Abstract
Members of the CELF family of RNA binding proteins have been implicated in alternative splicing regulation in developing heart. Transgenic mice that express a nuclear dominant-negative CELF protein specifically in the heart (MHC-CELFΔ) develop cardiac hypertrophy and dilated cardiomyopathy with defects in alternative splicing beginning as early as 3 weeks after birth. MHC-CELFΔ mice exhibit extensive cardiac fibrosis, severe cardiac dysfunction, and premature death. Interestingly, the penetrance of the phenotype is greater in females than in males despite similar levels of dominant-negative expression, suggesting that there is sex-specific modulation of splicing activity. The cardiac defects in MHC-CELFΔ mice are directly attributable to reduced levels of CELF activity, as crossing these mice with mice overexpressing CUG-BP1, a wild-type CELF protein, rescues defects in alternative splicing, the severity and incidence of cardiac hypertrophy, and survival. We conclude that CELF protein activity is required for normal alternative splicing in the heart in vivo and that normal CELF-mediated alternative splicing regulation is in turn required for normal cardiac function.
The importance of the posttranscriptional regulation of gene expression has been driven home in the last few years, as it is now known that the vast number of proteins thought to comprise the human proteome are generated from a much smaller number of genes (16). Pre-mRNA alternative splicing is an important mechanism for the generation of protein diversity (14). It is currently estimated that 60 to 74% of human genes undergo alternative splicing (17, 18). Tight regulation of alternative splicing is necessary for the appropriate temporal and spatial control of gene expression, and disruption of splicing regulation can cause or contribute to human disease (11).
Members of the CUG binding protein (CUG-BP) and the embryonic lethal abnormal vision type RNA binding protein 3 (ETR-3)-like factor (CELF) family of RNA binding proteins (also called BRUNOL proteins) regulate alternative splicing by binding to intronic elements within specific pre-mRNA targets (7, 10, 19, 21, 35, 38). Two CELF proteins, CUG-BP1 (also known as BRUNOL2) and ETR-3 (also known as BRUNOL3, CUG-BP2, or NAPOR), are expressed in the heart and are hypothesized to drive changes in alternative splicing during cardiac development (22). CUG-BP1 and ETR-3 have also been suggested to play key roles in the pathogenesis of a number of disorders in heart and skeletal muscle, including myotonic dystrophy, Duchenne and Becker muscular dystrophies, partial monosomy 10p, and familial arrhythmogenic right ventricular dysplasia (7, 23, 24, 35, 38, 40). These reports suggest an important role for CELF-mediated alternative splicing regulation in the myocardium.
We previously described CELFΔ, a truncated form of CELF4 (also known as BRUNOL4) that acts as a dominant negative to specifically repress the alternative splicing activity of the members of the CELF family in cultured cells without globally disrupting alternative splicing (6). Transient expression of CELFΔ in primary embryonic cardiomyocyte cultures demonstrated that endogenous CELF activity is required for the appropriate alternative splicing of CELF targets in heart muscle cells (22). To determine the role of CELF-mediated alternative splicing regulation in the heart in vivo, we generated transgenic mice that express CELFΔ specifically in the heart by using the α-myosin heavy chain promoter. A dominant-negative approach was chosen in lieu of a more traditional knockout approach for two reasons. First, in transient- transfection assays, CUG-BP1 and ETR-3 have very similar effects on the alternative splicing of known CELF target pre-mRNAs (10, 19), suggesting that these proteins have a high degree of functional redundancy. Second, both CUG-BP1 and ETR-3 are expressed in the cytoplasm as well as the nucleus in the heart (22) and several reports have implicated CELF proteins in the regulation of cytoplasmic RNA processing events, including deadenlyation and translation (12, 28, 32-34, 41). In knockout mice, it would be difficult to distinguish between the effects from loss of nuclear versus cytoplasmic CELF functions. Overexpression of a nuclear dominant-negative protein allows us to overcome both of these obstacles: we can repress the activity of all CELF proteins expressed in the heart, and we can target repression specifically to the nucleus, where splicing takes place.
Here, we report a striking cardiac phenotype in mice attributable to the inhibition of CELF splicing activity by the cardiac tissue (MHC-CELFΔ)-specific expression of a dominant-negative CELF protein. Animals expressing the MHC-CELFΔ transgene develop early-onset cardiac hypertrophy and dilated cardiomyopathy characterized by fibrosis, changes in gene expression associated with heart failure, severe cardiac dysfunction, and premature death. MHC-CELFΔ mice exhibit specific defects in the alternative splicing of pre-mRNA targets of CELF-mediated regulation, and both the splicing defects and gross pathological changes are rescued by the increased cardiac expression of CUG-BP1, a wild-type CELF protein. Our results indicate that CELF-mediated alternative splicing regulation is necessary for maintenance of normal cardiac structure and function.
MATERIALS AND METHODS
Generation of MHC-CELFΔ mice.
The nucleus-restricted dominant-negative CELF protein NLSCELFΔ was created by insertion of the strong nuclear localization signal from the simian virus 40 large T antigen in frame between the N-terminal Xpress epitope tag and the truncated CELF4 open reading frame of the previously described CELFΔ (6). The dominant-negative splicing activity of NLSCELFΔ was confirmed as previously described for CELFΔ (6). Nuclear localization of NLSCELFΔ was tested by nuclear/cytoplasmic fractionation (20) of transiently transfected cells, followed by Western blotting using an anti-Xpress antibody (Invitrogen). The cardiac tissue-specific dominant-negative transgene MHC-CELFΔ was created by subcloning the NLSCELFΔ open reading frame between the mouse α-myosin heavy chain promoter (provided by Robert Schwartz, Institute of Biosciences and Technology, Houston, TX) and the 3′ untranslated region of the bovine growth hormone amplified from the pcDNA3.1+ vector (Invitrogen). The expression cassette was excised with SacI and microinjected into the pronuclei of fertilized FVB oocytes in the Baylor College of Medicine Genetically Engineered Mouse Core Laboratory. Genotyping was performed by PCR amplification of mouse tails lysed in DirectPCR lysis reagent (Viagen Biotech, Inc.). A 493-bp MHC-CELFΔ transgene product (primers GCCCGGCACTCTTAGCAAAC and GGGAGGAGTACTTCACAAAG) and a 386-bp TSHβ internal control product (primers GTAACTCACTCATGCAAAGT and TCCTCAAAGATGCTCATTAG) were amplified in the same reaction using an annealing temperature of 55°C and 30 cycles of amplification and resolved by agarose gel electrophoresis.
Western blot analysis.
Hearts were homogenized in protein loading buffer (0.64 M Tris HCl [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate, 5% β-mercaptoethanol), and total protein samples were quantitated, resolved by polyacrylamide gel electrophoresis, transferred, and probed as previously described (19) using a monoclonal antibody against the Xpress epitope tag (Invitrogen). Equivalent loadings were confirmed by Ponceau S staining.
Alternative splicing.
Total RNA was extracted from the hearts of transgenic and wild-type littermates and subjected to reverse transcription (RT)-PCR analysis. Mtmr1 and Mef2A PCRs were performed as published previously (5). β1 integrin, Bin1, Rbm9, F1γ, and Mef2C were assessed using single-tube RT-PCRs with 0.5 μg RNA, 2.8 U avian myeloblastosis virus reverse transcriptase (Life Sciences, Inc.), and 0.4 U Platinum Taq (Invitrogen) per reaction in a 20-μl volume. Primers and conditions used are as follows: β1 integrin primers GGCAACAATGAAGCTATCG and CCTCATACTTCGGATTGAC, 57°C annealing temperature, 22 cycles; Bin1 primers TCAACACGTTCCAGAGCATC and GATCTCAGGGGTAGCAGCAG, 57°C annealing temperature, 25 cycles; Rbm9 primers CCAGCAACCAGAATGGATCT and ACCCGTGCTGTTGCATTATT, 57°C annealing temperature, 23 cycles; F1γ primers CCAGCAAGAACGCTTCTGA and TCAGTGGACCAAGTTTCTTCTG, 57°C annealing temperature, 22 cycles; Mef2C primers TCCACCTCGGCTCTGTAACT and CAGCTGCTCAAGCTGTCAAC, 57°C annealing temperature, 25 cycles. For SERCA2, RT-PCR analysis was performed as previously described (19) using forward primer GGCTCCATCTGCTTGTCCA and reverse primers GTGTCTGTGCTGTAGACCCAG and AGCGGTTACTCCAGTATT at a 55°C annealing temperature and 24 cycles of amplification.
Histology.
Hearts and lungs were removed from virgin female littermates and fixed overnight by immersion in 10% neutrally buffered formalin. Fixed organs were embedded in paraffin, sectioned, and stained with hematoxylin and eosin stain or Mason's trichrome stain.
Assessment of cardiac function.
M-mode echocardiography and 10-mHz Doppler ultrasound were performed on virgin female transgenic and wild-type littermates anesthetized with 1% isoflurane gas in oxygen (31). Measurements were performed blinded, and functional parameters were calculated using standard formulas (15).
Real time RT-PCR analysis.
Quantitative real time RT-PCR analysis was performed as previously described (37). Levels of marker gene expression were normalized against levels of glyceraldehyde-2-phosphate dehydrogenase (GAPDH) expression using the formula 2 to the power of [(number of cycles to reach set level of GAPDH) - (number of cycles to reach set level of marker gene)], setting the level for comparison within the linear range of amplification for all genes.
Statistics.
Data are reported as means ± standard errors of the means unless otherwise noted. Comparisons between groups were performed by analysis of variance with post hoc testing (Tukey) using SPSS software or with two-tailed t tests using Microsoft Excel. Differences were considered statistically significant at a P of ≤0.05.
RESULTS
MHC-CELFΔ mice exhibit defects in CELF-mediated alternative splicing.
CELFΔ is approximately 35 kDa, well below the size limit for passive diffusion through the nuclear pore complex (13, 30). To drive the localization of CELFΔ to the nucleus, where alternative splicing takes place, the simian virus 40 large T antigen nuclear localization signal was inserted in frame between the N-terminal epitope tag and the CELFΔ open reading frame to create NLSCELFΔ. In transfected cells, NLSCELFΔ is detectable only in the nucleus and represses the activity of the CELF family in a manner similar to the original CELFΔ (data not shown). To create mice that express this nuclear dominant-negative protein in the heart, a transgene was created (MHC-CELFΔ) (Fig. 1A) in which the expression of NLSCELFΔ is driven by the mouse α-myosin heavy chain (α-MHC) promoter.
FIG. 1.
MHC-CELFΔ transgenic mice exhibit defects in the alternative splicing of the CELF pre-mRNA target Mtmr1. (A) The MHC-CELFΔ transgene contains NLSCELFΔ behind the mouse α-myosin heavy chain promoter. (B) Expression of the NLSCELFΔ transgene protein was confirmed in MHC-CELFΔ mice by Western blotting using an antibody against the N-terminal Xpress epitope tag. Fifty micrograms of total protein lysate was loaded per lane from the hearts of the 10-day-old MHC-CELFΔ-11 pup and a 6-week-old MHC-CELFΔ-10 animal. Equivalent loadings were confirmed by Ponceau S staining (data not shown). (C) Mtmr1 contains two alternative exons, 2.1 and 2.2, which may be alternatively spliced such that they are both included (isoform C), only the first is included (isoform B) or both are skipped (isoform A). In the wild-type heart, the C isoform predominates in newborn and 3-week-old animals. In the transgenic heart, Mtmr1 alternative splicing is normal at birth but by 3 weeks, the C isoform is diminished and the B isoform is induced.
Three transgenic founders with germ line transmission were obtained (MHC-CELFΔ-10, -11, and -574). The MHC-CELFΔ-11 F0 female died from a postpartum uterine infection, however, and her line was subsequently lost. The only transgenic pup obtained from the MHC-CELFΔ-11 line died 10 days following birth, and dissection followed by Western blotting revealed an enlarged heart that expressed high levels of the transgene protein (Fig. 1B and data not shown). Transgenic animals from line MHC-CELFΔ-10 survived to maturity and expressed the NLSCELFΔ protein, though at levels lower than the pup from line MHC-CELFΔ-11 (Fig. 1B). The third line, MHC-CELFΔ-574, expressed the lowest levels of NLSCELFΔ of the three lines (data not shown). Except where otherwise noted, analyses were conducted on line MHC-CELFΔ-10, the higher expressing of the two surviving lines.
To confirm that the alternative splicing activity of the CELF family is disrupted in MHC-CELFΔ mice, the alternative splicing pattern of the myotubularin-related protein 1 (Mtmr1) transcripts in MHC-CELFΔ hearts was compared to that of wild-type littermates at different ages. The Mtmr1 gene has two alternative exons (2.1 and 2.2) that are alternatively spliced to give rise to three mRNA isoforms (Fig. 1C) and was recently identified as a target of CELF-mediated splicing regulation in humans, following a genomic screen for intronic sequences that contain U/G-rich motifs characteristic of CELF binding sites (10). Mtmr1 alternative splicing is conserved between mouse and human (5), and the mouse Mtmr1 gene also contains putative CELF binding sites in the introns adjacent to the alternative exons (see Table S1 in the supplemental material). The C isoform, which includes both alternative exons, predominates in the wild-type heart (Fig. 1C). In the transgenic heart, Mtmr1 alternative splicing was unperturbed at birth. By 3 weeks, however, there was a marked reduction in the amount of the C isoform in MHC-CELFΔ hearts, with a corresponding increase in the B isoform, which includes the 2.1 exon but skips the 2.2 exon (Fig. 1C). This suggests that for Mtmr1 splicing in the heart, CELF activity is important for the inclusion of exon 2.2.
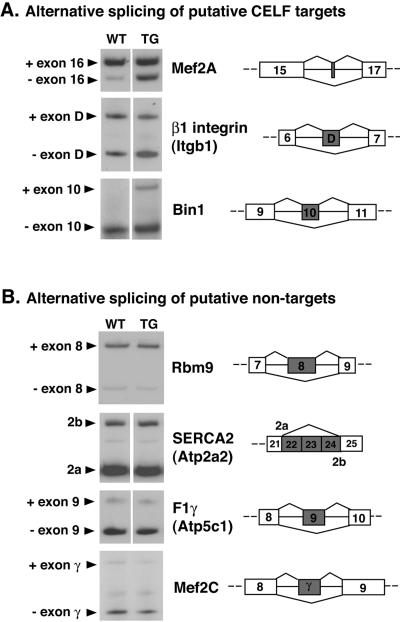
We hypothesized that the expression of NLSCELFΔ would specifically disrupt the alternative splicing of CELF target pre-mRNAs in vivo without globally disrupting alternative splicing. To confirm this, we identified three other putative CELF targets and four putative nontargets and compared their alternative splicing patterns in wild-type and MHC-CELFΔ hearts. Candidate targets contained U/G-rich motifs that could act as CELF binding sites in one or both introns adjacent to an alternatively spliced exon, whereas putative nontargets lacked any motifs resembling CELF binding sites in the flanking introns (see Table S1 in the supplemental material). At 3 weeks, all three of the putative targets identified showed different ratios of their alternative splice forms in MHC-CELFΔ hearts relative to the wild type whereas none of the four putative nontargets were affected (Fig. 2).
FIG. 2.
The alternative splicing patterns of putative CELF targets are affected in the hearts of MHC-CELFΔ mice. (A) Several mouse genes were identified as putative CELF targets by the presence of potential CELF binding sites in introns adjacent to alternatively spliced exons (see Table S1 in the supplemental material). The alternative splicing patterns of three putative targets were found to differ between the hearts of female wild-type and MHC-CELFΔ littermates at 3 weeks of age by RT-PCR. (B) Several alternatively spliced mouse genes that lack U/G-rich motifs resembling CELF binding sites in the introns adjacent to their alternative exons were also identified and were designated putative nontargets. The splicing patterns of four of these putative nontargets were identical in the hearts of 3-week-old wild-type and MHC-CELFΔ female littermates. Two or more individuals were evaluated for each putative target or nontarget with similar results. A representative example for each is shown. WT, wild type; TG, transgenic.
MHC-CELFΔ transgenic mice develop early-onset cardiac hypertrophy and dilated cardiomyopathy.
Hemizygous MHC-CELFΔ mice appeared normal at birth and were obtained at the expected Mendelian frequency. Attempts to generate homozygous MHC-CELFΔ mice failed, however, as the female transgenic mice did not tolerate pregnancy. From 16 F1 sibling crosses, eight females died during pregnancy, five died within a few days of delivery, one became severely distressed during pregnancy and was euthanized, and one failed to become pregnant. The females that died ranged from 8.5 to 16.5 weeks old, and all exhibited dramatic cardiac hypertrophy. The heart comprised 1.59% ± 0.12% of the body weight in the transgenic females that died (n = 11), almost three times that of age-matched, normal female littermates sacrificed for comparison (0.62% ± 0.06%, n = 6). Because normal littermates were virgins with lower body weights than their transgenic pregnant/postpartum counterparts (for pregnant females, body weights include unborn pups), this difference is likely to be underestimated. All subsequent data presented in this study are derived from hemizygous, nonparous animals.
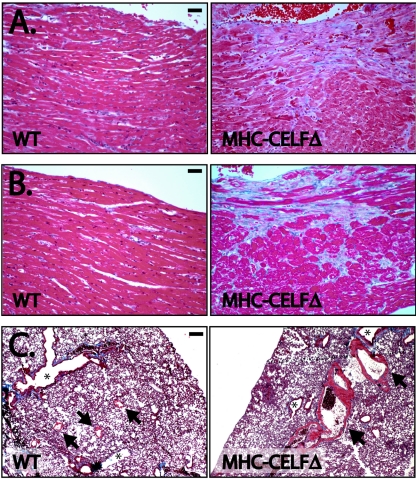
Cardiac hypertrophy in MHC-CELFΔ animals is not pregnancy dependent. Figure 3A shows the heart of a virgin 8-week-old MHC-CELFΔ female compared to that of her wild-type littermate. Coronal sections revealed mild cardiac enlargement that is primarily left sided with a globular left ventricular contour at 3 weeks following birth (Fig. 3B). At 9 weeks, transgenic hearts were more strikingly enlarged, exhibiting expansion of the chambers but without dramatic wall thinning, indicating both cardiac hypertrophy and dilation (Fig. 3C). Left atrial dilation was present at both 3 and 9 weeks of age, with thrombosis of the chamber at 9 weeks (Fig. 3C).
FIG. 3.
MHC-CELFΔ mice develop cardiac hypertrophy and dilated cardiomyopathy. (A) Hearts from wild-type (WT) and MHC-CELFΔ female littermates were isolated and photographed 8 weeks after birth. Bar represents 1 mm. Trichrome-stained coronal sections show progressive, predominantly left-sided, cardiac enlargement in the MHC-CELFΔ mice that is mild at 3 weeks (B) and more severe at 9 weeks (C). LV, left ventricle. Asterisk indicates left atrial thrombus. (D) Heart size was evaluated in wild-type and MHC-CELFΔ littermates over a time course of postnatal development. Each symbol represents an individual animal. (E) The mean heart size of the MHC-CELFΔ population is compared to that of the wild-type population at each time point. Values shown are the means ± the standard errors of the means. The number of animals in each group is shown. Asterisk indicates that the mean is significantly different from that of the wild type (P ≤ 0.05).
To characterize the progression of hypertrophy, the heart sizes of MHC-CELFΔ mice and their wild-type littermates were measured over a postnatal time course (Fig. 3D and E). Heart size was evaluated as the percentage of total body weight that is heart weight. This method was deemed valid because no differences were observed between the mean body weights of nonparous transgenic animals and their sex-matched wild-type littermates at any age evaluated (data not shown). Although there was considerable overlap in heart sizes between individuals in the wild-type and transgenic populations at 3 weeks, some individuals had slightly enlarged hearts (Fig. 3D) and there was already a small but significant increase in the mean heart sizes of female MHC-CELFΔ mice relative to their wild-type counterparts (Fig. 3E). By 6 weeks, cardiac hypertrophy was pronounced in transgenic males and females, and by 9 weeks, MHC-CELFΔ hearts reached their maximum mean size. It is interesting to note that the mean heart size of the transgenic males was not increased as much as that of transgenic females (see “Phenotypic penetrance is lower in MHC-CELFΔ males than females” below). The same differences between groups were seen when heart size was measured as a ratio of heart weight to tibia length (data not shown).
The development of cardiac hypertrophy is not due to the disruption of another gene at the integration site but depends on expression of the NLSCELFΔ protein. Just as the animal from the MHC-CELFΔ-11 line had severe hypertrophy and early death associated with very high levels of NLSCELFΔ expression, animals from the MHC-CELFΔ-574 line, which express the lowest levels of NLSCELFΔ protein, likewise develop cardiac hypertrophy that is less severe than that seen in the other lines. The mean heart size of MHC-CELFΔ-574 females at 9 weeks was 0.65% ± 0.02% of the body weight (n = 15), significantly greater than the 0.52% ± 0.01% of body weight (n = 34) observed for age-matched wild-type females (P ≤ 0.05).
Phenotypic penetrance is lower in MHC-CELFΔ males than females.
Premature death was commonly observed in MHC-CELFΔ animals. In the vast majority of cases, individuals appeared healthy and did not show overt signs of poor health (such as weight loss or lethargy) prior to death. Survival curves were plotted for transgenic mice out to 24 weeks of age (Fig. 4A). Strikingly, 91% of MHC-CELFΔ males but only 27% of MHC-CELFΔ females were still alive at 24 weeks of age. This is a dramatic increase in mortality in female mice, as the average life span of wild-type FVB mice in our vivarium is approximately 2 years (C. Beverly and J. Bondzinski, Baylor College of Medicine Center for Comparative Medicine, personal communication).
FIG. 4.
Penetrance of the MHC-CELFΔ phenotype is lower in males than in females, despite similar amounts of NLSCELFΔ protein expression. (A) Survival. Mortality is dramatically higher for MHC-CELFΔ females than males. The number of animals in each group is indicated. (B) Incidence of hypertrophy. By 6 weeks after birth, more than 80% of MHC-CELFΔ females have cardiac hypertrophy, whereas the incidence of hypertrophy remains low in transgenic males. An individual is considered to have cardiac hypertrophy if its heart is larger than 2 standard deviations above the mean heart size of sex- and age-matched wild-type mice. The number of animals in each group is indicated. (C) Splicing defects. All transgenic females, but only two-thirds of transgenic males have defects in Mtmr1 alternative splicing in the heart at 3 weeks of age. (D) NLSCELFΔ protein expression. Western blot probed with an antibody against the N-terminal Xpress epitope tag reveals similar levels of NLSCELFΔ expression in the hearts of male and female littermates at 6 weeks of age. From this litter, two of the three transgenic females but none of the transgenic males had pronounced cardiac hypertrophy. Fifty micrograms of total protein lysate was loaded per lane, and equivalent loadings were confirmed by Ponceau S staining (data not shown).
This difference in survival is not the only difference observed between the sexes in the MHC-CELFΔ population. As mentioned above, the mean heart size of transgenic males is not elevated as much as in transgenic females at any age (Fig. 4B). In fact, the hearts of many MHC-CELFΔ males remained unaffected. No more than 50% of MHC-CELFΔ males developed cardiac hypertrophy within the first 24 weeks, far fewer than MHC-CELFΔ females, 85% of whom developed hypertrophy by 6 weeks (Fig. 4B). This disparity is also seen between the sexes in the lower-expressing MHC-CELFΔ-574 line, as 60% of females but only 18% of males developed mild hypertrophy by 9 weeks (data not shown).
The observed differences between males and females are also evident at the level of alternative splicing. All of the female MHC-CELFΔ mice evaluated displayed defective Mtmr1 splicing by 3 weeks, but only a portion of the MHC-CELFΔ males exhibited splicing patterns that differed from the wild type (Fig. 4C). The dissimilar penetrance of the phenotype between MHC-CELFΔ males and females is not attributable to a difference in expression of the transgene. Western blot analysis demonstrated similar levels of NLSCELFΔ expression in male and female transgenic littermates, regardless of the presence or absence of cardiac hypertrophy in these individuals (Fig. 4D). Intriguingly, these results suggest that there is a sex-specific modulation of CELF-mediated alternative splicing activity.
MHC-CELFΔ transgenic mice exhibit severe pathology and cardiac dysfunction.
To characterize the health of the transgenic heart, we performed histological analysis on hearts from wild-type and MHC-CELFΔ females. Four days following birth, there were no pronounced differences between transgenic and wild-type hearts (data not shown). At 3 weeks following birth, trichrome staining revealed early fibrosis and myocyte degenerative changes, as evidenced by paler fibers indicative of myocytolysis (Fig. 5A). At 9 weeks, degenerative myocytolysis was more extensive, with continuing loss of myofibers and fibrosis (Fig. 5B). In addition to the evident cardiac damage, by 9 weeks, we saw secondary signs of cardiac dysfunction in the lungs of affected MHC-CELFΔ animals. Grossly, lungs looked gray and swollen (data not shown). Wet lung weights of MHC-CELFΔ females were more than twice those of age- and sex-matched wild-type littermates (283 ± 18 mg versus 131 ± 5 mg; n = 6 each). Trichrome-stained sections of lungs revealed dilated veins with muscular hypertrophy of the wall, indicative of chronically elevated left-ventricular-end diastolic pressure (Fig. 5C). The thrombosis observed in the dilated left atrium of the 9-week-old MHC-CELFΔ heart is also consistent with chronic left-heart dysfunction (Fig. 3C).
FIG. 5.
Histology reveals cardiac pathology in MHC-CELFΔ mice. Trichrome staining of sections demonstrates that myocytolysis and fibrosis are present at 3 weeks (A) and become more profound by 9 weeks (B). Bar represents 20 μm. (C) Trichrome-stained sections show dilated veins with muscular hypertrophy in the lungs of MHC-CELFΔ mice at 9 weeks, consistent with chronically increased left-ventricular-end diastolic pressure. Arrows indicate veins. Asterisks indicate airways. Bar represents 100 μm. WT, wild type.
To determine whether MHC-CELFΔ mice were undergoing heart failure, we used real-time RT-PCR analysis to measure the expression levels of three genes known to be upregulated in the failing heart: atrial natriuretic factor, B-type natriuretic peptide, and α-skeletal actin (4, 36). All three of these markers were upregulated in MHC-CELFΔ hearts by 3 weeks (see Fig. S1 in the supplemental material), prior to the onset of severe hypertrophy but consistent with the onset of splicing defects and the presence of scarring in the heart at this age (Fig. 1C and 5A).
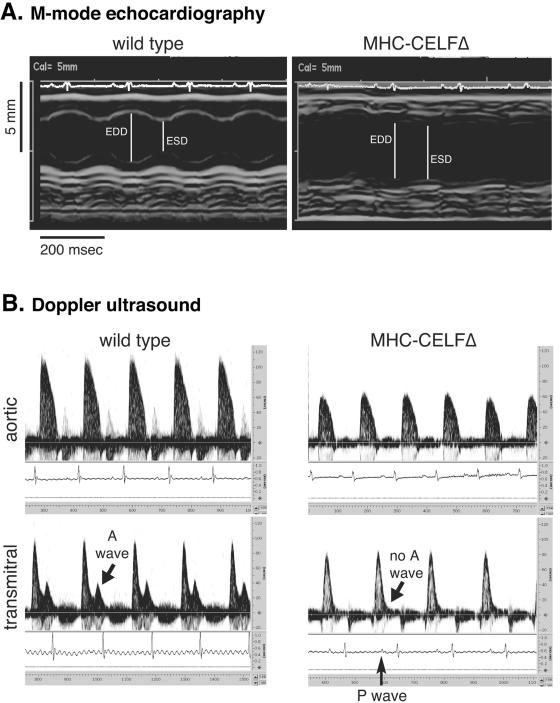
Cardiac function in female MHC-CELFΔ mice was compared to age- and sex-matched wild-type littermates using M-mode echocardiography and 10-mHz Doppler ultrasound. At 4 to 5 days following birth, the cardiac function of transgenic mice was indistinguishable from that of wild-type littermates when assessed by Doppler (data not shown). At 9 weeks, however, MHC-CELFΔ mice had profound cardiac dysfunction (Fig. 6 and Table 1). Under anesthesia, the transgenic mice had normal heart rates with no sign of persistent arrhythmia (Table 1). The beat-to-beat variability as measured by the standard deviation of the RR interval measuring the time between the R peaks in the electrocardiogram was also not different from the wild type (Table 1). Echocardiography confirmed that the left ventricles of the transgenic heart were enlarged without thinning of the ventricular wall (Fig. 6A). The end diastolic and end systolic dimensions were both significantly increased in MHC-CELFΔ hearts (Fig. 6 and Table 1). Some transgenic individuals also had a detectable increase in the thickness of the posterior wall, though the mean posterior wall thickness of transgenic hearts was not statistically different from that of the wild type (Table 1). Fractional shortening, an indicator of systolic function, was severely reduced in the transgenic heart, indicating impaired contraction. Doppler analysis confirmed poor systolic function. The peak aortic flow velocity and mean and peak aortic accelerations were significantly reduced in MHC-CELFΔ animals, whereas the isovolumic contraction time was significantly elevated (Table 1). Diastolic dysfunction was also evident. The peak early-filling velocity was reduced, whereas the isovolumic relaxation time and Tei index were both significantly increased in transgenic animals (Table 1). Interestingly, there was no easily discernible A wave on the transmitral Doppler in five out of the six MHC-CELFΔ mice evaluated, indicating that despite the presence of a clear electrical activation of the atria (see P waves on the electrocardiogram), there was no significant atrial contribution to left-ventricular filling (Fig. 6B). Together, these results show that MHC-CELFΔ mice have enlarged hearts that exhibit both ventricular wall thickening and dilation and have a severe systolic and diastolic defect.
FIG. 6.
MHC-CELFΔ mice have poor cardiac function. M-mode echocardiography (A) and 10-mHz Doppler ultrasound (B) were performed on nonparous wild-type and MHC-CELFΔ female littermates 9 weeks after birth. A typical example of each is shown. Echocardiograms reveal increased left-ventricular cavity size with decreased fractional shortening, whereas Doppler ultrasounds reveal impaired systolic and diastolic function. EDD, end diastolic dimension; ESD, end systolic dimension.
TABLE 1.
MHC-CELFΔ hearts are functionally impaired at 9 weeks
| Method of measurement (units) | Wild type (n = 6) | MLC-CELFΔa (n = 6) |
|---|---|---|
| M-mode echocardiography | ||
| Diastolic dimension (mm) | 3.33 ± 0.18 | 4.14 ± 0.16* |
| Systolic dimension (mm) | 2.05 ± 0.09 | 3.27 ± 0.22* |
| Posterior wall thickness (mm) | 0.86 ± 0.06 | 1.03 ± 0.05 |
| Fractional shortening | 0.38 ± 0.02 | 0.21 ± 0.03* |
| Vcfcb (s−1) | 2.17 ± 0.22 | 1.31 ± 0.22* |
| 10-mHz Doppler | ||
| Heart rate (beats/min) | 391 ± 19 | 366 ± 23 |
| SD of RR interval (ms) | 3.2 ± 1.1 | 1.8 ± 0.2* |
| Systolic parameters | ||
| Peak aortic flow velocity (cm/s) | 114.2 ± 2.8 | 62.5 ± 4.0* |
| Mean acceleration (cm/s2) | 7,984 ± 399 | 3,355 ± 280* |
| Peak acceleration (cm/s2) | 19,040 ± 1,420 | 6,980 ± 940* |
| Ejection time (ms) | 62.5 ± 0.8 | 59.3 ± 2.4 |
| Isovolumic contraction time (ms) | 11.6 ± 0.4 | 18.2 ± 3.6* |
| Diastolic parameters | ||
| Peak early-filling velocity (cm/s) | 97.7 ± 4.9 | 79.7 ± 9.4* |
| Isovolumic relaxation time (ms) | 14.4 ± 1.0 | 19.2 ± 3.33* |
| Tei indexc | 0.42 ± 0.02 | 0.69 ± 0.13* |
Asterisk indicates mean is significantly different from that of the wild type (P ≤ 0.05).
Vcfc, rate-corrected mean velocity of circumferential fiber shortening = (fractional shortening × square root of the RR interval)/ejection time.
Tei index = (isovolumic contraction time + isovolumic relaxation time)/ejection time.
The MHC-CELFΔ phenotype is rescued by the overexpression of CUG-BP1 in the heart.
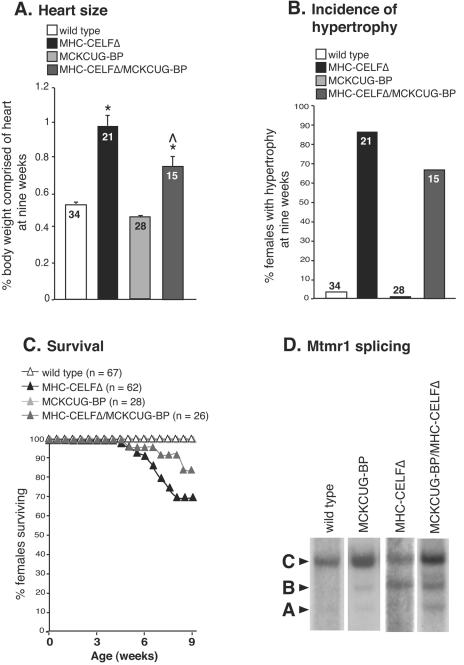
It is possible that the overexpression of exogenous NLSCELFΔ in the heart causes pathogenesis by a mechanism unrelated to its effects on CELF-mediated splicing. CELFΔ does not bind to RNA (39), making it unlikely that the effects on the splicing of CELF targets are due to a gain-of-function of the NLSCELFΔ protein acting directly on RNA. Both the defects in Mtmr1 alternative splicing and the development of cardiac hypertrophy occur with similar incidences in MHC-CELFΔ males, which supports the hypothesis that pathology in the transgenic heart can be attributed to the disruption of normal CELF-mediated alternative splicing. To confirm this hypothesis, we crossed MHC-CELFΔ males to female mice that overexpress CUG-BP1 in heart and skeletal muscle under the control of the muscle creatine kinase promoter (MCKCUG-BP). If the observed pathology in MHC-CELFΔ mice is in fact due to the repression of CELF activity, the effects should be alleviated in double-transgene offspring by the increase in wild-type CELF protein. Characterization of MCKCUG-BP mice is described in a separate report (15a). In the line of MCKCUG-BP mice used for these experiments, MCKCUG-BP-1036, transgenic animals overexpressed less than twofold the amount of endogenous CUG-BP1 protein found in the hearts of wild-type animals at 9 weeks (data not shown). Bitransgenic females were evaluated at 9 weeks, since a high proportion of MHC-CELFΔ females are affected and mean heart size has reached its peak level by this age (Fig. 3). Overexpression of CUG-BP1 partially rescued the effects of the dominant-negative protein but, alone, had no effect on heart size or survival compared to wild-type animals (Fig. 7). The mean heart size in MHC-CELFΔ/MCKCUG-BP double-transgene females was significantly smaller than that for MHC-CELFΔ females alone, although still greater than that of wild-type females (Fig. 7A). Likewise, the percentage of females that had cardiac hypertrophy was lower (Fig. 7B) and a greater fraction of MHC-CELFΔ/MCKCUG-BP females survived to 9 weeks (Fig. 7C).
FIG. 7.
The MHC-CELFΔ phenotype is rescued by overexpression of a wild-type CELF protein. (A) The mean heart sizes ± the standard errors of the means are shown for nonparous female animals at 9 weeks of age. The number of animals in each group is indicated. An asterisk indicates that the mean is significantly different from that of the wild type, and a caret indicates that the mean is significantly different from that of MHC-CELFΔ alone (P ≤ 0.05). (B) The female population overexpressing wild-type CUG-BP1 and NLSCELFΔ exhibits a lower incidence of cardiac hypertrophy than transgenic females that express the dominant-negative protein alone. Cardiac hypertrophy is defined as the heart size more than 2 standard deviations above the mean heart size of sex- and age-matched wild-type mice. (C) Overexpression of CUG-BP1 in the heart improves the survival rate of females expressing the dominant-negative protein. (D) At 9 weeks, the C isoform of Mtmr1 is the predominant splice form in wild-type mice. In MHC-CELFΔ mice, the C isoform is reduced and the level of the B isoform is elevated. Overexpression of CUG-BP1 increases the proportion of the C isoform.
To confirm that CUG-BP1 overexpression restores CELF-mediated alternative splicing in the MHC-CELFΔ heart, Mtmr1 splicing patterns were determined by RT-PCR analysis (Fig. 7D). In wild-type mice at 9 weeks, the C isoform predominated. The predominance of the C isoform was not affected by the overexpression of CUG-BP1 alone in the MCKCUG-BP-1036 line. In MHC-CELFΔ hearts at 9 weeks, however, the ratio of the C and B isoforms was approximately 50/50. Again, this suggests that CELF activity is important for inclusion of the second alternative exon. Inclusion of this exon improved in MHC-CELFΔ/MCKCUG-BP hearts as the C to B ratio was increased, though the proportion of total transcripts that were the C isoform was not completely restored to wild-type levels. Together, these results verify that defects in CELF-mediated splicing and the overt cardiac pathology in MHC-CELFΔ mice are both the result of reduced CELF activity in the hearts of these animals.
DISCUSSION
The present study demonstrates that CELF protein activity is critical for the appropriate alternative splicing of CELF targets in the heart in vivo and that normal CELF-mediated alternative splicing regulation is necessary for healthy cardiac function. It is important to note that α-MHC is transiently down-regulated in the ventricles during embryogenesis (27), so the MHC-CELFΔ transgene, which drives the expression of NLSCELFΔ under the control of the α-MHC promoter, would be expected to reach its highest level of expression and therefore its greatest efficacy shortly after birth. This is consistent with the onset of the MHC-CELFΔ phenotype. By all measures (alternative splicing patterns, histology, and functional tests), MHC-CELFΔ mice are indistinguishable from wild-type animals in the first 4 days after birth. By 3 weeks after birth, however, MHC-CELFΔ mice exhibit defects in alternative splicing, changes in gene expression, and scarring in the heart, and by 9 weeks, overt differences are observed in cardiac size and function.
In the past year, two mouse models have been reported that demonstrate the harmful effects of perturbing factors that regulate alternative splicing in the heart. Cardiac-specific loss of SC35 or ASF/SF2 causes dilated cardiomyopathy (8, 43). For ASF/SF2, loss leads to the aberrant regulation of CaMKII∂ alternative splicing and overexpression of the inappropriate CaMKII∂ splice form recapitulates the phenotype of ASF/SF2 ablation (43). In this report, reduced CELF activity in the hearts of MHC-CELFΔ mice was shown to affect the splicing of a natural pre-mRNA target, Mtmr1 (10), as well as the three putative CELF targets myocyte enhancer factor 2A (Mef2A), β1 integrin, and bridging integrator 1 (Bin1). With the exception of β1 integrin, the specific functional consequences of these alternative splicing events are unknown but these genes participate in diverse cellular functions that could contribute to pathogenesis. MTMR1 belongs to a family of phosphatases, and although its substrates are not known, aberrant ratios of Mtmr1 splice forms have been proposed to contribute to striated muscle defects in myotonic dystrophy (5, 15a). Mef2A belongs to a family of transcription factors critical for cardiogenesis that are involved in mediating the development of hypertrophic features in cardiac hypertrophy (1). Inclusion of the alternative exon D of β1 integrin alters the cytoplasmic domain of this cell-surface receptor and strengthens cytoskeleton-matrix linkages in muscle cells (3). Knockout of exon D in mice caused the increased expression of markers of ventricular dysfunction, suggesting that the increased adhesion of β1 integrin provided by the inclusion of exon D is important for cardiac contraction (2). Bin1 is an adaptor protein and putative tumor suppressor that is required in cardiac muscle (29). Though its precise function is not known, inclusion of the alternative exon 10 has been suggested to control its subcellular localization and is associated with impeded proliferation in cultured skeletal muscle cells (42). Additional targets that have not yet been identified may also be misspliced in the transgenic heart. Given that we have observed changes in alternative splicing for several pre-mRNAs, we feel that it is unlikely that the alternative splicing of any single target is fully responsible for the pathology in MHC-CELFΔ mice. Rather, it seems likely that the combined effects on the subset of CELF targets contribute to the observed cardiac dysfunction and pathology. Several targets of CELF regulation have been identified in skeletal muscle, and misregulation of these different CELF targets in the muscles of myotonic dystrophy patients is thought to explain distinct symptoms of the disease (7, 35, 38). MHC-CELFΔ mice will provide a useful tool in future studies to help delineate programs of CELF-mediated alternative splicing regulation in the heart.
Although the full complement of affected targets has not been elucidated, it is clear that the pathology observed in MHC-CELFΔ hearts can be attributed to a reduction in CELF splicing activity rather than an unrelated gain-of-function of the dominant-negative protein, as overexpression of CUG-BP1 in the hearts of these animals at least partially rescues both the splicing defects of the known target Mtmr1 and the overt signs of cardiac disease. The double-transgene animals overexpressing the dominant-negative protein and wild-type CUG-BP1 are not completely restored to a wild-type phenotype, however, perhaps because the levels of CUG-BP1 are insufficient to fully compensate for the extent of repression by NLSCELFΔ. The MCKCUG-BP-1036 line used for these experiments expresses levels of CUG-BP1 only slightly above normal endogenous levels in the first few weeks after birth. A second line that had similar levels of CUG-BP1 overexpression, MCKCUG-BP-1032, showed a similar level of rescue when crossed with MHC-CELFΔ animals (data not shown). Another possible explanation for the incomplete restoration of wild-type phenotype in the double-transgene animals is that we are only adding back a single CELF protein. ETR-3 is also expressed in the heart (19, 21, 22) and may exert some effects on splicing that do not overlap with CUG-BP1. Although CUG-BP1 and ETR-3 are highly conserved (the human proteins are 78% identical), they may interact with different subsets of transcripts or different protein partners to form distinct splicing complexes on their target RNAs. To date, their activities have been compared on only a few targets, predominantly by transient transfection assays in which the proteins are highly overexpressed (10, 19, 39). It would be interesting in the future to compare the effects of knocking out CUG-BP1 and ETR-3 activities independently.
The most surprising finding in this study was the pronounced phenotypic disparity between males and females despite similar levels of transgene expression. It is not uncommon for genetically modified mice to display sex differences in cardiac phenotypes, but in most cases, females fare better than males, with lower mortality, less severe hypertrophy, and mitigated pathology (9). MHC-CELFΔ mice are unusual because males are less likely than their female counterparts to exhibit splicing defects, develop cardiac hypertrophy, or die young. There are several possibilities that could explain the sex differences in MHC-CELFΔ mice. Males may have a greater tolerance for the misexpression of CELF-regulated splice variants than females. Alternatively, there may be female-specific pre-mRNA targets of CELF activity that contribute to the onset of pathogenesis or male-specific targets that offer some protective effect. The fact that there is a difference in penetrance not only for the gross features of the phenotype but also for Mtmr1 splicing suggests that there is a sex-specific modulation at the level of CELF splicing activity. It is not known what protein partners the CELF proteins require or what posttranslational modifications are important for their activity. Sex-specific differences in expression or activity of a cofactor or modifier could create differences in the levels of CELF activity between males and females. Sex-specific alternative splicing has been well described in Drosophila melanogaster, where the sex-specific expression of alternative splicing regulators controls sex determination (26). In vertebrates, tra2α, the mammalian homolog of a Drosophila splicing regulator, is upregulated in response to androgen stimulation (25). Emerging evidence also links levels of sex hormones to sex differences in cardiac pathology seen in mice (10). Sex-specific differences in cardiac CELF protein expression, modification, activity, and response to sex hormone pathways are now being investigated.
Supplementary Material
Acknowledgments
We thank Donnie Bundman, Thuy Pham, Motoaki Sano, and Gopal Singh for technical assistance, David Ladd for help with the statistical analysis, and Michael Schneider for helpful discussions.
This work was supported by grants from the National Institutes of Health to T.A.C. (RO1HL45653), C.H. (RO1HL22512), and G.T. (RO1AG17899). A.N.L. was supported by a postdoctoral NRSA fellowship from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (5F32AR008618-02) and a Development Grant from the Muscular Dystrophy Association (MDA3812).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akazawa, H., and I. Komuro. 2003. Roles of cardiac transcription factors in cardiac hypertrophy. Circ. Res. 92:1079-1088. [DOI] [PubMed] [Google Scholar]
- 2.Baudoin, C., M.-J. Goumans, C. Mummery, and A. Sonnenberg. 1998. Knockout and knockin of the β1 exon D define distinct roles for integrin splice variants in heart function and embryonic development. Genes Dev. 12:1202-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkin, A., S. Retta, O. Pletjushkina, F. Balzac, L. Silengo, R. Fassler, V. Koteliansky, K. Burridge, and G. Tarone. 1997. Muscle beta1D integrin reinforces the cytoskeleton-matrix link: modulation of integrin adhesive function by alternative splicing. J. Cell Biol. 139:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boluyt, M., L. O'Neill, A. Meredith, O. Bing, W. Brooks, C. Conrad, M. Crow, and E. Lakatta. 1994. Alterations in cardiac gene expression during the transition from stable hypertrophy to heart failure. Marked upregulation of genes encoding extracellular matrix components. Circ. Res. 75:23-32. [DOI] [PubMed] [Google Scholar]
- 5.Buj-Bello, A., D. Furling, H. Tronchere, J. Laporte, T. Lerouge, G. Butler-Browne, and J.-L. Mandel. 2002. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 11:2297-2307. [DOI] [PubMed] [Google Scholar]
- 6.Charlet-B., N., P. Logan, G. Singh, and T. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 7.Charlet-B., N., R. Savkur, G. Singh, A. Philips, E. Grice, and T. Cooper. 2002. Loss of the muscle-specific chloride channel in type I myotonic dystrophy lead to misregulated alternative splicing. Mol. Cell 10:45-53. [DOI] [PubMed] [Google Scholar]
- 8.Ding, J.-H., X. Xu, D. Yang, P.-H. Chu, N. Dalton, Z. Ye, J. Yeakley, H. Cheng, R.-P. Xiao, J. Ross, J. Chen, and X.-D. Fu. 2004. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in heart. EMBO J. 23:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, X.-J. 2004. Gender modulates cardiac phenotype development in genetically modified mice. Cardiovasc. Res. 63:510-519. [DOI] [PubMed] [Google Scholar]
- 10.Faustino, N., and T. Cooper. 2005. Identification of putative new splicing targets for ETR-3 using its SELEX sequences. Mol. Cell. Biol. 25:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faustino, N., and T. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 12.Good, P., Q. Chen, S. Warner, and D. Herring. 2000. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem. 275:28583-28592. [DOI] [PubMed] [Google Scholar]
- 13.Görlich, D. 1998. Transport into and out of the cell nucleus. EMBO J. 17:2721-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley, B. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 15.Hartley, C., G. Taffet, A. Reddy, M. Entman, and L. Michael. 2002. Noninvasive cardiovascular phenotyping in mice. ILAR J. 43:147-158. [DOI] [PubMed] [Google Scholar]
- 15a.Ho, T. H., D. Bundman, D. Armstrong, and T. Cooper. 2005. Transgenic mice expressing CUG-BP1 reproduce splicing misregulation observed in myotonic dystrophy. Hum. Mol. Genet. 14:1539-1547. [DOI] [PubMed]
- 16.Consortium International Human Genome Sequencing. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J., J. Castle, P. Garrett-Engele, Z. Kan, P. Loerch, C. Armour, R. Santos, E. Schadt, R. Stoughton, and D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 18.Kan, Z., E. Rouchka, W. Gish, and D. States. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladd, A., N. Charlet-B., and T. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd, A., and T. Cooper. 2004. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J. Cell Sci. 117:3519-3529. [DOI] [PubMed] [Google Scholar]
- 21.Ladd, A., N. Nguyen, K. Malhotra, and T. Cooper. 2004. CELF6, a member of the CELF family of RNA binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J. Biol. Chem. 279:17756-17764. [DOI] [PubMed] [Google Scholar]
- 22.Ladd, A., M. Stenberg, M. Swanson, and T. Cooper. 13 April 2005. A dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 10.1002/dvdy.20382. [DOI] [PubMed]
- 23.Li, D., L. Bachinski, and R. Roberts. 2001. Genomic organization and isoform-specific tissue expression of human NAPOR (CUGBP2) as a candidate gene for familial arrhythmogenic right ventricular dysplasia. Genomics 74:396-401. [DOI] [PubMed] [Google Scholar]
- 24.Lichtner, P., T. Attié-Bitach, S. Schuffenhauer, J. Henwood, P. Bouvagnet, P. Scambler, T. Meitinger, and M. Vekemans. 2002. Expression and mutation analysis of Brunol3, a candidate gene for heart and thymus developmental defects associated with partial monosomy 10p. J. Mol. Med. 80:431-442. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman, A., D. Friedlich, G. Harmison, B. Howell, C. Jordon, S. Breedlove, and K. Fischbeck. 2001. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem. Biophys. Res. Commun. 282:499-506. [DOI] [PubMed] [Google Scholar]
- 26.Lopez, A. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 27.Lyons, G., S. Schiaffino, D. Sassoon, P. Barton, and M. Buckingham. 1990. Developmental regulation of myosin gene expression in mouse cardiac muscle. J. Cell Biol. 111:2427-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, D., C. Houchen, S. Kennedy, B. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11:113-126. [DOI] [PubMed] [Google Scholar]
- 29.Muller, A. J., J. F. Baker, J. B. DuHadaway, K. Ge, G. Farmer, P. S. Donover, R. Meade, C. Reid, R. Grzanna, A. H. Roach, N. Shah, A. Peralta Soler, and G. C. Prendergast. 2003. Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol. Cell. Biol. 23:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nigg, E. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 31.Oh, H., G. Taffet, K. Youker, M. Entman, P. Overbeek, L. Michael, and M. Schneider. 2001. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc. Natl. Acad. Sci. USA 98:10308-10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paillard, L., V. Legagneux, D. Maniey, and H. Osborne. 2002. c-Jun ARE targets mRNA deadenlyation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J. Biol. Chem. 277:3232-3235. [DOI] [PubMed] [Google Scholar]
- 33.Paillard, L., V. Legagneux, and H. Osborne. 2003. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell 95:107-113. [DOI] [PubMed] [Google Scholar]
- 34.Paillard, L., F. Omilli, V. Legagneux, T. Bassez, D. Maniey, and H. Osborne. 1998. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 17:278-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philips, A., L. Timchenko, and T. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737-741. [DOI] [PubMed] [Google Scholar]
- 36.Ry, S., M. Andreassi, A. Clerico, A. Biagini, and D. Giannessi. 2001. Endothelin-1, endothelin-1 receptors and cardiac natriuretic peptides in failing human heart. Life Sci. 68:2715-2730. [DOI] [PubMed] [Google Scholar]
- 37.Sano, M., S. Wang, M. Shirai, F. Scaglia, M. Xie, S. Sakai, T. Tanaka, P. Kulkarni, P. Barger, K. Youker, G. Taffet, Y. Hamamori, L. Michael, W. Craigen, and M. Schneider. 2004. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 23:3559-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savkur, R., A. Phillips, and T. Cooper. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29:40-47. [DOI] [PubMed] [Google Scholar]
- 39.Singh, G., N. Charlet-B., J. Han, and T. Cooper. 2004. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 32:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sironi, M., R. Cagliani, G. Comi, U. Pozzoli, A. Bardoni, R. Giorda, and N. Bresolin. 2003. Trans-acting factors may cause dystrophin splicing misregulation in BMD skeletal muscle. FEBS Lett. 537:30-34. [DOI] [PubMed] [Google Scholar]
- 41.Timchenko, N., P. Iakova, Z.-J. Cai, J. Smith, and L. Timchenko. 2001. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 21:6927-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wechsler-Reya, R., K. Elliot, and G. Prendergast. 1998. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell. Biol. 18:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, X., D. Yang, J.-H. Ding, W. Wang, P.-H. Chu, N. Dalton, H.-Y. Wang, J. Bermingham, Z. Ye, F. Liu, M. Rosenfeld, J. Manley, J. Ross, J. Chen, R.-P. Xiao, H. Cheng, and X.-D. Fu. 2005. ASF/SF2 regulated CaMKII∂ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59-72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.