Abstract
Platelet-derived growth factor (PDGF) protein family members are potent mitogens and chemoattractants for mesenchymal cells. The classic PDGF ligands A and B are single-domain protein chains which are secreted as active dimers capable of activating their cognate PDGF receptors (PDGFRs). In contrast to PDGFs A and B, PDGF D contains an N-terminal complement subcomponent C1r/C1s, Uegf, and Bmp1 (CUB) domain and a C-terminal PDGF domain. PDGF D must undergo extracellular proteolytic processing, separating the CUB domain from the PDGF domain, before the PDGF domain can stimulate β-PDGFR-mediated cell signal transduction. Here, we report that prostate carcinoma cells LNCaP and PC3 autoactivate latent full-length PDGF D into its active form under serum-independent conditions and that this autoactivation is inhibited by PAI-1, a urokinase plasminogen activator (uPA)/tissue plasminogen activator (tPA) inhibitor. Interestingly, uPA, but not the closely related protease tPA, is capable of processing recombinant latent PDGF DD into the active form. We identify the uPA cleavage site between the CUB and PDGF domains of the full-length PDGF D by mutational analysis and show that PDGF D and uPA colocalize in human prostate carcinoma. This evidence provides a direct link between uPA- and PDGF D-mediated cell signaling, which may contribute to the progression of prostate cancer.
Platelet-derived growth factors (PDGFs) regulate a diverse array of cellular processes, including cell proliferation, transformation, migration, survival, and apoptosis of mesenchymal cells, in development as well as during pathogenesis (reviewed in references 31 and 42). For over 2 decades, PDGFs were thought to exist as the homodimers PDGF AA and BB or the heterodimer PDGF AB. These PDGF dimers are processed intracellularly and secreted as active dimers that readily activate PDGF receptors (PDGFRs). Recently, two new PDGF ligands (PDGF CC and DD) were discovered that have a unique two-domain structure with an N-terminal complement subcomponent C1r/C1s, Uegf, Bmp1 (CUB) domain and a C-terminal PDGF/vascular endothelial growth factor domain. PDGF CC and DD are secreted as full-length, latent dimers, and the proteolytic removal of the CUB domain is required for the growth factor domain of PDGF CC or DD to activate the PDGF receptors (4, 20, 21).
PDGFs exert their biological functions through the activation of dimeric receptors made up of two structurally similar protein-tyrosine kinase receptor subunits (αα-, αβ-, or ββ-PDGFR). While αα-PDGFR can be activated by PDGF AA, BB, or CC, ββ-PDGFR is activated by PDGF BB or DD. Although both α- and β-PDGFRs mediate strong mitogenic signals, studies suggest that β-PDGFR induces more potent transforming signals than α-PDGFR (3, 41). Consistently, increased β-PDGFR expression was shown to correlate with the development of many human tumors, including gliomas (30, 40), myelomonocytic leukemia (13), and osteosarcoma (37). Importantly, recent studies revealed a critical role for β-PDGFR in prostate cancer. A majority of prostate tumor tissues at both primary and metastasized sites express β-PDGFR as determined by immunohistochemical analysis. Interestingly, β-PDGFR staining is more prominent in endothelial cells and vascular smooth muscle cells adjacent to prostate tumor cells, suggestive of a role for β-PDGFR signaling in prostate cancer progression. PDGF BB, originally thought to be a sole ligand for β-PDGFR, has not been found in prostate cancer clinical samples, raising the possibility that PDGF D, a newly discovered ligand for β-PDGFR, may be responsible for β-PDGFR-mediated signal transduction during prostate cancer progression (10, 11, 18). To investigate the role of PDGF D/β-PDGFR in prostate cancer progression, we previously established a prostate carcinoma cell line, LNCaP, that overexpresses PDGF DD (38). Whereas PDGF DD was reported to be expressed as a latent dimer and processed into an active growth factor domain by a protease present in serum, we found that LNCaP cells autoactivate latent PDGF DD into the active PDGF domain, which can induce cell growth and migration in both autocrine and paracrine manners. More importantly, PDGF DD expression accelerates the early onset of prostate tumor growth and drastically enhances prostate carcinoma cell invasion into surrounding stromal cells in a SCID mouse model, demonstrating a potential oncogenic activity of PDGF DD in the development and/or progression of prostate cancer.
Since the extracellular proteolytic cleavage of PDGF DD is a key step to initiate PDGF D/β-PDGFR signaling, here we question which protease produced by prostate carcinoma cells is responsible for the processing of a latent PDGF DD into an active growth factor. To address this, we analyzed two prostate carcinoma cell lines, LNCaP and PC3, that both autoactivate latent full-length PDGF D into its active form under serum-independent conditions. Here, we identify urokinase plasminogen activator (uPA) as a protease capable of activating PDGF D, and this autoactivation is inhibited by PAI-1, an uPA/tissue plasminogen activator (tPA) inhibitor. We show that uPA, but not the closely related protease tPA, is capable of processing recombinant latent PDGF D into the active form and that the uPA cleavage site resides within the hinge region between the CUB and the growth factor domains, as determined by mutational analysis. Lastly, confocal microscopy assay reveals the colocalization of PDGF D and uPA in human prostate carcinoma, suggesting a direct link between uPA and PDGF D-mediated cell signaling during prostate cancer progression.
MATERIALS AND METHODS
Cell culture.
Human prostate carcinoma LNCaP and PC3 cells and resultant transfected cell lines were cultured in a humidified 5% CO2 incubator with RPMI 1640 medium supplemented with 5% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco BRL, Carlsbad, CA). The green monkey kidney cell line CV-1 was cultured in the same manner, except maintained in Dulbecco's modified Eagle medium (DMEM)-10% FBS.
Generation of stable cell lines overexpressing PDGF D.
Construction of the mammalian PDGF D expression vector pcDNA3-PDGF D:His and establishment of PDGF D-overexpressing LNCaP cell lines were previously described (38). PC3 cells transfected with pcDNA3-PDGF D:His or the corresponding empty vector pcDNA3-neo were selected using 350 μg/ml Geneticin (G418) in RPMI 1640 with 10% fetal bovine serum. Cells which survived selection were pooled together and are referred to herein as PC3-PDGF D or PC3-neo. Overexpression of PDGF D was confirmed by reverse transcription (RT)-PCR as described previously (primer pair is given below).
Production of rPDGF D protein using the vaccinia expression system.
Cloning of full-length PDGF D cDNA into the vaccinia virus expression vector pTF7-ECM1 was described previously (38). The expression vector pTF7-PDGF D:HIS was used to generate a stable, recombinant vaccinia virus which contains the full-length PDGF D cDNA under the control of a T7 promoter, using established protocols (12). This recombinant vaccinia virus expressing PDGF D is referred to as vPDGF D:HIS. vPDGF D:HIS was used to generate recombinant PDGF D (rPDGF D) conditioned medium. In short, the green monkey kidney cell line CV-1 was coinfected with vPDGF D:HIS and the control virus vTF7-3, which expresses the T7 RNA polymerase protein required for the expression of rPDGF D from vPDGF D:HIS. Coinfection was done in serum-free DMEM. After 30 min of infection, the cells were washed once with 1× phosphate-buffered saline (PBS) and then cultured in serum-free DMEM (Gibco BRL) for 24 h. Conditioned medium (CM) containing rPDGF D was collected, and cellular debris was pelleted by a 5-min centrifugation at 2,000 rpm. The presence of rPDGF D was confirmed by immunoblot analysis using our PDGF D growth domain antibody before use in further experiments.
Reagents.
Generation of custom-made antibody which recognizes the growth domain of PDGF D was described previously (38). uPA antibody which recognizes the beta chain was purchased from American Diagnostica (Stamford, CT). The proteases uPA and tPA were purchased from American Diagnostica and furin was purchased from R&D Biosystems (Minneapolis, MN). The protease inhibitors PAI-1, TAPI-1, and PDX-Portland were purchased from EMD Bioscience (San Diego, CA) and aprotinin and leupeptin were purchased from Sigma-Aldrich (St. Louis, MO). Spectrozyme PL plasmin activity assay (American Diagnostica) was used, per manufacturer's instructions, to assay for plasmin activity in CM.
RT-PCR.
The primer pair used to detect PDGF D expression was as follows: forward primer, 5′-GAACAGCTACCCCAGGAACC-3′, and reverse primer, 5′-CTTGTGTCCACACCATCGTC-3′. This primer pair amplifies a 185-bp product that represents part of the CUB coding region of the PDGF D protein. The primer pair for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as a positive control in all RT-PCR experiments with the primer pair comprised of forward primer 5′-ATCACCATCTTCCAGGAGCGA-3′ and reverse primer 5′-GCCAGTGAGCTTCCCGTTCA-3′, amplifying an approximately 500-bp product.
Construction of mutant PDGF DR247,249A and generation of rPDGF DR247,249A protein.
Full-length PDGF D cDNA, without the six-His C-terminus tag, was inserted into the pCR2.1-TOPO cloning vector (Invitrogen, La Jolla, CA) and used as a template in the QuikChange site-directed mutagenesis kit, per manufacturer's instructions (Stratagene, La Jolla, CA). The double mutation was confirmed by DNA sequencing (Elim Biopharmaceuticals, San Francisco, CA). Primers used to generate the site-directed mutation were as follows: sense primer, 5′-gacacccctcggtatGCaggcGCgtcataccatgaccg-3′, and antisense primer, 5′- cggtcatggtatgacGCgcctGCataccgaggggtgtc-3′ (mutations in uppercase). This plasmid, referred to as pPCR2.1-PDGF DR247,249A, was then digested with BclI and AflIII, and the ∼1.1-kb insert was gel purified and ligated into the NcoI-BamHI site of the vaccinia virus expression vector pT7F-ECM1. The resultant vaccinia virus expression plasmid pTF7-PDGF DR247,249A was then used to generate rPDGF DR247,249A using previously described coinfection/transfection protocols (12, 38).
PDGF D-mediated paracrine activation of β-PDGFR and ERKs.
LNCaP-PDGF D, LNCaP-neo, PC3-PDGF D, and PC3-neo cells were cultured in serum-free medium for 48 h. CM was collected and centrifuged at 2,000 rpm for 5 min to remove cell debris. Serum-starved NIH 3T3 cells were treated with indicated CM or positive and negative controls for 10 min and lysed in immunoprecipitation (IP)-lysis buffer (0.5% sodium deoxycholate, 1% Nonidet P-40, 50 mM Tris [pH 7.6], 2 mM EGTA, 2 mM EDTA, 150 mM NaCl, 2 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml of leupeptin, and 10 μg/ml aprotinin). The lysates were centrifuged for 20 min at 12,000 × g to remove debris, and protein concentrations were determined using the bicinchoninic acid protein assay kit, per manufacturer's instructions (Pierce Biotechnology, Rockford, IL). Next, 500 μg of lysate was used for immunoprecipitation with an anti-β-PDGFR antibody (Santa Cruz Biotech, Santa Cruz, CA) and protein G agarose beads (Pierce Biotechnology, Rockford, IL). Immunoprecipitates were washed five times with IP-lysis buffer and resolved by reducing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Tyrosine-phosphorylated β-PDGFR was detected by immunoblotting using an antiphosphotyrosine antibody (Upstate, Lake Placid, NY). Total levels of immunoprecipitated β-PDGFR were detected using the same antibody used for IP. To determine the total and activated levels of extracellular signal-regulating kinases 1 and 2 (ERK1/2), 50 μg of lysate from each sample was resolved by reducing SDS-PAGE and immunoblots were performed using anti-pERK1/2 or anti-ERK1/2 antibodies according to the manufacturer's instructions (Cell Signaling Technologies, Beverly, MA).
Ni-NTA concentration of rPDGF D.
rPDGF D expressed by vaccinia virus and PDGF D overexpressed by transformed cell lines contains a six-His tag at the carboxyl end of the full-length PDGF D. Therefore, a Ni affinity protocol can be used to immunoprecipitate the full-length PDGF D as well as the growth factor domain-only forms of rPDGF D. An appropriate amount of 10× binding buffer was added to either CM collected from LNCaP-PDGF, LNCaP-neo, PC3-PDGF D, or PC3-neo or CM containing virus-expressed rPDGF D to achieve the optimal pH of 6.5 and imidazole concentration of 1 mM. CM was incubated overnight at 4°C with up to 50 μl of a 50% Ni-nitrilotriacetic acid (NTA)-agarose bead slurry (QIAGEN, Valencia, CA). Beads were then collected by centrifugation for 1 min at 2,000 rpm and washed twice in a wash buffer consisting of 20 mM sodium phosphate buffer (pH 6.5), 300 mM NaCl, 1 mM imidazole, and 0.05% Tween 20. rPDGF D was stripped from the beads by incubating the beads in 10 mM EDTA at 4°C overnight.
Immunofluorescent double staining of uPA and PDGF D in human prostate tissue.
Hematoxylin and eosin (H&E) stained, as well as nonstained, slides of paraffin-embedded human prostate cancer specimens were obtained from the Wayne State University Pathology Research Core Facility. For immunofluorescent staining, slides were dewaxed with xylene and double probed with anti-GFD PDGF D rabbit polyclonal antibody and anti-uPA beta chain monoclonal antibody 3689 (American Diagnostica, Stamford, CT). Fluorescein isothiocyanate-conjugated anti-rabbit and Texas Red-conjugated anti-mouse secondary antibodies were used to detect PDGF D and uPA, respectively (Jackson Immunoresearch Laboratories, West Grove, PA). Confocal immunofluorescence microscopic analysis was performed using a Zeiss LSM 510 confocal microscopy system equipped with a C-Apochromat (numerical aperture = 1.2) 63× Korr objective lens (Carl Zeiss MicroImaging, Inc., Thornwood, NY). Images for figures were colored and resized with associated microscope software available through the Wayne State University Imaging Core Facility. H&E images were taken from corresponding serial slides at 10× and 40× magnifications using SPOT RT software v3.0 (Diagnostic Instruments, Sterling Heights, MI).
RESULTS
Human prostate carcinoma cell line PC3 processes PDGF D into its active form in a serum-independent manner.
Previously, we described the activation of full-length PDGF D (∼50-kDa monomer, ∼80-kDa dimer) by a protease produced by the human prostate carcinoma cell line LNCaP (38). To examine whether this was specific to LNCaP cells, the prostate carcinoma cell line PC3 was used to generate a PC3-PDGF D cell line that stably overexpresses PDGF D (see Materials and Methods). Very little PDGF D protein was detected in CM collected from PC3-PDGF D cells, whereas both full-length and activated growth factor domain forms of PDGF D were readily detected in CM from LNCaP-PDGF D cells (Fig. 1A), even though the expression levels of PDGF D in LNCaP-PDGF D and PC3-PDGF D cells were comparable at the RNA level (Fig. 1C). Taking advantage of the six-His tag engineered at the C terminus of PDGF D, CM was concentrated using Ni-NTA agarose beads and the concentrated sample was subjected to immunoblot analysis. Both full-length PDGF D and the ∼18-kDa form of the growth factor domain were detected in PC3-PDGF D CM (Fig. 1B). Interestingly, the majority of PDGF D protein was detected as a processed form in PC3-PDGF D whereas only a fraction of full-length PDGF D was processed into active forms in LNCaP-PDGF D cells. These results suggest that PDGF D is more efficiently processed into the growth factor domains in PC3 cells than those in the LNCaP cells.
FIG. 1.
PDGF D overexpression and activation by a protease secreted by LNCaP and PC3 cell lines. (A) PC3-neo, PC3-PDGF D, LNCaP-neo, and LNCaP-PDGF D cells were grown to 100% confluence in complete media, washed once with PBS, and then cultured in serum-free medium for 48 h. CM was collected, and proteins were resolved using SDS-PAGE under reducing conditions. (B) Three milliliters of CM from PC3-neo and PC3-PDGF D was concentrated using Ni-NTA agarose beads and resolved on a reducing SDS-PAGE gel (see Materials and Methods). Anti-PDGF D-growth domain (GD) antibody was used for immunoblot analysis. FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain monomer. (C) Semiquantitative RT-PCR analysis was performed to examine RNA levels of PDGF D in parental cell lines, as well as neo control and PDGF D overexpressing cell lines. The primer pair used detected both endogenously and exogenously expressed PDGF D.
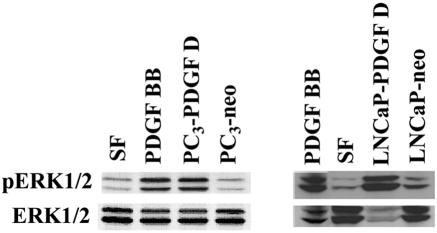
Paracrine activation of cell signaling by PDGF D produced from PC3 and LNCaP cells was confirmed by stimulating serum-starved NIH 3T3 cells with CM collected from these cells. Serum-free media with and without 25 ng/ml of PDGF BB were included as negative and positive controls, respectively. As shown in Fig. 2, CM from PC3-PDGF D and LNCaP-PDGF D, but not PC3-neo and LNCaP-neo, induced an increase in ERK1/2 activation in NIH 3T3 cells, indicating that PDGF D processed by PC3 and LNCaP cells is biologically active.
FIG. 2.
PC3-processed PDGF D is biologically active. Serum-starved NIH 3T3 cells were stimulated for 10 min with CM collected from PC3-PDGF D, PC3-neo, LNCaP-PDGF D, and LNCaP-neo. Serum-starved NIH 3T3 cells stimulated with serum-free (SF) medium or SF medium supplemented with 25 ng/ml PDGF BB served as negative and positive controls, respectively. Fifty micrograms of NIH 3T3 lysate was resolved by reducing SDS-PAGE and immunoblot analyzed using an antibody specific to active extracellular signal-regulated protein kinase (pERK1/2). To determine basal levels of ERK1/2, the same blot was stripped and immunoblotted with an antibody that recognizes both active and inactive forms of ERK1/2.
We previously showed that PDGF D can be processed, although inefficiently, by a protease at or near the LNCaP cell surface but not a soluble enzyme, as recombinant PDGF D (rPDGF D) was processed when incubated with LNCaP cells, but not in CM collected from LNCaP-PDGF D or LNCaP-neo cells. We also reported that LNCaP-PDGF D cells are slightly more effective in processing PDGF D than LNCaP-neo cells, indicating that PDGF D may induce positive feedback signaling to upregulate the protease expression/activity necessary for the PDGF D processing (38). To determine if the PC3-produced protease was similar to the LNCaP-produced protease, we incubated rPDGF D in the presence of PC3-neo and PC3-PDGF D cells or with CM collected from these two cell lines, followed by an assay for the presence of the growth factor domain of PDGF D. Full-length rPDGF D was effectively processed into the ∼18-kDa form of the growth domain when incubated in the presence of PC3-PDGF D or PC3-neo cells (Fig. 3A). A slight increase of rPDGF D processing was detected in the presence of PC3-PDGF D cells compared to PC3-neo. Additionally, the PC3-produced protease is present in a soluble form, as rPDGF D was processed into the ∼18-kDa form when incubated with CM collected from PC3-PDGF D or PC3-neo cells (Fig. 3B). These results demonstrate that both LNCaP and PC3 carcinoma cell lines produce a serum-independent protease which can activate PDGF D. Importantly, these data also show that PC3 cells produce high activity levels of protease, capable of processing PDGF D into its active form, whereas LNCaP produces low activity levels of protease that appear to be mainly cell surface associated.
FIG. 3.

rPDGF D is processed when incubated with PC3-PDGF D or PC3-neo cells or when mixed with PC3-PDGF D or PC3-neo conditioned media. (A) PC3-PDGF D and PC3-neo cells were grown to 100% confluence in a six-well plate, washed once with PBS, and then incubated in serum-free medium supplemented with rPDGF D (see Materials and Methods). CM was collected after the indicated time points and resolved on an SDS-PAGE gel under reducing conditions. PDGF D was detected through immunoblot analysis using anti-PDGF D-growth domain (GD) antibody. FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain monomer. (B) CM was collected from 48-h serum-starved PC3-PDGF D cells or PC3-neo cells and mixed with an equal amount of rPDGF D. Samples were incubated at 37°C for the indicated time points and then resolved on a reducing SDS-PAGE gel. Anti-PDGF D-GD antibody was used for immunoblot analysis.
Aprotinin and PAI-1 can inhibit PC3 or LNCaP cell processing of PDGF D.
To identify which class of protease is responsible for processing PDGF D, PC3-PDGF D and LNCaP-PDGF D cells were cultured in the presence of various protease inhibitors. As PDGF D was difficult to detect in CM collected from PC3-PDGF D cells, the initial inhibitor study presented in Fig. 4A was conducted with PC3-PDGF D cells coinfected with our recombinant vPDGF D and the vTF7-3 virus, which provides the T7 RNA polymerase required for expression of vPDGF D recombinant virus (12). As seen in Fig. 4A and B, the matrix metalloproteinase inhibitor TAPI-1, the cysteine protease inhibitor E64, and the trypsin-like cysteine protease inhibitor leupeptin had little effect on PDGF D processing in LNCaP and PC3 cells. Since furin was shown to mediate the intracellular processing of PDGF AA and BB, the furin inhibitor PDX-Portland was included in our study but was also found to have no effect on the processing of PDGF D (Fig. 4A) (33). Importantly, the serine protease inhibitor aprotinin significantly reduced the levels of processed PDGF D for both cell lines, suggesting that the serine protease(s) is responsible for PDGF D processing in these cells (Fig. 4A and B). Since the serine proteases uPA and tPA are often involved in human cancer development/progression, we tested whether the uPA/tPA inhibitor PAI-1 can suppress PDGF D processing. As shown in Fig. 4A, PAI-1 effectively suppressed the PDGF D processing at similar levels to aprotinin. These results suggest that PC3- and LNCaP-produced proteases, responsible for PDGF D processing, are a part of the uPA/tPA protease cascade.
FIG. 4.

PAI-1 inhibits LNCaP and PC3 processing of full-length PDGF D. (A) PC3-PDGF D cells were plated in a 48-well plate and grown to 100% confluence. Cells were then washed with PBS, coinfected with recombinant PDGF D virus and vTF7-3 for 30 min (see Materials and Methods), washed with PBS, and then cultured in serum-free medium with the indicated protease inhibitors or vehicle controls (EtOH or dimethyl sulfoxide [DMSO]) for 48 h. CM was then collected and resolved on a reducing SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with our PDGF D antibody. FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain (GD) monomer. (B) LNCaP-PDGF D cells were treated as described above, except infected with vTF7-3 virus control only. CM was collected and analyzed as described above. (C) PC3-PDGF D cells were plated in a six-well plate to 100% confluence, washed with PBS, and then incubated in serum-free medium with 10 μg/ml of aprotinin or 200 nM of PAI-1 for 48 h. Media were collected, and PDGF D was concentrated using Ni-NTA agarose beads. Samples were resolved on an SDS-PAGE gel under reducing conditions and transferred to a nitrocellulose membrane, and PDGF D was detected using anti-PDGF D-GD antibody.
To confirm the above results in the absence of vaccinia virus infection, PC3-PDGF D cells were plated to 100% confluence in a six-well plate in reduced serum medium (2.5% FBS) and allowed to attach overnight. Cells were then washed with PBS and incubated in serum-free medium for 2 h, followed by one more wash with PBS in an effort to reduce the levels of plasminogen/plasmin contamination from serum. Absence of plasmin contamination was confirmed by a plasmin activity assay (data not shown). Cells were then incubated for 48 h in the presence of 10 μg/ml aprotinin or 200 nM PAI-1. Consistent with rPDGF D processing (Fig. 4A), both aprotinin and PAI-1 effectively suppressed the processing of PDGF D in PC3-PDGF D cells as determined by immunoblot analysis of PDGF D protein concentrated using Ni-NTA agarose beads (Fig. 4C).
uPA is an activator of PDGF D.
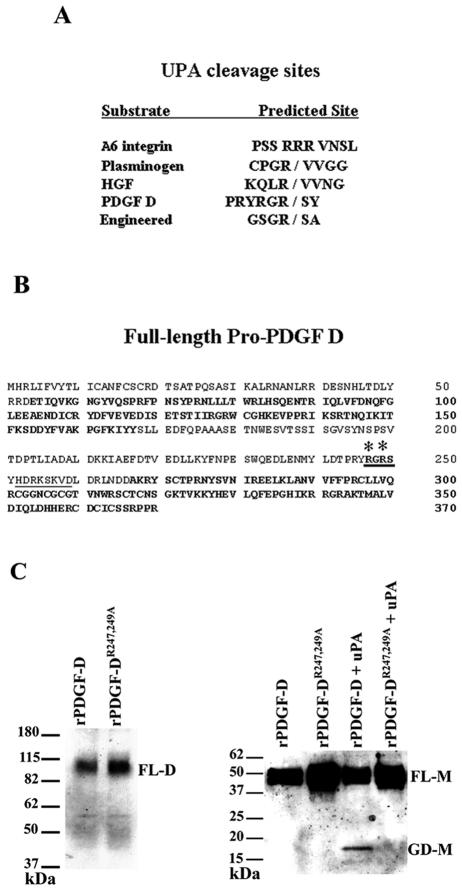
The above results suggest that uPA and/or tPA may be an activator of PDGF D. To determine which protease is an activator of PDGF D, we first surveyed a series of proteases, including uPA and tPA, for their ability to cleave rPDGF D into an active growth factor domain. Even though our inhibitor study did not indicate a role for furin in PDGF D processing, furin was also included in our survey, as computer predictions of the full-length PDGF D indicate a putative furin cleavage site in the hinge region of PDGF D (Fig. 7B, region is underlined) (35). Recombinant PDGF D was incubated with 10 nM of each enzyme and only uPA, but not tPA, was able to cleave rPDGF D into the ∼18-kDa form of the growth factor domain (Fig. 5A). Additionally, when rPDGF D was incubated with increased amounts of uPA, an increased amount of active PDGF D was generated in an uPA dose-dependent manner (Fig. 5B).
FIG. 7.
Mutational analysis identifies the uPA cleavage site in full-length PDGF D. (A) Comparison of uPA cleavage sites in engineered and natural uPA substrates. (B) Putative uPA cleavage site in the full-length PDGF D protein hinge region (cleavage site in bold and underlined). Asterisks indicate R247 and R249 mutated to alanines. The CUB domain and growth factor domain are in bold. The computer-predicted furin cleavage site is underlined. (C) Left panel, CV-1 cells were coinfected and transfected (see Materials and Methods) with vTF7-3 and either pTF7-PDGF DR247,249A or pTF7-PDGF D and cultured in serum-free medium for 24 h. CM was collected and resolved on an SDS-PAGE gel under nonreducing conditions to visualize the dimer form of full-length pro-PDGF D (∼80 kDa) using the anti-PDGF D-GD antibody. Right panel, rPDGF DR247,249A or rPDGF D was incubated at 37°C overnight with or without 10 nM uPA and then resolved on an SDS-PAGE gel under reducing conditions. PDGF D full-length (∼50 kDa) and growth domain (∼18 kDa) monomers were detected through immunoblot analysis using anti-PDGF D-GD antibody. FL-D, full-length PDGF D protein dimer; FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain monomer.
FIG. 5.
PDGF D is a substrate of uPA. (A) rPDGF D was incubated overnight with 10 nM of indicated protease, resolved on an SDS-PAGE gel under reducing conditions, and analyzed by immunoblotting using anti-PDGF D-growth domain (GD) antibody. FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain monomer. (B) rPDGF D was incubated with increasing nanomolar amounts of recombinant uPA and incubated at 37°C overnight. Samples were analyzed by immunoblotting using the anti-PDGF D-GD antibody. (C) Five milliliters of CM was collected from CV-1 cells infected with vaccinia virus-expressed rPDGF D and vTF7-3 or vTF7-3 only and was incubated with no uPA (lane 3 and lane 6) or 10 nM uPA (lane 4 and lane 7) overnight and then used to stimulate serum-starved NIH 3T3 cells for 10 min. In addition to the vTF7-3 control, additional negative controls included NIH 3T3 cells stimulated with serum-free (SF) medium (lane 1) or SF medium incubated overnight with 10 nM uPA (lane 5). β-PDGFR was immunoprecipitated by incubating 500 μg of lysate with anti-β-PDGFR antibody. Samples were resolved on an SDS-PAGE gel under reducing conditions, transferred to a nitrocellulose membrane, and then probed with an antiphosphotyrosine antibody. As a control of the immunoprecipitation efficiency, the membrane was stripped and reprobed with an anti-β-PDGFR antibody.
We next examined if uPA-digested rPDGF DD was biologically active. rPDGF DD incubated with 10 nM uPA was used to treat serum-starved NIH 3T3 cells, and then β-PDGFR was immunoprecipitated from NIH 3T3 lysate. Serum-free medium was included as a negative control, and serum-free medium supplemented with 25 ng/ml of PDGF BB was included as a positive control. Other controls included serum-free medium supplemented with 10 nM uPA and CM collected from CV-1 cells infected with only vTF7-3. The vTF7-3 controls were included to address the possibility that vaccinia virus infection of CV-1 cells induces the production of a latent growth factor independent of vPDGF D. As seen in Fig. 5C, only the PDGF BB-positive control and the rPDGF D plus uPA experimental sample induced phosphorylation of β-PDGFR, demonstrating that uPA processes full-length PDGF D into its biologically active growth factor.
Active uPA levels correlate with PDGF D activation in human prostate carcinoma cells.
Identifying uPA as a potential activator of PDGF D in human prostate carcinoma cells, we next examined the levels and activities of uPA in PC3 and LNCaP cells with and without PDGF D overexpression. uPA has a three-domain structure which includes (i) an N-terminal growth-factor-like domain (amino-terminal fragment) which mediates the association of uPA with its membrane-associated receptor, uPAR, (ii) a kringle domain which has been implicated in uPA-mediated chemotaxis, and (iii) the proteolytic/catalytic domain at the C terminus. uPA is secreted as a 54-kDa single-chain latent protease (sc-uPA) and activated when proteolytically cleaved between Lys158 and Ile159 to form the alpha and beta chains of two-chain uPA (tc-uPA). The alpha and beta chains of tc-uPA are held together by disulfide bonds, the 24-kDa alpha chain contains the amino-terminal fragment and the kringle domain, and the 30-kDa beta chain contains the catalytic domain required for uPA proteolytic activity. Immunoblot analysis of CM (Fig. 6A) and lysate (Fig. 6B) was performed under reducing conditions to detect sc-uPA (inactive) and the beta chain of tc-uPA (active). There was a significant amount of both soluble and cell-associated latent sc-uPA present in PC3-neo and PC3-PDGF D cells. Interestingly, levels of the active beta chain of tc-uPA were increased in CM collected from PC3-PDGF D cells (Fig. 6A) while the cell-associated beta chains of tc-uPA were comparable between PC3-PDGF D cells and PC3-neo cells (Fig. 6B). These results suggest that uPA activates PDGF D, which in turn regulates uPA expression and activity. At present, the molecular mechanism of this feedback signaling loop is unclear. However, this signaling loop for the uPA activation of PDGF D and PDGF D-induced cell-associated tc-uPA levels does not appear to involve the uPA receptor (uPAR), since the uPAR protein levels are similar among the four cell lines (data not shown). Consistent with the absence of PDGF D processing in LNCaP CM and inefficient processing of PDGF D at or near the LNCaP cell surface (38), uPA protein was undetected in LNCaP-neo and LNCaP-PDGF D CM and low levels of the active beta chain of tc-uPA were detected in LNCaP-neo and LNCaP-PDGF D cell lysate (Fig. 6B).
FIG. 6.

uPA expression in PC3-neo, PC3-PDGF, LNCaP-neo, and LNCaP-PDGF D cell lines. (A) Cells were grown to 100% confluence, washed with PBS, and cultured in serum-free medium for 48 h. CM was collected and resolved on an SDS-PAGE gel under denaturing conditions and then transferred to a nitrocellulose membrane. An antibody recognizing the beta chain of uPA was used for immunoblot analysis to detect the latent sc-uPA and the beta chain present in active, two-chain uPA. (B) Lysate was collected along with the CM used above, and 50 μg was resolved on an SDS-PAGE gel under reducing conditions, transferred to nitrocellulose, and probed with the same antibody used above.
Identification of the uPA cleavage sites in the PDGF D full-length protein.
When PDGF D was first identified, it was reported that full-length PDGF D was cleaved at either R247 or R249 of the hinge region of full-length PDGF D (20). Comparison of the R247/R249 region of PDGF D with the uPA cleavage sites in uPA natural substrates yielded no remarkable conservation (Fig. 7A) (7, 26). However, there was sequence similarity between the R247/R249 region of PDGF D and the preferred cleavage site of uPA identified by phage library display and confirmed in an engineered protein (Fig. 7A) (6, 14, 15). To determine whether this putative uPA cleavage site of PDGF D is critical for the uPA-mediated proteolytic cleavage of PDGF D, R247 and R249 were both mutated to alanines by site-directed mutagenesis and then the mutant PDGF DR247,249A was inserted into the vaccinia virus expression vector for further analysis (Fig. 7B). The mutant PDGF DR247,249A retains its ability to form a dimer (Fig. 7C, left panel), indicating that the double alanine mutation does not disrupt the dimeric structure of PDGF DD. Importantly, however, the double mutation disrupts uPA's ability to process PDGF D into the ∼18-kDa active growth domain form (Fig. 7C, right panel), indicating that uPA cleaves PDGF D at the R247/R249 site in the hinge region of full-length PDGF D.
PDGF D and uPA expression in human prostate carcinoma.
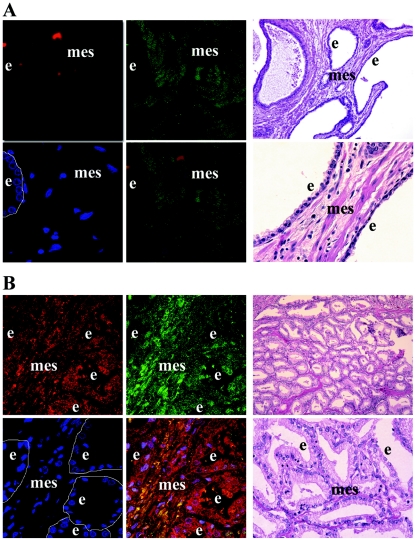
Previous histological studies reported uPA preferentially localized to the glandular epithelium of human prostate cancer (27, 39). There are no published histological data concerning PDGF D expression in the prostate gland. Therefore, to further investigate the expression of PDGF D and its possible colocalization with uPA in the prostate gland, we obtained slides of both normal prostate and human prostate carcinoma specimens from the Wayne State University Pathology Research Core Facility. Slides were coprobed with two antibodies, a monoclonal mouse antibody which recognizes the beta chain of uPA and our custom-made rabbit antibody which recognizes the growth domain of PDGF D. Confocal immunofluorescence microscopic analysis revealed that PDGF D was preferentially expressed in the mesenchyme of normal prostate tissue (Fig. 8A). Analysis of the prostate carcinoma sample shows increased and profuse PDGF D and, more interestingly, colocalization with its activator, uPA (Fig. 8B).
FIG. 8.
Localization of PDGF D and uPA in human prostate carcinoma. Microscopic immunofluorescence analysis of uPA (Texas Red) and PDGF D (fluorescein isothiocyanate; green) expression in samples of the human prostate was performed at 63× magnification. Cell nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; blue). Yellow in the merged panel represents the colocalization of uPA and PDGF D. Images of H&E staining of corresponding serial slides taken at 10× and 40× magnification were used for histological analysis of tissues. Epithelial cells lining the ducts of the prostate are outlined in the DAPI panels. e, epithelium; mes, mesenchyme. (A) Normal human prostate specimen. (B) Human prostate carcinoma specimen.
DISCUSSION
Members of the platelet-derived growth factor family have long been known as potent stimulators of cell proliferation, migration, and transformation. PDGF ligands are typically secreted by tumor cells or inflammatory cells, which then activate the PDGF receptors on the cell surfaces of surrounding mesenchymal cells, mediating cell-cell communication. PDGFR correlation to poor prognosis in prostate cancer and the therapeutic value of PDGFR-antagonistic chemotherapeutic drugs are now being established. Both α- and β-PDGFR expression levels are upregulated in prostatic intraepithelial neoplasia and adenocarcinoma. Interestingly, even though increased expression of especially the β-PDGFR protein is often found in advanced stages of prostate cancer with phenotypic changes typical to β-PDGFR activation, its cognate ligand PDGF B expression has not been reported in prostate cancer clinical samples (10, 11, 18). This raises the possibility that PDGF D, a newly discovered ligand for β-PDGFR, may be responsible for β-PDGFR-mediated signal transduction during prostate cancer progression. In phase I and phase II clinical trials, the PDGFR antagonistic drugs ST1571 (also known as Gleevac or imatinib mesylate) and SU101 have shown promise in reducing prostate-specific antigen levels, tumor progression, and resultant bony pain (8, 17, 23). Similar to the clinical studies, a mouse model of prostate cancer bone metastasis exhibits increased β-PDGFR phosphorylation in prostate tumor xenographs as well as in the surrounding mouse bone endothelial cells and vascular smooth muscle cells (36). Accordingly, administration of ST1571 either alone or in combination with other cancer therapy drugs results in a decrease of PDGFR phosphorylation, significant reduction of tumor growth and angiogenesis, and reduced destruction of host bone in prostate cancer murine models (16, 29).
To better understand the roles of PDGF/β-PDGFR signaling in prostate cancer progression, we have investigated the effects of PDGF D expression, a newly discovered ligand for β-PDGFR, in prostate carcinoma cells. Whereas PDGF AA, PDGF AB, and PDGF BB are processed intracellularly and secreted as active dimers that can readily activate PDGFRs, PDGF CC and PDGF DD are secreted as latent dimers. Thus, the proteolytic cleavage of PDGF DD is an essential step in regulating its activity for β-PDGFR-mediated signal transduction. PDGF D is produced in many secretory organs, including the adrenal and salivary glands, and its expression is often upregulated in many human tumors (19, 20). Yet, a protease responsible for PDGF D processing in the circulation or at the tumor site was unknown. This present study identifies uPA as an activator of PDGF D in human prostate carcinoma cells. Our data are in good agreement with the recent paper by Fredriksson et al., which focuses on the tPA activation of PDGF CC but also reports that tPA does not activate PDGF DD (9). The levels of active uPA expressed by PC3 correlate with the efficiency of PDGF D processing, suggesting uPA as a critical regulator of PDGF-D/β-PDGFR signaling for prostate cancer progression.
uPA has been localized to the sites of cell-cell contact, as well as the leading edge of migrating cells, and is well correlated with cancer metastasis (reviewed in references 2 and 32). Increased serum levels of uPA are also associated with prostate cancer development and are elevated even more in patients with metastasis to the bone (24, 27). The most well-known substrate of uPA is plasminogen, and until recently, it was believed that its pathogenic effects involved the aberrant activation of plasminogen into the active protease plasmin. Plasmin degrades the extracellular matrix of the cell, allowing for the release of growth factors and other matrix degrading enzymes, such as the metalloproteases. However, recent studies suggest that uPA can induce cell migration and proliferation independent of plasmin generation. The plasmin-independent uPA activity is thought to induce cell proliferation/migration by proteolytically activating growth factors (hepatocyte growth factor, vascular endothelial growth factor) or by interacting with an as-yet-unidentified cell surface receptor other than uPAR (25, 34).
Our finding further supports the direct role of uPA in the regulation of growth factor signaling critical for cancer progression. Considering potent roles for β-PDGFR signaling in cell growth, migration, transformation, and angiogenesis, uPA activation of PDGF D has a significant implication in coordinating the oncogenic activities of uPA and PDGF D at the tumor site. In fact, our previous study showed that PDGF DD stimulates prostate cancer cell proliferation in an autocrine manner and also induces the cell proliferation and migration of prostate fibroblast cells in a paracrine manner. More importantly, PDGF D drastically enhances tumor cell interactions with the surrounding stroma in a mouse tumor model (38), demonstrating a potential oncogenic activity of PDGF D in prostate cancer progression. Now, in this study, we show that PDGF D and its activator, uPA, can colocalize in human prostate carcinoma; therefore, uPA and PDGF D interaction can be biologically significant in a pathological context. Interestingly, recent studies showed a synergistic effect of uPA with PDGF BB signaling in inducing human smooth muscle cell migration and proliferation that is reliant on the uPA catalytic domain but independent of plasmin generation (5, 28). It would be of particular importance to examine whether PDGF DD expressed in smooth muscle cells is also involved in the synergistic signaling loop between uPA and PDGF BB signaling.
Active uPA protein is found in both CM and cell lysate collected from PC3 cells, which correlates with our observations that PDGF D is activated when incubated with either PC3 cells or CM. In contrast, very low levels of active uPA are detected only in LNCaP cell lysate, which correlates with our observation that PDGF D is inefficiently activated in the presence of LNCaP cells, but not in CM collected from LNCaP. Interestingly, when PDGF D signaling is constitutively activated by PC3 cells, active tc-uPA levels significantly increase. Although we do not understand the molecular mechanisms of this feedback signaling loop at present, this may be critical both for uPA localization to the leading edge of migrating tumor cells which are often found in vivo and for uPA activation of PDGF D at or near the cell surface. Recent studies report that localized PDGF BB expression in tumor cells, compared to diffuse PDGF BB expression, has a profound impact on the recruitment of pericytes to the new vasculature developing within the tumor (1, 22). Our preliminary histological data reveal that PDGF D in normal prostate is preferentially expressed in the mesenchyme of the gland and that the expression of PDGF D is increased and more profuse in prostate carcinoma. Interestingly, colocalization of uPA and PDGF D appears to be preferentially localized to the border of the carcinoma. Therefore, localized activation of PDGF DD by uPA at prostate tumor-stromal cell contacts could contribute to vascularization of the tumor and the migration and invasion of cancer cells.
Acknowledgments
This work is supported by National Institutes of Health National Cancer Institute grant CA64139 (to H.-R.C.K.).
We are grateful to S. Sheng and R. Fridman for many thoughtful discussions and constructive comments. We thank M. Schober and W. Huang for their assistance with the immunoprecipitation experiments. We are grateful to L. Schuger for pathological analysis of the prostate tissue and assistance with the resultant figure. We also thank Pam Osenkowski and Steve Singson for their assistance with the vaccinia virus system and the Kim Lab for assistance in preparing the manuscript.
REFERENCES
- 1.Abramsson, A., P. Lindblom, and C. Betsholtz. 2003. Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J. Clin. Investig. 112:1142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreasen, P. A., L. Kjoller, L. Christensen, and M. J. Duffy. 1997. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int. J. Cancer 72:1-22. [DOI] [PubMed] [Google Scholar]
- 3.Bejcek, B. E., R. M. Hoffman, D. Lipps, D. Y. Li, C. A. Mitchell, P. W. Majerus, and T. F. Deuel. 1992. The v-sis oncogene product but not platelet-derived growth factor (PDGF) A homodimers activate PDGF alpha and beta receptors intracellularly and initiate cellular transformation. J. Biol. Chem. 267:3289-3293. [PubMed] [Google Scholar]
- 4.Bergsten, E., M. Uutela, X. Li, K. Pietras, A. Ostman, C. H. Heldin, K. Alitalo, and U. Eriksson. 2001. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat. Cell Biol. 3:512-516. [DOI] [PubMed] [Google Scholar]
- 5.Carlin, S. M., M. Roth, and J. L. Black. 2003. Urokinase potentiates PDGF-induced chemotaxis of human airway smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 284:L1020-L1026.12576295 [Google Scholar]
- 6.Coombs, G. S., R. C. Bergstrom, E. L. Madison, and D. R. Corey. 1998. Directing sequence-specific proteolysis to new targets. The influence of loop size and target sequence on selective proteolysis by tissue-type plasminogen activator and urokinase-type plasminogen activator. J. Biol. Chem. 273:4323-4328. [DOI] [PubMed] [Google Scholar]
- 7.Demetriou, M. C., M. E. Pennington, R. B. Nagle, and A. E. Cress. 2004. Extracellular alpha 6 integrin cleavage by urokinase-type plasminogen activator in human prostate cancer. Exp. Cell Res. 294:550-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckhardt, S. G., J. Rizzo, K. R. Sweeney, G. Cropp, S. D. Baker, M. A. Kraynak, J. G. Kuhn, M. A. Villalona-Calero, L. Hammond, G. Weiss, A. Thurman, L. Smith, R. Drengler, J. R. Eckardt, J. Moczygemba, A. L. Hannah, D. D. Von Hoff, and E. K. Rowinsky. 1999. Phase I and pharmacologic study of the tyrosine kinase inhibitor SU101 in patients with advanced solid tumors. J. Clin. Oncol. 17:1095-1104. [DOI] [PubMed] [Google Scholar]
- 9.Fredriksson, L., H. Li, C. Fieber, X. Li, and U. Eriksson. 2004. Tissue plasminogen activator is a potent activator of PDGF-CC. EMBO J. 23:3793-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fudge, K., D. G. Bostwick, and M. E. Stearns. 1996. Platelet-derived growth factor A and B chains and the alpha and beta receptors in prostatic intraepithelial neoplasia. Prostate 29:282-286. [DOI] [PubMed] [Google Scholar]
- 11.Fudge, K., C. Y. Wang, and M. E. Stearns. 1994. Immunohistochemistry analysis of platelet-derived growth factor A and B chains and platelet-derived growth factor alpha and beta receptor expression in benign prostatic hyperplasias and Gleason-graded human prostate adenocarcinomas. Mod. Pathol. 7:549-554. [PubMed] [Google Scholar]
- 12.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golub, T. R., G. F. Barker, M. Lovett, and D. G. Gilliland. 1994. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77:307-316. [DOI] [PubMed] [Google Scholar]
- 14.Ke, S. H., G. S. Coombs, K. Tachias, D. R. Corey, and E. L. Madison. 1997. Optimal subsite occupancy and design of a selective inhibitor of urokinase. J. Biol. Chem. 272:20456-20462. [DOI] [PubMed] [Google Scholar]
- 15.Ke, S. H., G. S. Coombs, K. Tachias, M. Navre, D. R. Corey, and E. L. Madison. 1997. Distinguishing the specificities of closely related proteases. Role of P3 in substrate and inhibitor discrimination between tissue-type plasminogen activator and urokinase. J. Biol. Chem. 272:16603-16609. [DOI] [PubMed] [Google Scholar]
- 16.Kim, S. J., H. Uehara, S. Yazici, R. R. Langley, J. He, R. Tsan, D. Fan, J. J. Killion, and I. J. Fidler. 2004. Simultaneous blockade of platelet-derived growth factor-receptor and epidermal growth factor-receptor signaling and systemic administration of paclitaxel as therapy for human prostate cancer metastasis in bone of nude mice. Cancer Res. 64:4201-4208. [DOI] [PubMed] [Google Scholar]
- 17.Ko, Y. J., E. J. Small, F. Kabbinavar, A. Chachoua, S. Taneja, D. Reese, A. DePaoli, A. Hannah, S. P. Balk, and G. J. Bubley. 2001. A multi-institutional phase ii study of SU101, a platelet-derived growth factor receptor inhibitor, for patients with hormone-refractory prostate cancer. Clin. Cancer Res. 7:800-805. [PubMed] [Google Scholar]
- 18.Langley, R. R., D. Fan, R. Z. Tsan, R. Rebhun, J. He, S. J. Kim, and I. J. Fidler. 2004. Activation of the platelet-derived growth factor-receptor enhances survival of murine bone endothelial cells. Cancer Res. 64:3727-3730. [DOI] [PubMed] [Google Scholar]
- 19.LaRochelle, W. J., M. Jeffers, J. R. Corvalan, X. C. Jia, X. Feng, S. Vanegas, J. D. Vickroy, X. D. Yang, F. Chen, G. Gazit, J. Mayotte, J. Macaluso, B. Rittman, F. Wu, M. Dhanabal, J. Herrmann, and H. S. Lichenstein. 2002. Platelet-derived growth factor D: tumorigenicity in mice and dysregulated expression in human cancer. Cancer Res. 62:2468-2473. [PubMed] [Google Scholar]
- 20.LaRochelle, W. J., M. Jeffers, W. F. McDonald, R. A. Chillakuru, N. A. Giese, N. A. Lokker, C. Sullivan, F. L. Boldog, M. Yang, C. Vernet, C. E. Burgess, E. Fernandes, L. L. Deegler, B. Rittman, J. Shimkets, R. A. Shimkets, J. M. Rothberg, and H. S. Lichenstein. 2001. PDGF-D, a new protease-activated growth factor. Nat. Cell Biol. 3:517-521. [DOI] [PubMed] [Google Scholar]
- 21.Li, X., A. Ponten, K. Aase, L. Karlsson, A. Abramsson, M. Uutela, G. Backstrom, M. Hellstrom, H. Bostrom, H. Li, P. Soriano, C. Betsholtz, C. H. Heldin, K. Alitalo, A. Ostman, and U. Eriksson. 2000. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nat. Cell Biol. 2:302-309. [DOI] [PubMed] [Google Scholar]
- 22.Lindblom, P., H. Gerhardt, S. Liebner, A. Abramsson, M. Enge, M. Hellstrom, G. Backstrom, S. Fredriksson, U. Landegren, H. C. Nystrom, G. Bergstrom, E. Dejana, A. Ostman, P. Lindahl, and C. Betsholtz. 2003. Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes Dev. 17:1835-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathew, P., P. F. Thall, D. Jones, C. Perez, C. Bucana, P. Troncoso, S. J. Kim, I. J. Fidler, and C. Logothetis. 2004. Platelet-derived growth factor receptor inhibitor imatinib mesylate and docetaxel: a modular phase I trial in androgen-independent prostate cancer. J. Clin. Oncol. 22:3323-3329. [DOI] [PubMed] [Google Scholar]
- 24.Miyake, H., I. Hara, K. Yamanaka, S. Arakawa, and S. Kamidono. 1999. Elevation of urokinase-type plasminogen activator and its receptor densities as new predictors of disease progression and prognosis in men with prostate cancer. Int. J. Oncol. 14:535-541. [DOI] [PubMed] [Google Scholar]
- 25.Mukhina, S., V. Stepanova, D. Traktouev, A. Poliakov, R. Beabealashvilly, Y. Gursky, M. Minashkin, A. Shevelev, and V. Tkachuk. 2000. The chemotactic action of urokinase on smooth muscle cells is dependent on its kringle domain. Characterization of interactions and contribution to chemotaxis. J. Biol. Chem. 275:16450-16458. [DOI] [PubMed] [Google Scholar]
- 26.Naldini, L., E. Vigna, A. Bardelli, A. Follenzi, F. Galimi, and P. M. Comoglio. 1995. Biological activation of pro-HGF (hepatocyte growth factor) by urokinase is controlled by a stoichiometric reaction. J. Biol. Chem. 270:603-611. [DOI] [PubMed] [Google Scholar]
- 27.Ohta, S., H. Fuse, Y. Fujiuchi, O. Nagakawa, and Y. Furuya. 2003. Clinical significance of expression of urokinase-type plasminogen activator in patients with prostate cancer. Anticancer Res. 23:2945-2950. [PubMed] [Google Scholar]
- 28.Padro, T., R. M. Mesters, B. Dankbar, H. Hintelmann, R. Bieker, M. Kiehl, W. E. Berdel, and J. Kienast. 2002. The catalytic domain of endogenous urokinase-type plasminogen activator is required for the mitogenic activity of platelet-derived and basic fibroblast growth factors in human vascular smooth muscle cells. J. Cell Sci. 115:1961-1971. [DOI] [PubMed] [Google Scholar]
- 29.Pietras, K., K. Rubin, T. Sjoblom, E. Buchdunger, M. Sjoquist, C.-H. Heldin, and A. Ostman. 2002. Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res. 62:5476-5484. [PubMed] [Google Scholar]
- 30.Plate, K. H., G. Breier, C. L. Farrell, and W. Risau. 1992. Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas. Lab. Investig. 67:529-534. [PubMed] [Google Scholar]
- 31.Rosenkranz, S., and A. Kazlauskas. 1999. Evidence for distinct signaling properties and biological responses induced by the PDGF receptor alpha and beta subtypes. Growth Factors 16:201-216. [DOI] [PubMed] [Google Scholar]
- 32.Sheng, S. 2001. The urokinase-type plasminogen activator system in prostate cancer metastasis. Cancer Metastasis Rev. 20:287-296. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried, G., A.-M. Khatib, S. Benjannet, M. Chretien, and N. G. Seidah. 2003. The proteolytic processing of pro-platelet-derived growth factor-A at RRKR86 by members of the proprotein convertase family is functionally correlated to platelet-derived growth factor-A-induced functions and tumorigenicity. Cancer Res. 63:1458-1463. [PubMed] [Google Scholar]
- 34.Stepanova, V. V., and V. A. Tkachuk. 2002. Urokinase as a multidomain protein and polyfunctional cell regulator. Biochemistry (Moscow) 67:109-118. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, N. A., W. J. Van De Ven, and J. W. Creemers. 2003. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 17:1215-1227. [DOI] [PubMed] [Google Scholar]
- 36.Uehara, H., S. J. Kim, T. Karashima, D. L. Shepherd, D. Fan, R. Tsan, J. J. Killion, C. Logothetis, P. Mathew, and I. J. Fidler. 2003. Effects of blocking platelet-derived growth factor-receptor signaling in a mouse model of experimental prostate cancer bone metastases. J. Natl. Cancer Inst. 95:458-470. [DOI] [PubMed] [Google Scholar]
- 37.Uren, A., M. S. Merchant, C. J. Sun, M. I. Vitolo, Y. Sun, M. Tsokos, P. B. Illei, M. Ladanyi, A. Passaniti, C. Mackall, and J. A. Toretsky. 2003. Beta-platelet-derived growth factor receptor mediates motility and growth of Ewing's sarcoma cells. Oncogene 22:2334-2342. [DOI] [PubMed] [Google Scholar]
- 38.Ustach, C. V., M. E. Taube, N. J. Hurst, Jr., S. Bhagat, R. D. Bonfil, M. L. Cher, L. Schuger, and H. R. Kim. 2004. A potential oncogenic activity of platelet-derived growth factor d in prostate cancer progression. Cancer Res. 64:1722-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Veldhuizen, P. J., R. Sadasivan, R. Cherian, and A. Wyatt. 1996. Urokinase-type plasminogen activator expression in human prostate carcinomas. Am. J. Med. Sci. 312:8-11. [DOI] [PubMed] [Google Scholar]
- 40.Westermark, B., C. H. Heldin, and M. Nister. 1995. Platelet-derived growth factor in human glioma. Glia 15:257-263. [DOI] [PubMed] [Google Scholar]
- 41.Yu, J., T. F. Deuel, and H. R. Kim. 2000. Platelet-derived growth factor (PDGF) receptor-alpha activates c-Jun NH2-terminal kinase-1 and antagonizes PDGF receptor-beta-induced phenotypic transformation. J. Biol. Chem. 275:19076-19082. [DOI] [PubMed] [Google Scholar]
- 42.Yu, J., C. Ustach, and H. R. Kim. 2003. Platelet-derived growth factor signaling and human cancer. J. Biochem. Mol. Biol. 36:49-59. [DOI] [PubMed] [Google Scholar]