Abstract
Glutamate-cysteine ligase catalytic subunit (GCLC) is regulated transcriptionally by Nrf1 and Nrf2. tert-Butylhydroquinone (TBH) induces human GCLC via Nrf2-mediated trans activation of the antioxidant-responsive element (ARE). Interestingly, TBH also induces rat GCLC, but the rat GCLC promoter lacks ARE. This study examined the role of Nrf1 and Nrf2 in the transcriptional regulation of rat GCLC. The baseline and TBH-mediated increase in GCLC mRNA levels and rat GCLC promoter activity were lower in Nrf1 and Nrf2 null (F1 and F2) fibroblasts than in wild-type cells. The basal protein and mRNA levels and nuclear binding activities of c-Jun, c-Fos, p50, and p65 were lower in F1 and F2 cells and exhibited a blunted response to TBH. Lower c-Jun and p65 expression also occurs in Nrf2 null livers. Levels of other AP-1 and NF-κB family members were either unaffected (i.e., JunB) or increased (i.e., Fra-1). Overexpression of Nrf1 and Nrf2 in respective cells restored the rat GCLC promoter activity and response to TBH but not if the AP-1 and NF-κB binding sites were mutated. Fra-1 overexpression lowered endogenous GCLC expression and rat GCLC promoter activity, while Fra-1 antisense had the opposite effects. In conclusion, Nrf1 and Nrf2 regulate rat GCLC promoter by modulating the expression of key AP-1 and NF-κB family members.
Glutathione (GSH) is the main nonprotein thiol in mammalian cells that participates in many critical cellular functions, including antioxidant defense and cell growth (14, 24, 28). The synthesis of GSH from its constituent amino acids involves two ATP-requiring enzymatic steps: the formation of γ-glutamylcysteine from glutamate and cysteine and the formation of GSH from γ-glutamylcysteine and glycine. The first step of GSH biosynthesis is rate limiting and catalyzed by glutamate-cysteine ligase (GCL, also known as γ-glutamylcysteine synthetase), while the second step is catalyzed by GSH synthetase (14). The GCL enzyme is composed of a catalytic (GCLC, Mr of ∼73,000) and a modifier (GCLM, Mr of ∼30,000) subunit which are encoded by different genes and dissociate under reducing conditions (7, 27, 35). The catalytic subunit exhibits all of the catalytic activity of the isolated enzyme as well as feedback inhibition by GSH (27). The modifier subunit is enzymatically inactive but plays an important regulatory function by lowering the Km of GCL for glutamate and raising the Ki for GSH (7, 8). GCL is a major determinant of the overall GSH synthesis capacity, and changes in GCL activity can result from regulation at multiple levels affecting only the catalytic or modifier subunit or both (14). Both human GCLC and GCLM promoters have been cloned (4, 5, 16, 18, 34). Antioxidant-response element (ARE, also known as electrophile response element, EpRE) and activator protein 1 (AP-1) are two cis-acting elements present in the promoter of both human GCL subunits that have been implicated in their transcriptional regulation by oxidants and β-naphthoflavone (5, 14, 16, 18).
Nrf1 and Nrf2, members of the cap ‘n’ collar-basic leucine zipper proteins (CNC-bZIP), are important in the transcriptional regulation of human and mouse GCL subunits and GSH levels. Nrf2 is known to bind and trans activate the ARE present in the human GCLC and GCLM promoters in response to β-naphthoflavone, pyrrolidine dithiocarbamate, and tert-butylhydroquinone (TBH) (4, 34). Nrf1 knockout mice die in utero, but fetal hepatocytes and embryonal fibroblasts from these animals have lower GSH levels and are more susceptible to oxidative stress than wild-type (WT) mice (3, 11). Mice lacking Nrf2 also exhibit lower GSH levels and are more susceptible to acetaminophen-induced liver injury than WT mice (2). Nrf1 and Nrf2 knockouts have lower GCLC expression (2, 3), and overexpression of Nrf1 and Nrf2 has been shown to induce the human GCLC promoter activity (19, 34). Interestingly, TBH also induces the expression of rat GCLC and the activity of the rat GCLC promoter even though the promoter lacks ARE (36, 37). Instead, AP-1 appears to be essential for TBH-mediated induction of the rat GCLC gene (36). The aim of this work was to determine whether Nrf1 and Nrf2 are important in the transcriptional regulation of rat GCLC, and if so, to determine the molecular mechanisms. These studies were carried out in Nrf1 and Nrf2 knockout (F1 and F2) and WT fibroblasts, and results were confirmed in Nrf2 knockout mouse livers. We found that Nrf1 and Nrf2 are important for the rat GCLC promoter activity despite the absence of ARE. The mechanism involves the dependence of key family members of AP-1 and NF-κB on Nrf1 and Nrf2 for their expression. This dependency has not been previously reported and may be an important determinant in the phenotype of the cells and mice that lack Nrf1 or Nrf2.
MATERIALS AND METHODS
Materials.
Cell culture media and fetal bovine serum were obtained from Gibco BRL Life Technologies (Grand Island, NY). The luciferase assay system was obtained from Promega (Madison, WI). All restriction enzymes were obtained from either Promega or Invitrogen (Carlsbad, CA). [32P]dCTP and [γ-32P]ATP (3,000 Ci/mmol) were purchased from New England Nuclear (DuPont, Boston, MA). A total RNA isolation kit was obtained from Invitrogen. All other reagents were of analytical grade and were obtained from commercial sources.
Nrf2 knockout mouse.
Nrf2 knockout mice were generated and maintained as described previously (1). Five-month-old knockout male mice and wild-type littermates were sacrificed, and livers were snap-frozen for RNA isolation as described below. Animals were treated humanely, and all procedures were in compliance with our institutions' guidelines for the use of laboratory animals.
Cell culture and TBH treatment.
Nrf1 and Nrf2 knockout (F1 and F2) and WT fibroblasts were grown in Dulbecco's minimal essential medium/F12 medium supplemented with 10% fetal bovine serum as previously described (1, 11). Prior to treatment with TBH, the amount of serum was reduced to 0.5% for 3 h. Cells were then treated with TBH (5 to 60 μM) for 1 to 32 h for various assays as described below.
Recombinant plasmids, transfection, and luciferase assays.
Rat GCLC promoter-luciferase constructs were previously described (36, 37). Nrf1 and Nrf2 expression plasmids were previously described (1, 11). pCI-Fra-1 and pCI-antisense Fra-1 expression vectors were kind gifts from D. Chalbos (23). Jun2-luciferase (Jun2-LUC) was kindly provided by Ze'ev Ronai (Ruttenberg Cancer Center, Mount Sinai School of Medicine). This contains one of the key AP-1 binding sites of the human c-Jun promoter and is known to be trans activated by c-Jun (31). Full-length human c-Jun promoter (−1780/+731) was kindly provided by W. Vedeckis (33). Mouse c-Fos promoter (−954 to +3, relative to translational start site) was obtained by cloning using forward primer 5′-CACGAATTTATGAATGAACCCAGTAC-3′ (−954 to −929) and reverse primer 5′-CATGGTCGAAGTTTGGGGAAAGC-3′ (−20 to +3) with the wild-type fibroblast genomic DNA as we described previously (36). Sequence was confirmed in both directions as described previously (36) and agreed with the sequence of GenBank accession number AF332140 and mouse genomic DNA sequence. The mouse c-Fos promoter fragment (−954 to +3) was cloned into the promoterless pGL-3 basic vector, creating the recombinant plasmid −954/+3 c-Fos-LUC. The NF-κB-driven luciferase construct (NF-κB-LUC, which contains five repeats of the consensus NF-κB binding sequence) and vector control were obtained from Stratagene (La Jolla, CA).
To study the transcriptional activity of the rat GCLC promoter, F1, F2, or WT cells (5 × 105 cells in 2 ml medium) were transiently transfected with 1.5 μg GCLC promoter luciferase construct −595/+2-LUC or promoterless pGL-3 enhancer vector (as a negative control) using the Superfect transfection reagent (QIAGEN, Valencia, CA) as we described previously (36). In some experiments, cells were cotransfected with Nrf1, Nrf2, Fra-1, or Fra-1 antisense expression vectors as previously described (1, 11, 23). To control for transfection efficiency, cells were cotransfected with the Renilla phRL-TK vector (Promega), which is a plasmid containing the Renilla luciferase gene driven by herpes simplex virus-thymidine kinase promoter. After 11 h, cells were harvested and lysed in 200 μl of reporter lysis buffer (Luciferase Assay System, Promega). Aliquots of the cell lysates were sequentially assessed for firefly and Renilla luciferase activities using a TD-20/20 Luminometer (Promega). The luciferase activity driven by the rat GCLC promoter construct was normalized to Renilla luciferase activity. Each experiment was done with triplicate samples.
The effect of TBH on rat GCLC promoter activity was examined by measuring luciferase activity driven by the rat GCLC promoter luciferase gene constructs in transfected F1, F2, or WT cells treated with TBH (60 μM) during the last 8 h of the transfection. To confirm the importance of NF-κB and AP-1 binding in mediating the effect of TBH on rat GCLC promoter activity, rat GCLC promoter construct −595/+2-LUC mutated in the NF-κB and/or AP-1 binding sites was generated using a GeneTailor site-directed mutagenesis system (Invitrogen). Rat GCLC promoter constructs mutated in either the putative NF-κB site (−378 to −369) (from 5′-GGGAACACCC-3′ to 5′-GGGAACTCTC-3′ [underlining shows nucleotide changes]), the AP-1 site (−356 to −346) (from 5′-CCTGACGGCCC-3′ to 5′-CCTAATGGCCC-3′), or both were subcloned into the pGL-3 promoter-luciferase vector (Promega). F1 and F2 cells transfected with either native or mutant rat GCLC promoter constructs were treated with TBH (60 μM) during the last 8 h of the transfection. In some experiments, cells were cotransfected with Nrf2, Fra-1, Fra-1 antisense expression vector, or empty vector for 12 h prior to treatment with TBH for another 8 h.
To evaluate the effect of Nrf2 overexpression on reporter activity driven by Jun2-LUC, NF-κB-LUC, c-Jun promoter, or c-Fos promoter, F2 cells were cotransfected with Nrf2 or empty expression vector and one of these constructs for 16 h. Reporter activity was measured as described above.
Northern blot analysis and real-time PCR.
Total RNA was extracted, and Northern hybridization analysis was performed using specific rat GCLC cDNA and c-Jun probes as described previously (36). Northern hybridization analysis was also performed using specific c-Fos, Fra-1, p50, and p65 cDNA probes designed according to their published sequences (6, 9, 20, 26). These cDNA probes were obtained by reverse transcription and PCR using a one-step reverse transcription-PCR kit (Clontech). To ensure equal loading of RNA samples and transfer in each of the lanes, prior to hybridization, membranes were rinsed with ethidium bromide and photographed, and the same membranes were also rehybridized with a 32P-labeled β-actin cDNA probe as described previously (36). Autoradiography and densitometry (Gel Documentation System, Scientific Technologies, Carlsbad, CA, and NIH Image 1.60 software program) were used to quantitate relative RNA. Results of Northern blot analysis were normalized to β-actin.
Synthesis of cDNAs from total RNA and real-time reverse transcription-PCR were done to assess the mRNA levels of c-Jun, p65, and GCLC in the livers of Nrf2 knockout mice and wild-type littermates as we described previously (13). GCLC primers were described previously (13). Sequences of the c-Jun and p65 primers were as follows: c-Jun forward, 5′CAGCAACTTTCCTGACCCAGAGGA-3′; c-Jun reverse, 5′-AGACTCCGCTAGCACTCACGTTGG-3′; p65 forward, 5′-CTCTGGGGCGGCACGTAC-3′; and p65 reverse, 5′-CATCCCACCTGTTCCCCTTGG-3′. Aliquots of cDNA were amplified in an Opticon PCR machine (MJ Research) using Dynamo HS (MJ Research) in duplicates in 20-μl reaction volumes. PCR cycling conditions consist of 95°C for 15 min and 45 cycles of 95°C for 30 s, 60°C for 30 s, and 68°C for 45 s. Expression levels were calculated relative to 18S rRNA levels as endogenous control. Relative expression was calculated as 2(CT), where CT is the cycle threshold.
Western blot analysis.
Total cell lysates were extracted from F1, F2, and WT cells after various treatments and subjected to Western blot analysis as described previously (15). Membranes were probed with antibodies directed against c-Jun, c-Fos, phospho-c-Jun, JunB, JunD, Fra-1, Fra-2, JAB1, p50, p65, RelB, and c-Rel (Santa Cruz Biotechnology, Santa Cruz, CA). To ensure equal loading, protein gels were stained with Coomassie blue and/or membranes were stripped and reprobed with antiactin antibodies (Santa Cruz Biotechnology). A horseradish peroxidase-conjugated secondary antibody was used. Blots were developed by enhanced chemiluminescence.
EMSA and supershift assay.
Electrophoretic mobility shift assays (EMSAs) for AP-1, NF-κB, and ARE were done as described previously (36, 38). Five to 15 μg of nuclear protein from WT, F1, or F2 cells treated with TBH (60 μM for 6 h) or vehicle control (dimethyl sulfoxide [DMSO]) were preincubated with 2 μg of poly(dI-dC) in a buffer containing 10 mM HEPES (pH 7.6), 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2, and 10% glycerol for 10 min on ice. 32P-end-labeled double-stranded DNA fragments containing consensus AP-1 (5′-CGCTTGATGACTCAGCCGGAA-3′) or NF-κB binding sites (5′-AGTTGAGGGGACTTTCCCAGGC-3′) or an ARE consensus site (5′-GTTTCTGCTTAGTCATTGTCTTC-3′ [underlining indicates the ARE binding site]) were then added with or without a 100-fold excess of unlabeled specific probe. Mixtures were incubated for 20 min on ice, loaded on a 4% nondenaturing polyacrylamide gel, and subjected to electrophoresis in 50 mM Tris, 45 mM borate, and 0.5 mM EDTA (pH 8.0). Gels were dried and subjected to autoradiography. Further confirmation of the identity of the binding proteins was done by antibody supershift assays for c-Jun, c-Fos, p50, p65, Nrf1, and Nrf2 (Biotechnology, Lake Placid, NY, or Santa Cruz Biotechnology) as we described previously (36, 38). To see whether Nrf1 or Nrf2 binds to the AP-1 and NF-κB sites of the rat GCLC promoter, EMSA with supershift was done using DNA fragment 5′-CCCCTGACGGCCCCGCCCACGAC-3′ for the AP-1 site (−358 to −336 of rat GCLC, with the AP-1 site underlined) and 5′-CCGGGAACACCCACGGCCTCAAC-3′ for the NF-κB site (−380 to −358 of the rat GCLC, with the NF-κB site underlined). The ARE consensus site was used as a positive control for Nrf1 and Nrf2 supershift analysis.
GSH levels.
GSH level was determined by the recycling method of Tietze (30).
ChIP assay.
To see whether c-Jun, c-Fos, Fra-1, and Nrf2 bind to the AP-1 site of the rat GCLC promoter in an endogenous chromatin configuration, a chromatin immunoprecipitation (ChIP) assay was carried out following the ChIP assay kit protocol provided by Upstate (Waltham, MA). H4IIE cells (for the rat GCLC AP-1 site) or wild-type fibroblasts (for binding to ARE) were treated with TBH (60 μM) or vehicle control (DMSO) for 6 h and processed for ChIP assay as per protocol except for minor modifications. Briefly, proteins were cross-linked to DNA by treating cells with 1% formaldehyde at 37°C for 10 min. After fixation, cells were washed with ice-cold phosphate-buffered saline, resuspended, lysed in sodium dodecyl sulfate lysis buffer (Upstate), and centrifuged for 5 min at 1,000 rpm. Cell lysates were sonicated at 25 to 30% power with five 10-s pulses using a Sonic Dimembrator model F60 (Fisher Scientific, Pittsburgh, PA) to fragment the chromatin to about 1 kb or less. The sonicated cell lysates were spun in a microcentrifuge at 13,000 × g for 10 min at 4°C. The supernatant containing the soluble chromatin sample was treated with 1 μl of 5 M NaCl and heated at 65°C for 4 h to reverse the protein-DNA cross-links. Twenty microliters of the reversed soluble chromatin sample was removed and used as input control (total chromatin fraction) for final PCR. The remaining chromatin solution was split into equal fractions and subjected to immunoprecipitation in the presence or absence of specific antibodies. Antibodies used for immunoprecipitation were anti-c-Jun, c-Fos, Fra-1, and Nrf2 antibodies (Santa Cruz Biotechnology). For PCR of the rat GCLC promoter region across the AP-1 site, an equal amount of template DNA from the antibody-treated, no-antibody, and input samples was amplified using forward primer 5′-CCAGTATTCTCTTGGGAACCAAG-3′ (bp −437 to −413 relative to the ATG start codon) and reverse primer 5′-CACGGGCTTCCTACTTGCGAC-3′ (bp −234 to −213 relative to the ATG start codon). Input DNA was PCR amplified for 20 to 35 cycles to determine the linear range. PCR of the mouse GCLM promoter region across the ARE site (CTGCTTAGTCA, −340 to −328 bp relative to the ATG start codon) was done using the forward primer 5′-AACGGTTACGAAGCACTTTCTCGG-3′ (bp −454 to −433 relative to the ATG start codon) and the reverse primer 5′-ACTCCGCGCGGCCACAGCCCGGTG-3′ (bp −170 to −146 relative to the ATG start codon). All PCR products were run on 8% acrylamide gels and stained with ethidium bromide for 15 to 30 min.
Statistical analysis.
Data are given as the mean ± the standard error of the mean (SEM). Statistical analysis was performed using analysis of variance (ANOVA) followed by Fisher's test for multiple comparisons. For changes in mRNA levels, ratios of individual genes to β-actin densitometric values were compared; for changes in protein levels, ratios of individual proteins to actin densitometric values were compared. Significance was defined by a P value of <0.05.
RESULTS
GCL expression at baseline and in response to TBH.
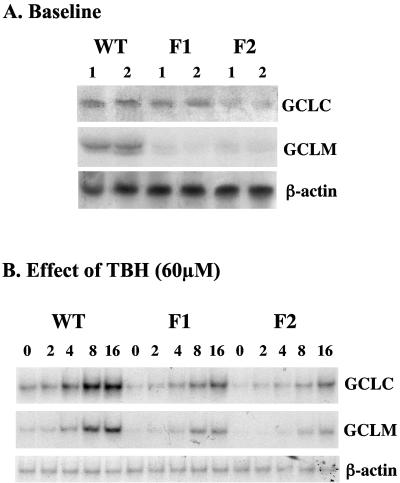
We first compared the steady-state mRNA levels of GCLC and GCLM at baseline and in response to TBH treatment in WT, F1, and F2 cells. At baseline, the GCLC mRNA levels are 40% and 81% lower in F1 and F2 cells, respectively, than in WT cells (F1 = 60% ± 10% and F2 = 19% ± 4% of WT, P < 0.05 versus WT) (Fig. 1A). Similarly, the GCLM mRNA levels are 70% and 86% lower in F1 and F2 cells, respectively, than in WT cells (F1 = 30% ± 10% and F2 = 14% ± 2% of WT, P < 0.05 versus WT) (Fig. 1A). TBH induces maximal GCLC and GCLM expression at 60 μM (data not shown) and in a time-dependent fashion (Fig. 1B). The GCLC mRNA level increased by 1.6- and 2.7-fold after 8 and 16 h, respectively, of TBH treatment in WT cells. Although both GCLC and GCLM mRNA levels increased with time in the F1 and F2 cells after TBH treatment, the response was significantly blunted compared to WT cells (Fig. 1B).
FIG. 1.
GCL subunit expression at baseline and in response to TBH in WT, F1, and F2 cells. RNA (25 μg/lane) samples from WT, F1, and F2 cells at baseline (A) or treated with 60 μM TBH for 0 to 16 h (B) were analyzed by Northern blot analysis with a 32P-labeled GCLC cDNA probe as described in Materials and Methods. The same membranes were then sequentially rehybridized with 32P-labeled GCLM and β-actin cDNA probes.
Rat GCLC promoter activity at baseline and in response to TBH.
We next compared the rat GCLC promoter activity in these three types of cells. The rat GCLC promoter construct −595/+2-LUC was chosen for these studies because it contains the maximum promoter activity of the cloned rat GCLC promoter (37). Furthermore, we had previously shown that acetaldehyde and TBH induced this promoter construct which contains both AP-1 and NF-κB sites (36, 37). Figure 2 shows that the rat GCLC promoter activity in WT cells is nearly doubled in response to TBH treatment. In comparison, the basal rat GCLC promoter activity is 30% and 60% lower in F1 and F2 cells, respectively. Furthermore, TBH treatment induced the GCLC promoter activity only 30 to 40% (Fig. 2).
FIG. 2.
Effect of TBH on rat GCLC promoter activity in WT, F1, and F2 cells. WT, F1, and F2 cells were transiently transfected with the rat GCLC promoter-luciferase construct −595/+2-LUC (labeled GCLC-LUC) or pGL-3 enhancer vector and treated with TBH (60 μM for 8 h) or vehicle control (DMSO) as described in Materials and Methods. Results represent the mean ± SEM from three independent experiments performed in triplicate. Data are expressed as relative luciferase activity to that of pGL-3 enhancer vector control in WT cells, which is assigned a value of 1.0. *, P < 0.05 versus WT GCLC-LUC; †, P < 0.05 versus respective control and WT GCLC-LUC plus TBH (ANOVA followed by Fisher's test).
Role of Nrf1 and Nrf2 in the expression of AP-1 and NF-κB family members.
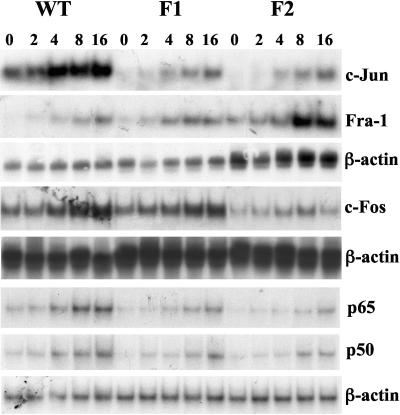
We have previously shown that both the AP-1 and NF-κB sites of the rat GCLC promoter may be involved in mediating the induction of acetaldehyde on GCLC promoter (37) and that the basal expression of rat GCLC as well as induction by TBH require c-Jun (36). This prompted us to examine whether Nrf1 and Nrf2 may influence the expression and/or DNA binding and trans-activating activities of AP-1 and NF-κB. Figure 3 shows that the steady-state protein levels of many of the AP-1 family members are lower in F1 and F2 cells than in WT cells. The most affected are c-Jun and c-Fos levels, which are 70% to 85% lower in F1 and F2 cells than in WT cells. However, JunB and JunD levels are essentially unchanged, while Fra-1 and JAB1 levels are markedly increased, especially in F2 cells (Fig. 3). Lower c-Jun levels resulted in lower levels of phosphorylated c-Jun in both F1 and F2 cells (Fig. 3). Figure 4 shows that NF-κB family members are similarly affected, with p50 and p65 levels markedly lower in F1 and F2 cells than in WT cells, whereas c-Rel levels are increased, especially in F2 cells.
FIG. 3.
Steady-state protein levels of the AP-1 family members in WT, F1, and F2 cells. Total cell lysates (40 μg/lane) from WT, F1, and F2 cells were subjected to Western blot analysis using anti-c-Fos, c-Jun, phospho-c-Jun (p-c-Jun), JunB, JunD, Fra-1, Fra-2, and JAB1 antibodies as described in Materials and Methods. The same membranes were stripped and probed with antibodies against actin to ensure equal protein loading. The right panels show densitometric changes expressed as percentages of WT. *, P < 0.05 versus WT. Representative blots are shown.
FIG. 4.
Steady-state protein levels of the NF-κB family members in WT, F1, and F2 cells. Total cell lysates (40 μg/lane) from WT, F1, and F2 cells were subjected to Western blot analysis using anti-p50, p65, RelB, and c-Rel antibodies as described in Materials and Methods. The same membranes were stripped and probed with antibodies against actin to ensure equal protein loading. The right panels show densitometric changes expressed as percentages of WT. *, P < 0.05 versus WT. Representative blots are shown.
Effect of TBH on mRNA levels and nuclear binding activities of AP-1 and NF-κB family members.
We next examined the effect of TBH on the mRNA levels of some of the AP-1 and NF-κB family members. Baseline mRNA levels of c-Jun, c-Fos, p50, and p65 are all lower in F1 and F2 cells than in WT cells (Fig. 5). In contrast, baseline Fra-1 mRNA levels are much higher in F1 and especially F2 cells. In WT cells, TBH treatment led to a time-dependent induction of c-Jun, c-Fos, Fra-1, p50, and p65. This effect on c-Jun, c-Fos, p50, and p65 is significantly blunted in F1 and F2 cells (Fig. 5). Interestingly, the effect on Fra-1 is significantly enhanced, especially in F2 cells (Fig. 5).
FIG. 5.
Effect of TBH on c-Jun, Fra-1, c-Fos, p65, and p50 mRNA levels in WT, F1, and F2 cells. RNA (25 μg/lane) samples from WT, F1, and F2 cells treated with 60 μM TBH for 0 to 16 h were analyzed by Northern blot analysis with 32P-labeled c-Jun, c-Fos, or p65 cDNA probes as described in Materials and Methods. The same membranes were then sequentially rehybridized with 32P-labeled Fra-1 and β-actin cDNA probes, β-actin cDNA probe, or p50 and β-actin cDNA probes, respectively.
EMSA with supershift was next performed to determine if there is less AP-1 and NF-κB nuclear binding activity in F1 and F2 cells. Figure 6 shows that baseline AP-1 nuclear binding activity is lower in F1 and F2 cells and the response to TBH treatment is significantly blunted compared to WT cells. Supershift confirmed decreased c-Jun and c-Fos binding in F1 and F2 cells. Similarly, baseline NF-κB nuclear binding activity is also lower in F1 and F2 cells than in WT cells, and the response to TBH is also blunted. Supershift also confirmed decreased p50 and p65 binding (Fig. 7).
FIG. 6.
Effect of TBH on electrophoretic mobility shift and supershift assays for AP-1 binding. Nuclear protein extracts (10 μg) were obtained from WT, F1, and F2 cells treated with TBH (60 μM for 0, 4, or 8 h), and EMSA was done as described in Materials and Methods using a consensus AP-1 probe. Panel A shows supershift analysis using anti-c-Jun antibodies, and panel B shows supershift analysis using anti-c-Fos antibodies. The arrows to the right point to complexes that were supershifted in the presence of specific antibodies. Representative EMSAs are shown.
FIG. 7.
Effect of TBH on electrophoretic mobility shift and supershift assays for NF-κB binding. Nuclear protein extracts (10 μg) were obtained from WT, F1, and F2 cells treated with TBH (60 μM for 0, 4, or 8 h), and EMSA was done as described in Materials and Methods using a consensus NF-κB probe. Panel A shows supershift analysis using anti-p65 antibodies, and panel B shows supershift analysis using anti-p50 antibodies. The arrows to the right point to complexes that were supershifted in the presence of specific antibodies.
AP-1 and NF-κB expression in the livers of Nrf2 knockout mice.
To confirm that Nrf2 modulates the expression of AP-1 and NF-κB family members in vivo, we measured the mRNA levels of c-Jun and p65 in the livers of Nrf2 knockout mice and wild-type littermates. Figure 8 shows that, consistent with our findings in the F2 fibroblasts, the mRNA levels of c-Jun and p65 are more than 80% lower in the knockout mouse livers. These animals also have lower expression of GCLC, as previously described (2, 13).
FIG. 8.
Real-time PCR analysis of hepatic c-Jun, p65, and GCLC mRNA levels in Nrf2 knockout mice and wild-type littermates. RNA was extracted from the livers of wild-type and Nrf2 knockout mice and subjected to real-time PCR as described in Materials and Methods. Results represent means ± standard deviation from three animals each relative to wild-type mice. Expression levels were calculated relative to 18S rRNA levels as endogenous control. *, P < 0.005 versus wild type.
Overexpression of Nrf2 in F2 cells.
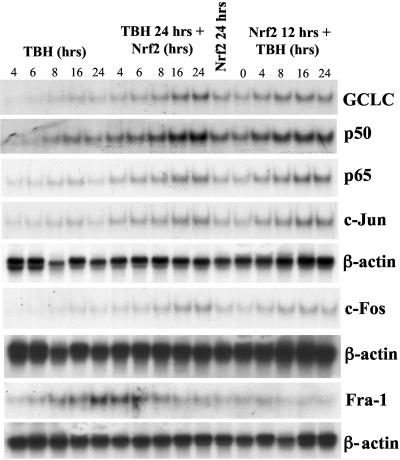
The effect of overexpression of Nrf2 in F2 cells was next examined. Figure 9 shows that compared to untransfected F2 cells, Nrf2 overexpression alone induced GCLC, p50, p65, c-Jun, and c-Fos expression (Fig. 9, Nrf2 24 hrs and Nrf2 12 hrs + TBH 0 hrs columns). TBH treatment further increased the mRNA levels of these genes. In contrast, Nrf2 overexpression led to a time-dependent suppression of Fra-1 expression (Fig. 9, middle panels of TBH 24 hrs + Nrf2 column).
FIG. 9.
Effect of Nrf2 overexpression and TBH treatment on the steady-state mRNA levels of GCLC, p50, p65, c-Jun, c-Fos, and Fra-1 in F2 cells. F2 cells were treated with either 60 μM TBH for 4 to 24 h (lanes 1 to 5 from the left), 60 μM TBH for 24 h plus Nrf2 expression vector during the last 4 to 24 h of the TBH treatment (lanes 6 to 10 from the left), Nrf2 expression vector alone for 24 h (lane 11 from the left), or Nrf2 expression vector for 12 h followed by 60 μM TBH for 0 to 24 h (lanes 12 to 16 from the left). RNA (25 μg/lane) samples following these treatments were analyzed by Northern blot analysis with 32P-labeled GCLC, p50, p65, c-Jun, c-Fos, and Fra-1 cDNA probes as described in Materials and Methods. β-Actin was used for housekeeping control.
Nrf2 overexpression alone also induced the rat GCLC promoter activity and, in conjunction with TBH treatment, resulted in a synergistic induction of the rat GCLC promoter activity (Fig. 10A). However, this induction is blocked significantly if the AP-1 site or the NF-κB site is mutated and completely prevented if both sites are mutated (Fig. 10A).
FIG. 10.
Effect of Nrf2/Nrf1 overexpression and/or TBH treatment on rat GCLC, human c-Jun, and mouse c-Fos promoter activities and c-Jun and NF-κB-dependent reporter activities. (A) F2 cells were transiently transfected with the rat native or mutant GCLC promoter-luciferase construct −595/+2-LUC (GCLC-LUC) or pGL-3 enhancer vector and treated with TBH (60 μM for 8 h) plus empty vector, Nrf2 expression vector, or TBH plus Nrf2 expression vector as described in Materials and Methods. To assess the importance of the AP-1 and NF-κB binding sites, these sites were mutated by two bases as described in Materials and Methods. The effect of Nrf2 overexpression and TBH was examined in the mutant constructs containing only the mutated AP-1 site (AP-1mut), only the mutated NF-κB site (NFκBmut), or both. Results represent means ± SEM from four independent experiments performed in triplicates. Data are expressed as luciferase activity relative to that of pGL-3 enhancer vector, which is assigned a value of 1.0. *, P < 0.05 versus native GCLC-LUC construct; **, P < 0.05 versus native GCLC-LUC construct and treatment with either TBH or Nrf2 expression vector; †, P < 0.05 versus native construct treated with Nrf2 expression vector and TBH (ANOVA followed by Fisher's test). (B) F2 cells were cotransfected with c-Jun or c-Fos promoter constructs (c-Jun-LUC or c-Fos-LUC), c-Jun- or NF-κB-dependent constructs (Jun2-LUC or NFκB-LUC), and Nrf2 expression vector or empty vector control as described in Materials and Methods. Results represent means ± SEM from three to four independent experiments performed in triplicates. Data are expressed relative to activities without Nrf2 (empty vector control). *, P < 0.05 versus without Nrf2. (C) F1 cells were transiently transfected with the rat native or mutant GCLC promoter-luciferase construct −595/+2-LUC (GCLC-LUC) or pGL-3 enhancer vector and treated with empty vector or Nrf1 expression vector as described in Materials and Methods. Results represent means ± SEM from four independent experiments performed in triplicates. Data are expressed as luciferase activity relative to that of pGL-3 enhancer vector, which is assigned a value of 1.0. *, P < 0.05 versus native GCLC-LUC construct; †, P < 0.05 versus native construct and native construct treated with Nrf1 expression vector (ANOVA followed by Fisher's test).
Nrf2 overexpression in F2 cells induced the reporter activities driven by c-Jun and c-Fos promoters as well as c-Jun- and NF-κB-dependent constructs Jun2-LUC and NF-κB-LUC, respectively (Fig. 10B), which further supports the notion that the effect of Nrf2 on the rat GCLC promoter is exerted at the level of AP-1 and NF-κB.
Overexpression of Nrf1 in F1 cells.
Similar to the effect of Nrf2 overexpression, Nrf1 overexpression in F1 cells also induced the rat GCLC promoter activity, but the effect is blocked completely if the AP-1 site or the NF-κB site is mutated (Fig. 10C). In fact, the GCLC promoter activity is lower when either site is mutated.
Nrf1 and Nrf2 binding to AP-1 and NF-κB sites of rat GCLC promoter.
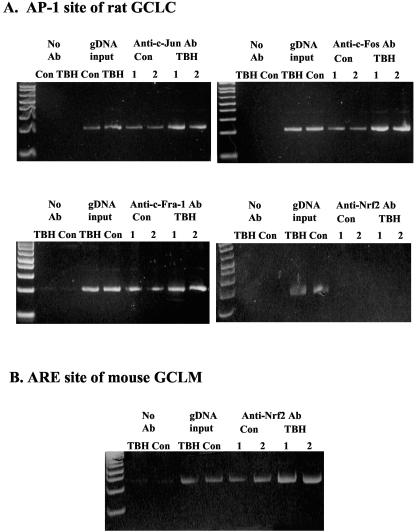
To see whether Nrf1 and Nrf2 might bind to the AP-1 and NF-κB sites of the rat GCLC promoter, EMSA with supershift analysis was carried out using DNA fragments containing either the AP-1 or the NF-κB site. Figure 11 shows that Nrf1 and Nrf2 bind to the consensus ARE site but not the AP-1 or NF-κB site, while strong binding occurred with c-Jun and p50, as demonstrated by the supershift analysis.
FIG. 11.
EMSA and supershift analysis of the rat GCLC AP-1 and NF-κB sites. WT cells were treated with TBH (60 μM for 8 h) or vehicle control and subjected to EMSA with supershift analysis for the AP-1 site at −356 or the NF-κB site at −378. Supershift analysis was performed using antibodies directed against c-Jun, Nrf1, and Nrf2 for the AP-1 site (A) and p50, Nrf1, and Nrf2 for the NF-κB site (B). Note that supershift occurred only with anti-c-Jun antibodies for the AP-1 site and anti-p50 antibodies for the NF-κB site. As a positive control, TBH treatment induced Nrf1 and Nrf2 binding to the ARE site of the mouse GCLM (C). Arrows in panel C point to the Nrf1 and Nrf2 supershifts.
Role of Fra-1 in GCLC expression and rat GCLC promoter activity.
We next examined whether Fra-1 induction in F2 cells might play a role in the reduced expression of GCLC and rat GCLC promoter activity. Figure 12A shows that overexpression of Fra-1 in F2 cells lowered the endogenous GCLC mRNA levels (70% of control by densitometry), while overexpression with Fra-1 antisense increased the endogenous GCLC mRNA levels (196% of control by densitometry).
FIG. 12.
Effect of Fra-1 or Fra-1 antisense overexpression on the endogenous GCLC expression (A) or rat GCLC promoter activity (B). (A) F2 cells were treated with Fra-1 or Fra-1 antisense expression vector for 12 h, and RNA (25 μg/lane) samples following these treatments were analyzed by Northern blot analysis with a 32P-labeled GCLC cDNA probe as described in Materials and Methods. β-Actin was used for housekeeping control. (B) F2 cells were transiently transfected with the rat GCLC promoter-luciferase construct −595/+2-LUC (GCLC-LUC) and treated with TBH (60 μM for 8 h) plus empty vector, Nrf2 expression vector, Fra-1 expression vector, Fra-1 antisense expression vector (Fra-1 AS), or a combination of Nrf2 and TBH, Fra-1 and TBH, or Fra-1 AS and TBH as described in Materials and Methods. Results represent means ± SEM from six independent experiments performed in triplicates. Data are expressed as luciferase activity relative to that of pGL-3 enhancer vector, which is assigned a value of 1.0. *, P < 0.01 versus GCLC-LUC construct; †, P < 0.05 versus GCLC-LUC construct; **, P < 0.005 versus GCLC-LUC construct and treatment with either TBH, Nrf2, or Fra-1 AS expression vectors (ANOVA followed by Fisher's test).
Similar to their effect on the endogenous GCLC gene, Fra-1 overexpression also reduced the rat GCLC promoter activity, while Fra-1 antisense increased the promoter activity. Fra-1 overexpression completely prevented the ability of TBH to induce the rat GCLC promoter, while Fra-1 antisense greatly enhanced the ability of TBH to induce the promoter. In fact, Fra-1 antisense treatment resulted in a level of induction similar to that resulting from Nrf2 overexpression, either alone or in combination with TBH.
Influence of Nrf1, Nrf2, TBH, and Fra-1 on GSH levels.
Table 1 summarizes the GSH levels in WT, F1, and F2 cells treated with TBH, Nrf1, Nrf2, Fra-1, or Fra-1 antisense overexpression vectors. Consistent with previous reports (1, 19), F1 and F2 cells have lower GSH levels than WT cells, which can be restored by Nrf1 and Nrf2 overexpression, respectively. TBH treatment increased the GSH levels in all three cell types, with the largest increase in WT cells. Fra-1 overexpression decreased and Fra-1 antisense increased GSH levels in all three cell types.
TABLE 1.
Effect of TBH, Nrf1, Nrf2, Fra-1, and Fra-1 antisense treatments on cell GSH levels
| Treatment | Cell GSH level, nmol/mg proteina
|
||
|---|---|---|---|
| WT | F1 | F2 | |
| Control | 46.9 ± 2.7 | 37.9 ± 0.8† | 25.3 ± 1.0† |
| TBH | 87.1 ± 1.5* | 67.7 ± 1.7** | 41.7 ± 1.6** |
| Fra-1 | 36.3 ± 1.2** | 32.3 ± 1.3* | 16.8 ± 0.5** |
| Fra-1 antisense | 80.7 ± 6.2** | 53.4 ± 3.9* | 44.6 ± 5.1* |
| Nrf1 | 60.9 ± 2.6** | ||
| Nrf2 | 47.1 ± 7.6* | ||
Results represent the mean ± SE from three experiments done in triplicate. WT, F1, and F2 cells were treated with TBH (60 μM for 8 h), Fra-1, Fra-1 antisense, Nrf1, or Nrf2 expression vectors for 12 h as described in Materials and Methods. *, P < 0.05 versus controls; **, P < 0.01 versus controls; †, P < 0.05 versus WT control by ANOVA followed by Fisher's test.
Protein binding to the endogenous rat GCLC AP-1 site.
ChIP assay was used to further evaluate protein binding to the rat GCLC AP-1 site in the endogenous chromatin configuration. Figure 13 shows that following TBH treatment, there is increased c-Jun, c-Fos, and Fra-1 binding to the rat GCLC AP-1 site. Nrf2 does not bind to the rat GCLC AP-1 site, but TBH treatment induced Nrf2 binding to the ARE site of mouse GCLM.
FIG. 13.
Effect of TBH treatment on protein binding to the rat GCLC AP-1 site in vivo. ChIP assay was used to assess transcription factor binding to the AP-1 site of rat GCLC in an endogenous chromatin configuration as described in Materials and Methods. Note that TBH treatment increased c-Jun, c-Fos, and Fra-1 binding to the rat GCLC AP-1 site. Nrf2 does not bind to this site, but increased binding to the ARE site of mouse GCLM can be seen following TBH treatment.
DISCUSSION
CNC-bZIP proteins are a subfamily of bZIP transcription factors characterized by the presence of a 45-amino-acid homology region known as the CNC domain (3, 21). There are six known CNC-bZIP family members in mice: p45NFE2, Nrf1, Nrf2, Nrf3, Bach1, and Bach2 (3). Bach2 and p45NFE2 have tissue-specific expression, whereas Nrf1 and Nrf2 are ubiquitously expressed (3, 21). CNC-bZIP factors form heterodimers with other bZIP proteins and regulate transcription via an extended AP-1-like site called the NFE2 site (TGCTGACTCAT) (3, 17), which resembles the consensus ARE element [5′-(A/G)T(G/C)A(C/T)NNNGC(A/G)-3′] (4). Indeed, both Nrf1 and in particular Nrf2 have been shown to trans activate ARE present in the promoter of genes encoding enzymes involved in phase II detoxification and antioxidant defense (2, 3, 4, 11, 19, 32, 34).
GSH is the most abundant nonprotein thiol that is important in numerous cellular processes, including antioxidant defense, storage of cysteine, maintenance of the redox state, and proliferation (14). As the rate-limiting enzyme in GSH synthesis, GCL activity is a major determinant of the GSH synthesis capacity. Because of its importance, regulation of GCL has been a topic of extensive research (reviewed in reference 14). Many of the treatments and conditions known to affect GCL activity exert their effect at the transcriptional level, affecting both GCLC and GCLM in a coordinated manner (14). In particular, critical ARE elements have been identified in both of the human GCLC and GCLM promoters that mediate the coordinated induction by β-naphthoflavone, pyrrolidine dithiocarbamate, and TBH (4, 34). Nrf2 has been identified as the key transcription factor, possibly in complexes with other Jun or Maf proteins, that trans activates the human GCLC and GCLM promoters via binding to ARE in response to these treatments (4, 34). The importance of Nrf2 in GCL expression has been further demonstrated using Nrf2 knockout mice, which exhibit increased susceptibility to acetaminophen-induced liver injury attributed to decreased GCL expression and GSH levels (2). Nrf2 overexpression increased human GCLC and GCLM promoter activity (10, 34) and restored GCL subunit expression and GSH levels in Nrf2 knockout fibroblasts (1). A similar role for Nrf1 has also been demonstrated, as fetal hepatocytes from Nrf1 knockout mice have lower GCLC expression (3) and overexpression of Nrf1 also induced the human GCLC promoter activity via ARE (10, 19). ARE elements have also been reported in the mouse GCLC and GCLM promoters (12).
The rat GCL subunits are regulated similarly to the human GCL subunits (14). TBH induces the expression of both GCL subunits in human and rat (34, 36). Interestingly, the 1.8-kb 5′-flanking region of the rat GCLC does not contain any consensus ARE element, but the reporter activity driven by a recombinant rat GCLC-luciferase construct is induced by TBH treatment (36). This prompted us to examine whether Nrf2 and Nrf1 are important in the transcriptional regulation of the rat GCLC gene as they are in the human and mouse genes.
Using embryonal fibroblasts from Nrf1 and Nrf2 knockout mice, we first established that the endogenous GCLC and GCLM expression is lower than in wild-type fibroblasts, as in previous reports (2, 3). TBH exerted a time-dependent induction in the expression of both genes, but the response is markedly diminished in the F1 and F2 cells, with the F2 cells more significantly impaired than F1 cells. Comparable to the response of the endogenous genes, basal rat GCLC promoter activity is lower in F1 and F2 cells than WT cells, with F2 cells exhibiting the lowest activity. TBH treatment increased the rat GCLC promoter activity in all three cell types, but while it doubled the GCLC promoter activity in WT cells, it increased the GCLC activity in F1 and F2 cells only by about 30%. We had previously shown that the rat GCLC promoter is induced by treatment with acetaldehyde and TBH (36, 37). Consensus NF-κB and AP-1 binding sites are present in the rat GCLC −595/+2 promoter fragment (37). Increased nuclear binding to both sites occurred after treatment with acetaldehyde (37), while TBH-mediated increased GCLC promoter activity was blocked by treatment with dominant negative c-Jun (36). These findings led us to investigate whether Nrf1 and Nrf2 might influence the expression and trans-activating activity of AP-1 and NF-κB.
To our surprise, we found dramatic changes in the steady-state protein levels of several AP-1 and NF-κB family members in fibroblasts lacking either Nrf1 or Nrf2. Changes in the protein levels are not uniform within each family. Among AP-1 family members, c-Fos and c-Jun levels are reduced by 70 to 85%, JunB protein levels are essentially unchanged, JunD levels are unchanged in F1 cells but slightly reduced in F2 cells, Fra-2 levels are slightly increased in F1 cells, and Fra-1 and JAB1 levels are markedly induced, especially in F2 cells. Among the NF-κB family members, p50 and p65 are reduced by 60 to 70%, RelB levels are less affected, and c-Rel levels are actually induced. The molecular mechanism for changes in the protein levels of c-Jun, c-Fos, Fra-1, p50, and p65 is at the pretranslational level, as the mRNA levels of these genes are lower (or higher in the case of Fra-1) in the F1 and F2 cells than in WT cells. To make sure our findings in the fibroblasts are relevant in vivo, we also measured the mRNA levels of c-Jun and p65 in the livers of Nrf2 knockout and wild-type mice. Both c-Jun and p65 mRNA levels are significantly decreased in the knockout livers, supporting an important role of Nrf2 in the expression of AP-1 and NF-κB family members.
TBH treatment resulted in a time-dependent induction of c-Jun, c-Fos, p50, and p65, with the highest induction seen in WT cells and the most severely blunted induction seen in F2 cells. These changes also translated to nuclear binding activity, as TBH-mediated induction of increased AP-1 and NF-κB nuclear binding is significantly blunted in both F1 and F2 cells, with the F2 cells exhibiting the lowest nuclear binding activity of the three. However, the opposite effect was observed with Fra-1, as TBH-mediated Fra-1 induction is most prominent in F2 cells. Using EMSA and supershift, we found that Nrf2 does not bind to either of these sites. ChIP assay confirmed that TBH treatment induced binding of c-Jun, c-Fos, and Fra-1, but not Nrf2, to the rat GCLC AP-1 site.
Transfection of F2 cells with Nrf2 expression vector increased the basal expression of GCLC, p50, p65, c-Jun, and c-Fos, decreased the basal expression of Fra-1, and restored the ability of the cell to respond to TBH. This further demonstrates that changes in the expression of these genes is causally related to lack of Nrf2. Nrf2 overexpression also induced the rat GCLC promoter activity and together with TBH exerted a synergistic effect on the rat GCLC promoter activity. This is comparable to the synergistic induction of GCLC, p50, p65, c-Jun, and c-Fos mRNA levels in cells treated with both Nrf2 expression vector and TBH. The mechanism(s) of Nrf2- and TBH-mediated increase in rat GCLC promoter activity requires intact AP-1 and NF-κB binding sites, as mutation of either site significantly blocked the induction and mutation of both sites completely prevented the increase. Similarly, transfection of F1 cells with Nrf1 also increased the basal rat GCLC promoter activity, although to a lesser extent than overexpression of Nrf2 in F2 cells. This effect is also blocked by mutation of either the AP-1 site or the NF-κB site. The fact that mutation of either the AP-1 or NF-κB site was able to exert such a significant inhibitory effect also suggests that AP-1 and NF-κB may cooperate to achieve a full TBH-mediated induction. These results also show that Nrf2 and Nrf1 regulate the rat GCLC promoter activity, albeit via an indirect mechanism. The most plausible mechanism involves the ability of Nrf2 and Nrf1 to modulate the expression of key AP-1 and NF-κB family members so that in their absence, reduced expression of these proteins resulted in reduced rat GCLC promoter activity, which is corrected when Nrf2 or Nrf1 is overexpressed. This is also consistent with our previous finding that basal rat GCLC expression is significantly diminished by treatment with dominant negative c-Jun (36). However, our present study employed mouse fibroblasts to study a rat promoter construct. Whether Nrf1 and Nrf2 also regulate AP-1 and NF-κB family members in a similar fashion in rat cells remains to be explored.
Since Fra-1 expression is markedly induced in F2 cells, we also examined the role that this induction may play in the reduced expression of GCLC. Fra-1 belongs to the Fos family proteins, and although they are all capable of forming heterodimers with the Jun family members, the contribution of individual family members to transcriptional activation may be different (29). Fra-1 lacks the C-terminal transactivation domain present in c-Fos, and it has been shown to function as a negative regulator of AP-1 activity in certain cell types (23, 29). We found this to be true also for GCLC in fibroblasts, as overexpression of Fra-1 further reduced the endogenous GCLC expression and the rat GCLC promoter activity, while the opposite was true for treatment with Fra-1 antisense. Interestingly, Fra-1 antisense treatment and Nrf2 expression vector resulted in comparable levels of GCLC induction, suggesting increased Fra-1 expression in the F2 cells contributes significantly to the reduced GCLC expression and rat GCLC promoter activity in these cells.
Cell GSH levels correlated closely with GCLC expression. Consistent with previous reports, F1 and F2 cells have reduced cell GSH levels and overexpression with Nrf1 or Nrf2 restored the levels (1, 19). TBH treatment increased GSH levels in all three cells, with the effect most prominent in WT cells. Consistent with the role of a negative regulator of GCLC, Fra-1 overexpression reduced while Fra-1 antisense increased cell GSH levels.
The ability of Nrf1 and Nrf2 to modulate the expression of AP-1 and NF-κB family members has not been previously reported. The question is how the absence of Nrf1 or Nrf2 leads to reduced levels of some and increased levels of other family members. Much more additional work will be required to address this. One possibility is that Nrf1 and Nrf2 trans activate the promoter of c-Jun, c-Fos, p50, and p65 but act as repressors for Fra-1, JAB1, and c-Rel. Nrf2 sites are present in the promoter region of mouse c-Jun (accession number U60582) and c-Fos (accession number AF332140) genes, making this possibility an attractive hypothesis. Indeed, Nrf2 overexpression induced the promoter activity of c-Jun and c-Fos and reporter activity driven by c-Jun and NF-κB-dependent constructs. The exact molecular mechanism remains to be defined. It should be noted that there are published reports examining differential gene expression in Nrf2-deficient tissues, and many of the Nrf2-dependent genes do not have any potential ARE sequence (12, 25). Interestingly, AP-4 is listed as one of the Nrf2-dependent genes (12), and one of the genes highly induced by TBH in an Nrf2-dependent manner (9.4-fold), proliferin, lacks an ARE and has been shown to be induced by acetaldehyde (22). We have previously shown that acetaldehyde induces both NF-κB and AP-1 (37). Collectively, these data and our new data show that Nrf2 can regulate gene expression in an ARE-independent manner, possibly involving both AP-1 and NF-κB. While the present work focused on the altered expression of c-Jun, c-Fos, Fra-1, p50, and p65, we cannot exclude the possibility that increased expression of JAB1 and c-Rel may also contribute to the phenotype observed. This will also need to be addressed in the future.
In summary, we have uncovered a highly novel action of Nrf1 and Nrf2, namely, their ability to modulate the expression of AP-1 and NF-κB family members. It is through this mechanism that both Nrf1 and Nrf2 regulate the activity of the rat GCLC promoter despite the absence of ARE. These findings further illustrate the complex cross-talks among the different families of transcription factors and suggest many of the biological functions of Nrf1 and Nrf2 may be related to their ability to modulate AP-1 and NF-κB expression.
Acknowledgments
This work was supported by NIH grants DK-45334 (S.C.L.) and CA-91907 (J.Y.C.).
WT, F1, and F2 mouse embryo fibroblasts were provided by the Cell Culture Core of the USC Liver Disease Research Center (P30 DK48522) via J.Y.C. originally.
REFERENCES
- 1.Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Chan, K., X. D. Han, and Y. W. Kan. 2001. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA 98:4611-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L., M. Kwong, R. Lu, D. Ginzinger, C. Lee, L. Leung, and J. Y. Chan. 2003. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell. Biol. 23:4673-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erickson, A. M., Z. Nevarea, J. J. Gipp, and R. T. Mulcahy. 2002. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. J. Biol. Chem. 277:30730-30737. [DOI] [PubMed] [Google Scholar]
- 5.Galloway, D. C., D. G. Blake, A. G. Shepherd, and L. I. McLellan. 1997. Regulation of human γ-glutamylcysteine synthetase: co-ordinate induction of the catalytic and regulatory subunits in HepG2 cells. Biochem. J. 328:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh, S., A. M. Gifford, L. R. Riviere, P. Tempst, G. P. Nolan, and D. Baltimore. 1990. Cloning of the p50 DNA binding subunit of NF-kappa B: homology to rel and dorsal. Cell 62:1019-1029. [DOI] [PubMed] [Google Scholar]
- 7.Huang, C., M. E. Anderson, and A. Meister. 1993. Amino acid sequence and function of the light subunit of rat kidney gamma-glutamylcysteine synthetase. J. Biol. Chem. 268:20578-20583. [PubMed] [Google Scholar]
- 8.Huang, C., L. Chang, M. E. Anderson, and A. Meister. 1993. Catalytic and regulatory properties of the heavy subunit of rat kidney gamma-glutamylcysteine synthetase. J. Biol. Chem. 268:19675-19680. [PubMed] [Google Scholar]
- 9.Huo, L., and T. L. Rothstein. 1996. Isolation and characterization of murine fra-1: induction mediated by CD40 and surface Ig is protein kinase C dependent. J. Immunol. 157:3812-3818. [PubMed] [Google Scholar]
- 10.Jeyapaul, J., and A. K. Jaiswal. 2000. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 59:1433-1439. [DOI] [PubMed] [Google Scholar]
- 11.Kwong, M., Y. W. Kan, and J. Y. Chan. 1999. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. J. Biol. Chem. 274:37491-37498. [DOI] [PubMed] [Google Scholar]
- 12.Lee, J. M., M. J. Calkins, K. Chan, Y. W. Kan, and J. A. Johnson. 2003. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 278:12029-12038. [DOI] [PubMed] [Google Scholar]
- 13.Leung, L., M. Kwong, S. Hou, C. Lee, and J. Y. Chan. 2003. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 278:48021-48029. [DOI] [PubMed] [Google Scholar]
- 14.Lu, S. C. 1999. Regulation of hepatic glutathione synthesis: current concepts and controversies. FASEB J. 13:1169-1183. [PubMed] [Google Scholar]
- 15.Lu, S. C., Z. Z. Huang, H. Yang, and H. Tsukamoto. 1999. Effect of thioacetamide on the hepatic expression of γ-glutamylcysteine synthetase subunits in the rat. Toxicol. Appl. Pharmacol. 159:161-168. [DOI] [PubMed] [Google Scholar]
- 16.Moinova, H. R., and R. T. Mulcahy. 1998. An electrophile responsive element (EpRE) regulates β-naphthoflavone induction of the human β-glutamylcysteine synthetase regulatory subunit gene. J. Biol. Chem. 273:14683-14689. [DOI] [PubMed] [Google Scholar]
- 17.Motohashi, H., T. O'Connor, F. Katsuoka, J. D. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Mulcahy, R. T., M. A. Wartman, H. H. Bailey, and J. J. Gipp. 1997. Constitutive and β-naphthoflavone-induced expression of the human γ-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 272:7445-7454. [DOI] [PubMed] [Google Scholar]
- 19.Myhrstad, M. C. W., C. Husberg, P. Murphy, O. Nordström, R. Blomhoff, J. Ø. Moskaug, and A. B. Kolstø. 2001. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochim. Biophys. Acta 1517:212-219. [DOI] [PubMed] [Google Scholar]
- 20.Nolan, G. P., S. Ghosh, H. C. Liou, P. Tempst, and D. Baltimore. 1991. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell 64:961-969. [DOI] [PubMed] [Google Scholar]
- 21.Novotny, V., E. E. Prieschl, R. Csonga, G. Fabjani, and T. Baumruker. 1998. Nrf1 in a complex with fosB, c-jun, junD and ATF2 forms the AP1 component at the TNF alpha promoter in stimulated mast cells. Nucleic Acids Res. 26:5480-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parfett, C. L. 2003. Combined effects of tumor promoters and serum on proliferin mRNA induction: a biomarker sensitive to saccharin, 2,3,7,8-TCDD, and other compounds at minimal concentrations promoting C3H/10T1/2 cell transformation. J. Toxicol. Environ. Health Part A 66:1943-1966. [DOI] [PubMed] [Google Scholar]
- 23.Philips, A., C. Teyssier, F. Galtier, C. Rivier-Covas, J. M. Rey, H. Rochefort, and D. Chalbos. 1998. FRA-1 expression level modulates regulation of activator protein-1 activity by estradiol in breast cancer cells. Mol. Endocrinol. 12:973-985. [DOI] [PubMed] [Google Scholar]
- 24.Poot, M., H. Teubert, P. S. Rabinovitch, and T. J. Kavanagh. 1995. De novo synthesis of glutathione is required for both entry into and progression through the cell cycle. J. Cell. Physiol. 163:555-560. [DOI] [PubMed] [Google Scholar]
- 25.Rangasamy, T., C. Y. Cho, R. K. Thimmulappa, L. Zhen, S. S. Srisuma, T. W. Kensler, M. Yamamoto, I. Petrache, R. M. Tuder, and S. Biswal. 2004. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Investig. 114:1248-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renz, M., M. Neuberg, C. Kurz, R. Bravo, and R. Muller. 1985. Regulation of c-fos transcription in mouse fibroblasts: identification of DNase I-hypersensitive sites and regulatory upstream sequences. EMBO J. 4:3711-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seelig, G. F., R. P. Simondsen, and A. Meister. 1984. Reversible dissociation of gamma-glutamylcysteine synthetase into two subunits. J. Biol. Chem. 259:9345-9347. [PubMed] [Google Scholar]
- 28.Suthanthiran, M., M. E. Anderson, V. K. Sharma, and A. Meister. 1990. Glutathione regulates activation-dependent DNA synthesis in highly purified normal human T lymphocytes stimulated via the CD2 and CD3 antigens. Proc. Natl. Acad. Sci. USA 87:3343-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, T., H. Okuno, T. Yoshida, T. Endo, N. Nishina, and H. Iba. 1991. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 19:5537-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tietze, F. 1969. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal. Biochem. 27:502-522. [DOI] [PubMed] [Google Scholar]
- 31.van Dam, H., M. Duyndam, R. Rottier, A. Bosch, L. de Vries-Smits, P. Herrlich, A. Zantema, P. Angel, and A. J. van der Eb. 1993. Heterodimer formation of c-Jun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 12:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venugopal, R., and A. K. Jaiswal. 1998. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17:3145-3156. [DOI] [PubMed] [Google Scholar]
- 33.Wei, P., N. Inamdar, and W. V. Vedeckis. 1998. Transrepression of c-jun gene expression by the glucocorticoid receptor requires both AP-1 sites in the c-jun promoter. Mol. Endocrinol. 12:1322-1333. [DOI] [PubMed] [Google Scholar]
- 34.Wild, A. C., H. R. Moinova, and R. T. Mulcahy. 1999. Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274:33627-33636. [DOI] [PubMed] [Google Scholar]
- 35.Yan, N., and A. Meister. 1990. Amino acid sequence of rat kidney gamma-glutamylcysteine synthetase. J. Biol. Chem. 265:1588-1593. [PubMed] [Google Scholar]
- 36.Yang, H., Y. Zeng, T. D. Lee, Y. Yang, X. Ou, L. Chen, M. Haque, R. Rippe, and S. C. Lu. 2002. Role of AP-1 in the coordinate induction of rat glutamate-cysteine ligase and glutathione synthetase by tert-butylhydroquinone. J. Biol. Chem. 277:35232-35239. [DOI] [PubMed] [Google Scholar]
- 37.Yang, H. P., Z. Z. Huang, J. H. Wang, X. P. Ou, and S. C. Lu. 2001. Cloning and characterization of the 5′-flanking region of the rat glutamate-cysteine ligase catalytic subunit. Biochem. J. 357:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, H. P., M. R. Sadda, V. Yu, Y. Zeng, T. D. Lee, X. P. Ou, L. X. Chen, and S. C. Lu. 2003. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor α: role of NF-κB and AP-1. J. Biol. Chem. 278:50887-50896. [DOI] [PubMed] [Google Scholar]