Abstract
The oncogenic TLS-ERG fusion protein is found in human myeloid leukemia and Ewing's sarcoma as a result of specific chromosomal translocation. To unveil the potential mechanism(s) underlying cellular transformation, we have investigated the effects of TLS-ERG on both gene transcription and RNA splicing. Here we show that the TLS protein forms complexes with RNA polymerase II (Pol II) and the serine-arginine family of splicing factors in vivo. Deletion analysis of TLS-ERG in both mouse L-G myeloid progenitor cells and NIH 3T3 fibroblasts revealed that the RNA Pol II-interacting domain of TLS-ERG resides within the first 173 amino acids. While TLS-ERG repressed expression of the luciferase reporter gene driven by glycoprotein IX promoter in L-G cells but not in NIH 3T3 cells, the fusion protein was able to affect splicing of the E1A reporter in NIH 3T3 cells but not in L-G cells. To identify potential target genes of TLS-ERG, the fusion protein and its mutants were stably expressed in both L-G and NIH 3T3 cells through retroviral transduction. Microarray analysis of RNA samples from these cells showed that TLS-ERG activates two different sets of genes sharing little similarity in the two cell lines. Taken together, these results suggest that the oncogenic TLS-ERG fusion protein transforms hematopoietic cells and fibroblasts via different pathways.
In acute myelogenous leukemia, chronic myelogenous leukemia in BLAST crisis, and certain myelodysplastic syndromes, the TLS (translocation liposarcoma) gene is fused to the ERG (ets-related gene) through a recurrent t(16;21) chromosomal translocation (18). Interestingly, the same t(16;21) rearrangement and the resultant TLS-ERG chimeric fusion protein were also reported in Ewing's sarcoma (36). The TLS-ERG fusion protein retains the N-terminal domain of TLS, but the C-terminal domain of TLS is replaced by the DNA-binding domain of ERG. Previous studies have demonstrated that TLS-ERG fusion protein is capable of transforming mouse cell lines (19) as well as normal human hematopoietic cells (28).
The TLS gene was originally cloned as a fusion partner with the CHOP gene in human myxoid liposarcoma (9, 33). TLS belongs to a family of closely related proteins that include the Ewing's sarcoma protein EWS (11) and the TATA-binding protein-associated factor TAFII68 (3). EWS is known to interact with the transcription coactivator CBP/p300 (35). TLS has been reported to be a target of the BCR/ABL oncoprotein and binds to DNA in a phosphorylation-dependent manner (29, 30). In addition, transient-expression experiments revealed that TLS binds to RNA polymerase II (Pol II) through the N-terminal domain of TLS and interacts with splicing factors through the C-terminal domain of TLS (8, 42, 43).
TLS-ERG was originally speculated to act as a chimeric transcription factor leading to transformation through deregulation of gene transcription (31), but accumulating evidence suggests that TLS-ERG and the related EWS-FLI-1 fusion proteins may lead to cellular abnormalities by deregulating both gene transcription and RNA splicing (20, 22, 40, 42). TLS has been proposed to function as an adaptor molecule linking gene transcription by RNA Pol II with RNA processing by splicing factors, whereas the TLS-ERG fusion protein is thought to disrupt this linkage by binding to RNA Pol II but failing to recruit splicing factors to the sites of active transcription (42). Interestingly, transformation assays with L-G myeloid progenitor cells and with NIH 3T3 fibroblasts suggested that there might exist at least two transforming subdomains within the N-terminal region of the TLS-ERG fusion protein (19), yet it is unclear whether these two transforming subdomains affect the same set of genes or deregulate two distinct sets of genes in different cellular backgrounds (hematopoietic cells versus fibroblasts). It is important to address this question, as similar TLS and EWS fusion proteins have been found in many types of cancer, and the cells may be transformed differently depending on the histogenetic background from which the tumor originates.
In this report, we studied the TLS-ERG fusion protein in both mouse L-G myeloid progenitors and NIH 3T3 fibroblasts to mimic hematopoietic and nonhematopoietic cells. The differences between gene transcription and RNA splicing in these two unrelated lineages of cells were further analyzed by deletion mutants of TLS-ERG. We found that TLS-ERG and its mutants indeed behaved differently in L-G and NIH 3T3 cells when tested for their transactivation potential and the ability to interfere with RNA splicing. Our observations were further supported by DNA microarray experiments showing that different sets of genes are affected by the same TLS-ERG construct in L-G and NIH 3T3 cells. These findings suggest that TLS-ERG fusion protein transforms hematopoietic and nonhematopoietic cells via different pathways.
MATERIALS AND METHODS
Plasmids.
LNCX retroviral constructs expressing hemagglutinin (HA) epitope-tagged HA-TLS-ERG, HA-TLS-ERGΔ1-173, HA-TLS-ERGΔ174-265, and HA-TLS-ERGΔETS were described previously (19). The point mutation R367L was introduced into TLS-ERG by the Gene Editor site-directed mutagenesis kit (Promega). HA-tagged wild-type ERG was generated by PCR. For transient expression, DNA fragments encoding HA-tagged TLS-ERG, its mutants, and ERG were released from the LNCX retroviral vector and cloned into the unique HindIII site of the pCR3 vector (Invitrogen). Firefly luciferase reporter pGL3-GPIX was generated by cloning the promoter region of the human glycoprotein IX gene into the KpnI-BglII sites of the pGL3-basic vector (2). pGL3-ESET was generated by cloning the promoter region of mouse ESET gene into the KpnI-BglII sites of the pGL3-basic vector (4). cDNAs encoding splicing factors TASR-1 and TASR-2 were cloned into the EcoRI-KpnI sites of the pFlag-CMV-5a vector (Sigma) with the Flag epitope tagged at the C-terminal end of the protein. The pCS3-MT-E1A splicing reporter has been previously described (17).
Antibodies.
The mouse monoclonal horseradish peroxidase-conjugated anti-HA antibody (3F10) was purchased from Roche Applied Science (Indianapolis, IN). The mouse monoclonal horseradish peroxidase-conjugated anti-Flag antibody (M2) was from Sigma. The rabbit polyclonal anti-Pol II (C21) was from Santa Cruz Biotechnology (Santa Cruz, CA). The mouse monoclonal anti-Pol II antibodies 8WG16 and H5 were from Covance (Berkeley, CA). The mouse monoclonal anti-SC35 and anti-TLS antibodies were from PharMingen (San Diego, CA), the mouse hybridoma clone m104 was purchased from the American Type Culture Collection (Rockville, MD) and used for collecting an antibody recognizing a family of classic SR proteins including SC35. The rabbit polyclonal anti-TLS antibody was raised against the N-terminal 165 amino acids (17).
Cell culture and retroviral infection.
Human cervix carcinoma HeLa cells and mouse fibroblast NIH 3T3 cells were cultured in Dulbecco's modified Eagle medium plus 10% fetal calf serum. Human acute myeloid leukemia YNH-1 cells and mouse myeloid progenitor L-G cells were cultured in RMPI 1640 medium supplemented with 10% fetal calf serum and 50 μM β-mercaptoethanol plus 10 ng/ml recombinant human granulocyte colony-stimulating factor (G-CSF; for YNH-1 cells) or 1 ng/ml recombinant mouse interleukin-3 (for L-G cells). BOSC23 retrovirus packaging cells were purchased from the American Type Culture Collection and maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum plus 0.025 mg/ml mycophenolic acid (Sigma) and 2.176 μg/ml aminopterin (Sigma). For retroviral infection, BOSC23 packaging cells were transfected with pLNCX constructs by the calcium phosphate precipitation method. After 48 h, the supernatant was collected and used to infect L-G and NIH 3T3 cells. Infected cells were selected in medium containing 1 mg/ml G418, and G418-resistant clones were pooled for this study.
Immunoprecipitation and Western blot analysis.
Cells (2 × 107 HeLa cells, 10 × 107 L-G cells, or NIH 3T3 cells from one 15-cm plate) were collected and lysed with 5 ml NP-40 lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40). The resultant nuclear pellet was resuspended in 0.5 ml lysis buffer X (50 mM Tris, pH 7.4, 270 mM NaCl, 0.5% Triton X-100) supplemented with protease inhibitors and phosphatase inhibitors to prepare the nuclear extract. For immunoprecipitation, 4 μl of 8WG16, 25 μl of anti-SC35, or 50 μl of anti-TLS was incubated with 45 μl of protein A/G agarose for 3 h at 4°C in 0.2 ml buffer A (10 mM Tris, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100), and the antibody-protein A/G-agarose complex was then incubated with 0.2 ml nuclear extract overnight at 4°C on a rotating wheel. After one wash with buffer X and two washes with radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), the immunoprecipitates were mixed with 40 μl of SDS sample buffer and denatured at 96°C for 5 min. After separation by SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane, the proteins were blotted with appropriate antibodies and visualized by the ECL Western blotting analysis system (Amersham Pharmacia Biotech, Piscataway, NJ).
Immunostaining.
Retrovirus-transduced NIH 3T3 cells harboring HA-TLS-ERG or its mutants were seeded onto cover glass slides (20 by 20 mm). Twenty-four hours later, the cells were fixed by 2% paraformaldehyde for 5 min. Following two washes in phosphate-buffered saline (PBS), the cells were blocked with 3% normal goat serum for 1 h and then incubated with 1:1,000-diluted 3F10 anti-HA at 4°C overnight. After three washes, the cells were incubated with a Cy3-conjugated goat anti-mouse immunoglobulin G (IgG; Jackson ImmunoResearch Labs, West Grove, PA) diluted 1:200 in PBS for 1 h and then incubated with 1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) to counterstain the DNA. The cells were washed three times with PBS, mounted with DAKO fluorescent mounting medium, and visualized with a Zeiss immunofluorescence microscope.
Transfection and luciferase assay.
NIH 3T3 cells (65% confluent) in a 3.5-cm well or 1 × 107 L-G cells were transfected with 1.5 μg pGL3-GPIX luciferase reporter, 250 ng pCR3-HA-TLS-ERG construct, and 20 ng pRL-SV40 Renilla luciferase control. NIH 3T3 cells were transfected using the TransIT-TKO transfection reagent from Mirus (Madison, WI) according to the manufacturer's instructions. L-G cells were transfected using a Nucleofector from Amaxa, Inc. (Gaithersburg, MD) in 0.1 ml solution V using program T20. After 24 h, cells were lysed and assayed using the Promega dual-luciferase assay system. Transfection was repeated at least three times, and the luciferase activity was normalized to the internal Renilla luciferase control.
E1A pre-mRNA splicing assay.
For in vivo splicing of E1A pre-mRNA in NIH 3T3 cells, 1.5 μg pCS3-MT-E1A and 1.5 μg pFlag-CMV-5a-TASR-1 plus 1.5 μg pCR3-HA-TLS-ERG constructs were used for transfection of cells in a 3.5-cm well. For L-G cells, 1 μg pCS3-MT-E1A and 1 μg pFlag-CMV-5a-TASR plus 1 μg pCR3-HA-TLS-ERG were introduced into 2 × 107 cells by nucleofection. After 24 h, total RNA from half of the cells was isolated using an RNeasy column (QIAGEN) and eluted with 40 μl H2O. Nuclear extracts from the remaining cells were prepared for Western blotting with the mouse monoclonal M2 anti-Flag antibody. For hybridization, 20 μl of total RNA was mixed with 1.5 × 106 cpm of 32P-labeled RNA probes antisense to the E1A genomic sequence. After overnight incubation, excessive RNA probes were digested with RNase A plus T1 supplied with the RNase protection assay system (PharMingen). The protected antisense E1A RNA fragments were isolated and separated on a 6% denaturing gel as previously described (42).
DNA microarray analysis.
Total RNAs from retrovirus-transduced L-G and NIH 3T3 cells stably expressing HA-tagged TLS-ERG and its mutants were isolated for DNA array analysis at the University of Washington Center for Expression Array. Target labeling and hybridization to Affymetrix GeneChips (mouse genome 430, version 2.0, array) were carried out with minor modifications from procedures recommended by the manufacturer. The chips were scanned using the GeneChip Scanner, and the CHP files were generated using Affymetrix GCOS 1.1 software. The expression settings for scaling were set for all probe sets with a target value of 250, and normalization was also set for all probe sets. Default values were used for all other parameters. Gene expression in cells harboring HA-TLS-ERGΔETS was used as the baseline control for comparison analysis.
To identify genes that showed either an increase or decrease over the baseline control, the comparison CHP files were first filtered using the change P value to determine a “call” of no change, increase, decrease, marginal increase, or marginal decrease. Only probe sets with a “call” of increase or decrease were analyzed using the detection P value to determine a “call” of present, absent, or marginal. Probe sets with a “call” of present and a signal log ratio of −1.0 (i.e., twofold decrease) and lower or an signal log ratio of +1.0 (i.e., twofold increase) and higher were selected. Using the NetAffx Analysis Center on the Affymetrix website, the selected probe sets were then uploaded as a batch query to obtain gene annotations. The annotated genes were searched with the NetAffx Analysis Center's GeneOntology (GO) browser for assignment into various biological pathways. The number of probe sets shared by TLS-ERG and its mutants was determined using the intersection tool.
RESULTS
Endogenous TLS interacts with both RNA Pol II and SR splicing factors.
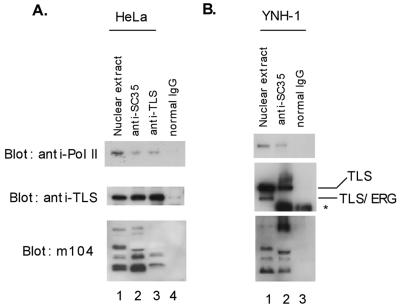
In previous studies, we showed that the overexpressed TLS N-terminal domain interacts with RNA Pol II and the overexpressed TLS C-terminal domain interacts with the SR family of splicing factors. To test whether endogenous TLS can function as an adaptor molecule coupling gene transcription and RNA splicing under physiological conditions, we performed coimmunoprecipitation experiments using nuclear extract prepared from HeLa cells. The expression of endogenous RNA Pol II, TLS, and SR proteins was confirmed by Western blotting with the H5 antibody recognizing the hyperphosphorylated RNA Pol IIo, a rabbit polyclonal antibody against TLS, and the m104 antibody recognizing a family of SR proteins (Fig. 1A, lane 1). The anti-SC35 antibody could immunoprecipitate SR proteins, as confirmed in our study (Fig. 1A, lane 2, bottom panel). Together with these SR proteins, the anti-SC35 antibody also brought down both endogenous TLS and hyperphosphorylated RNA Pol IIo (Fig. 1A, lane 2, top and middle panels). For reciprocal immunoprecipitation, a mouse monoclonal anti-TLS antibody was able to bring down RNA Pol IIo and several SR proteins (Fig. 1A, lane 3). As a negative control, normal mouse IgG failed to immunoprecipitate SR proteins, TLS, or Pol IIo (Fig. 1A, lane 4).
FIG. 1.
Detection of RNA Pol II-TLS-SR protein complexes. (A) HeLa nuclear extract (lane 1) was immunoprecipitated with the mouse monoclonal anti-SC35 (lane 2), a mouse monoclonal antibody against the N-terminal region of TLS (lane 3), or a nonspecific mouse IgG as a negative control (lane 4). The nuclear extract and the immunoprecipitates were blotted with the H5 anti-RNA Pol IIo (top panel), a rabbit polyclonal anti-TLS (middle panel), and the m104 antibody recognizing a family of SR proteins (bottom panel). (B) A similar immunoprecipitation experiment was carried out with the t(16;21) acute leukemia cell line YNH-1 expressing endogenous TLS-ERG. Note the absence of TLS-ERG from the anti-SC35 immunoprecipitate. The position of the IgG band is indicated by an asterisk.
To investigate the interactions of SR proteins with TLS and Pol II in TLS-ERG-positive tumor cells, we incubated the anti-SC35 antibody with nuclear extract from the YNH-1 acute myeloid leukemia cell line (41). One of the TLS alleles in these YNH-1 cells is fused to the ERG gene as a result of the t(16;21) translocation, and full-length TLS protein is expressed from the remaining intact allele (41). Since the anti-TLS antibody used in the Western blotting was raised against the N-terminal domain of TLS, it can detect the existence of both TLS and the TLS-ERG fusion protein in the YNH-1 nuclear extract (Fig. 1B, lane 1). The anti-SC35 antibody brought down both full-length TLS and Pol IIo, but the TLS-ERG fusion protein was absent from the anti-SC35 immunoprecipitate (Fig. 1B, lane 2), confirming that the association with SR proteins is mediated through the C-terminal domain of TLS. In the negative-control experiment with normal mouse IgG, none of these proteins was detectable from the immunoprecipitate (Fig. 1B, lane 3). Since a direct interaction between Pol II and the classic SR family of proteins has not been demonstrated (16), these results provide evidence for the existence of Pol II-TLS-SR multiprotein complexes in vivo and support the adaptor role of TLS in recruiting SR proteins to the Pol II basal transcription machinery.
Amino acids 1 to 173 of TLS mediate interaction with RNA Pol II.
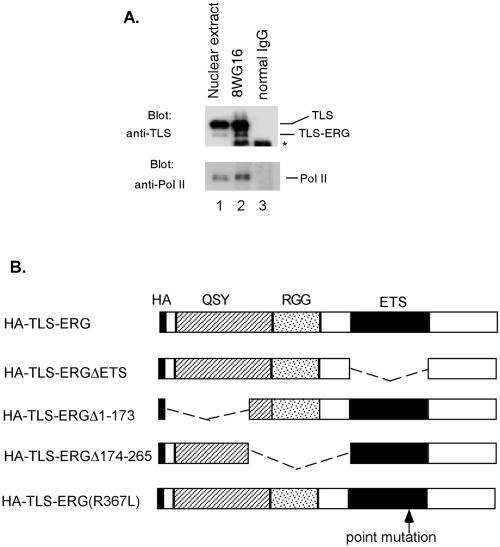
Our previous studies of Pol II interaction were carried out with cells overexpressing epitope-tagged TLS-ERG fusion protein (42). To investigate the association of endogenous TLS-ERG fusion protein with Pol II under physiological conditions, YNH-1 nuclear extract (Fig. 2A, lane 1) was treated with the anti-Pol II antibody 8WG16 (Fig. 2A, lane 2) or the normal mouse IgG (Fig. 2A, lane 3). Western blotting showed that both full-length TLS and the TLS-ERG leukemia fusion protein associate with Pol II in vivo, demonstrating that Pol II interaction was mediated through the N-terminal domain of TLS.
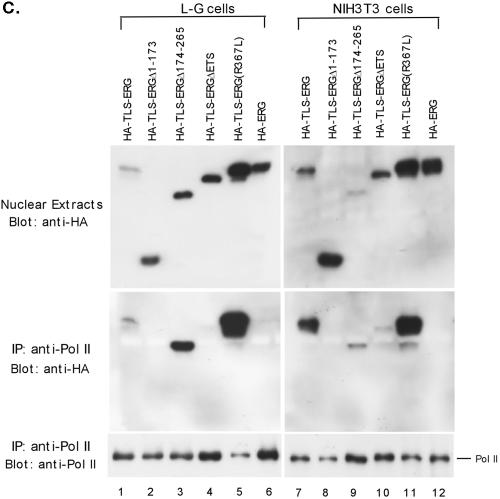
FIG. 2.
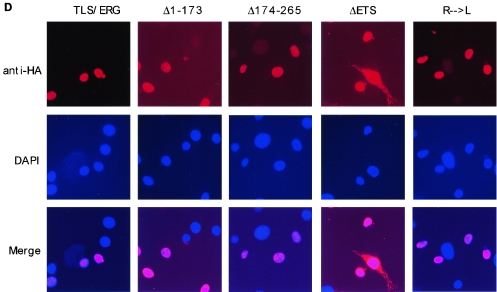
Interaction of RNA Pol II with the first 173 amino acids of TLS-ERG. (A) YNH-1 nuclear extract (lane 1) was incubated with the anti-Pol II antibody 8WG16 (lane 2) or normal mouse IgG as a negative control (lane 3). The position of the IgG band is indicated by an asterisk. (B) Schematic of HA-TLS-ERG and its mutants with different domains deleted or mutated. HA, hemagglutinin epitope; QSY, glutamine-, serine-, and tyrosine-rich domain; RGG, region with multiple Arg-Gly-Gly repeats; ETS, ets DNA-binding domain. (C) Nuclear extracts from L-G cells (lanes 1 to 6) and NIH 3T3 cells (lanes 7 to 12) expressing HA-TLS-ERG or its mutants were blotted with the 3F10 anti-HA antibody (top panels). The nuclear extracts were incubated with 8WG16, and the immunoprecipitates (IP) were blotted with the anti-HA antibody (middle panels) or the C21 anti-Pol II antibody (bottom panels). (D) NIH 3T3 cells expressing HA-TLS-ERG or its mutants were stained with a Cy3-conjugated anti-HA antibody (top panel), the nuclei were indicated by DAPI staining (middle panel), and the two images were merged to show subcellular localization of the HA-tagged protein.
The structure of TLS-ERG is shown in the schematic (Fig. 2B). In an earlier study, we identified two distinct functional subdomains within the TLS sequence of the fusion protein. The subdomain that resides between amino acids 1 and 173 (exons 1 to 5) is required for the transformation of NIH 3T3 cells, and the subdomain between amino acids 174 and 265 (exons 6 and 7) is essential for the transformation of L-G cells (19). We also found that the ETS domain of ERG is required for transformation of both L-G and NIH 3T3 cells. For this study, we used these three TLS-ERG deletion mutants and an additional mutant that replaces the arginine at position 367 with leucine (R367L) (Fig. 2B). This point mutation is known to abolish the DNA binding and transactivating ability of ERG (27).
To determine the N-terminal sequence of TLS-ERG that is responsible for interaction with Pol II, HA-tagged TLS-ERG as well as three deletion mutants were stably expressed in L-G cells as well as in NIH 3T3 cells after retroviral infection and G418 selection. The expression of HA-tagged proteins in L-G and NIH 3T3 cells was confirmed by Western blotting with a mouse monoclonal anti-HA antibody (Fig. 2C, upper panels). Nuclear extracts from the infected cells were immunoprecipitated with the 8WG16 anti-Pol II antibody that was known to coimmunoprecipitate the TLS-ERG fusion protein. After washes, the precipitates were blotted with the anti-HA antibody to detect the coimmunoprecipitated mutants. HA-tagged TLS-ERG was clearly brought down by the anti-Pol II antibody from both L-G and NIH 3T3 cells (Fig. 2C, lanes 1 and 7, middle panels). Deletion of amino acids 1 to 173 from TLS-ERG completely abolished interaction of the fusion protein with Pol II in both types of cells (Fig. 2C, lanes 2 and 8, middle panels). Even though deletion of amino acids 174 to 265 had little effect on the coimmunoprecipitation (Fig. 2C, lanes 3 and 9, middle panels), deletion of the ETS domain from the fusion protein impaired its interaction with Pol II (Fig. 2C, lanes 4 and 10, middle panels). Point mutant R367L retained the ability to associate with Pol II (Fig. 2C, lanes 5 and 11, middle panels), and wild-type HA-ERG did not possess such an ability (Fig. 2C, lanes 6 and 12, middle panels). These results suggest that the Pol II-binding domain is located within the first 173 amino acids of TLS-ERG, a subdomain that is required for transformation of NIH 3T3 cells but not L-G cells.
To investigate why deletion of the ETS domain from TLS-ERG had an adverse effect on its association with Pol II, we carried out immunostaining experiments with NIH 3T3 cells that stably express HA-TLS-ERG and its mutants. As these cells represent pooled G418-resistant clones, it became clear that not all of the cells expressed the epitope-tagged proteins. The immunostaining results also revealed that HA-TLS-ERG and its mutants, with the exception of HA-TLS-ERGΔETS, are exclusively localized in the nucleus (Fig. 2D). Interestingly, deletion of the ETS domain resulted in partial cytoplasmic localization of the mutant HA-TLS-ERGΔETS among some of the cells. We speculate that deletion of the ETS domain may also have changed the subnuclear localization of the mutant to a site that is less likely to encounter Pol II for association.
TLS-ERG inhibits reporter gene expression in L-G cells but not in NIH 3T3 cells.
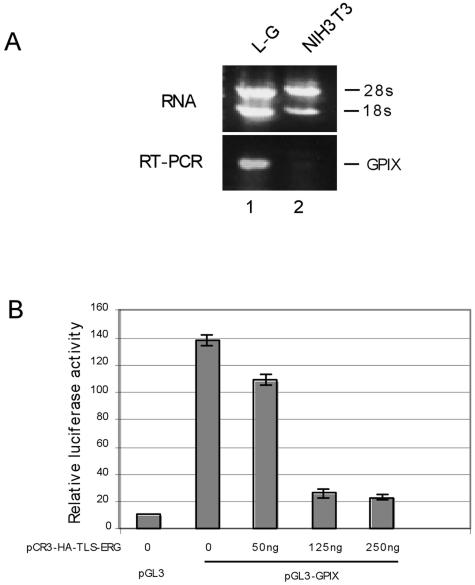
As a chimeric transcription factor, TLS-ERG was thought to contribute to transformation by transcriptional deregulation of target genes. To determine how this fusion protein affects transcription in different cellular contexts and how subdomains within TLS-ERG contribute to such an effect, we cloned the glycoprotein IX (GPIX) promoter to drive expression of the luciferase reporter gene. GPIX is a cell surface protein that mediates adhesion of platelets (26). Previous studies of the GPIX promoter have identified an ETS binding site, recognizable by ERG protein, to be important to transcription of the GPIX gene (2). Through reverse transcription (RT)-PCR analysis of GPIX mRNA, we found that expression of the GPIX gene is detectable in L-G myeloid progenitor cells but not in NIH 3T3 fibroblasts (Fig. 3A).
FIG. 3.
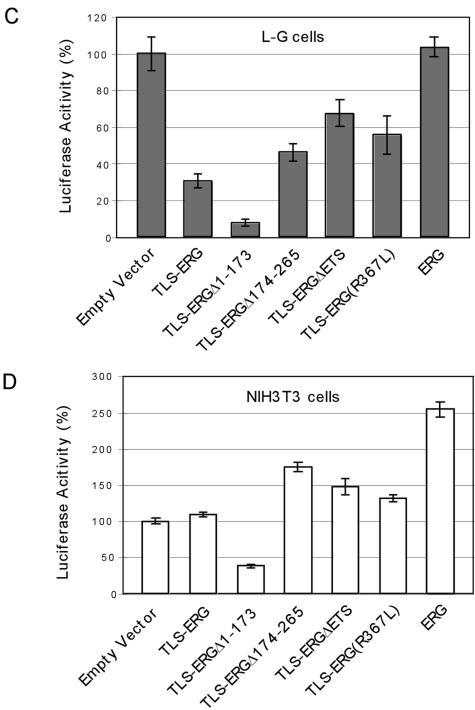
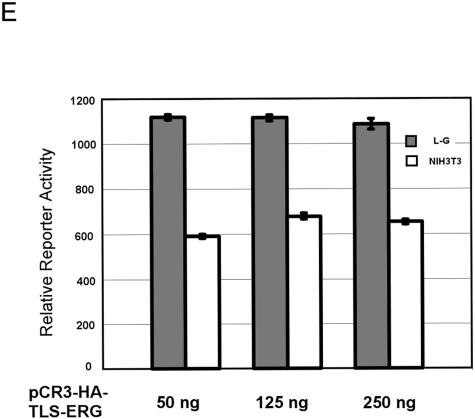
Effects of TLS-ERG and its mutants on GPIX promoter in L-G and NIH 3T3 cells. (A) Expression of the mouse GPIX gene was examined by RT-PCR in L-G (lane 1) and NIH 3T3 (lane 2) cells. (B) pGL3 or pGL3-GPIX firefly luciferase reporter construct (1.5 μg) was introduced into L-G cells with increasing amounts of pCR3-HA-TLS-ERG as indicated. The total amount of the DNA was kept constant by the addition of pCR3 empty vector. (C) pGL3-GPIX (1.5 μg) was cotransfected into L-G cells with 250 ng of pCR3-HA-TLS-ERG construct expressing TLS-ERG or its mutants. (D) pGL3-GPIX (1.5 μg) was cotransfected into NIH 3T3 cells with 250 ng of pCR3-HA-TLS-ERG construct expressing TLS-ERG or its mutants. (E) pGL3-ESET firefly luciferase reporter construct (1.5 μg) was introduced into L-G and NIH 3T3 cells with increasing amounts of pCR3-HA-TLS-ERG as indicated. All transfections were carried out independently at least three times, and the promoter activity was normalized to that of the Renilla luciferase control.
In L-G cells, the cloned GPIX promoter generated a luciferase activity that is more than 10-fold greater than that of the promoterless reporter construct (Fig. 3B, compare columns 1 and 2). This reflected the fact that the GPIX gene is constitutively activated in these L-G cells. When cotransfected with pCR3-HA-TLS-ERG, the GPIX promoter activity was severely repressed by coexpression of TLS-ERG in a dose-dependent manner (Fig. 3B, columns 3 to 5), suggesting that the TLS-ERG fusion protein might compete with endogenous transactivators for access to the GPIX promoter.
While TLS-ERG was able to repress luciferase expression driven by the GPIX promoter, deletion of the first 173 amino acids from the fusion protein resulted in an even more severe repression of the luciferase reporter gene (Fig. 3C, compare columns 1 to 3). TLS-ERG repression of the reporter was somewhat lessened by deletion of amino acids 174 to 265, by deletion of the ETS domain, or by the R367L mutation (Fig. 3C, columns 4 to 6). The fact that mutations within the ETS domain of TLS-ERG did not completely restore the GPIX promoter activity in L-G cells suggests that TLS-ERG may exert its effect on the reporter gene through an additional mechanism. Coexpression of ERG did not affect the promoter activity (Fig. 3C, column 7).
In contrast to L-G cells, the GPIX promoter was relatively weak in NIH 3T3 cells and coexpression of TLS-ERG fusion protein had little effect on promoter activity (Fig. 3D, compare columns 1 and 2). However, the TLS-ERG mutant lacking the first 173 amino acids also caused repression of promoter activity in NIH 3T3 cells (Fig. 3D, column 3). This may reflect the fact that amino acids 1 to 173 mediate interaction with RNA Pol II, mutants that lack Pol II-interacting ability but retain DNA-binding ability can function in a dominant-negative manner. Cotransfection with the TLS-ERG mutant lacking amino acids 174 to 265 or the ETS domain caused slight increases in luciferase activity, as did cotransfection with the R367L mutant (Fig. 3D, columns 4 to 6). Wild-type ERG was able to transactivate the GPIX promoter and generated a luciferase activity that is 2.5-fold that of the empty vector control in NIH 3T3 cells (Fig. 3D, column 7). These results showed that amino acids 174 to 265 in TLS-ERG might represent a subdomain of transcriptional repression regardless of cellular background.
To ensure that the effects of TLS-ERG on the GPIX promoter construct are gene-specific and not the results of nonspecific squelching of coactivators in L-G cells or squelching of corepressors in NIH 3T3 cells, we performed similar cotransfection of pCR3-HA-TLS-ERG with the pGL3-ESET reporter. The ESET promoter in this reporter construct does not have an ETS binding site and is constitutively activated in various cell types (4). When the amount of pCR3-HA-TLS-ERG was increased from 50 ng to 250 ng in the cotransfection, we did not observe any effect on luciferase activity driven by the ESET promoter in either L-G or NIH 3T3 cells (Fig. 3E). It is therefore unlikely that the transiently expressed TLS-ERG protein in the cotransfection would have a discernible squelching effect under our experimental conditions.
TLS-ERG influences E1A splicing in NIH 3T3 cells but not in L-G cells.
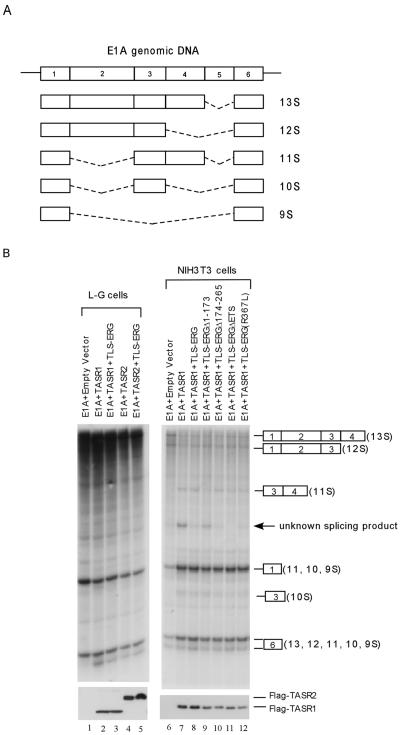
Since TLS associates with both RNA Pol II and SR splicing factors and the TLS-ERG fusion protein uncouples such an association, we investigated whether TLS-ERG and its mutants can affect RNA splicing in L-G myeloid progenitor cells and NIH 3T3 fibroblasts. To this end, the adenovirus E1A gene was used as a splicing reporter because alternative splicing of E1A pre-mRNA transcripts has been well characterized (5, 39). The five different E1A splicing isoforms designated 13S, 12S, 11S, 10S, and 9S are illustrated in Fig. 4A. Transfection of the E1A splicing reporter into L-G cells generated 13S, 12S, and 9S, as detected by RNase protection assay (Fig. 4B, lane 1). The TLS-associated SR protein TASR-1 was previously reported to promote 11S and 10S, whereas TASR-2 promoted 9S in HeLa cells and TLS-ERG was able to interfere with TASR-mediated E1A splicing (42). In L-G cells, both TASR-1 and TASR-2 slightly increased splicing products containing exon 6 (Fig. 4B, compare lane 1 with lanes 2 and 4). However, TLS-ERG did not have an appreciable effect on E1A splicing mediated by TASR proteins in L-G myeloid progenitor cells (Fig. 4B, lanes 3 and 5).
FIG. 4.
Effects of TLS-ERG and its mutants on TASR-mediated E1A pre-mRNA splicing in L-G and NIH 3T3 cells. (A) Diagram of E1A genomic DNA and different splicing isoforms. Numbers indicate individual exons, and dashed lines represent spliced sequences. (B) pCS3-MT-E1A splicing reporter was cotransfected into L-G cells with an empty vector (lane 1) or plasmid expressing Flag-TASR-1 (lanes 2 and 3) or Flag-TASR-2 (lanes 4 and 5) in the presence or absence of TLS-ERG. In NIH 3T3 cells, the pCS3-MT-E1A splicing reporter was cotransfected with an empty vector (lane 6) or plasmids expressing Flag-TASR-1 (lane 7) plus TLS-ERG or its mutants (lanes 8 to 12). In vivo alternative splicing of E1A pre-mRNA was analyzed by an RNase protection assay. Protected E1A RNA fragments are shown on the right, with exons designated by numerals in boxes. Nuclear extracts from the transfected cells were blotted with an anti-Flag antibody to show comparable expression of Flag-tagged TASR proteins (bottom panels).
In NIH 3T3 cells, coexpression of TASR-1 with the E1A splicing reporter promoted the 11S isoform as well as an unknown splicing product (Fig. 4B, compare lanes 6 and 7). This unknown splicing product was significantly inhibited by the TLS-ERG fusion protein (Fig. 4B, compare lanes 7 and 8). While TLS-ERGΔ1-173 did not affect this unknown splicing product, the mutant protein instead appeared to decrease splicing to the 11S isoform (Fig. 4B, lane 9). TLS-ERGΔ174-265 retained the ability to inhibit this unknown splicing product (Fig. 4B, lane 10). To our surprise, TLS-ERGΔETS and TLS-ERG(R367L) also had an inhibitory effect on this unknown splicing product (Fig. 4B, lanes 11 to 12), suggesting that retention of the first 173 amino acids (the same domain that mediates interaction with RNA Pol II) of TLS-ERG is sufficient to interfere with TASR-1-mediated E1A splicing in NIH 3T3 cells.
TLS-ERG alters expression of different sets of genes in L-G and NIH 3T3 cells.
Since our cotransfection experiments with the GPIX promoter and E1A splicing reporters suggest that transformation of L-G myeloid progenitor cells and NIH 3T3 fibroblasts may be mediated through different mechanisms, we set out to investigate whether TLS-ERG affects a common set of genes or alters two sets of unrelated genes in different cellular backgrounds (hematopoietic cells versus fibroblasts). To compare the effects of the TLS-ERG fusion protein on gene expression in L-G and NIH 3T3 cells, we carried out DNA microarray experiments with RNAs from these two types of cells harboring retroviral HA-TLS-ERG and its deletion mutants. With the Affymetrix GeneChip (mouse genome 430, version 2.0, array) that covers up to 45,000 probe sets corresponding to 39,000 transcripts, global gene expression was profiled and compared between L-G myeloid progenitor cells and NIH 3T3 fibroblasts. To exclude genes of less importance to transformation, we used cells harboring HA-TLS-ERGΔETS as the baseline control for microarray analysis, since this mutant was unable to transform either L-G or NIH 3T3 cells (19).
After comparison analysis with the TLS-ERGΔETS control, our microarray data revealed that 942 genes were up- or down-regulated at least twofold in L-G cells and 1,622 genes were affected at least twofold in NIH 3T3 cells by TLS-ERG (Fig. 5A; see also http://www.vams.edu/pathology/research/default.asp). To examine the accuracy of the microarray data, we carried out RT-PCR analysis of 20 transcripts identified in the microarray. Changes in expression for 19 of these transcripts were confirmed by RT-PCR (data not shown), indicating that the microarray data generally reflected levels of gene expression in retrovirus-transduced L-G and NIH 3T3 cells.
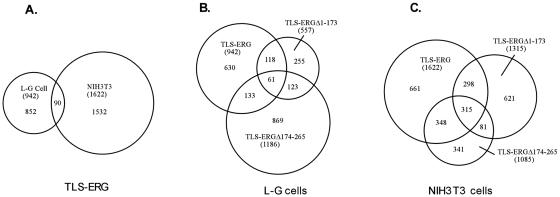
FIG. 5.
Venn diagrams of genes affected by TLS-ERG and its mutants in L-G and NIH 3T3 cells. (A) RNAs from L-G and NIH 3T3 cells stably expressing HA-TLS-ERG were used to analyze gene expression patterns on the Affymetrix GeneChips (mouse genome 430, version 2.0, array). Probe sets that differed from the HA-TLS-ERGΔETS baseline control by at least twofold were selected. The sizes of the Venn circles are proportional to the total number of modulated probe sets. Venn numbers correspond to overlapped probe sets between L-G and NIH 3T3 cells. RNAs from L-G (B) and NIH 3T3 (C) cells stably expressing HA-TLS-ERG, HA-TLS-ERGΔ1-173, or HA-TLS-ERGΔ174-265 were analyzed by the mouse genome 430, version 2.0, array using HA-TLS-ERGΔETS as the baseline control. The total number of probe sets that were affected twofold or higher by each construct is included in parentheses. The number of probe sets shared by the mutants is also indicated.
In NIH 3T3 cells, TLS-ERG appeared to regulate genes that are also targets of the EWS-ETS family of fusion proteins (such as EWS-FLI-1, EWS-ERG, and EWS-ETV1) found in Ewing's sarcoma. It was noticed that the target genes shared by TLS-ERG and EWS-ETS included the well-characterized ubiquitin-conjugating enzyme E2-C (1), cyclin D1 (10, 15), uridine phosphorylase (12), matrix metalloproteinase 3, and inhibitor of DNA binding 1 (13). In L-G cells, however, TLS-ERG appeared to regulate a different set of genes than the one regulated in NIH 3T3 cells. We identified 90 genes that were up- or down-regulated by TLS-ERG in both L-G and NIH 3T3 cells (Fig. 5A), and the ones with a change of at least fourfold are listed in Table 1. None of the listed genes in Table 1 has been found to be the target of EWS-ETS fusion proteins, suggesting that TLS-ERG mediates transformation of hematopoietic and nonhematopoietic cells through different pathways. This notion is also supported by significant divergence observed in microarray data from EWS-FLI-1-expressing mouse NIH 3T3 cells and primary human fibroblasts (13, 25).
TABLE 1.
Genes similarly regulated in both L-G and NIH 3T3 cells by TLS-ERGa
| Probe set identification no. | Gene title | Gene name | Fold change in cell typeb:
|
|
|---|---|---|---|---|
| L-G | NIH 3T3 | |||
| 1417689_a_at | Membrane-associated protein 17 | Map17 | −9 | −6 |
| 1419709_at | Stefin A3 | Stfa3 | 34 | 8 |
| 1425675_s_at | CEA-related cell adhesion molecule 1 | Ceacam1 | 4 | 4 |
| 1426284_at | Keratin 20 | Krt20 | 42 | 37 |
| 1427747_a_at | Lipocalin 2 | Len2 | 56 | 74 |
| 1428781_at | RIKEN cDNA 1110014F24 gene | 1110014F24Rik | 7 | 5 |
| 1429700_at | RIKEN cDNA 3110040M04 gene | 3110040M04Rik | 12 | 5 |
| 1448617_at | CD53 antigen | Cd53 | 7 | 4 |
| 1450009_at | Lactotransferrin | Ltf | 97 | 7 |
| 1450494_x_at | CEA-related cell adhesion molecule 1 | Ceacam1 | 5 | 4 |
Complete raw data can be found at http://www.vams.edu/pathology/research/default.asp.
Change is calculated relative to the TLS/ERGΔETS control. Listed are genes with a change of at least four-fold.
Since both TLS-ERG and TLS-ERGΔ1-173 can transform L-G myeloid progenitor cells (19), we examined whether these two constructs indeed regulate genes that are not shared with the nontransforming TLS-ERGΔ174-265. With a minimum of twofold change as the criteria, we identified 118 genes commonly affected by TLS-ERG and TLS-ERGΔ1-173 but not by TLS-ERGΔ174-265 in L-G cells (Fig. 5B). As shown in Table 2, expression of 17 of these genes were affected at least fourfold in L-G cells expressing TLS-ERG and TLS-ERGΔ1-173 compared to L-G cells expressing TLS-ERGΔ174-265. It was also noticed that the G-CSF receptor was up-regulated and granzyme B was down-regulated by TLS-ERG and TLS-ERGΔ1-173 but not by TLS-ERGΔ174-265 (see http://www.vams.edu/pathology/research/default.asp), as reported previously (19).
TABLE 2.
Genes regulated in L-G cells by TLS-ERG and TLS-ERGΔ1-173a
| Probe set identification no. | Gene title | Gene name | Fold change in cells expressingb:
|
|||
|---|---|---|---|---|---|---|
| TLS-ERG | TLS-ERGΔ1-173 | TLS-ERGΔ174-265 | TLS-ERGΔETS | |||
| 1415960_at | Myeloperoxidase | Mpo | 478 | 18 | NCc | 1 |
| 1417323_at | RIKEN cDNA 5430413I02 gene | 5430413I02Rik | 6 | 6 | NC | 1 |
| 1419394_s_at | S100 calcium binding protein A8 (calgranulin A) | S100a8 | 388 | 6 | NC | 1 |
| 1420463_at | Cytokine-dependent hematopoietic cell linker | Clnk | 12 | 13 | NC | 1 |
| 1421732_at | Glutamine repeat protein 1 | Glrp1 | 5 | 4 | NC | 1 |
| 1424111_at | Insulin-like growth factor 2 receptor | Igf2r | 5 | 5 | NC | 1 |
| 1426433_at | Myc target 1 | Myct1 | 6 | 23 | NC | 1 |
| 1427381_at | Immunoresponsive gene 1 | Irg1 | 23 | 9 | NC | 1 |
| 1427604_a_at | ATPase, class II, type 9A | Atp9a | 5 | 13 | NC | 1 |
| 1435761_at | Stefin A1 | Stfa1 | 79 | 34 | NC | 1 |
| 1438855_x_at | Tumor necrosis factor alpha-induced protein 2 | Tnfaip2 | 26 | 23 | NC | 1 |
| 1439568_at | Gene regulated by estrogen in breast cancer protein | Greb1 | −60 | −9 | −4 | 1 |
| 1440837_at | BG144448 | −28 | −7 | −3 | 1 | |
| 1447584_s_at | AI642973 | 7 | 30 | NC | 1 | |
| 1448756_at | S100 calcium binding protein A9 (calgranulin B) | S100a9 | 294 | 9 | −3 | 1 |
| 1449074_at | RIKEN cDNA 1700019N12 gene | 1700019N12Rik | 4 | 4 | NC | 1 |
| 1452340_at | RIKEN cDNA 6820424L24 gene | 682042L24Rik | 11 | 14 | NC | 1 |
Complete raw data can be found at http://www.vams.edu/pathology/research/default.asp.
Change is calculated relative to the TLS/ERGΔETS control. Listed are genes with a change of at least fourfold in both TLS-ERG and TLS-ERGΔ1-173 samples over TLS-ERGΔ174-265.
NC, no change in expression level.
In NIH 3T3 cells, both TLS-ERG and TLS-ERGΔ174-265 possess the transformation potential, whereas TLS-ERGΔ1-173 is nontransforming (19). Analysis of microarray data revealed that 348 genes may be related to transformation of fibroblasts, since they were found to be affected at least twofold by both TLS-ERG and TLS-ERGΔ174-265 but not by the nontransforming TLS-ERGΔ1-173 (Fig. 5C). 19 of the genes that were affected at least fourfold by the transforming TLS-ERG and TLS-ERGΔ174-265 are listed in Table 3, and none of the 19 genes is significantly affected by TLS-ERG in L-G myeloid progenitor cells (see http://www.vams.edu/pathology/research/default.asp).
TABLE 3.
Genes regulated in NIH 3T3 cells by TLS-ERG and TLS-ERGΔ174-265a
| Probe set identification no. | Gene title | Gene name | Fold change in cells expressingb:
|
|||
|---|---|---|---|---|---|---|
| TLS-ERG | TLS-ERGΔ1-173 | TLS-ERGΔ174-265 | TLS-ERGΔETS | |||
| 1416101_a_at | Histone 1, H1c | Hist1h1c | −5 | −1.4 | −4 | 1 |
| 1416645_a_at | Alpha fetoprotein | Afp | 8 | NCc | 12 | 1 |
| 1417426_at | Proteoglycan, secretory granule | Prg1 | 28 | NC | 23 | 1 |
| 1417851_at | Chemokine (C-X-C motif) ligand 13 | Cxcl13 | 8 | NC | 17 | 1 |
| 1417940 | RAD51-associated protein 1 | Rad51ap1 | 4 | NC | 4 | 1 |
| 1420773_at | Deubiquitinating enzyme 1 | Dub1 | 9 | NC | 4 | 1 |
| 1420855_at | Elastin | Eln | −6 | 3 | −5 | 1 |
| 1423093_at | Inner centromere protein | INCenp | 4 | NC | 4 | 1 |
| 1425039_at | Integrin beta-like 1 | Itgb11 | −4 | −1.9 | −4 | 1 |
| 1428142_at | Ets variant gene 5 | Etv5 | −4 | NC | −4 | 1 |
| 1429659_at | SMC2-structural maintenance of chromosomes 2-like 1 (yeast) | Smc211 | 15 | NC | 17 | 1 |
| 1442077_at | RIKEN cDNA 2310076G05 gene | 2310076G05Rik | 24 | NC | 20 | 1 |
| 1448859_at | Chemokine (C-X-C motif) ligand 13 | Cxcl13 | 6 | NC | 8 | 1 |
| 1449877_s_at | Kinesin family member C1 | Kifc1 | 4 | 2 | 4 | 1 |
| 1451679_at | RIKEN cDNA 6530401D17 gene | 6530401D17Rik | 20 | 3 | 11 | 1 |
| 1453202_at | RIKEN cDNA E330016A19 gene | E330016A19Rik | 4 | NC | 4 | 1 |
| 1453748_a_at | Kinesin family member 23 | Kif23 | 4 | NC | 4 | 1 |
| 1455609_at | RIKEN cDNA C030025P15 gene | C030025P15Rik | 6 | NC | 5 | 1 |
| 1458374_at | Expressed sequence C79407 | C79407 | 4 | NC | 4 | 1 |
Complete raw data can be found at http://www.vams.edu/pathology/research/default.asp.
Change is calculated relative to the TLS/ERGΔETS control. Listed are genes with a change of at least fourfold in both TLS-ERG and TLS-ERGΔ174-265.
NC, no change in expression level.
To identify potential common targets that are associated with cellular transformation regardless of cell type, the 118 probe sets from L-G cells (affected twofold or more by transforming TLS-ERG and TLS-ERGΔ1-173) and the 348 probe sets from NIH 3T3 cells (affected twofold or more by transforming TLS-ERG and TLS-ERGΔ174-265) were compared side by side. We found that only one single probe set, corresponding to the transcription factor 19 (Tcf19) (23), was present in both groups. Further analysis of our array data revealed that transformation of L-G cells was accompanied by a 2-fold decrease of Tcf19, whereas transformation of NIH 3T3 cells resulted in a 2.5-fold increase of Tcf19.
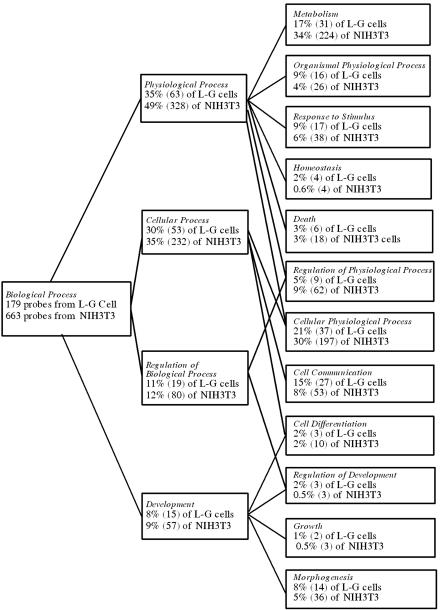
To examine biological processes that might be differentially affected by the TLS-ERG fusion protein during transformation of these two types of cells, we analyzed the 179 probe sets identified from L-G cells (shared by the transforming TLS-ERG and TLS-ERGΔ1-173) and the 663 probe sets from NIH 3T3 cells (shared by the transforming TLS-ERG and TLS-ERGΔ174-265) using the GO browser on the Affymetrix website. As shown in Fig. 6, transformation of L-G cells is associated with a significantly higher percentage of genes involved in organismal physiological process, homeostasis, cell communication, development regulation, and growth. On the other hand, transformation of NIH 3T3 cells is associated with a higher percentage of genes involved in metabolism and regulation of physiological process. These results suggest that transformation by TLS-ERG may be mediated through different mechanisms by affecting distinct sets of genes in hematopoietic and nonhematopoietic cells.
FIG. 6.
Biological processes affected by the transforming HA-TLS-ERG and its mutants in L-G and NIH 3T3 cells. One hundred seventy-nine probe sets shared by the transforming TLS-ERG and TLS-ERGΔ1-173 in L-G cells and 663 probe sets shared by the transforming TLS-ERG and TLS-ERGΔ174-265 in NIH 3T3 cells were analyzed using the Affymetrix GO browser. After the analysis, subgroups of the probe sets (with the number in parentheses) were assigned to the specific biological processes indicated in italics. As a gene product can potentially be involved in more than one biological process, a probe set may thus be assigned to more than one subgroup. Some of the probe sets were not assigned due to lack of information regarding their gene products.
DISCUSSION
In this study, we have demonstrated coimmunoprecipitation of Pol II, TLS, and SR proteins in an in vivo context. Since all three proteins (Pol II, TLS, and SR) interact with each other, we believe that the existence of Pol II-TLS-SR multiprotein complexes can better account for our results than the alternative explanation proposing separate Pol II-SR and TLS-SR complexes inside cells. Our study therefore provides further support to the notion that TLS and the related EWS and TAFII68 proteins function as bridges connecting gene transcription with RNA splicing. As TLS-ERG still binds to Pol II but no longer recruits splicing factors, the fusion protein is potentially capable of deregulating several cellular processes, including gene transcription, RNA splicing, and/or DNA-binding-independent antiapoptotic activity (20, 22, 34, 40, 42).
It was originally thought that specific fusion genes are only associated with specific tumor types (24), but such a belief has been challenged by the findings that the ETV6-NTRK fusion protein is present in infantile fibrosarcoma, acute myeloid leukemia, and secretory breast carcinoma (14, 21, 38). The fact that the TLS-ERG fusion protein is found in both human myeloid leukemia and sarcoma provides additional support to the notion that a specific fusion gene can be associated with multiple types of tumor.
How does expression of the TLS-ERG fusion protein lead to two distinct tumor phenotypes? It may be argued that TLS-ERG utilizes the same oncogenic mechanism in both leukemia and sarcoma and it is the cell lineage in which the translocation occurs that determines the tumor type. On the other hand, it is also possible that TLS-ERG activates different oncogenic pathways in the transformation of hematopoietic and nonhematopoietic cells. To differentiate these possibilities, we used two cell lines of different lineages in this study: the mouse L-G myeloid progenitor cells and NIH 3T3 fibroblasts. In accordance with previous studies (19), transformation of L-G cells is defined as proliferation in the presence of G-CSF instead of differentiation into neutrophils and transformation of NIH 3T3 cells is defined as anchorage-independent colony formation in soft agar.
Through analysis of TLS-ERG mutants, we identified that the first 173 amino acids of TLS comprise the subdomain that mediates interaction with RNA Pol II. Deletion of this Pol II-interacting subdomain appears to cause transcriptional repression of the GPIX promoter construct in both L-G and NIH 3T3 cells. We speculate that the repression is caused, at least in part, by the unmasking of a potential repression domain between amino acids 174 and 265. In support of such a contention, deletion of amino acids 174 to 265 appears to modestly relieve the repression on the GPIX promoter construct in both L-G and NIH 3T3 cells.
In L-G myeloid progenitor cells, both TLS-ERG and TLS-ERGΔ1-173 could repress the GPIX promoter construct and cause cellular transformation. The nontransforming TLS-ERGΔ174-265 and TLS-ERGΔETS did not repress the promoter construct or transform the cells. In NIH 3T3 fibroblasts, TLS-ERG and TLS-ERGΔ174-265 could cause cellular transformation but neither protein showed a significant effect on the transcription of the luciferase reporter gene driven by the GPIX promoter. These results suggest that transformation of L-G cells is primarily associated with deregulation of gene transcription. Since the GPIX gene is not expressed in NIH 3T3 cells, the GPIX luciferase construct may not be an ideal reporter to predict whether transformation of NIH 3T3 cells is dependent on transcriptional deregulation.
When the effect of TLS-ERG on E1A splicing was assayed in NIH 3T3 cells, however, the transforming TLS-ERG and TLS-ERGΔ174-265 were able to inhibit an unknown splicing product and the nontransforming TLS-ERGΔ1-173 did not possess such an ability. It should be pointed out that interference with such a splicing product appears necessary but not sufficient to induce transformation of NIH 3T3 fibroblasts. Other potential functions of TLS-ERG, such as the antiapoptotic activity recently reported to not require DNA-binding ability of the structurally homologous EWS-FLI-1 fusion protein (34), may also play a role in transformation of NIH 3T3 cells.
We carried out DNA microarray experiments in an attempt to identify TLS-ERG target genes that are critical to transformation of L-G myeloid progenitors and NIH 3T3 fibroblasts. The microarray experiments measure changes in mRNA steady-state level, which reflects the combined contributions from gene transcription, pre-mRNA splicing, and mRNA stability. How TLS-ERG regulates some of these target genes is currently under further investigation.
Our microarray analysis reveals that TLS-ERG can influence a unique set of genes in L-G myeloid progenitors. In NIH 3T3 cells, however, the TLS-ERG fusion protein appears to affect many genes that have been reported to be targets of the structurally related EWS-ETS fusion proteins including EWS-FLI-1 and EWS-ERG. Among the common targets of TLS-ERG, EWS-FLI-1, and EWS-ERG is the gene encoding uridine phosphorylase (13). Ectopic expression of uridine phosphorylase has recently been reported to support anchorage-independent growth of NIH 3T3 cells (12). Since uridine phosphorylase is also induced by the nontransforming HA-TLS-ERGΔ174-265, it is not clear whether this TLS-ERG target gene alone plays a critical role in the transformation of NIH 3T3 fibroblasts. It is probably more likely that transformation of NIH 3T3 fibroblasts by TLS-ERG requires contributions from a battery of deregulated genes.
Based on studies from our group and others, one can conclude that the structurally related TLS and EWS fusion proteins transform NIH 3T3 fibroblasts through a similar mechanism that include activation of a common set of genes such as uridine phosphorylase. However, our study also indicates that the same TLS-ERG fusion protein most likely utilizes a different pathway in a different cellular background such as that of the L-G myeloid progenitor cells. This notion is also supported by studies of EWS-FLI-1 in different cellular backgrounds (13, 25, 37, 44) and may have implications for how we should proceed with the investigation of TLS and EWS fusion proteins, as most experiments have been done in the surrogate NIH 3T3 cells. Target genes identified in NIH 3T3 fibroblasts may not be the ones that are actually affected in the cancer itself. In this regard, the study of cells derived from the original cancer should serve as a better alternative to identify target genes using techniques such as RNA interference (6, 7, 32). Ideally, this should be complemented by introduction of the fusion gene into stem cells that are capable of differentiation into precursor cells from which the corresponding cancer arises.
Acknowledgments
J.Z. was supported by an award from the National Scholarship Fund of China. H.A.C. was supported by a Veterans Affairs Merit Review Award. L.Y. was supported by National Institutes of Health grants 1RO1CA090941 and 1RO1AR051455.
We thank Daniel Y. Wu and Danilo A. Perrotti for critical reading of the manuscript.
REFERENCES
- 1.Arvand, A., H. Bastians, S. M. Welford, A. D. Thompson, J. V. Ruderman, and C. T. Denny. 1998. EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 17:2039-2045. [DOI] [PubMed] [Google Scholar]
- 2.Bastian, L. S., M. Yagi, C. Chan, and G. J. Roth. 1996. Analysis of the megakaryocyte glycoprotein IX promoter identifies positive and negative regulatory domains and functional GATA and Ets sites. J. Biol. Chem. 271:18554-18560. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti, A., Y. Lutz, D. J. Heard, P. Chambon, and L. Tora. 1996. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro- oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15:5022-5031. [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn, M. L., H. A. Chansky, A. Zielinska-Kwiatkowska, Y. Matsui, and L. Yang. 2003. Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim. Biophys. Acta 1629:8-14. [DOI] [PubMed] [Google Scholar]
- 5.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 6.Chansky, H. A., F. Barahmand-Pour, Q. Mei, W. Kahn-Farooqi, A. Zielinska-Kwiatkowska, M. Blackburn, K. Chansky, E. U. Conrad III, J. D. Bruckner, T. K. Greenlee, and L. Yang. 2004. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J. Orthop. Res. 22:910-917. [DOI] [PubMed] [Google Scholar]
- 7.Chansky, H. A., M. Blackburn, W. Kahn-Farooqi, D. Benedetti, and L. Yang. 2004. Trans. 50th Annu. Meet. Orthop. Res. Soc., vol. 29, p. 0058.
- 8.Chansky, H. A., M. Hu, D. D. Hickstein, and L. Yang. 2001. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 61:3586-3590. [PubMed] [Google Scholar]
- 9.Crozat, A., P. Aman, N. Mandahl, and D. Ron. 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363:640-644. [DOI] [PubMed] [Google Scholar]
- 10.Dauphinot, L., C. De Oliveira, T. Melot, N. Sevenet, V. Thomas, B. E. Weissman, and O. Delattre. 2001. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene 20:3258-3265. [DOI] [PubMed] [Google Scholar]
- 11.Delattre, O., J. Zucman, B. Plougastel, C. Desmaze, T. Melot, M. Peter, H. Kovar, I. Joubert, P. de Jong, G. Rouleau, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162-165. [DOI] [PubMed] [Google Scholar]
- 12.Deneen, B., H. Hamidi, and C. T. Denny. 2003. Functional analysis of the EWS/ETS target gene uridine phosphorylase. Cancer Res. 63:4268-4274. [PubMed] [Google Scholar]
- 13.Deneen, B., S. M. Welford, T. Ho, F. Hernandez, I. Kurland, and C. T. Denny. 2003. PIM3 proto-oncogene kinase is a common transcriptional target of divergent EWS/ETS oncoproteins. Mol. Cell. Biol. 23:3897-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eguchi, M., M. Eguchi-Ishimae, A. Tojo, K. Morishita, K. Suzuki, Y. Sato, S. Kudoh, K. Tanaka, M. Setoyama, F. Nagamura, S. Asano, and N. Kamada. 1999. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood 93:1355-1363. [PubMed] [Google Scholar]
- 15.Fuchs, B., C. Y. Inwards, and R. Janknecht. 2004. Vascular endothelial growth factor expression is up-regulated by EWS-ETS oncoproteins and Sp1 and may represent an independent predictor of survival in Ewing's sarcoma. Clin. Cancer Res. 10:1344-1353. [DOI] [PubMed] [Google Scholar]
- 16.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 17.Hallier, M., A. Lerga, S. Barnache, A. Tavitian, and F. Moreau-Gachelin. 1998. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 273:4838-4842. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa, H., K. Shimizu, Y. Hayashi, and M. Ohki. 1994. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 54:2865-2868. [PubMed] [Google Scholar]
- 19.Ichikawa, H., K. Shimizu, R. Katsu, and M. Ohki. 1999. Dual transforming activities of the FUS (TLS)-ERG leukemia fusion protein conferred by two N-terminal domains of FUS (TLS). Mol. Cell. Biol. 19:7639-7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaishankar, S., J. Zhang, M. F. Roussel, and S. J. Baker. 1999. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene 18:5592-5597. [DOI] [PubMed] [Google Scholar]
- 21.Knezevich, S. R., D. E. McFadden, W. Tao, J. F. Lim, and P. H. Sorensen. 1998. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 18:184-187. [DOI] [PubMed] [Google Scholar]
- 22.Knoop, L. L., and S. J. Baker. 2001. EWS/FLI alters 5′-splice site selection. J. Biol. Chem. 276:22317-22322. [DOI] [PubMed] [Google Scholar]
- 23.Ku, D. H., C. D. Chang, J. Koniecki, L. A. Cannizzaro, L. Boghosian-Sell, H. Alder, and R. Baserga. 1991. A new growth-regulated complementary DNA with the sequence of a putative trans-activating factor. Cell Growth Differ. 2:179-186. [PubMed] [Google Scholar]
- 24.Ladanyi, M., and J. A. Bridge. 2000. Contribution of molecular genetic data to the classification of sarcomas. Hum. Pathol. 31:532-538. [DOI] [PubMed] [Google Scholar]
- 25.Lessnick, S. L., C. S. Dacwag, and T. R. Golub. 2002. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 1:393-401. [DOI] [PubMed] [Google Scholar]
- 26.Lopez, J. A. 1994. The platelet glycoprotein Ib-IX complex. Blood Coagul. Fibrinolysis 5:97-119. [PubMed] [Google Scholar]
- 27.Matsui, Y., H. A. Chansky, F. Barahmand-Pour, A. Zielinska-Kwiatkowska, N. Tsumaki, A. Myoui, H. Yoshikawa, L. Yang, and D. R. Eyre. 2003. COL11A2 collagen gene transcription is differentially regulated by EWS/ERG sarcoma fusion protein and wild-type ERG. J. Biol. Chem. 278:11369-11375. [DOI] [PubMed] [Google Scholar]
- 28.Pereira, D. S., C. Dorrell, C. Y. Ito, O. I. Gan, B. Murdoch, V. N. Rao, J. P. Zou, E. S. Reddy, and J. E. Dick. 1998. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. Proc. Natl. Acad. Sci. USA 95:8239-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotti, D., S. Bonatti, R. Trotta, R. Martinez, T. Skorski, P. Salomoni, E. Grassilli, R. V. Lozzo, D. R. Cooper, and B. Calabretta. 1998. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 17:4442-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrotti, D., A. Iervolino, V. Cesi, M. Cirinna, S. Lombardini, E. Grassilli, S. Bonatti, P. P. Claudio, and B. Calabretta. 2000. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol. Cell. Biol. 20:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad, D. D., M. Ouchida, L. Lee, V. N. Rao, and E. S. Reddy. 1994. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene 9:3717-3729. [PubMed] [Google Scholar]
- 32.Prieur, A., F. Tirode, P. Cohen, and O. Delattre. 2004. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 24:7275-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabbitts, T. H., A. Forster, R. Larson, and P. Nathan. 1993. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 4:175-180. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan, R., Y. Fujimura, J. P. Zou, F. Liu, L. Lee, V. N. Rao, and E. S. Reddy. 2004. Role of protein-protein interactions in the antiapoptotic function of EWS-Fli-1. Oncogene 23:7087-7094. [DOI] [PubMed] [Google Scholar]
- 35.Rossow, K. L., and R. Janknecht. 2001. The Ewing's sarcoma gene product functions as a transcriptional activator. Cancer Res. 61:2690-2695. [PubMed] [Google Scholar]
- 36.Shing, D. C., D. J. McMullan, P. Roberts, K. Smith, S. F. Chin, J. Nicholson, R. M. Tillman, P. Ramani, C. Cullinane, and N. Coleman. 2003. FUS/ERG gene fusions in Ewing's tumors. Cancer Res. 63:4568-4576. [PubMed] [Google Scholar]
- 37.Staege, M. S., C. Hutter, I. Neumann, S. Foja, U. E. Hattenhorst, G. Hansen, D. Afar, and S. E. Burdach. 2004. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res. 64:8213-8221. [DOI] [PubMed] [Google Scholar]
- 38.Tognon, C., S. R. Knezevich, D. Huntsman, C. D. Roskelley, N. Melnyk, J. A. Mathers, L. Becker, F. Carneiro, N. MacPherson, D. Horsman, C. Poremba, and P. H. Sorensen. 2002. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2:367-376. [DOI] [PubMed] [Google Scholar]
- 39.Wang, J., and J. L. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:335-346. [PMC free article] [PubMed] [Google Scholar]
- 40.Welford, S. M., S. P. Hebert, B. Deneen, A. Arvand, and C. T. Denny. 2001. DNA binding domain-independent pathways are involved in EWS/FLI1-mediated oncogenesis. J. Biol. Chem. 276:41977-41984. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, K., H. Hamaguchi, K. Nagata, M. Kobayashi, F. Tanimoto, and M. Taniwaki. 1997. Establishment of a novel human acute myeloblastic leukemia cell line (YNH-1) with t(16;21), t(1;16) and 12q13 translocations. Leukemia 11:599-608. [DOI] [PubMed] [Google Scholar]
- 42.Yang, L., L. J. Embree, and D. D. Hickstein. 2000. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol. Cell. Biol. 20:3345-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273:27761-27764. [DOI] [PubMed] [Google Scholar]
- 44.Zwerner, J. P., J. Guimbellot, and W. A. May. 2003. EWS/FLI function varies in different cellular backgrounds. Exp. Cell Res. 290:414-419. [DOI] [PubMed] [Google Scholar]