Abstract
In Saccharomyces cerevisiae, Sum1p is a promoter-specific repressor. A single amino acid change generates the mutant Sum1-1p, which causes regional silencing at new loci where wild-type Sum1p does not act. Thus, Sum1-1p is a model for understanding how the spreading of repressive chromatin is regulated. When wild-type Sum1p was targeted to a locus where mutant Sum1-1p spreads, wild-type Sum1p did not spread as efficiently as mutant Sum1-1p did, despite being in the same genomic context. Thus, the SUM1-1 mutation altered the ability of the protein to spread. The spreading of Sum1-1p required both an enzymatically active deacetylase, Hst1p, and the N-terminal tail of histone H4, consistent with the spreading of Sum1-1p involving sequential modification of and binding to histone tails, as observed for other silencing proteins. Furthermore, deletion of the N-terminal tail of H4 caused Sum1-1p to return to loci where wild-type Sum1p acts, consistent with the SUM1-1 mutation increasing the affinity of the protein for H4 tails. These results imply that the spreading of repressive chromatin proteins is regulated by their affinities for histone tails. Finally, this study uncovered a functional connection between wild-type Sum1p and the origin recognition complex, and this relationship also contributes to mutant Sum1-1p localization.
In eukaryotes, specialized chromatin structures create metastable transcriptional domains and define chromosomal elements, including centromeres and telomeres. Some of these chromatin structures can propagate along chromosomes, whereas other chromatin modifications are restricted to one or a few nucleosomes. To explore how the spreading of repressive chromatin is regulated, we have studied a nonspreading locus-specific repressor protein, Sum1p, and a mutant version of this protein, Sum1-1p, which spreads along a chromosome and achieves regional silencing (37, 43). The mutant SUM1-1 phenotype is conferred by a single amino acid change (6), suggesting that a very slight alteration can tip the balance between spreading and not spreading. Operating at this balance point must be important for precise regulation of where and when repressive chromatin structures spread.
Repression mechanisms are often categorized as locus-specific or regional. Locus-specific repressors are typically DNA-binding proteins that associate with operator sequences near the promoters of target genes. These repressors recruit other corepressor complexes, such as histone deacetylases, and interact with core transcription factors to mediate repression. Regional silencing, in contrast, represses transcription throughout a chromosomal domain in a promoter-independent fashion. Silencing proteins typically associate with nucleosomes and propagate along chromosomes to form a specialized chromatin structure, known as heterochromatin or silenced chromatin, which is inhibitory to transcription. Although at first glance these two mechanisms of repression seem completely distinct, recent work reveals shared mechanistic features, as described below in more detail (7, 9, 37).
Sir protein-mediated silencing in Saccharomyces cerevisiae (reviewed in reference 35) provided the context for the discovery and subsequent investigations of mutant Sum1-1p. Sir-mediated silencing occurs at the donor cassettes for mating type switching, HMR and HML, and also at telomeres. Silencing is mediated by regulatory sites known as silencers, which consist of binding sites for three DNA-binding proteins, origin recognition complex (ORC), Rap1p, and Abf1p. The assembly of silenced chromatin at HM loci involves two distinguishable steps (18, 27, 36). First, the Sir proteins assemble at the silencer via interactions with silencer binding proteins. Then, Sir2p, Sir3p, and Sir4p spread from the silencer. The NAD+-dependent deacetylase activity of Sir2p is not required for assembly of Sir proteins at the silencer but is required for their spreading (18, 36). Sir3p and Sir4p bind the tails of histones H3 and H4, and at least Sir3p has a higher affinity for deacetylated tails (5, 16). These observations inspire a “sequential deacetylation” model for the propagation of silenced chromatin (36). In this model, Sir2p associated with a silencer deacetylates the neighboring nucleosome, creating new high-affinity binding sites for Sir3p and Sir4p. Since Sir2p is found in complex with Sir4p, the binding of Sir3p and Sir4p to the newly deacetylated nucleosome recruits additional Sir2p, which can then deacetylate the next nucleosome.
Sum1p is a locus-specific repressor of genes normally expressed at midsporulation (49) and of genes involved in NAD+ biosynthesis (2). Sum1p binds directly to a DNA sequence upstream of repressed genes (32, 49) and is implicated in the repression of at least 48 genes. For about half of these genes, repression also requires two corepressor proteins, Hst1p and Rfm1p (28), which are found in a complex with Sum1p (28, 33, 37). Hst1p is a deacetylase related to Sir2p (43). Rfm1p, a 35-kDa protein with coiled-coil motifs, is required for the association of Hst1p with Sum1p (28).
A mutant form of Sum1p, known as Sum1-1p, spreads and achieves regional silencing by a mechanism similar to that of Sir proteins (37, 43). The SUM1-1 mutation, T988I (6), restores silencing in strains lacking Sir proteins (23, 24) by redirecting Sum1-1p to HM loci, where it spreads to form repressive chromatin. Like the wild-type Sum1p, Sum1-1p associates with Hst1p and Rfm1p (28, 37, 43), and these proteins are required for deacetylation of nucleosomes in the silenced domain (37).
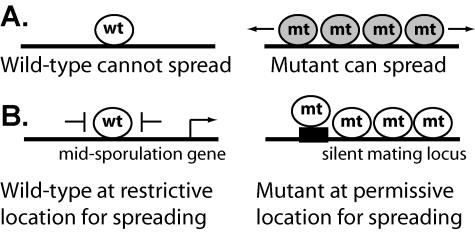
The single most striking effect of the SUM1-1 mutation is its ability to convert a site-specific repressor into a protein capable of spreading over multiple nucleosomes. There are at least two models to explain the ability of mutant Sum1-1p to spread (Fig. 1). In one model (Fig. 1A), the primary effect of the SUM1-1 mutation is to change the ability of Sum1p to spread, perhaps by increasing the stability of Sum1-1p-containing chromatin. In this scenario, the observed change in the location of Sum1-1p compared to that of Sum1p would be a secondary effect of the increased ability to spread. In the other model (Fig. 1B), the primary effect of the mutation is to change the location of Sum1p by altering the affinity of the protein for a DNA-associated protein or DNA itself. In this model, both wild-type Sum1p and mutant Sum1-1p are fully capable of spreading, but features at the loci where wild-type Sum1p normally acts, such as boundary elements, prevent spreading. Only when Sum1-1p fortuitously ends up at the silent mating type loci, where there are no such boundaries, can it spread. To test these models, we investigated the extent to which the SUM1-1 mutation alters the location of Sum1p and compared the spreading abilities of mutant and wild-type Sum1p.
FIG. 1.
Models for the conversion of a repressor to a silencing protein. (A) The ability to spread changes. (B) The location changes.
MATERIALS AND METHODS
Plasmids and yeast strains.
Plasmids generated for this study are described in Table 1. To construct the SUM1 and SUM1-1 integrating plasmids, pLR376 and pLR377, the entire SUM1-containing inserts were excised from plasmids DMC283 and DMC326 (6) and ligated into pRS404, a TRP1 integrating plasmid lacking replication or centromere sequences (41). The hst1-N291A allele in plasmid pLR100 was generated by site-specific mutagenesis of plasmid pLP316 (3). The original codon for asparagine 291 (AAT) was changed to alanine (GCT). The hmrSum1 allele in plasmid pLR337 was generated by site-directed mutagenesis of plasmid pJR759 (24) to modify two sequences in the E silencer. The sequence AAAAACCCATCAAC, containing the Rap1p binding site, was replaced with the sequence CACTAATTTGTGACA, containing a Sum1p binding site, and the Abf1p binding site, ATCATAAAATACGA, was mutated to ATCAGGAAAATACGA to disrupt binding of Abf1p. The hmrSum1X2 allele in plasmid pLR391 was generated by site-directed mutagenesis of plasmid pLR337 to modify the I silencer. The sequence TTTAATTTTTAA AAAAACAAA TTTAATTGACC between the Abf1p and ORC binding sites was modified to TTTAATTTTTAA GTGTCACAAATTAG TTTAATTGACC to create a Sum1p binding site. Plasmid pLR39 containing 7myc-SUM1 in pRS415 was generated as previously described (37). The ARS1015 and ARS1013 plasmids (48) contained yeast intergenic regions, iYJR003C and iYJL038C, cloned into a yeast shuttle vector with CEN4 and URA3 but no other autonomously replicating sequence (ARS).
TABLE 1.
Plasmids generated for this study
| Plasmid | Description | Vector description | Vector reference |
|---|---|---|---|
| pLR39 | 7myc-SUM1 in pRS415 | CEN/ARS LEU2 | 41 |
| pLR100 | hst1-N291A in YEp352 | 2μm URA3 | 17 |
| pLR337 | hmrSum1 in pRS316 | CEN/ARS URA3 | 41 |
| pLR376 | SUM1-1 in pRS404 | Integrating TRP1 | 41 |
| pLR377 | SUM1 in pRS404 | Integrating TRP1 | 41 |
| pLR378 | hmrSum1×2 in pRS316 | CEN/ARS URA3 | 41 |
Strains used in this study were all derived from W303-1a (Table 2). The hst1Δ::KanMX, sum1Δ::LEU2, 7myc-SUM1-1, 3myc-SUM1, sir2Δ::HIS3, sir2Δ::TRP1, sir2Δ::LEU2 (37), SUM1-1 (6), hhf1Δ::HIS3, hhf2Δ4-19::TRP1 (20), orc2-1 (13), and orc5-1 alleles (26) were previously described. The rfm1Δ::LEU2 allele was generated by one-step gene conversion (34) with a PCR product amplified from plasmid pRS415 using primers 5′-AATTTATTAGACAAC AGGAAGGTGTTATAAGAAAGTGCGAGATTGTACTGAGAGTGCACC and 5′-TATTTCT CTCTATTTATATTTATTTACTTCTTCAAAGAAGCTGTGCGGTATTTCACACCG. The hst1-N291A and 7myc-SUM1 alleles were generated by a serial gene replacement approach, in which HST1 or SUM1 was first replaced with URA3, which was then replaced with the hst1-N291A allele from plasmid pLR100 or the 7myc-SUM1 allele from plasmid pLR39. The chromosomal TRP1::SUM1 and TRP1::SUM1-1 alleles were generated using plasmid pLR376 or pLR377 linearized within the TRP1 gene.
TABLE 2.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1b | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| MC89 | W303 MATα SUM1-1 | D. Shore |
| TD4 | MATahis4-519 leu2-3,112 trp1 ura3-52 | G. Fink |
| LRY134 | W303 MATα hst1Δ::KanMX | |
| LRY142 | W303 MATα sum1Δ::LEU2 | |
| LRY200 | W303 MATα SUM1-1 hst1Δ::KanMX | |
| LRY273 | W303 MATα SUM1-1 sir2Δ::TRP1 | |
| LRY312 | W303 MATα SUM1-1 orc5-1 | |
| LRY316 | W303 MATα orc5-1 | |
| LRY325 | W303 MATα orc2-1 | |
| LRY459 | W303 MATα 7myc-SUM1-1 sir2Δ::HIS3 | |
| LRY464 | W303 MATα 3myc-SUM1 sir2Δ::HIS3 | |
| LRY466 | W303 MATα 3myc-SUM1 | |
| LRY526 | W303 MATα 7myc-SUM1-1 sir2Δ::HIS3 hst1Δ::KanMX | |
| LRY529 | W303 MATα 7myc-SUM1-1 | |
| LRY532 | W303 MATα sum1Δ::LEU2 sir2Δ::HIS3 | |
| LRY576 | W303 MATα 7myc-SUM1-1 sir2Δ::HIS3 orc5-1 | |
| LRY1021 | MATahis4 | P. Schatz |
| LRY1222 | W303 MATα 7myc-SUM1-1 hhf1Δ::HIS3 sir2Δ::LEU2 | |
| LRY1229 | W303 MATα 7myc-SUM1-1 hhf1Δ::HIS3 hhf2Δ4-19::TRP1 sir2Δ::LEU2 | |
| LRY1238 | W303 MATα 7myc-SUM1-1 sir2Δ::HIS3 hst1-N291A | |
| LRY1258 | W303 MATα sir2Δ::HIS3 7myc-SUM1 | |
| LRY1261 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 TRP1::SUM1 | |
| LRY1269 | W303 MATα sir2Δ::HIS3 7myc-SUM1 TRP1::SUM1-1 | |
| LRY1291 | W303 MATα 7myc-SUM1-1 sir2Δ::HIS3 rfm1Δ::LEU2 | |
| LRY1295 | W303 MATα sir2Δ::HIS3 | |
| LRY1341 | W303 MATα 3myc-SUM1 rfm1Δ::LEU2 |
This study employed the original SUM1-1 allele (23), the protein encoded by which differs from that of the published SUM1 sequence (1) by seven amino acids (A87D, F241L, M615L, L638S, T704I, T748I, and G782N) in addition to the mutation at threonine 988 that confers the SUM1-1 phenotype. The additional amino acid changes in protein encoded by the SUM1-1 allele did not contribute in any apparent way to the mutant phenotype, since a mutant protein with the single amino acid change, Sum1p-T988I, behaved identically to Sum1-1p (see also reference 6). Specifically, Sum1p-T988I associated with HM loci, where it spread. In addition, the association of Sum1p-T988I with midsporulation promoters was reduced, and the midsporulation genes were derepressed in a strain bearing the SUM1-T988I allele (data not shown).
Chromatin IP.
Chromatin immunoprecipitations (IP) were performed as previously described (37) using 10 optical density equivalents of cells and 3 μl anti-Myc tag (06-549; Upstate Biotechnology). Cells were treated with 1% formaldehyde for up to 3 h to cross-link proteins to DNA. The DNA was sheared by sonication to an average size of 600 to 700 bp in all experiments. For quantitative real-time PCR, a standard curve was prepared using input DNA. The standard curve and immunoprecipitated samples were amplified with primers for a control locus (ATS1) and the locus of interest in separate reactions, which enabled the relative amount of each locus in the IP sample to be determined. ATS1 was selected as a control locus because it is relatively far from ORC binding sites, which, as discovered here, are associated with Sum1-1p. Reported values represent averages of at least two independent IP experiments, each analyzed in two separate PCRs. Sequences of the oligonucleotides used for PCR are in Table 3.
TABLE 3.
Primers used to analyze chromatin IP
| Region | Primer 1 | Primer 2 |
|---|---|---|
| HMR-E | CTAAATCGCATTTCTTTTCGTCCAC | TAACAAAAACCAGGAGTACCTGCGC |
| HMR-a1 | GTGGCATTACTCCACTTCAAGTAAG | CAAGAGCAAGACGATGGGG |
| HMR-I | TGTCACCAACATTTTCGTATATGGCG | CTACCACATTATCAATCCTTGCATCCAG |
| HML-E | CCAGAAGATAATTTAGAAGACAAGTAGCG | TTTGGCCCCCGAAATCG |
| HML-I | TGAAAGCCCGACGTTTGC | GCTGTTACGGAGATGCAAAGC |
| SMK1 | GCGACGAGGCGTGAGGGTAG | CATAGGCTCCCTTGCCCAGG |
| SPR3 | TAGGTCGTTTCCTACCTCATTGATG | AATGATAGTCGAGGTCAGACACACTT |
| ATS1 | GGTAACGCAGCCGTTTGAGC | CCTCATCGTGCCCCAGTCC |
| ARS309 | CGTGGCAACATCTTCTCCG | GTACTGGCTCGTCCATTCCC |
| ARS1 | GAAGGCAAGAGAGCCCCG | TTGTAGCGTATGCGCCTGTG |
| HMR-1 | AGATTCATATATCTTCAAGGGGAACTTCTTGTAC | TAGTTTCTTAAGTACTACCGGATTAGAGGTTTG |
| HMR-2 (HMR-E) | ||
| HMR-3 | TCCGCCATACTACAAATATCATCC | TACCAACCCATCCGCCG |
| HMR-4 (HMR-a1) | ||
| HMR-5 (HMR-I) | ||
| HMR-6 | GACACCCAGGTTGCCGC | TGGTGGCCCATGCCTTG |
| HMR-7 | GCAGCTTACTCCCAAGAGTGC | CAACATGGTGTTCCAAAGCAC |
| SMK1-1 | TTATCGGCCTGGGCCTTG | GGGCTTTGTCCGCCTACAT |
| SMK1-2 | GGTGACCATTGCGACTGTGC | GTGGCGCCGAATTCTACC |
| SMK1-3 | GCGACGAGGCGTGAGGGTAG | GGAGCACCGAGGTTGGAG |
| SMK1-4 | CCTGGGCAAGGGAGCCTA | TTAGCTCACGGATGGCCC |
| SMK1-5 | GAGATAGTGACCAGCTCGCCCT | CGGTGGATGACATCTGCAC |
| YJL038C-1 | TCTATTTCGAGCGACTCTCTCTCA | GAGAAGGTGCCTATTATCGAGGGAC |
| YJL038C-2 | GCAACAACATTCACCATAGGTGC | GCTATGCTATGTATTCAATCCCCCT |
| YJL038C-3 | TGGCGCCACAACCTAGTCA | CAAGAGATTTACTTCGAGCAATTTTGG |
| YJL038C-4 | CGAAAGTACCGGAACCAAACTGG | TCAACACAGACAGAACTCTGCGAG |
| YJL038C-5 | CAAGAAGGGCGGGGAATTG | CGACGCCTCCAATGCTGG |
Mating assays.
For patch mating assays, cells were grown for 8 h on solid yeast extract-peptone-dextrose (YPD) medium or minimal medium without uracil to retain plasmids. The yeast cells were subsequently replica plated to mating-type tester lawns (LRY1021 or TD4) on minimal medium. Plates were incubated at 30°C and scanned after 2 or 3 days. For quantitative mating assays, overnight cultures of cells were diluted and plated on YPD (to determine total cell number) and on minimal plates with approximately 2 × 107 mating-type tester cells to determine the number of mating events by the number of prototrophic colonies. The fraction of cells that mated was calculated by dividing the number of colonies on the mating plate by the number of colonies on YPD.
Microarray expression studies.
Total RNA was isolated from logarithmically growing cells as described previously (39). cDNA and then cRNA were generated, labeled, and hybridized to Affymetrix S98 yeast arrays, as recommended by the manufacturer.
RNA blots.
RNA was separated on 1% agarose-formaldehyde gels and transferred to Zeta Probe membranes (Bio-Rad) (38). Probes were generated by PCR using total yeast genomic DNA. The probes were labeled using the RediPrime II kit (Amersham). The amount of mRNA was normalized to ACT1 mRNA using a Storm phosphorimager.
RESULTS
Sum1p and Sum1-1p were localized to different sites.
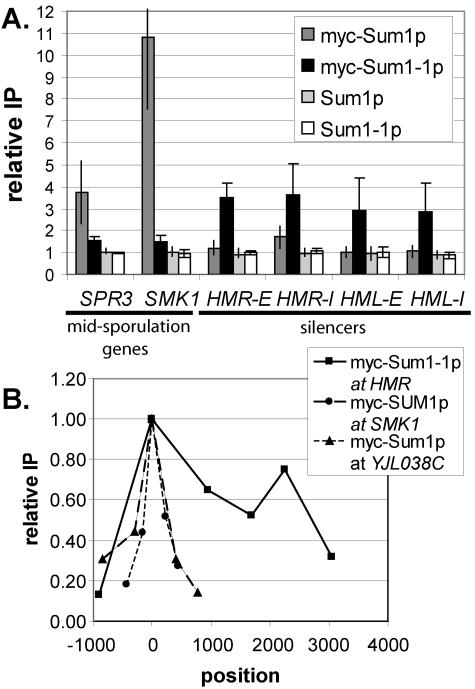
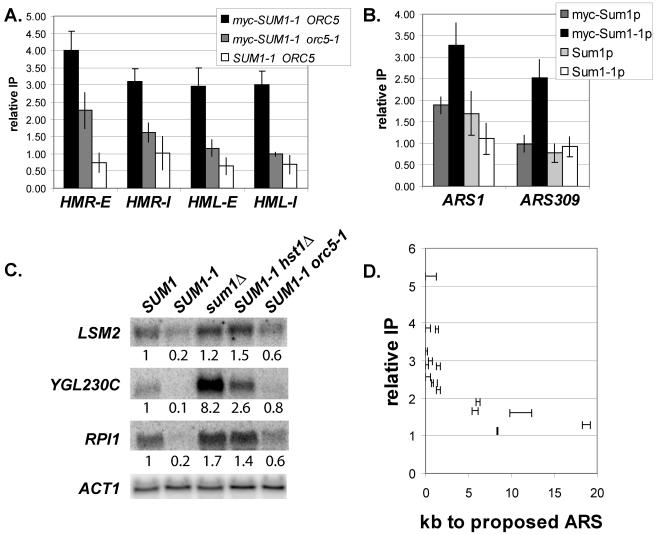
Previous results based on qualitative chromatin IP analyses are consistent with the relocalization of mutant Sum1-1p from midsporulation promoters to the HM loci (37). To accurately assess the distribution of wild-type and mutant Sum1p at HM loci and midsporulation promoters, quantitative real-time PCR analysis of immunoprecipitated DNA was performed (Fig. 2A). Wild-type Sum1p associated strongly with the promoters of two genes repressed by Sum1p (SMK1 and SPR3) but did not precipitate significantly with any of the four silencers (HMR-E, HMR-I, HML-E, and HML-I). In contrast, mutant Sum1-1p had a slight association with the promoters of Sum1p-repressed genes and associated robustly with all four silencers. Therefore, the SUM1-1 mutation reduced but did not completely eliminate the association of mutant Sum1-1p with the sites of action of wild-type Sum1p. Furthermore, mutant Sum1-1p accumulated at new locations where wild-type Sum1p was not found. This change in distribution was consistent with the SUM1-1 mutation acting solely by altering the localization of the protein (Fig. 1B) and could result from mutant Sum1-1p having increased affinity for a protein or DNA sequence associated with the HM loci. However, these data do not exclude models in which the accumulation of mutant Sum1-1p at new loci is a secondary effect of an increased ability to spread.
FIG. 2.
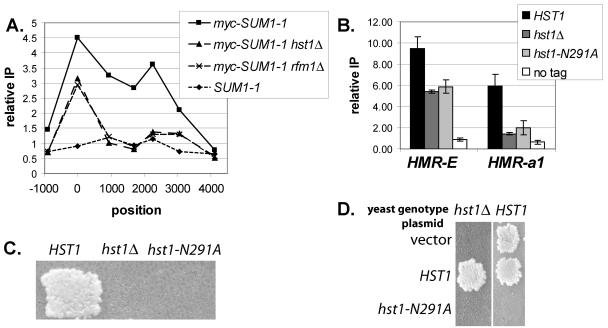
(A) Wild-type Sum1p and mutant Sum1-1p associated with different genomic locations. DNA coprecipitated with myc-Sum1p or myc-Sum1-1p was quantified by real-time PCR. The y axis represents the relative enrichment of the specified regions compared to the ATS1 promoter, which is not regulated by Sum1p or Sum1-1p. If the recovery of a particular region is not enhanced compared to ATS1, the relative enrichment is 1. Strain genotypes were myc-SUM1 (LRY466), myc-SUM1-1 (LRY529), SUM1 (W303), and SUM1-1 (MC89). (B) Sum1-1p was more broadly distributed than Sum1p. The x axis represents the position along the chromosome (in base pairs) of the center of each PCR product. The site of recruitment (Sum1p binding site for SMK1 and YJL038C or HMR-E silencer for Sum1-1p) was set to zero. The y axis represents the relative enrichment at each position, compared to the maximum enrichment, which was set to 1.0. The data points for each locus were connected to aid visualization and are not meant to imply anything about the level of protein association in the regions between data points. The strain genotype was myc-SUM1 (LRY466) or myc-SUM1-1 sir2Δ (LRY459).
Wild-type Sum1p did not spread at repressed promoters.
To investigate the two models of SUM1-1 action further, we examined the ability of wild-type Sum1p to spread. The distribution of Sum1p was examined at several genes normally repressed by Sum1p, including SMK1, YJL038C, CRR1, and YSW1. (Genes repressed by Sum1p were identified by their induction in the absence of Sum1p [28] and their association with Sum1p in chromatin IP studies [25].) In all cases, there was a single, sharp peak of Sum1p association with chromatin, centering on the predicted Sum1p binding site (Fig. 2B). Association of Sum1p dropped to less than 50% of the maximum level within 500 bp of the binding site. This narrow distribution of Sum1p was observed at genes that were shown (28) to require Hst1p for repression (e.g., YJL038C) and those that did not (e.g., SMK1). In contrast, mutant Sum1-1p was distributed across at least 2,300 bp at the HMR locus. Thus, wild-type Sum1p did not appear to spread at its normal sites of action as efficiently as mutant Sum1-1p spread at HMR. Although these results could not prove that wild-type Sum1p never spreads, Sum1p clearly did not spread at these four examined loci. Thus, the abilities of Sum1p and Sum1-1p to spread at their sites of action were different.
Wild-type Sum1p did not spread efficiently at HMR.
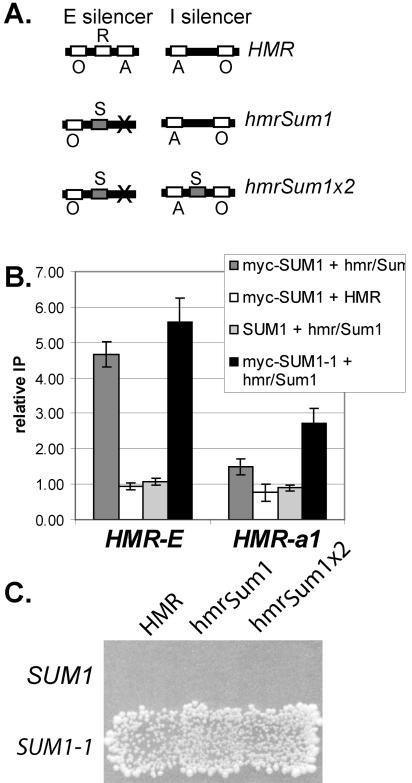
To determine whether wild-type Sum1p had an intrinsic ability to spread that was constrained at the sites where it normally binds (Fig. 1B), wild-type Sum1p was recruited to HMR. At this locus there should not be any factors that limit spreading, since mutant Sum1-1p and Sir proteins are able to spread. The binding site for Sum1p (32) was inserted into the HMR silencer in place of the Rap1p binding site, which is not required for Sum1-1p-mediated silencing (37). In addition, the Abf1p binding site was mutated to help prevent the silencer from initiating Sir-mediated silencing. Thus, a silencer was constructed that should bind wild-type Sum1p and which will no longer be silenced by Sir proteins due to the absence of the Rap1p and Abf1p binding sites (Fig. 3A, hmrSum1). The I silencer, which is still intact in this plasmid, cannot initiate Sir-mediated silencing on its own (4, 36). This modified HMR locus was transformed into sir2Δ yeast on a CEN plasmid, and the ability of Sum1p to spread was assessed by chromatin IP (Fig. 3B). As expected, Sum1p associated with the mutated silencer sequence but not the wild-type silencer, indicating that the inserted Sum1 binding site was functional. However, wild-type Sum1p associated only to a small degree with the a1 gene at hmrSum1. In contrast, the mutant Sum1-1p did spread to the a1 gene on this same plasmid. Thus, within the limits of this assay, wild-type Sum1p did not spread as well at HMR as mutant Sum1-1p.
FIG. 3.
Wild-type Sum1p did not spread efficiently at HMR. (A) Modified silencers used in this study. The binding sites for ORC (O), Rap1p (R), Abf1p (A), and Sum1p (S) at the E and I silencers are represented. (B) Association of myc-Sum1p with wild-type and modified silencers. Strains of the genotype myc-SUM1 sir2Δ (LRY464), SUM1 sir2Δ (LRY1295), or myc-SUM1-1 sir2Δ (LRY459) were transformed with plasmids bearing wild-type HMR (pJR759) or hmrSum1 (pLR337). Real-time PCR analysis, performed as described in the legend to Fig. 2, automatically accounts for the difference in copy number between the plasmid-based silencer and the chromosomal ATS1 promoter, since it involves a ratio of ratios (HMRIP/ATS1IP/HMRinput/ATS1input). (C) Mating assay for HMR silencing. Yeast cells of genotype SUM1 sir2Δ (LRY1295) or myc-SUM1-1 sir2Δ (LRY 459) were transformed with plasmids containing wild-type HMR (pJR759), hmrSum1 (pLR337), or hmrSum1x2 (pLR391). Yeast cells were replica plated to a mating tester lawn (TD4) on minimal medium supplemented with leucine and tryptophan.
To determine whether wild-type Sum1p could spread at all, even if only in a small fraction of cells, we employed a mating assay, which has the sensitivity to detect rare instances of silencing. In these MATα cells, expression of a1 mRNA from HMR prevents mating. However, if HMR were silenced in a very small fraction of cells, these cells would mate and be detected as individual colonies on medium that selects for diploids. When the modified hmrSum1 locus was transformed into yeast bearing wild-type Sum1p, mating was not detected (Fig. 3C, first row, second column). In addition, quantitative mating assays revealed that the fraction of SUM1 cells that mated was not significantly different from the fraction of sum1Δ cells that mated (2 × 10−6 for SUM1 and 3 × 10−6 for sum1Δ). In contrast, mutant Sum1-1p was much more successful at silencing this same plasmid (second row), and the fraction of mating-competent cells was roughly 104 times greater (4.3 × 10−2) with mutant Sum1-1p than with wild-type Sum1p in a quantitative mating assay. Thus, mutant Sum1-1p had an enhanced ability to spread compared to wild-type Sum1p.
Both the E and I silencers that flank HMR are bound by ORC, and Sum1-1p acts through ORC (37, 43). Thus, two sites of recruitment flanking the silenced locus may be necessary for stable spreading, and the single wild-type Sum1p binding site in the hmrSum1 allele may not be sufficient. To determine whether recruitment of Sum1p to two sites flanking HMR could promote spreading, a second Sum1p binding site was inserted at the HMR-I silencer (hmrSum1X2) (Fig. 3A). Silencing of this hmrSum1X2 plasmid was not detected in a patch mating assay (Fig. 3C, third column), and the quantitative mating assay revealed that the fraction of cells that mated in the presence of Sum1p (1 × 10−5) was just slightly greater than the fraction that mated in a sum1Δ strain (2.5 × 10−6). Chromatin IP analysis verified that Sum1p did associate robustly with the second binding site (data not shown). In contrast, mutant Sum1-1p successfully silenced hmrSum1X2 (Fig. 3C). Furthermore, since the fraction of SUM1-1 cells that mated was essentially the same on all three HMR alleles (4.5 × 10−2 for hmrSum1X2, 4.3 × 10−2 for hmrSum1, and 3.8 × 10−2 for HMR), the insertion of the two Sum1p binding sites did not alter any features of the plasmid necessary for silencing. Thus, wild-type Sum1p did not spread as efficiently as mutant Sum1-1p at HMR.
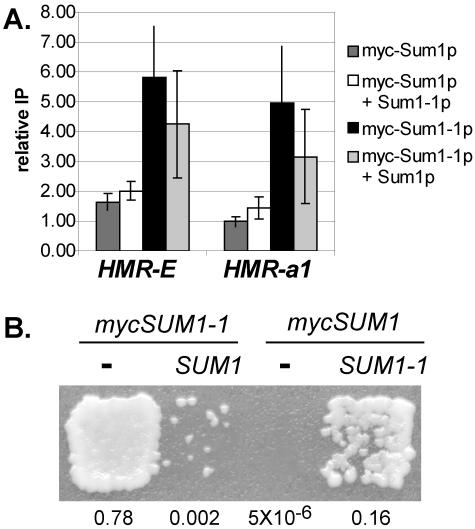
It was possible that wild-type Sum1p had the ability to spread but was recruited to HMR in a way that limited its ability to initiate the formation of silenced chromatin. Therefore, an experiment was designed which eliminated the requirement for Sum1p to initiate silenced chromatin yet would still allow an evaluation of its ability to spread. The mutant and wild-type proteins were coexpressed, and the ability of wild-type Sum1p to be incorporated into Sum1-1p-silenced chromatin was examined. If wild-type Sum1p had the ability to spread in the same way as mutant Sum1-1p, then both wild-type and mutant proteins should make up the silenced chromatin. However, when tagged wild-type Sum1p was coexpressed with untagged mutant Sum1-1p, wild-type Sum1p did not associate with the E silencer or the a1 gene significantly more than it did when expressed alone (Fig. 4A). Therefore, wild-type Sum1p was not incorporated into the Sum1-1p-silenced chromatin to a significant extent. The Sum1-1p-silenced chromatin was functional in these cells, since they were able to mate (Fig. 4B, fourth patch). Interestingly, the ability of Sum1-1p to silence HMR was diminished when wild-type Sum1p was coexpressed (Fig. 4B), and this effect was greater when mutant Sum1-1p was tagged than when wild-type Sum1p was tagged. Immunoblots revealed that tagged wild-type and mutant Sum1p were expressed at roughly equivalent levels (data not shown). These observations suggested that wild-type Sum1p, rather than participating in the formation of silenced chromatin, actually disrupted the spreading of mutant Sum1-1p. It also appeared that the myc tag partially compromised the function of the protein. Taken together, these experiments demonstrated that mutant Sum1-1p had a much greater ability to spread than wild-type Sum1p, consistent with model A and not model B (Fig. 1).
FIG. 4.
Sum1p interfered with Sum1-1p-mediated silencing. (A) Association of Sum1p and Sum1-1p with HMR in strains coexpressing wild-type and mutant proteins. Strain genotypes were myc-SUM1 sir2Δ (LRY1258), myc-SUM1 SUM1-1 sir2Δ (LRY1269), myc-SUM1-1 sir2Δ (LRY459), and myc-SUM1-1 SUM1 sir2Δ (LRY1261). (B) Mating assay of the same yeast used in panel A. Yeast cells were replica plated to a mating tester lawn (LRY1021) on minimal medium. The fraction of mating-competent cells determined in a separate quantitative mating assay is indicated below each strain.
Spreading of mutant Sum1-1p required deacetylation by Hst1p and the histone H4 tail.
To understand how the SUM1-1 mutation increased the ability of Sum1p to spread, it was necessary to understand the process by which mutant Sum1-1p spreads. Since Sum1-1p-mediated silencing involves a protein deacetylase highly related to Sir2p and results in deacetylation of nucleosomes (37, 43), we tested whether spreading of Sum1-1p, like spreading of Sir proteins, required an active deacetylase. In the case of the Sir proteins, spreading occurs by a sequential deacetylation mechanism (18, 36). Sir2p deacetylates a nucleosome, thereby creating new binding sites for Sir3p and Sir4p, which in turn recruit additional Sir2p to the newly deacetylated nucleosomes. If Sum1-1p spreads by a similar sequential deacetylation mechanism, then three predictions can be made. First, spreading of Sum1-1p should require the deacetylase activity of Hst1p. Second, enzymatically inactive Hst1p should have a dominant-negative phenotype, since incorporation of enzymatically inactive Hst1p into a growing chromatin structure should prevent further spreading of the repressed domain. Finally, histone tails should be required for spreading of Sum1-1p. These three predictions were tested.
To test the first prediction, that deacetylation by Hst1p is required for the spreading of Sum1-1p, HST1 was deleted and the distribution of Sum1-1p at HMR was assessed by chromatin immunoprecipitation (Fig. 5A). Sum1-1p still associated with the E silencer at HMR in the absence of Hst1p, indicating that Hst1p was not required for the stability of Sum1-1p or its recruitment to the silencer. However, Sum1-1p did not spread to internal sites at HMR in the absence of Hst1p. The distribution of mutant Sum1-1p was also examined in the absence of Rfm1p, a protein that is required for the association of Hst1p with wild-type Sum1p and for Sum1-1p-mediated silencing (28). In the absence of Rfm1p, Hst1p should not be recruited to HMR, and hence, Sum1-1p should not spread. Consistent with this prediction, Sum1-1p did not spread in the absence of Rfm1p (Fig. 5A). Immunoblot analysis demonstrated that the amount of Sum1-1p did not change in the absence of Hst1p or Rfm1p (data not shown).
FIG. 5.
Spreading of Sum1-1p required deacetylase activity. (A) Distribution of myc-Sum1-1p at HMR locus. DNA coprecipitated with myc-Sum1-1p was quantified by semiquantitative PCR using [32P]dCTP. The y axis represents the relative enrichment of the specified regions compared to the SSC1 promoter. The x axis represents the position along the chromosome (in base pairs) of the center of each PCR product. The HMR-E silencer was set to zero, and the HMR-I silencer was at 2,225. Strain genotypes were myc-SUM1-1 sir2Δ (LRY459), myc-SUM1-1 hst1Δ sir2Δ (LRY526), myc-SUM1-1 rfm1Δ sir2Δ (LRY1291), and SUM1-1 sir2Δ (LRY273). (B) Association of myc-Sum1-1p with HMR in hst1 strains. The strain genotype was myc-SUM1-1 sir2Δ (LRY459), myc-SUM1-1 hst1Δ sir2Δ (LRY526), myc-SUM1-1 hst1-N291A sir2Δ (LRY1238), or SUM1-1 sir2Δ (LRY273). (C) Mating of the same yeast strains used in panel B. Yeast cells were replica plated to a mating tester lawn (LRY1021) on minimal medium. (D) Mating of myc-SUM1-1 hst1Δ sir2Δ (LRY526) or myc-SUM1-1 sir2Δ (LRY459) yeast strains transformed with vector (YEp352), HST1 (pLP316), or hst1-N291A (pLR100). Yeast cells were replica plated to a mating tester lawn (TD4) on minimal medium supplemented with leucine and tryptophan. These conditions require that the plasmid be retained for the diploids to grow on the selective plate.
The lack of spreading by Sum1-1p in the absence of Hst1p or Rfm1p could indicate either a requirement for deacetylase activity or a structural requirement for Hst1p that is independent of the enzymatic activity. Therefore, a mutation was created in the presumptive active site of Hst1p (N291A) which is identical to a characterized mutation in the highly conserved active site of Sir2p, the paralog of Hst1p. The sir2 mutation, sir2-N345A, disrupts enzymatic activity in vitro (19) but is unlikely to cause structural changes (30). The association of Sum1-1p with HMR was evaluated in HST1, hst1Δ, and hst1-N291A strains. In the presence of mutant Hst1p-N291A, Sum1-1p did associate with the E silencer but not the a1 gene at HMR, indicating that spreading did not occur (Fig. 5B). In contrast, in the presence of wild-type Hst1p, Sum1-1p associated with both the E silencer and the a1 gene. Consistent with the chromatin immunoprecipitation results, mating did not occur in the presence of the hst1-N291A mutation, suggesting that HMR was no longer silenced (Fig. 5C). Thus, the deacetylase activity of Hst1p was required for the spreading of Sum1-1p.
A second prediction of the sequential deacetylation model is that the hst1-N291A allele should have a dominant-negative phenotype, since incorporation of enzymatically inactive Hst1p into a growing chromatin structure should prevent further spreading of the chromatin. To test this prediction, plasmids bearing either wild-type or mutant HST1 were transformed into a SUM1-1 HST1 sir2Δ strain, and mating was tested under conditions that required retention of the plasmid. Robust mating occurred in the presence of the wild-type HST1 plasmid, but mating did not occur in the presence of the hst1-N291A plasmid (Fig. 5D, second column). Thus, enzymatically inactive Hst1p had a dominant-negative effect on Sum1-1p-mediated silencing, as predicted by the sequential deacetylation model. The ability of Hst1p-N291A to disrupt Sum1-1p-mediated silencing also demonstrates that this mutant protein was stably expressed.
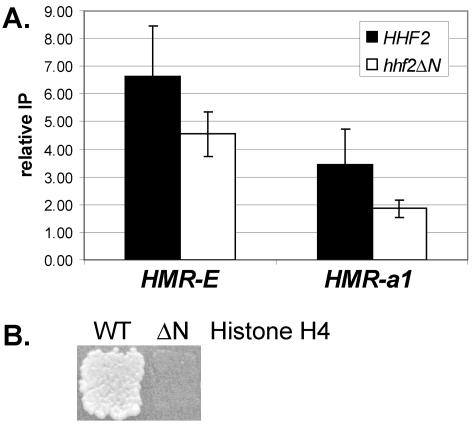
A third prediction of the sequential deacetylation model is that histone tails should be required for the spreading of Sum1-1p. To test this prediction, a SUM1-1 sir2Δ strain was constructed in which amino acids 4 to 19 of histone H4 were deleted. This truncation of histone H4 was previously shown to reduce silencing by Sir proteins (20). To determine whether the histone H4 tail deletion affected the spreading of Sum1-1p, chromatin immunoprecipitation was conducted (Fig. 6A). The association of Sum1-1p with the a1 gene was reduced in the absence of the histone H4 tail, consistent with the tail of histone H4 being required for spreading of Sum1-1p. In addition, mating did not occur in the presence of the histone H4 tail deletion (Fig. 6B), indicating that HMR was no longer silenced. In conclusion, all three predictions of the sequential deacetylation model were met for the spreading of Sum1-1p. It was therefore highly likely that Sum1-1p spread by this mechanism.
FIG. 6.
Sum1-1p-mediated silencing required the N-terminal tail of histone H4. (A) Association of Myc-Sum1-1p with HMR in the absence of the N-terminal tail of histone H4. Strain genotypes were myc-SUM1-1 HHF2 hhf1Δ sir2Δ (LRY1222) and myc-SUM1-1 hhf2Δ4-19 hhf1Δ sir2Δ (LRY1229). (B) Mating of same yeast strains used in panel A. Yeast cells were replica plated to a mating tester lawn (LRY1021) on minimal medium.
Mutant Sum1-1p accumulated near ORC binding sites.
The experiments above established that the SUM1-1 mutation altered both the localization and spreading ability of Sum1p. To understand how a single amino acid substitution could confer both effects, we next investigated the mechanism by which mutant Sum1-1p was recruited to chromatin. Previous studies suggested that mutant Sum1-1p is recruited to silencers through interactions with the origin recognition complex, ORC (37, 43). This conclusion was based on the dependence of Sum1-1p-mediated silencing on the ORC binding sites at silencers and also on qualitative chromatin IP studies showing reduced association of Sum1-1p with silencers in an orc5-1 mutant strain. orc5-1 is a temperature-sensitive allele that reduces Orc5p levels even at permissive temperatures. To assess more accurately the dependence of Sum1-1p on ORC for association with silencers, quantitative real-time PCR analysis of chromatin IP samples was performed. The association of Sum1-1p with all four silencers was significantly reduced in an orc5-1 temperature-sensitive strain (Fig. 7A). These experiments were conducted in cells grown at the permissive temperature, at which some residual Orc5p is present in the cell, which could explain the low-level association of Sum1-1p with the silencers in the orc5-1 strain.
FIG. 7.
myc-Sum1-1p accumulated near ORC binding sites. (A) Association of myc-Sum1-1p with silencers under conditions of reduced ORC function. Strain genotypes were myc-SUM1-1 sir2Δ (LRY459), myc-SUM1-1 orc5-1 sir2Δ (LRY576), and SUM1-1 sir2Δ (LRY273). Cultures were grown at the permissive temperature of 25°C. (B) Association of myc-Sum1p and myc-Sum1-1p with two ORC binding sites. The strain genotype was myc-SUM1 (LRY466), myc-SUM1-1 (LRY529), SUM1 (W303), or SUM1-1 (MC89). Two separate immunoprecipitations were conducted in which all four strains were cross-linked for 2.5 or 3 h. (C) RNA from cells of the genotypes SUM1 (W303), SUM1-1 (MC89), sum1Δ (LRY142), SUM1-1 hst1Δ (LRY200), and SUM1-1 orc5-1 (LRY312) was analyzed for LSM2, YGL230C, RPI1, or ACT1 mRNA. Cultures were grown at the permissive temperature of 25°C. The relative amount of each mRNA is indicated below each blot. The amount of mRNA from each gene was normalized to ACT1, and then the expression level in the SUM1 strain was set to 1.0. (D) Proximity of Sum1-1p-repressed promoters to predicted ORC binding sites. For each repressed gene, the distance from the 5′ end of the open reading frame to the nearest predicted ORC binding site was plotted as a range on the x axis. The y axis represents the relative enrichment of the promoter compared to ATS1 in a Sum1-1p chromatin IP performed using myc-SUM1-1 (LRY529) cells.
The observation that ORC is involved in the recruitment of Sum1-1p to silencers led us to examine the relationship between ORC and Sum1-1p in more detail. If ORC is indeed involved in recruiting Sum1-1p to silencers, then it may also recruit Sum1-1p to other origins of replication, of which there are approximately 400 in the yeast genome (48). To determine whether Sum1-1p does associate with other ORC binding sites, the association of wild-type and mutant Sum1p was examined at the well-characterized origin ARS1 and at ARS309, which is unusual in that the sequence to which ORC binds differs from the consensus sequence (44). Mutant Sum1-1p was enriched at both origins, whereas wild-type Sum1p was not (Fig. 7B), consistent with mutant Sum1-1p being recruited to chromatin at least in part through ORC. The enrichment of Sum1-1p at these origins was convincingly detected only when cross-linking was carried out for longer times (2 to 3 h rather than 20 to 30 min), suggesting that the interaction between Sum1-1p and the coprecipitated DNA is indirect, perhaps through ORC. The same requirement for longer cross-linking times was not seen at HMR, perhaps because spreading of Sum1-1p is more extensive, which could contribute to increased immunoprecipitation by stabilizing the association of Sum1-1p with chromatin and by providing more sites for antibody binding.
If mutant Sum1-1p is recruited to origins throughout the genome, then it would be expected to repress nearby genes. Indeed, SUM1-1 cells grow slowly compared to wild-type cells, whereas sum1Δ cells grow at the same rate as wild-type cells, suggesting that other genes, perhaps near ORC binding sites, may be silenced by Sum1-1p. To determine whether Sum1-1p-mediated silencing is initiated at other ORC binding sites, we identified additional genes repressed by Sum1-1p and then asked whether those genes were near ORC binding sites. Microarray expression analysis was used to compare the expression profiles of SUM1, SUM1-1, and sum1Δ cells. In two separate experiments, 16 genes were repressed at least threefold in SUM1-1 cells compared to SUM1 cells and were not also repressed in sum1Δ cells, indicating that repression was not an effect of the loss of wild-type SUM1. Four of these repressed genes, LSM2, PAM16, KAR1, and VAS1, are essential for life in S. cerevisiae (1), and their reduced expression may explain, in part, the slow growth of SUM1-1 strains. Only 1 of the 16 genes, YGL230C, was derepressed in sum1Δ cells, whereas the expression of the others was unchanged. None of these genes showed significant association with wild-type Sum1p in a genome-wide localization of Sum1p (25), although three of them were significantly associated with Sum1p when examined individually (Table 4). Since there were no genes that were both associated with Sum1p and derepressed in sum1Δ cells, we conclude that these genes were not normal targets of wild-type Sum1p. RNA hybridization experiments confirmed that these genes were repressed in SUM1-1 cells compared to expression in wild-type cells (Fig. 7C, lanes 1 and 2; also data not shown). Furthermore, these genes were silenced by a mechanism similar to that operating at HMR, since repression of these genes was reduced in hst1Δ and orc5-1 strains (Fig. 7C, lanes 4 and 5).
TABLE 4.
Sum1-1p-repressed genes
| ORFa | Gene | Essential? | Sum1-1p IPb | Sum1p IP | No. of kb to pro-ARS |
|---|---|---|---|---|---|
| YBL026W | LSM2 | Yes | 5.26 | No | 0-1.2 |
| YBR242W | No | 3.88 | No | 0-0.62 | |
| YHR127W | No | 3.84 | Yes | 1.2-1.5 | |
| YGL230C | No | 3.27 | Minimal | 0-0.29 | |
| YBR268W | MRPL37 | No | 3.01 | No | 0.32-0.86 |
| YMR274C | RCE1 | No | 2.89 | Minimal | 0-0.34 |
| YIL119C | RPI1 | No | 2.88 | Yes | 1.2-1.8 |
| YNL211C | No | 2.58 | No | 0-0.62 | |
| YIR043C | No | 2.42 | No | 1.4 | |
| YIR030C | DCG1 | No | 2.41 | Yes | 0.73-0.93 |
| YJL104W | PAM16 | Yes | 2.22 | No | 1.2-1.7 |
| YNL188W | KAR1 | Yes | 1.9 | No | 5.9-6.4 |
| YIL149C | MLP2 | No | 1.7 | No | 5.3-6.1 |
| YGR094W | VAS1 | Yes | 1.6 | No | 9.8-12 |
| YLR068W | FYV7 | No | 1.3 | No | 18-19 |
| YIL015W | BAR1 | No | 1.15 | No | 8.3-8.5 |
Open reading frame.
Relative enrichment compared to ATS1.
To determine those genes that were directly silenced by Sum1-1p, chromatin IP was employed. Eleven of the genes were significantly associated with Sum1-1p, whereas five were slightly associated with Sum1-1p, with relative enrichments less than two (Table 4). If ORC were indeed responsible for recruiting Sum1-1p to chromatin, the 11 strongly associated genes should be closer to ORC binding sites than the 5 less strongly associated genes and should also be closer, on average, than genes not influenced by Sum1-1p. To determine the nearest ORC binding site, we used a study in which ORC and MCM proteins were localized throughout the genome to map proposed origins of replication (pro-ARSs) (48). This study did not pinpoint the exact location of each pro-ARS but narrowed it to a region of approximately 1 kb. Using this data, we found that the 5′ ends of all 11 genes that were strongly associated with Sum1-1p were less than 1.8 kb from the nearest pro-ARS (Fig. 7D; Table 4). In contrast, the five genes weakly associated with Sum1-1p were 5.3 to 19 kb from the nearest pro-ARS. Thus, the association of Sum1-1p with a promoter correlated with proximity to an ORC binding site (P = 0.00073).
Truncation of histone H4 caused Sum1-1p to associate with midsporulation promoters.
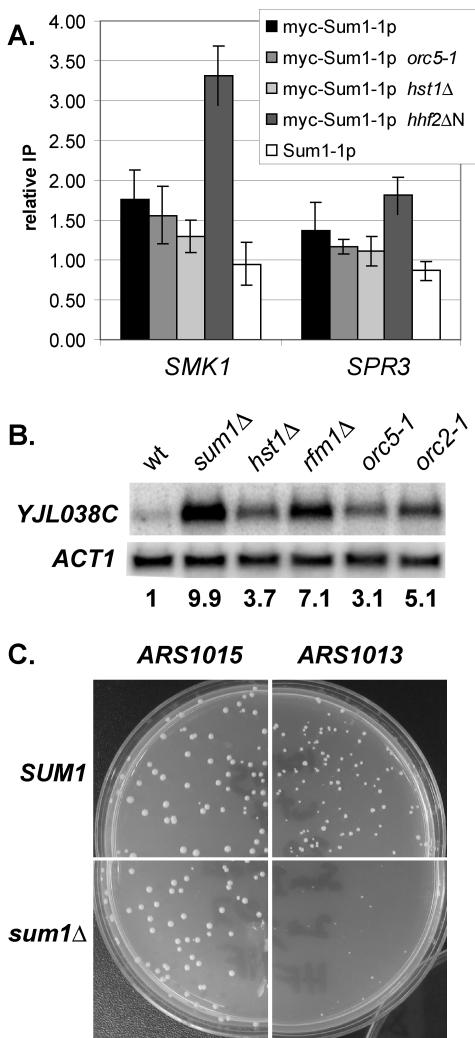
To resolve the puzzle of how a single amino acid change in Sum1p alters both the localization of the protein and its spreading ability, we considered whether an increased affinity for any of the proteins known to act with Sum1-1p could affect both localization and spreading of Sum1-1p. If mutant Sum1-1p has an increased affinity for another protein and is consequently drawn away from midsporulation promoters, then the absence of that binding partner might allow the mutant Sum1-1p to return to the midsporulation genes. Therefore, the association of Sum1-1p with the midsporulation genes SMK1 and SPR3 was tested in strains bearing mutations in genes known to be important for Sum1-1p-mediated silencing (Fig. 8A). Deletion of the N-terminal tail of histone H4 increased the association of mutant Sum1-1p with midsporulation promoters, suggesting that the Sum1-1p mutation may result in an increased affinity for histone H4 tails. In contrast, mutation of HST1, ORC5 (Fig. 8A), or RFM1 (data not shown) did not increase the association. This observation suggests that although mutant Sum1-1p accumulates at and near ORC binding sites to a greater extent than does wild-type Sum1p, this accumulation is not due to an increased affinity for ORC.
FIG. 8.
(A) Association of Myc-Sum1-1p with midsporulation promoters. The strain genotype was myc-SUM1-1 sir2Δ (LRY459), myc-SUM1-1 orc5-1 sir2Δ (LRY576), myc-SUM1-1 hst1Δ sir2Δ (LRY526), myc-SUM1-1 hhf2Δ4-19 hhf1Δ sir2Δ (LRY1229), or SUM1-1 sir2Δ (LRY273). (B) RNA from cells of the genotype SUM1 (W303), sum1Δ (LRY142), hst1Δ (LRY134), rfm1Δ (LRY1341), orc5-1 (LRY316), or orc2-1 (LRY325) was analyzed for YJL038C mRNA. Cultures were grown at the permissive temperature of 25°C. The relative amount of each mRNA is indicated below the blot. The amount of mRNA from each gene was normalized to ACT1, and then the expression level in the SUM1 strain was set to 1.0. (C) Yeast cells of the genotypes SUM1 (W303) and sum1Δ (LRY142) were transformed with plasmids bearing ARS1015 or ARS1013. The resultant transformed colonies are shown.
Wild-type Sum1p may interact with ORC.
The reversion of mutant Sum1-1p to midsporulation genes in the absence of histone H4 tails but not when ORC function was reduced was inconsistent with Sum1-1p having an increased affinity for ORC. An alternative model is that both the wild-type and mutant forms of Sum1p have a low affinity for ORC, and an additional interaction that mutant Sum1-1p is able to make with histone tails leads to the stabilization of Sum1-1p near ORC binding sites. Indeed, several lines of evidence supported an interaction between wild-type Sum1p and ORC, as outlined below.
If Sum1p and ORC do interact, their binding sites might colocalize in the genome. To quantify the extent of colocalization between ORC and Sum1p, we identified 406 intergenic regions that both were found to contain probable ARS elements (based on genome-wide ORC and MCM binding measurements (48) and were included as microarray probes in a genome-wide protein localization survey of 113 transcription factors (including Sum1p) (25). Of these 406, 66 (16.3%) were found to also bind Sum1p at the level of P values of <0.05. This is a 3.3-fold enrichment over what would be expected by chance, which is a significant (Fisher's exact test; P < 10−18) departure from randomness. In fact, Sum1p is the most highly enriched for colocalization with ARS elements among all 113 transcription factors tested, being more than five orders of magnitude more significantly enriched than even the next most enriched factor. Similar, though slightly weaker, enrichments were found when using only ORC binding data instead of both ORC and MCM; this suggests that Sum1p may have the greatest affinity for ORC in the context of ARS elements, as opposed to other sites of ORC binding.
There are two ways in which ORC and Sum1p could interact functionally. ORC could play a role in the Sum1p-mediated repression of some genes, or Sum1p could affect replication at some origins. To determine whether ORC plays a role in Sum1p-mediated repression, RNA was isolated from orc5-1 and orc2-1 yeast and analyzed for the expression of nine Sum1p-repressed genes near ORC binding sites. One of these genes, YJL038C, was clearly induced in both orc5-1 and orc2-1 strains (Fig. 8B), consistent with ORC participating in Sum1p-mediated repression.
To determine whether Sum1p plays a role in replication, we compared the stability of plasmids bearing one of two ARS elements as the sole origin of replication (48). ARS1015 is not associated with a Sum1p binding site, whereas ARS1013, which is located near YJL038C, is associated with a Sum1p binding site. Transformation of ARS1015 into wild-type or sum1Δ cells yielded robust colonies. In contrast, colonies were slightly smaller when ARS1013 was transformed into wild-type yeast and extremely small in sum1Δ yeast (Fig. 8C). These results are consistent with Sum1p facilitating replication at ARS1013 but not ARS1015. Thus, the clustering of ORC and Sum1p binding sites, the derepression of YJL038C in orc mutant strains, and the reduced stability of ARS1013 in the absence of Sum1p all suggest a functional interaction between wild-type Sum1p and ORC.
The proposed contribution of Sum1p to replication, at least at a subset of origins, suggests that the lower growth rate of SUM1-1 strains may be due to replication defects rather than repression of essential genes, as proposed above. To test this idea, we compared the growth rates of a double orc5-1 SUM1-1 mutant strain with orc5-1 and SUM1-1 single-mutant strains. If SUM1-1 caused a defect in replication, the double orc5-1 SUM1-1 mutant should have even more severely compromised replication and grow poorly if at all. In contrast, if the slow growth of SUM1-1 strains were due to decreased expression of an essential gene, the orc5-1 mutation should relieve this repression and result in an increased growth rate. The double orc5-1 SUM1-1 mutant strain grew at roughly the same rate as an orc5-1 single-mutant strain and faster than a SUM1-1 strain, and the same result was seen with orc2-1 mutant strains (data not shown). Thus, it is more likely that the poor growth of SUM1-1 strains is due to the reduced expression of essential genes rather than a replication defect.
DISCUSSION
A mutation affected the localization and spreading ability of a eukaryotic repressor.
This study revealed that the SUM1-1 mutation alters two properties of the protein—it changes both its location in the genome and its ability to spread. The altered localization of Sum1-1p was demonstrated by chromatin IP studies (Fig. 2A) and is consistent with previous observations (37, 43). The wild-type and mutant proteins clearly had different spreading abilities at the locations where they acted (Fig. 2B). However, to determine whether the difference in spreading was due to a change in protein function or was a property of its genomic location, it was necessary to compare the abilities of the mutant and wild-type proteins to spread at the same location. To do this, the wild-type Sum1p was recruited to the silent mating type locus, HMR, by inserting a Sum1p binding site into the silencer. Although wild-type Sum1p did bind to this modified silencer, it did not spread as efficiently as mutant Sum1-1p (Fig. 3). In addition, wild-type Sum1p was not efficiently incorporated into Sum1-1p-containing silenced chromatin, and wild-type Sum1p even appeared to disrupt the ability of mutant Sum1-1p to spread efficiently (Fig. 4). Therefore, wild-type Sum1p behaved rather differently than Sum1-1p and had, at best, a limited capacity to spread beyond its binding site. The ability of wild-type Sum1p to disrupt, but not eliminate, silencing by mutant Sum1-1p may also explain why the SUM1-1 allele appeared recessive in some studies but dominant in others (23, 24).
A difference in spreading ability between wild-type and mutant Sum1p is also supported by increased telomeric silencing in a SUM1-1 strain (6). This increase in silencing occurs in the presence, but not the absence, of Sir proteins and is probably due to mutant Sum1-1p becoming incorporated into Sir-silenced chromatin, thereby enhancing silencing. In this case, silencing is initiated by the Sir proteins, and therefore the enhanced silencing seen in a SUM1-1 strain compared to a SUM1 strain must be due to an increased ability of the protein to spread.
Having determined that the SUM1-1 mutation affects both the location and spreading ability of the protein, we considered how a single amino acid substitution could alter these two properties. Often, a single amino acid change affects a single function of a protein, for example, the affinity of the protein for another protein or catalysis in the active site. In this case, since no enzymatic activity is known for Sum1p, it is probable that the mutation alters the affinity of the protein for a binding partner, and this alteration causes the observed changes in spreading and location. To identify the particular binding partner for which mutant Sum1-1p had higher affinity, the association of mutant Sum1-1p with midsporulation genes was assessed in various mutant backgrounds. Sum1-1p returned to the midsporulation genes when the N-terminal tail of histone H4 was truncated but not when ORC5, HST1, or RFM1 was mutated (Fig. 8A). The most parsimonious interpretation of this data is that the changes in location and spreading ability resulted from an increased affinity for the N-terminal tail of histone H4, although an indirect effect of the histone H4 truncation cannot be ruled out. An increased interaction between mutant Sum1-1p and histone tails is consistent with our observations that in chromatin IP experiments, more nonspecific DNA precipitates with mutant Sum1-1p than with wild-type Sum1p (data not shown). Since histones are found throughout the genome, mutant Sum1-1p might be at virtually any position in the genome in a fraction of cells in the population. Furthermore, the dependence of Sum1-1p on the Hst1p deacetylase for spreading suggests that Sum1-1p has a higher affinity for deacetylated than acetylated histone tails, perhaps explaining why a slight enrichment for Sum1-1p was observed at repressed genes that are not close to ORC binding sites (Table 4).
The spreading of mutant Sum1-1p.
Three important conclusions emerged regarding the mechanism by which Sum1-1p spreads: the deacetylase activity of Hst1p was required for the spreading of Sum1-1p, enzymatically inactive Hst1p had a dominant-negative effect on Sum1-1p-mediated silencing, and the tail of histone H4 was required for Sum1-1p-mediated silencing. These results are all consistent with Sum1-1p spreading by a sequential deacetylation mechanism, much as the Sir proteins do. It is interesting that other types of repressive chromatin, such as silenced chromatin at the mating type locus in Schizosaccharomyces pombe, also propagate through the sequential modification of, and specific binding to, histones (15). It is therefore likely that this is a general mechanism by which specialized chromatin propagates.
The requirement for the N-terminal tail of histone H4 for Sum1-1p to spread (Fig. 6) implied that alterations in the affinity of the protein for the tail of histone H4 could modulate the extent of spreading of the Sum1 protein. Furthermore, since the SUM1-1 mutation appeared to increase the affinity of Sum1-1p for this histone tail (Fig. 8A), this change in affinity was the probable mechanism by which the SUM1-1 mutation increased the ability of the protein to spread. Therefore, one way in which the extent of spreading can be regulated is through the modulation of interactions between chromatin proteins and histone tails. In this view, amino acid substitution, as occurs in the SUM1-1 mutation, is a genetic surrogate for modulating the affinity of a protein for histone tails by posttranslational modifications, such as acetylation, methylation, and phosphorylation, all of which occur frequently on histone tails. Thus, these posttranslational modifications probably play a key role in regulating the spreading of chromatin proteins by impacting the affinity of chromatin proteins for nucleosomes. Indeed, changes in the modification status of histones in the vicinity of silenced domains do alter the extent to which Sir proteins spread (11, 22, 29, 42).
Some chromatin-associated proteins that do not spread also have the ability to modify histones and specifically bind to those modified histones. For example, the yeast Ssn6-Tup1 and mammalian SMRT/N-CoR corepressor complexes both associate with histone deacetylases and bind to unacetylated histone tails, yet neither spreads extensively (8, 9, 12, 47, 50). In these cases, the ability to generate and bind to a specific histone modification is thought to stabilize the association of these protein complexes with chromatin. It is possible that wild-type Sum1p has a similar ability to bind to histones and that the affinity of this interaction is not sufficient to allow spreading. Thus, the ability to modify histones and specifically bind to those modified histones serves at least two purposes—stabilizing the association of a protein complex with a promoter or facilitating the spreading of proteins along the chromosome. The strength of the interaction with histone tails is important in determining which of these modes of action predominates. Thus, promoter-specific repressor complexes and silencing complexes that spread are mechanistically related.
Recruitment of mutant Sum1-1p to chromatin.
This study also investigated the mechanism by which mutant Sum1-1p is recruited to particular sites in the genome. Three observations indicate that Sum1-1p accumulated near ORC binding sites. First, quantitative PCR analysis of chromatin IP samples revealed that the association of Sum1-1p with silencers was significantly reduced in an orc5-1 temperature-sensitive strain (Fig. 7A). Second, mutant Sum1-1p associated with two generic ORC binding sites, ARS1 and ARS309 (Fig. 7B). Finally, genes that were repressed in a Sum1-1p-dependent manner were near ORC binding sites (Fig. 7D). Thus, Sum1-1p joins a growing list of repressive proteins, including yeast Sir1p (14, 45) and Drosophila HP1 (31, 40), that are recruited to chromatin at least in part through ORC. In addition, the apparent functional interaction between wild-type Sum1p and ORC, as suggested by the higher-than-expected frequency of Sum1p binding sites near ORC binding sites, the derepression of YJL038C in orc mutant strains (Fig. 8B), and the decreased function of ARS1013 in sum1Δ cells (Fig. 8C), implies a fundamental link between DNA replication, chromatin structure, and transcriptional repression.
What causes Sum1-1p to accumulate near ORC binding sites? One possibility is that the SUM1-1 mutation increases the affinity of the protein for ORC, as suggested by an observed two-hybrid interaction between ORC5 and mutant SUM1-1 but not wild-type SUM1 (43). However, the orc5-1 mutation did not increase the association of mutant Sum1-1p with midsporulation promoters, whereas deletion of the N-terminal tail of histone H4 did (Fig. 8A). Therefore, it was more likely that Sum1-1p was drawn away from midsporulation genes by an increased affinity for histone tails rather than for ORC. In addition, an increased affinity for ORC would not change the spreading ability of the protein and hence cannot be the sole effect of the SUM1-1 mutation. An alternative model is that both wild-type and mutant Sum1p have a low affinity for ORC. The additional interaction that mutant Sum1-1p is able to make with histone tails in the vicinity of ORC binding sites would provide an additional attachment point, leading to the accumulation of Sum1-1p but not Sum1p near ORC binding sites. It is curious that the proposed increased affinity of Sum1-1p for histone tails does not also strengthen the association of Sum1-1p with midsporulation promoters. Perhaps Sum1-1p has a higher affinity for the particular histone tail modification pattern found near ORC binding sites compared to the pattern found near midsporulation genes. Alternatively, the same Sum1-1p molecule may not be able to bind to histone tails and DNA simultaneously.
Evolution of silencing complexes.
The SUM1-1 mutation provides a fascinating opportunity to explore the evolution of repressive chromatin. In essence, Sum1-1p represents the de novo evolution of a new type of silencing protein. A single nucleotide change in the SUM1 gene has given rise to a protein that can form silenced chromatin, whereas the wild-type gene cannot. It is not hard to imagine that similar types of mutations have occurred during the course of evolution, giving rise to novel expression patterns and phenotypes. In fact, Sir-mediated silencing itself is evolutionarily related to Sum1p-mediated repression and could have arisen from a promoter-specific repression complex. The deacetylases Sir2p and Hst1p are paralogs that arose in a genome duplication which occurred in the evolution of Saccharomyces (10, 21, 46). Hence, in an ancestor of Saccharomyces, there was only one Sir2p/Hst1p protein, and the two distinct functions that exist today most likely evolved after the genome duplication. In addition, Sir3p and Orc1p are paralogs (10), suggesting that the silencing-specific function of Sir3p could also have arisen after the genome duplication. Finally, the apparent functional interaction between Sum1p and ORC suggests that an ancestral interaction between a Sum1-like complex and ORC could have been elaborated in the development of the Sir silencing apparatus.
Acknowledgments
We thank Kelley Wallace, Charity Kirk, Michael Kobor, and Jane Zhu for technical assistance and Pei Zhou and members of the Rusche and Rine labs for helpful discussions.
This research was supported by a postdoctoral fellowship from the American Cancer Society (PF-01-116-01-GMC) to L.N.R. and grants from the National Institutes of Health to J.R. (GM31105) and L.R. (GM073991).
REFERENCES
- 1.Balakrishnan, R., K. R. Christie, M. C. Costanzo, K. Dolinski, S. S. Dwight, S. R. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. Nash, R. Oughtred, M. Skrzypek, C. L. Theesfeld, G. Binkley, C. Lane, M. Schroeder, A. Sethuraman, S. Dong, S. Weng, S. Miyasato, R. Andrada, D. Botstein, and J. M. Cherry. 2005. Saccharomyces Genome Database. [Online.] http://www.yeastgenome.org.
- 2.Bedalov, A., M. Hirao, J. Posakony, M. Nelson, and J. A. Simon. 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:7044-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 4.Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz, and K. Nasmyth. 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41:41-48. [DOI] [PubMed] [Google Scholar]
- 5.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 6.Chi, M. H., and D. Shore. 1996. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16:4281-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 8.Davie, J. K., D. G. Edmondson, C. B. Coco, and S. Y. Dent. 2003. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278:50158-50162. [DOI] [PubMed] [Google Scholar]
- 9.Davie, J. K., R. J. Trumbly, and S. Y. Dent. 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 11.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 13.Foss, M., F. J. McNally, P. Laurenson, and J. Rine. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262:1838-1844. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, K. A., J. Rine, and C. A. Fox. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 16.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 18.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 20.Kayne, P. S., U.-J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 21.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32:370-377. [DOI] [PubMed] [Google Scholar]
- 23.Klar, A. J., S. N. Kakar, J. M. Ivy, J. B. Hicks, G. P. Livi, and L. M. Miglio. 1985. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics 111:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurenson, P., and J. Rine. 1991. SUM1-1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics 129:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 26.Loo, S., C. A. Fox, J. Rine, R. Kobayashi, B. Stillman, and S. Bell. 1995. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell. 6:741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, K., M. A. Vega-Palas, and M. Grunstein. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 30.Min, J., J. Landry, R. Sternglanz, and R. M. Xu. 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105:269-279. [DOI] [PubMed] [Google Scholar]
- 31.Pak, D. T., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum, J. Marr, P. Romanowski, and M. R. Botchan. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311-323. [DOI] [PubMed] [Google Scholar]
- 32.Pierce, M., K. R. Benjamin, S. P. Montano, M. M. Georgiadis, E. Winter, and A. K. Vershon. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 23:4814-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothstein, R. 1991. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast, p. 281-301. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed]
- 35.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 36.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 39.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shareef, M. M., R. Badugu, and R. Kellum. 2003. HP1/ORC complex and heterochromatin assembly. Genetica 117:127-134. [DOI] [PubMed] [Google Scholar]
- 41.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32:378-383. [DOI] [PubMed] [Google Scholar]
- 43.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theis, J. F., and C. S. Newlon. 1997. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl. Acad. Sci. USA 94:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381:251-253. [DOI] [PubMed] [Google Scholar]
- 46.Wolfe, K. 2004. Evolutionary genomics: yeasts accelerate beyond BLAST. Curr. Biol. 14:R392-R394. [DOI] [PubMed] [Google Scholar]
- 47.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 48.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 49.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]