. 18:1761-1763. [DOI] [PubMed] [Google Scholar]
- 25.Le Goff, C., O. Zemlyanko, S. Moskalenko, N. Berkova, S. Inge-Vechtomov, M. Philippe, and G. Zhouravleva. 2002. Mouse GSPT2, but not GSPT1, can substitute for yeast eRF3 in vivo. Genes Cells 7:1043-1057. [DOI] [PubMed] [Google Scholar]
- 26.Le Goff, X., M. Philippe, and O. Jean-Jean. 1997. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol. 17:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miccoli, L., A. Beurdeley-Thomas, G. De Pinieux, F. Sureau, S. Oudard, B. Dutrillaux, and M. F. Poupon. 1998. Light-induced photoactivation of hypericin affects the energy metabolism of human glioma cells by inhibiting hexokinase bound to mitochondria. Cancer Res. 58:5777-5786. [PubMed] [Google Scholar]
- 28.Otto, A. I., L. Riou, C. Marionnet, T. Mori, A. Sarasin, and T. Magnaldo. 1999. Differential behaviors toward ultraviolet A and B radiation of fibroblasts and keratinocytes from normal and DNA-repair-deficient patients. Cancer Res. 59:1212-1218. [PubMed] [Google Scholar]
- 29.Patino, M. M., J. J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622-626. [DOI] [PubMed] [Google Scholar]
- 30.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1997. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol. 17:2798-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like Ψ+ determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips-Jones, M. K., L. S. Hill, J. Atkinson, and R. Martin. 1995. Context effects on misreading and suppression at UAG codons in human cells. Mol. Cell. Biol. 15:6593-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salas-Marco, J., and D. M. Bedwell. 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24:7769-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Stansfield, I., L. Eurwilaichitr, Akhmaloka, and M. F. Tuite. 1996. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 20:1135-1143. [DOI] [PubMed] [Google Scholar]
- 36.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 38.Urbero, B., L. Eurwilaichitr, I. Stansfield, J. P. Tassan, X. Le Goff, M. Kress, and M. F. Tuite. 1997. Expression of the release factor eRF1 (Sup45p) gene of higher eukaryotes in yeast and mammalian tissues. Biochimie 79:27-36. [DOI] [PubMed] [Google Scholar]
- 39.Weimer, T., J. Salfeld, and H. Will. 1987. Expression of the hepatitis B virus core gene in vitro and in vivo. J. Virol. 61:3109-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]